Summary
The N‐end rule pathway of targeted protein degradation is an important regulator of diverse processes in plants but detailed knowledge regarding its influence on the proteome is lacking.
To investigate the impact of the Arg/N‐end rule pathway on the proteome of etiolated seedlings, we used terminal amine isotopic labelling of substrates with tandem mass tags (TMT‐TAILS) for relative quantification of N‐terminal peptides in prt6, an Arabidopsis thaliana N‐end rule mutant lacking the E3 ligase PROTEOLYSIS6 (PRT6).
TMT‐TAILS identified over 4000 unique N‐terminal peptides representing c. 2000 protein groups. Forty‐five protein groups exhibited significantly increased N‐terminal peptide abundance in prt6 seedlings, including cruciferins, major seed storage proteins, which were regulated by Group VII Ethylene Response Factor (ERFVII) transcription factors, known substrates of PRT6. Mobilisation of endosperm α‐cruciferin was delayed in prt6 seedlings. N‐termini of several proteases were downregulated in prt6, including RD21A. RD21A transcript, protein and activity levels were downregulated in a largely ERFVII‐dependent manner. By contrast, cathepsin B3 protein and activity were upregulated by ERFVIIs independent of transcript.
We propose that the PRT6 branch of the pathway regulates protease activities in a complex manner and optimises storage reserve mobilisation in the transition from seed to seedling via control of ERFVII action.
Keywords: Arabidopsis thaliana, cruciferin, N‐end rule, N‐terminomics, protease, quantitative proteomics, TAILS, tandem mass tag (TMT)
Short abstract
See also the Commentary on this article by https://doi.org/10.1111/nph.15156.
Introduction
The transitions from dormant seed to photosynthetically active plant are key steps in the life cycle of plants (Holdsworth et al., 2008; Wu, 2014; de Wit et al., 2016). Dependent on the light environment following germination, a seedling may undergo skotomorphogenesis (hypocotyl elongation in the dark) or photomorphogenesis (opening of the apical hook and development of the photosynthetic apparatus). In both cases, mobilisation of seed storage reserves fuels growth until plants become fully photoautotrophic (Penfield et al., 2006b; Theodoulou & Eastmond, 2012). Seed reserves comprise starch, lipids in the form of triacylglycerol (TAG) and specialised seed storage proteins (SSPs), but the relative proportions differ considerably between species (Baud et al., 2008). In oilseed plants, such as Arabidopsis, TAG is the most abundant storage reserve but the endosperm and embryo of Arabidopsis seeds also contain numerous protein storage vacuoles (PSVs). Arabidopsis has two major classes of SSP: the 12S globulins (cruciferins) and 2S albumins (napins) which are synthesised as precursors during seed maturation and accumulate in PSVs after processing (Herman & Larkins, 1999; Baud et al., 2008). Following imbibition, catabolism of lipid and protein reserves is initiated in endosperm cells adjacent to the radical tip (Mansfield & Briarty, 1996). Tissue‐specific analysis of abscisic acid (ABA) signalling has shown that mobilisation of embryo and endosperm lipid reserves is under distinct hormonal control (Penfield et al., 2004, 2006a).
Numerous genetic studies have provided valuable insight into the control of germination and seedling establishment (Holdsworth et al., 2008). Previously, we identified PROTEOLYSIS6 (PRT6) as a positive regulator of germination in Arabidopsis (Holman et al., 2009). prt6 null alleles exhibit a range of phenotypes related to germination and seedling establishment: germination of prt6 is hypersensitive to inhibition by ABA and insensitive to nitric oxide (NO), prt6 seedling establishment is hypersensitive to sucrose, and hypocotyls and endosperm of prt6 seedlings retain oil bodies for several days following germination (Holman et al., 2009; Gibbs et al., 2014a). PRT6 encodes a ubiquitin E3 ligase belonging to the N‐end rule pathway of targeted protein degradation, which is a specialised subset of the ubiquitin proteasome system (Bachmair et al., 1986; Garzón et al., 2007; Varshavsky, 2011; Gibbs et al., 2014b, 2016). The N‐end rule relates the half‐life of a protein to its amino terminal (Nt) residue and has three branches, the Arg/N‐end rule and the Ac/N‐end rule, which target free and acetylated N‐termini, respectively, and the recently defined Pro/N‐end rule pathway (Supporting Information Fig. S1; Hwang et al., 2010; Varshavsky, 2011; Chen et al., 2017). In eukaryotes, proteins are synthesised with Met at the N‐terminus but can become Arg/N‐end rule substrates following cleavage by nonprocessive endopeptidases, if the new Nt is large or bulky (a so‐called destabilising residue). Arabidopsis has two characterised E3 ligases that recognise different types of destabilising residues. PROTEOLYSIS1 (PRT1) recognises aromatic Nt amino acids, whereas PRT6 is specific for basic Nt residues (Potuschak et al., 1998; Stary et al., 2003; Garzón et al., 2007; Graciet et al., 2010; Mot et al., 2018). As well as primary destabilising residues revealed by endopeptidase cleavage, PRT6 substrates can be generated via enzymatic modification of secondary and tertiary destabilising residues (Figs 1, S1). Five Arabidopsis transcription factors belonging to Group VII of the Ethylene Response Factor (ERFVII) family, namely HYPOXIA RESPONSIVE1 (HRE1), HRE2, RELATED TO APETALA2.2 (RAP2.2), RAP2.3 and RAP2.12, are N‐end rule substrates (Gibbs et al., 2011, 2015; Licausi et al., 2011). These proteins are substrates by virtue of a Cys residue at position 2: following N‐terminal Met excision (NME), Cys2 is oxidised by specific oxidases, which enables Nt arginylation, catalysed by arginyltransferase enzymes, ATE1 and ATE2. The sequential reactions of NME, Cys oxidation and arginylation produce an Nt degradation signal (N‐degron) for PRT6. The stability of these Met‐Cys initiating transcription factors is controlled by oxygen availability and action on exposed Cys‐2, thereby providing a mechanism by which oxygen status is sensed and transduced by the Arg/N‐end rule pathway in plants (Gibbs et al., 2011, 2015; Licausi et al., 2011; Weits et al., 2014; Mendiondo et al., 2016; White et al., 2017). Hypoxia responsive genes, such as ALCOHOL DEHYDROGENASE (ADH), PYRUVATE DECARBOXYLASE (PDC) and HAEMOGLOBIN1, are ectopically expressed in prt6 alleles (Choy et al., 2008; Gibbs et al., 2011; Riber et al., 2015). The Arg/N‐end rule also acts as a sensor of NO, which is required in addition to O2 for the degradation of ERFVII proteins in plants and G‐protein regulators in mammals (Hu et al., 2005; Gibbs et al., 2014a, 2015).
Figure 1.
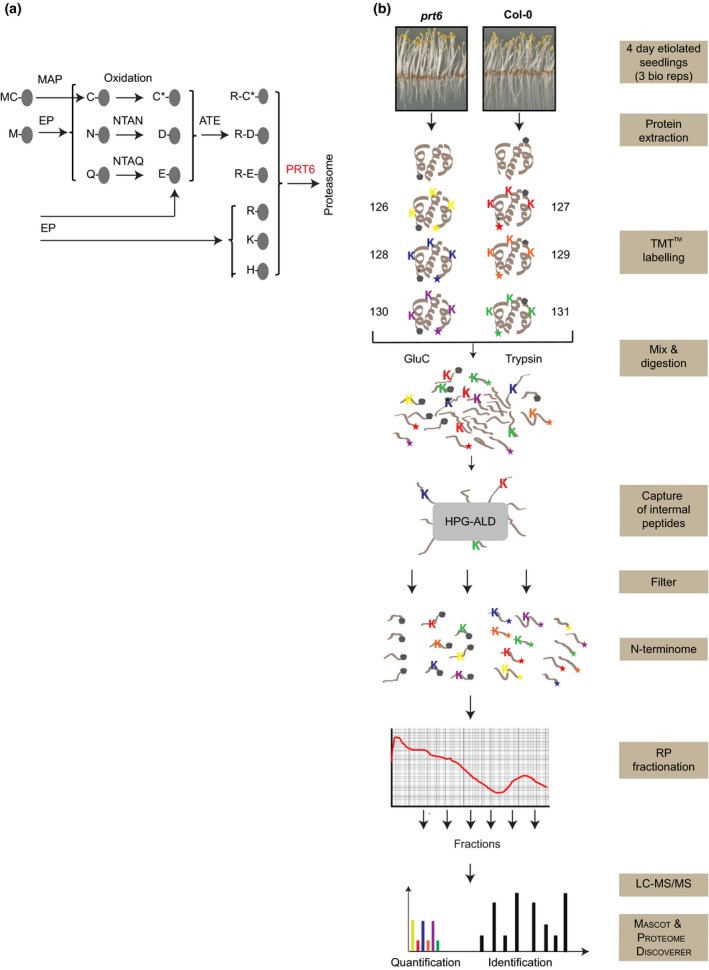
Identification and quantitation of N‐terminal peptides with TMT™‐TAILS. (a) The PRT6 branch of the Arg/N‐end rule. Substrates are generated by the action of endopeptidases (EP) or by methionine aminopeptidase (MAP)‐dependent excision of Met1 from proteins initiating Met‐Cys. PRT6, PROTEOLYSIS6 E3 ligase; ATE, arginyl tRNA transferase; NTAN1, asparagine‐specific N‐terminal amidase; NTAQ1, glutamine‐specific N‐terminal amidase. Amino acids are indicated with single letter codes; C*, oxidised cysteine. (b) Schematic representation of the TAILS workflow. Primary amines of proteins with free N‐termini (star) and lysine (K) side‐chain amines of proteins were labelled with 6‐plex TMT reagents (three biological replicates per genotype). After combining labelled samples from WT and prt6‐5 plants, the sample was divided into two, proteins were digested with either GluC or trypsin, and internal peptides were removed via hyperbranched polyglycerol aldehyde (HPG‐ALD) polymer binding of the free N‐terminal amine group. The unbound peptides (highly enriched for N‐terminal peptides) were fractionated by reversed‐phase (RP) chromatography, then analysed by high‐accuracy LC‐MS/MS. Mascot and ProteomeDiscoverer™ were used for protein identification and quantification. Grey pentagons represent naturally blocked (acetylated) N‐termini.
Genetic approaches in Arabidopsis have revealed further roles for the PRT6 branch of the Arg/N‐end rule pathway in leaf development and senescence (Yoshida et al., 2002; Graciet et al., 2009), quiescence under submergence (Riber et al., 2015), plant–pathogen interactions (Gravot et al., 2016; de Marchi et al., 2016) and photomorphogenesis (Choy et al., 2008; Abbas et al., 2015). The Arg/N‐end rule also plays roles in gametophyte development, starch accumulation and senescence in the moss Physcomitrella patens (Schuessele et al., 2016). With the exception of germination, gas sensing and photomorphogenesis, which are ERFVII‐dependent (Gibbs et al., 2014a,b; Abbas et al., 2015), the mechanisms underlying N‐end rule loss of function phenotypes have not been identified. In this study, we set out to determine the impact of the PRT6 E3 ligase on the proteome. We hypothesised that substrates would be stabilised in the prt6 mutant and therefore increased in abundance relative to the wild type, as would proteins acting downstream of PRT6 substrates such as transcription factors. Quantitative proteomics techniques, in particular N‐terminome analysis (Huesgen & Overall, 2012; Tsiatsiani et al., 2012), offer an opportunity to analyse the N‐end rule in this way: enrichment of N‐terminal peptides not only simplifies the proteome but also provides information about protein cleavage events that can be used to identify and validate potential N‐end rule substrates (Kleifeld et al., 2010, 2011). The N‐terminome is also a useful resource for protein annotation (Hartmann & Armengaud, 2014; Lange et al., 2014; Willems et al., 2017). Previously, we achieved efficient enrichment of Nt peptides from roots of Arg/N‐end rule mutants, using terminal amine isotopic labelling of substrates (TAILS) coupled with dimethyl labelling (Zhang et al., 2015). Here, we incorporate tandem mass tag (TMT™) labelling into the TAILS workflow to quantify the impact of the Arg/N‐end rule on etiolated seedlings. We identified and quantified c. 4000 Nt peptides. Of these, Nt peptides corresponding to 146 protein groups exhibited significantly altered abundance in prt6 seedlings. Surprisingly, we detected increased levels of SSP N‐termini in prt6, notably representing all four major cruciferins. We provide evidence that this reflects delayed mobilisation in Arg/N‐end rule mutants, due to increased stability of the ERFVII transcription factors. Our N‐terminomics data set also revealed that several proteases were differentially regulated in prt6, and subsequent validation showed that protease accumulation and activity are subject to complex regulation by the ERFVIIs. Collectively, our studies reveal that the Arg/N‐end rule serves to co‐ordinate the mobilisation of seed storage reserves and to regulate the abundance and activities of several proteases following germination.
Materials and Methods
N‐end rule mutant alleles and transgenic lines
prt6‐1, prt6‐5 and ate1/2 are well‐characterised Arabidopsis thaliana L. Heynh. null T‐DNA alleles, described by Holman et al. (2009) and Graciet et al. (2009). Higher order mutants are described by Gibbs et al. (2011, 2014a) and Abbas et al. (2015). X‐GUS lines are described by Garzón et al. (2007).
Plant growth and seedling treatments
Seeds were raised from plants grown under long day conditions (16 h : 18 h; 23°C : 18°C); all genotypes to be compared were raised in the same cabinet. Seeds were harvested, sieved (< 425 µm; Endecotts, London, UK ) and stored at room temperature. After ripened seeds were surface‐sterilised and plated on nylon mesh (Sefar NITEX, 03‐110/47; Heiden, Switzerland) on 0.5× Murashige and Skoog (MS) medium containing 0.5% (w/v) sucrose. After 2–3 d dark chilling at 4°C, plates were exposed to light for 6 h to induce germination, then wrapped in foil and incubated in a vertical position at 22°C for 4 d. Etiolated seedlings were harvested under green light; note that mutant and wild type (WT) seedlings grown under these conditions were at the same developmental stage.
TMT labelling and enrichment of N‐termini by TAILS
TMT‐TAILS was performed according to Klein et al. (2015) and Prudova et al. (2016), with modifications and MS as described in Methods S1.
TAILS MS data analysis
Raw data were searched against the TAIR10 database using Mascot v.2.4 (Matrix Science, London, UK) and Proteome Discoverer™ v.1.4.1.14 as described by Zhang et al. (2015), employing Top 10 peaks filter node and percolator nodes and reporter ions quantifier with semi‐ArgC or semi‐GluC enzyme specificity with a maximum of one missed cleavage. Carbamidomethylation (+57.021 Da) of cysteine and TMT isobaric labelling (+229.162 Da) of lysine were set as static modifications while TMT (+229.162 Da) labelling of the peptide N‐termini, the acetylation of the peptide (+42.011) N‐termini and methionine oxidation (+15.996) were considered dynamic. Mass tolerances were set to 10 ppm for MS and 0.06 Da for MS/MS. For quantification, integration window tolerance was set to 0.0075 Da. Each reporting ion was divided by the sum of total ions. Ratios were normalised by the medians of pre‐TAILS samples (Methods S1; Lange et al., 2014) searched with ArgC or GluC specificity. Statistical significance of quantification was assessed with an unpaired two‐sample Student's t‐test on 4 df. Data were log transformed and statistically significant results (P > 0.05) were further restricted to those with more than two‐fold change. No correction for multiplicity was applied. The statistical software package R 3.2.2 was used for all analyses. MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2016) with the dataset identifier PXD006450.
SDS‐PAGE and immunoblotting
Proteins were extracted in modified RIPA buffer containing 50 mM HEPES‐KOH pH 7.8, 100 mM KCl, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 10% (v/v) glycerol, 1% (v/v) IGEPAL, 0.5% (w/v) deoxycholate, 0.1% (w/v) sodium dodecyl sulphate (SDS), 1 mM Na4VO3, 1 mM phenylmethylsulfonylfluoride, 1× proteinase inhibitor cocktail (Roche), 1× phosphostop (Roche) and 50 μM MG‐132. Proteins were separated in precast 4–12% Bis‐Tris gels, using 1× SDS MES buffer and stained with Coomassie Brilliant Blue or transferred to polyvinylidene fluoride using iblot dry blotting system (ThermoFisher, Waltham, MA, USA). Detailed information is given in Methods S2. Primary antibodies were: Brassica napus Cruciferin (Wan et al., 2007), 1 : 10 000–20 000; OLE1 (anti‐rS3; D'Andréa et al., 2007), 1 : 5000–10 000; Anti N‐terminal AtCathB3 (kind gift of Dr Patrick Gallois, University of Manchester), 1 : 1000; Arabidopsis RD21 (residues 137–150; LPESIDWRKKGAVAC; Kaschani et al., 2009; kind gift of Prof. Carol Mackintosh, Dundee), 1 : 1000; and PDC and ADH (Agrisera, Vännäs, Sweden), 1 : 10 000 and 1 : 3000. For analysis of dry seeds and dissected endosperm, identical numbers of similar size seeds or endosperm were collected using a dissecting microscope, processed in parallel and identical amount of extracts were loaded, as indicated in the figure legends.
Real time quantitative reverse‐transcription PCR (RT‐qPCR)
Four‐day‐old etiolated seedlings without endosperm or seed coat were harvested under green light and RNA were extracted using an RNeasy Plant Mini Kit (Qiagen) and treated with RQ1 RNase‐free DNase (Promega). A Transcriptor First Strand cDNA Synthesis Kit (Roche) and anchored ‐oligo(dT)18 were used for cDNA synthesis for a two‐step RT‐PCR. Faststart Essential DNA Green Master (Roche) was used for real‐time PCR using a Lightcycler®96. Relative quantification was done using both ACT2 (At3g18780.2) and TUB4 (At5g44340.1) as references. Student's t‐test was used to calculate P values; error bars are shown as standard errors. Primers used are given in Table S1.
Activity‐based protein profiling
Activity‐based protein profiling (ABPP) was carried out as described by Lu et al. (2015). Band intensities were quantified using ImageJ. Student's t‐test was used to calculate P‐values.
Results
The seedling N‐terminome: identification of Nt peptides by TMT‐TAILS
prt6 RNA is expressed at a low level throughout the plant (Schmid et al., 2005; Winter et al., 2007; Zhang et al., 2015) and prt6 alleles exhibit phenotypes throughout development, including the transition from dark‐grown seedlings to light (Abbas et al., 2015). Etiolated seedlings were selected for analysis because PRT6 is active at this developmental stage, as demonstrated by stabilisation of the artificial Arg/N‐end rule substrate, R‐GUS, in the prt6 mutant background (Fig. S2). Labelling of proteins with TMTsixplex™ reagents was used in combination with TAILS to identify and quantify Nt peptides in seedlings of Col‐0 and the null mutant, prt6‐5 (Graciet et al., 2009). The experimental workflow is presented in Fig. 1.
The full N‐terminome dataset for etiolated seedlings is presented in Table S2. A total of 2396 protein groups were identified, with < 20% overlap between the two proteases used in the TAILS workflow (Fig. 2a). The combined GluC and Trypsin TAILS data sets comprised 5004 unique peptides for which location information was available. Of these, 32% were acetylated, 55% had free N‐termini and the remainder represented internal peptides not removed by the hyperbranched polyglycerol aldehyde polymer (Fig. 2b). In total, 4337 unique Nt peptides representing 3648 unique N‐termini were identified. More unique peptides were identified in the tryptic digest (2997, compared to 2007 for GluC), whereas GluC yielded a higher percentage of free Nt peptides, with a lower proportion of acetylated peptides due to lack of the basic residues (Biniossek & Schilling, 2012). The majority of acetylated Nt peptides were acetylated at Met1 or at residue 2, and therefore are probably the result of co‐translational Nt acetylation, either at the original N‐terminus or following NME by Met amino peptidases (Fig. 2c,e). We also detected free N‐termini generated by NME (Fig. 2f); both these and the acetylated peptides conformed to the established specificity of Met aminopeptidases (Bonissone et al., 2013). Of 2740 free Nt peptides, 43 corresponded to unmodified protein N‐termini initiating with Met (Fig. 2d). The remainder of the non‐acetylated peptides putatively generated by a post‐translational cleavage event were classified as ‘neo’ Nt peptides. In total, 2332 peptides initiated at residue 3 or beyond, relative to the predicted translation start (‘other’) (Fig. 2g). As we observed previously (Zhang et al., 2015), peptides with destabilising residues were underrepresented in the N‐terminome.
Figure 2.
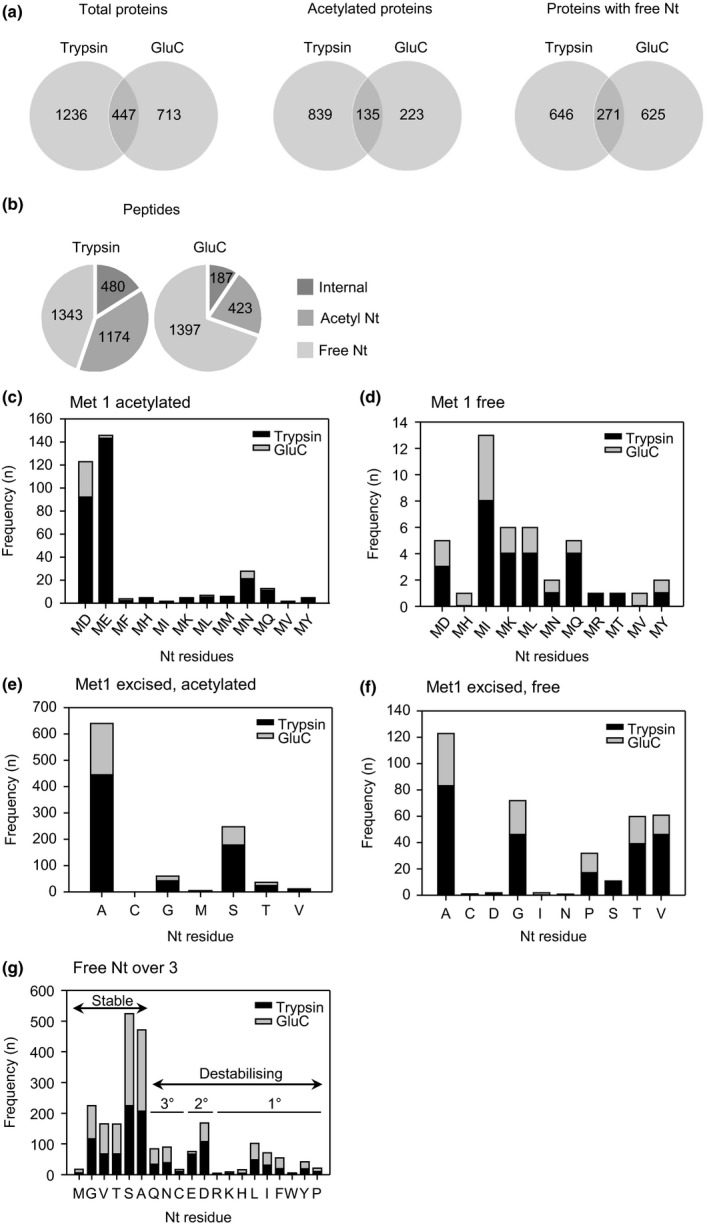
Analysis of protein groups and N‐terminal peptides identified by TMT™‐TAILS. Peptides were enriched by TMT‐TAILS, using two different proteases, trypsin and GluC. (a) Venn diagrams showing overlap in protein groups identified with the N‐terminal peptide datasets from two different proteases. (b) Numbers of unique peptides with location information identified in different categories (free N‐terminal (Nt), acetylated Nt and non‐Nt (internal) peptides) following enrichment by TAILS. When an N‐terminal peptide matched to more than one protein group, positional information was derived for the master protein defined by ProteomeDiscoverer. (c) Analysis of first and second residues of Nt peptides with Met 1 acetylated. (d) Analysis of first and second residues of Nt peptides with free Met 1. (e) Nt peptides resulting from N‐terminal methionine excision followed by Nt acetylation. (f) Free Nt peptides resulting from N‐terminal methionine excision. (g) Occurrence of different Nt‐amino acid residues in free Nt peptides which initiate at amino acid residues ≥ 3, relative to the protein encoded by the published open reading frame (ORF). Met, Gly, Val, Thr, Ser and Ala are stabilising residues. Primary, secondary and tertiary destabilising residues are indicated on the graph.
TMT‐TAILS identifies protein N‐termini with altered abundance in prt6
Peptide abundance was quantified and normalised with three biological replicates of Col‐0 and prt6. The majority of peptides were of similar abundance in Col‐0 and prt6 seedlings (Fig. S3; Table S3). However, Nt peptides corresponding to 45 protein groups exhibited significantly increased abundance in prt6 (defined as two‐fold at P < 0.05; Table 1). Sixteen groups are represented only by ‘original’ N‐termini (i.e. Met 1 or residue 2, relative to the TAIR10 gene model), 27 were identified only from N‐termini generated by endopeptidase cleavage and two were represented by both the original N‐terminus and a new N‐terminus generated by cleavage. Whilst the abundance of Nt peptides may not accurately reflect the abundance of the full‐length protein, three classes of protein of particular interest with regard to the known physiological functions of PRT6 were identified and selected for further study. First, an Nt peptide derived from the ABA receptor component, PYR1‐like 2 (PYL2), was upregulated in prt6 (Fig. 3a). PYL2 is unlikely to be a PRT6 substrate, because the peptide did not bear an Nt destabilising residue but appeared to have been generated by NME followed by acetylation at position 2. Analysis of transcript abundance by RT‐qPCR indicated that PYL2 expression was increased in prt6 relative to Col‐0 and that this increase was dependent upon the ERFVII transcription factors, RAP2.12, RAP2.2 and RAP2.3, but not HRE1 and HRE2 (Fig. 3b). Eight proteins encoded by genes known to be transcriptionally upregulated by hypoxia showed increased abundance in prt6; these included proteins encoded by ‘core’ hypoxia responsive genes, ADH, HAEMOGLOBIN1 and ACC OXIDASE 1 (Mustroph et al., 2009), and also two proteins belonging to the adenine nucleotide α‐hydrolase superfamily, which are homologous to the hypoxia‐responsive universal stress protein, HRU1 (Gonzali et al., 2015). Remarkably, seed storage proteins represented the largest category of proteins with greater abundance in prt6 compared to Col‐0 (Table 1). All four major 12S globulins (cruciferins) were represented by several neo‐Nt peptides in the prt6‐up data set.
Table 1.
N‐terminal peptides with increased abundance in seedlings of the Arabidopsis thaliana prt6 mutant
| AGI code | Description | Synonyms | Peptide | Start | Finish | Log2 fold change |
|---|---|---|---|---|---|---|
| Seed storage proteins | ||||||
| AT1G03880.1 | Cruciferin 2 | CRU2, CRB, At12S3 | gEGQGQGQSQGFR | 117 | 129 | 5.08 |
| qGQGQSQGFR | 120 | 129 | 5.02 | |||
| gQGQGQSQGFR | 119 | 129 | 4.75 | |||
| qGQSQGFR | 122 | 129 | 4.65 | |||
| gQGQSQGFR | 121 | 129 | 3.93 | |||
| eGQGQGQSQGFR | 118 | 129 | 3.80 | |||
| gLEETLcTMR | 270 | 279 | 3.63 | |||
| gQGQGQSQGFRD | 119 | 130 | 3.26 | |||
| nLDDPSDAD | 283 | 291 | 3.02 | |||
| gLEETLcTmR | 270 | 279 | 2.65 | |||
| aLEPSQIIkSE | 37 | 47 | 2.61 | |||
| AT1G03890.1 | RmlC‐like cupins superfamily protein | At12S2 | aPFPNAcHFSQ | 30 | 40 | 4.34 |
| aPFPNAcHFS | 30 | 39 | 3.26 | |||
| eAPFPNAc | 29 | 36 | 2.89 | |||
| gIEETYcTAkIHENIDDPER | 271 | 290 | 2.75 | |||
| pETFAEVEGSSGR | 113 | 125 | 2.17 | |||
| aPFPNAcH | 30 | 37 | 2.00 | |||
| sLAPAQATkFE | 43 | 53 | 1.41 | |||
| AT1G52690.1 | Late embryogenesis abundant protein (LEA) family protein | LEA7 | aSHQEQSYkAGETR | Ac‐2 | 15 | 2.85 |
| AT2G28490.1 | RmlC‐like cupins superfamily protein | gEGEGGGEWGGGGEGGGGGR | 63 | 82 | 3.68 | |
| AT3G15670.1 | Late embryogenesis abundant protein (LEA) family protein | tAQSAkE | 75 | 81 | 1.48 | |
| aSNQQSYkAGETR | Ac‐2 | 14 | 1.40 | |||
| AT3G22640.1 | Cupin family protein | PAP85 | qEEEEDmSENVHkVVSR | 364 | 380 | 4.23 |
| qEEEEDMSENVHkVVSR | 364 | 380 | 4.14 | |||
| eEEEDMSENVHkVVSR | 365 | 380 | 3.52 | |||
| ePPQQGEQEGPR | 33 | 44 | 2.96 | |||
| eEEEDmSENVHkVVSR | 365 | 380 | 2.64 | |||
| AT4G27170.1 | Seed storage albumin 4 | SESA, At12S4 | gQQHQPEQVR | 125 | 134 | 2.28 |
| AT4G28520.1 | Cruciferin 3 | CRU3, CRC, At12S1 | gQPWEGQGQQGQQGFR | 175 | 190 | 4.84 |
| vGVSVARYVIE | 71 | 81 | 4.81 | |||
| gQQGQQGFR | 182 | 190 | 4.71 | |||
| eILYcTGGQGR | 403 | 413 | 4.55 | |||
| eGQGQQGQQGFR | 179 | 190 | 4.28 | |||
| dNLDVLQATE | 40 | 49 | 4.15 | |||
| qQGQQGFR | 183 | 190 | 4.02 | |||
| gQGQQGQQGFR | 180 | 190 | 3.16 | |||
| qQGQPWEGQGQQGQQGFR | 173 | 190 | 3.00 | |||
| tIcSMRSHE | 338 | 346 | 2.98 | |||
| nLDNLDVLQATE | 38 | 49 | 2.67 | |||
| qGQQGQQGFR | 181 | 190 | 2.54 | |||
| gQGQQGQQGFR | 180 | 190 | 2.37 | |||
| gLEETIcSMR | 334 | 343 | 2.19 | |||
| sVNSYTLPILE | 366 | 376 | 2.18 | |||
| gLEETIcSmR | 334 | 343 | 1.98 | |||
| rQSLGVPPQLQNE | 24 | 36 | 1.91 | |||
| gVPPQLQNE | 28 | 36 | 1.59 | |||
| gQGQQGQQGFRD | 180 | 191 | 1.09 | |||
| aMVLPkYNMNANE | 391 | 403 | 1.09 | |||
| AT5G44120.3 | Cruciferin 1 | CRU1, CRA1, At12S4 | gLEETIcSARcTDNLDDPSR | 283 | 302 | 5.22 |
| tDNLDDPSR | 294 | 302 | 5.16 | |||
| sGVSFARYIIE | 70 | 80 | 4.66 | |||
| aLEPSHVLkSE | 43 | 53 | 3.65 | |||
| eTFQDSSEFQPR | 114 | 125 | 3.63 | |||
| qGQQGQQFPNE | 25 | 35 | 3.58 | |||
| gQQFPNEcQLDQLNALEPSHVLkSEAGR | 29 | 56 | 3.55 | |||
| cTDNLDDPSR | 293 | 302 | 3.38 | |||
| gLEETIcSAR | 283 | 292 | 2.90 | |||
| tTLTHSSGPA | 453 | 462 | 2.77 | |||
| gNNPQGQVWLQGRE | 195 | 208 | 2.77 | |||
| qQGQQFPNEcQLDQLNALEPSHVLkSEAGR | 27 | 56 | 2.77 | |||
| qQFPNEcQLDQLNALEPSHVLkSEAGR | 30 | 56 | 2.56 | |||
| gQQGQQFPNE | 26 | 35 | 2.46 | |||
| qQGQQFPNE | 27 | 35 | 2.12 | |||
| fEGQGQSQR | 126 | 134 | 2.00 | |||
| aETFQDSSEFQPR | 113 | 125 | 1.70 | |||
| dGEAQIQIVNDNGNR | 358 | 372 | 1.69 | |||
| qGQQFPNE | 28 | 35 | 1.54 | |||
| tTLTHSSGPAS | 453 | 463 | 1.40 | |||
| sGDTIATTPGVAQW | 147 | 160 | 1.22 | |||
| Hypoxia‐responsive | ||||||
| AT1G43800.1 | Plant stearoyl‐acyl‐carrier‐protein desaturase family protein | FTM, SAD6 | gTIAADEkR | 248 | 256 | 2.71 |
| AT1G77120.1 | Alcohol dehydrogenase 1 | ADH1 | aVGLGAAEGAR | 205 | 215 | 2.09 |
| sTTGQIIRckAAVAWE | Ac‐2 | 17 | 1.64 | |||
| AT2G16060.1 | Haemoglobin 1 | HB1 | mESEGkIVF | Ac‐1 | 9 | 3.26 |
| AT2G19590.1 | ACC oxidase 1 | AtACO1 | lQDDQVPGLE | 192 | 201 | 2.41 |
| AT2G47710.1 | Adenine nucleotide alpha hydrolases‐like superfamily protein | aTGDGkSVmVVGVDDSEQSTY | Ac‐2 | 22 | 1.96 | |
| AT3G11930.3 | Adenine nucleotide alpha hydrolases‐like superfamily protein | aEEQAATAmETSAVEkQPE | Ac‐2 | 20 | 1.25 | |
| AT3G21720.1 | Isocitrate lyase | ICL | iMEEEGR | 11 | 17 | 2.30 |
| aVSEHINR | 223 | 230 | 1.09 | |||
| AT5G19550.1 | Aspartate aminotransferase 2 | ASP2 | aDSPAITESR | 89 | 98 | 1.34 |
| Other | ||||||
| AT1G06680.1 | Photosystem II subunit P‐1 | PSBP‐1 | aQQSHEDDNSAVSR | 42 | 55 | 4.11 |
| kAQQSHEDDNSAVSR | 41 | 55 | 3.02 | |||
| AT1G07600.1 | Metallothionein 1A | MT1A, ATMT‐2, ATMT‐Q, LSR4 | ADSNcGcGSSckcGD | 2 | 16 | 1.04 |
| AT1G14950.1 | Polyketide cyclase/dehydrase and lipid transport superfamily protein | aTSGTYVTEVPLkGSAkNHY | Ac‐2 | 21 | 1.66 | |
| aTSGTYVTEVPLkGSAkN | Ac‐2 | 19 | 1.29 | |||
| AT1G17810.1 | Beta‐tonoplast intrinsic protein | BETA‐TIP | eATHPDSIR | 16 | 24 | 4.40 |
| dEATHPDSIR | 15 | 24 | 2.18 | |||
| aDEATHPDSIR | 14 | 24 | 1.95 | |||
| AT1G23870.1 | Trehalose‐phosphatase/synthase 9 | TPS9 | tVPGIISELDGGYSDGSSDVNSSNSSR | 32 | 58 | 1.98 |
| AT1G48130.1 | 1‐Cysteine peroxiredoxin 1 | PER1 | pGITLGDTVPNLE | 2 | 14 | 1.08 |
| AT1G54870.1 | NAD(P)‐binding Rossmann‐fold superfamily protein | ChlADR | iEEIDEPR | 185 | 192 | 2.22 |
| AT1G64970.1 | Gamma‐tocopherol methyltransferase | G‐TMT, VTE4, TMT1 | aATSTEALR | 53 | 61 | 1.11 |
| AT1G65090.2 | Unknown protein | sQTmEEYQSNESEDkR | Ac‐2 | 17 | 1.81 | |
| AT1G69410.1 | Eukaryotic elongation factor 5A‐3 | ELF5A‐3 | sDDEHHFESSDAGASkTYPQ | Ac‐2 | 21 | 1.08 |
| AT2G17200.1 | Ubiquitin family protein | DSK2 | gGEGDSSQPQSGEGEAVAVN | 2 | 21 | 1.64 |
| AT2G23240.1 | Plant EC metallothionein‐like protein | AtMT4b | aDTGkGSASAScNDR | 2 | 16 | 2.54 |
| AT2G26040.1 | PYR1‐like 2 | PYL2 | sSSPAVkGLTDE | Ac‐2 | 13 | 1.05 |
| AT2G30950.1 | FtsH extracellular protease family | VAR2, FTSH2 | dEQGVSSSR | 83 | 91 | 1.05 |
| AT2G38400.2 | Alanine:glyoxylate aminotransferase 3 | AGT3 | dSDEFQAR | 35 | 42 | 1.92 |
| AT3G13120.1 | Ribosomal protein S10p/S20e family protein | dTLDPTPE | 60 | 67 | 3.23 | |
| AT3G21380.1 | Mannose‐binding lectin superfamily protein | aAATMSWDDGkH | Ac‐2 | 13 | 3.21 | |
| aAATmSWDDGkH | Ac‐2 | 13 | 2.68 | |||
| AT3G51100.1 | Unknown protein | nEGSSEEVTR | 2 | 11 | 1.00 | |
| AT3G57560.1 | N‐Acetyl‐l‐glutamate kinase | NAGK | tVSTPPSIATGNAPSPDYR | 51 | 69 | 1.28 |
| AT3G58450.1 | Adenine nucleotide alpha hydrolases‐like superfamily protein | mETYVDAIGEDTAATTTTAETAANkN | Ac‐1 | 26 | 1.58 | |
| AT3G61870.1 | Unknown | aGGEFGILEGR | 75 | 85 | 1.03 | |
| AT4G12420.1 | Cupredoxin superfamily protein | SKU5 | aDPYSFYNFE | 21 | 30 | 1.25 |
| AT4G26870.1 | Class II aminoacyl‐tRNA and biotin synthetases superfamily protein | sSNYGDVTTNE | 53 | 63 | 1.86 | |
| AT5G10160.1 | Thioesterase superfamily protein | eIPIELR | 61 | 67 | 2.64 | |
| AT5G47110.1 | Chlorophyll A‐B binding family protein | LIL3:2 | aSSDNGTTSPVVE | 43 | 55 | 1.52 |
| sSDNGTTSPVVE | 44 | 55 | 1.29 | |||
| AT5G51545.1 | Low PSII accumulation2 | LPA2 | qNSQIESDTTEDPSR | 32 | 46 | 1.70 |
| AT5G53460.1 | NADH‐dependent glutamate synthase 1 | GLT1 | cGVGFVAE | 117 | 124 | 1.02 |
| AT5G58290.1 | Regulatory particle triple‐A ATPase 3 | RPT3 | aSAAVASmVLDPkASPALMD | Ac‐2 | 21 | 2.12 |
Peptides listed are more than two‐fold increased in abundance in prt6‐5, compared to Col‐0, at P < 0.05. The start and finish amino acid positions are defined with respect to TAIR10 gene models. Residues with modifications Nt‐TMT, side‐chain Lys TMT or other (e.g. oxidised Met) are indicated in lower case; full details are given in Supporting Information Table S3. Ac, N‐terminal acetylation.
Figure 3.
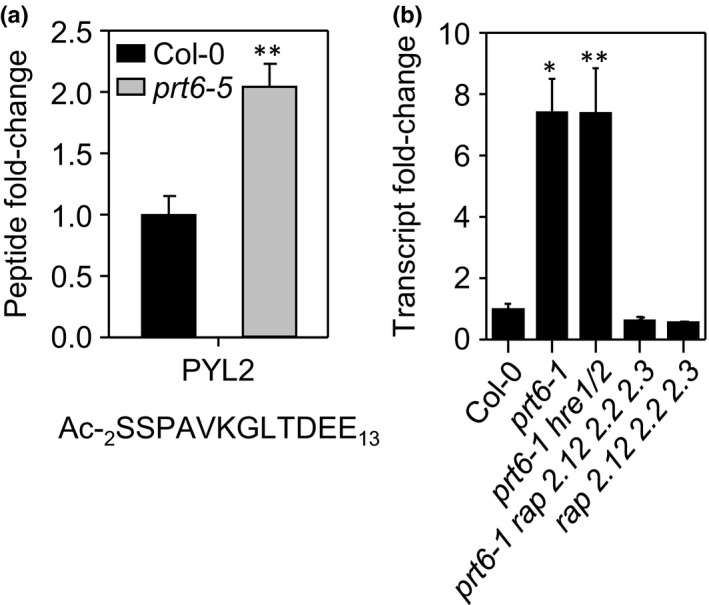
Abscisic acid (ABA) receptor component, PYL2, is regulated by the Arg/N‐end rule in an ERF‐dependent manner in etiolated Arabidopsis thaliana seedlings. (a) Relative abundance of Nt peptide corresponding to amino acids 2–13 of PYL2 in Col‐0 and prt6‐5. (b) PYL2 transcript abundance in single and combination N‐end rule and erfVII mutants, relative to Col‐0. Values are means ± SE (n = 3); *, P < 0.05; **, P < 0.01.
Increased abundance of seed storage proteins in prt6 seedlings requires RAP‐type ERFVII transcription factors
Cruciferins are highly abundant in embryo and endosperm of seeds but are mobilised following germination and are not normally present in 4‐d‐old seedlings. Multiple cruciferin‐derived Nt peptides were identified but only one had a primary destabilising residue. Several neo Nt peptides had secondary or tertiary destabilising residues; these did not bear the enzymatic modifications (arginylation, deamidation) required for degradation (Table S2), arguing against cruciferins being novel Arg/N‐end rule substrates and implying that cruciferin abundance is controlled directly or indirectly by stabilisation of an N‐end rule substrate in prt6 seedlings. Since RAP2.12, RAP2.2 and RAP2.3 control the transition from dormancy to germination and seedling response to ABA (Gibbs et al., 2014b; Papdi et al., 2015), we tested whether they also underpin the role of the Arg/N‐end rule in regulating storage reserve mobilisation. Proteins extracted from 4‐d‐old seedlings of mutants lacking PRT6 and different combinations of ERFVIIs were analysed by immunoblotting, using antisera towards the α‐subunit of cruciferin (Wan et al., 2007). Hypoxia marker proteins, ADH and pyruvate decarboxylase (PDC), both known to be regulated transcriptionally by the Arg/N‐end rule (Gibbs et al., 2011), were also tested and the oil body structural protein, Oleosin 1 (Ole1) was included because prt6 exhibits an oil body retention phenotype (Holman et al., 2009). Signals with all four antisera were increased in both the prt6‐1 and prt6‐5 null mutants and the prt6‐1 hre1 hre2 triple mutant relative to WT (Figs 4, S4). However, abundances of Ole1, ADH and PDC in quadruple prt6‐1 rap2.12 rap2.2 rap2.3 and sextuple prt6‐1 rap2.12 rap2.2 rap2.3 hre1 hre2 (hereafter, ‘prt6 erf VII ’) mutant seedlings were comparable to those in WT. Abundance of α‐cruciferin in the sextuple mutant was also similar to that in Col‐0, but was reproducibly lower in the quadruple mutant, suggesting possible feedback regulation by HRE1 and/or HRE2 (Figs 4, S4). The rap2.12 rap2.2 rap2.3 triple mutant had a surprisingly high level of α‐cruciferin but all proteins (including α‐cruciferin) were present at wild type amounts in plants lacking all five ERFVII transcription factors (erf VII). Taken together, removal of PRT6 function is associated with increased abundance of the storage protein cruciferin, which can be attributed to the action of PRT6 on different members of the ERFVII transcription factor family.
Figure 4.
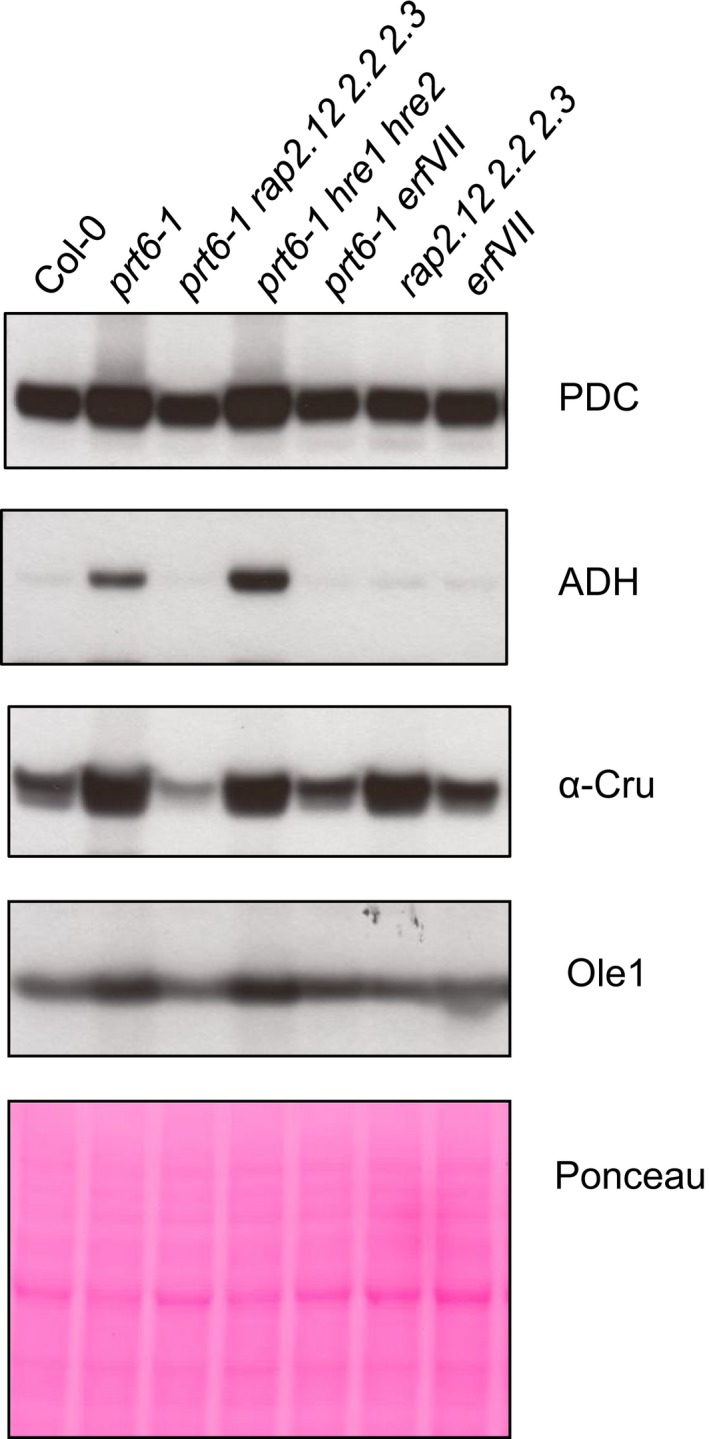
Increased abundance of proteins in prt6 seedlings requires RAP‐type ERFVII transcription factors. Proteins were extracted from 4‐d‐old etiolated seedlings of Arabidopsis thaliana N‐end rule and erfVII combination mutants and subjected to immunoblotting (25 µg per lane) with antisera towards pyruvate decarboxylase (PDC), alcohol dehydrogenase (ADH), cruciferin α subunit (α‐Cru) and oleosin1 (Ole1). Representative of three independent experiments.
Mobilisation of cruciferin is aberrant and delayed in Arg/N‐end rule mutants
Examination of the protein profiles of dry and imbibed seeds indicated that prt6‐5 and wild type seeds contain similar amounts of storage proteins (Fig. 5a), and therefore the difference in cruciferin abundance between the two genotypes is established following germination. The polypeptide pattern of prt6‐5 seeds was consistent with correct processing of seed storage proteins during maturation. In agreement with this, neo‐Nt peptides corresponding to the α‐ and β‐subunit N‐termini of Cru1/At12S4 and Cru3/At12S1 (as defined by Higashi et al., 2006) were identified in 4‐d‐old etiolated seedlings, as were peptides corresponding to the β‐subunit N‐termini of Cru2/At12S3 and At12S2 (Figs 5b, S5). During germination, the α‐subunits of cruciferins are degraded successively from the C‐terminus (Higashi et al., 2006; Li et al., 2007). However, numerous different neo‐Nt peptides derived from the α‐subunits of the four cruciferins were observed in the N‐terminome dataset, indicative of aberrant degradation (Figs 5b, S5). To determine whether the presence of seed storage proteins detected in prt6 seedlings was a result of delayed mobilisation, seed coat and endosperm were dissected from 4‐d‐old etiolated seedlings and analysed separately by immunoblotting. ADH, α‐cruciferin and Ole1 exhibited increased abundance in intact prt6‐5 seedlings. Whilst endosperms of germinated prt6‐5 seeds contained α‐cruciferin and Ole1, these proteins were not detected in wild type endosperm (Fig. 5c). ADH was strongly upregulated in whole prt6‐5 seedlings but absent from seed coat and endosperm in both mutant and wild type. These findings demonstrated that removal of PRT6 function inhibits seed reserve mobilisation.
Figure 5.
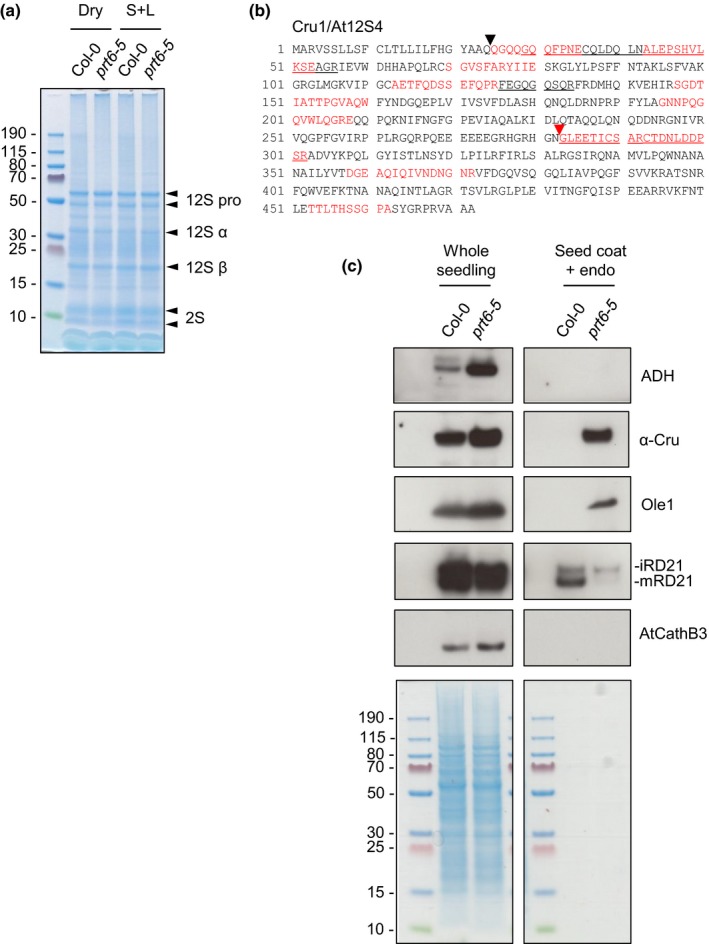
Mobilisation of 12S seed storage proteins is impaired in prt6 endosperm. (a) Quick Coomassie blue‐stained gel of proteins extracted from dry and germinating Arabidopsis thaliana seeds (protein extracted from five seeds was loaded in each lane). Dry, dry seed; S + L, seeds after 48 h of stratification on agar plus 6 h of light. The arrows indicate positions of the 12S cruciferin pro‐protein (pro), α and β subunits, and 2S albumins (napins). (b) Amino acid sequence of CRU1/At12S4; peptides identified by TMT™‐TAILS are indicated in red and/or underlined (some peptide sequences overlap). Black arrow indicates Nt of the α‐subunit generated by removal of residues 1–24; red arrow indicates Nt of the β‐subunit generated by proteolytic processing. (c) Immunoblots of 4‐d‐old seedlings, with endosperm and seed coat attached (loading equivalent to eight seedlings) and 15 dissected endosperms plus seed coat, probed with antisera towards alcohol dehydrogenase (ADH), cruciferin α subunit (α‐Cru), oleosin 1 (Ole1), RD21A (identifies both intermediate and mature forms, iRD21 and mRD21, respectively) and AtCathB3. The panel below shows the corresponding Quick Commassie blue‐stained gel; positions of molecular weight markers (kDa) are indicated to the left of the panel.
Multiple protein groups exhibit reduced abundance in the N‐terminome of prt6 seedlings
Peptides representing 101 protein groups were significantly reduced in abundance in prt6 seedlings relative to Col‐0 (Table 2). As analysis of gene ontogeny terms was uninformative, proteins were categorised manually. N‐termini of various proteins associated with the apoplast and cell wall were downregulated in prt6, including xylan‐modifying enzymes, β‐galactosidases and a prolyl 4‐hydroxylase that modifies extensin proteins. Numerous chloroplast proteins were also represented in the prt6 downregulated dataset, most notably proteins involved in Chl biosynthesis, consistent with the known role for the Arg/N‐end rule pathway in regulating tetrapyrrole synthesis as part of photomorphogenesis (Abbas et al., 2015). Other proteins with reduced abundance in the prt6 N‐terminome included a disparate group of enzymes involved in carbon metabolism and, interestingly, all enzymes of the S‐adenosyl methionine (SAM) cycle. Finally, N‐terminal peptides of seven proteases were decreased in abundance in prt6.
Table 2.
N‐terminal peptides with decreased abundance in seedlings of the Arabidopsis thaliana prt6 mutant
| AGI code | Description | Synonyms | Peptide | Start | Finish | Log2 fold change |
|---|---|---|---|---|---|---|
| Proteases and inhibitors | ||||||
| AT1G47128.1 | Granulin repeat cysteine protease family protein | RD21A | dELPESIDWR | 135 | 144 | −1.02 |
| AT3G14067.1 | Subtilase family protein | SASP | sAGNSGPNPE | 318 | 327 | −1.05 |
| aGNSGPNPE | 319 | 327 | −1.01 | |||
| AT3G45010.1 | Serine carboxypeptidase‐like 48 | Scp148 | gSGGSPSVQDFGH | 89 | 101 | −1.47 |
| AT4G34980.1 | Subtilisin‐like serine protease 2 | SLP2 | aVGSNEGDR | 444 | 452 | −1.20 |
| AT4G36195.1 | Serine carboxypeptidase S28 family protein | tAVTPESADR | 327 | 336 | −1.56 | |
| AT4G36880.1 | Cysteine proteinase1 | CP1, RDL1 | gkEVPETVDWR | 142 | 152 | −1.30 |
| AT4G39090.1 | Papain family cysteine protease | RD19A | aAGYAPAR | 311 | 318 | −1.01 |
| AT3G12490.2 | Cystatin B | CYSB, AtCYS6 | dVPANQNSGEVESLAR | 42 | 57 | −1.84 |
| C metabolism | ||||||
| AT1G32710.1 | Cytochrome c oxidase, subunit Vib family protein | sSAQMDPHDkMR | Ac‐2 | 13 | −1.03 | |
| AT1G53310.1 | Phosphoenolpyruvate carboxylase 1 | PPC1 | nLAEEVQIAYR | 106 | 116 | −1.29 |
| AT2G30970.1 | Aspartate aminotransferase 1 | ASP1 | sTILEDPE | 326 | 333 | −1.42 |
| AT3G23940.1 | Dihydroxy acid dehydratase | DHAD | mTVTGQTLAQNLE | 392 | 404 | −1.44 |
| AT3G55410.1 | 2‐Oxoglutarate dehydrogenase, E1 component | gEVSQQDIDR | 536 | 545 | −1.09 | |
| AT5G14740.1 | Carbonic anhydrase 2 | CA2 | sFPLDGNNSTDFIE | 233 | 246 | −1.51 |
| AT5G39410.1 | Saccharopine dehydrogenase | gFDSIPAE | 153 | 160 | −1.40 | |
| Lipid metabolism | ||||||
| AT1G30120.1 | Pyruvate dehydrogenase E1 beta | lSSQDVPTPYAGTLE | 373 | 387 | −1.01 | |
| AT1G55020.1 | Lipoxygenase 1 | LOX1 | tLEDVPGHGR | 129 | 138 | −1.01 |
| AT3G06650.1 | ATP‐citrate lyase B‐1 | ACLB‐1 | vSGAHNTIVTAR | 417 | 428 | −1.55 |
| AT3G16170.1 | AMP‐dependent synthetase and ligase family protein | AAE13 | nQFQDDSFE | 155 | 163 | −1.75 |
| AT5G43590.1 | Acyl transferase/acyl hydrolase/lysophospholipase superfamily protein | sLDGGGVR | 13 | 20 | −1.23 | |
| AT5G46290.3 | 3‐Ketoacyl‐acyl carrier protein synthase I | KAS1 | dVDAYYE | 77 | 83 | −1.04 |
| One‐carbon metabolism and ethylene | ||||||
| AT1G05010.1 | Ethylene‐forming enzyme | ACCO, ACO4 | mESFPIINLEkLNGEER | Ac‐1 | 17 | −1.50 |
| AT3G09820.1 | Adenosine kinase 1 | AK1 | lPYmDYIFGNE | 213 | 223 | −1.09 |
| AT3G59970.3 | Methylenetetrahydrofolate reductase 1 | MTHFR | aLDLVNHIR | 131 | 139 | −1.59 |
| AT4G01850.1 | S‐Adenosylmethionine synthetase 2 | SAM‐2 | gTGLIPDkE | 324 | 332 | −1.06 |
| AT4G13940.1 | S‐Adenosyl‐l‐homocysteine hydrolase | HOG1 | sFTNQVIAQLE | 412 | 422 | −1.75 |
| AT5G14780.1 | Formate dehydrogenase | FDH | vENALGIR | 59 | 66 | −1.33 |
| AT5G17920.1 | Methioine synthase | ATMS1 | lQAFTGAYAE | 219 | 228 | −1.49 |
| aGIGPGVYDIHSPR | 691 | 704 | −1.09 | |||
| Photosynthesis and Chl biosynthesis | ||||||
| AT4G27440.1 | Protochlorophyllide oxidoreductase B | PORB | akEPTYSAE | 181 | 189 | −1.78 |
| aLFPPFQkY | 325 | 333 | −1.72 | |||
| iASTGLFRE | 310 | 318 | −1.40 | |||
| nAAVYFPTAkEPTYSAE | 173 | 189 | −1.30 | |||
| iASTGLFR | 310 | 317 | −1.16 | |||
| AT5G08280.1 | Hydroxymethylbilane synthase | HEMC, RUGOSA | sLNHEETR | 292 | 299 | −1.24 |
| AT5G54190.1 | Protochlorophyllide oxidoreductase A | PORA | aIATSTPSVTkSSLDR | Ac‐71 | 86 | −1.19 |
| nAAVYQPTANQPTFTAE | 177 | 193 | −1.09 | |||
| AT5G64040.2 | Photosystem I reaction centre subunit PSI‐N | PSAN | gVIDEYLER | 87 | 95 | −1.45 |
| ATCG00120.1 | ATP synthase subunit alpha | ATPA | qSQSAPLTVEE | 423 | 433 | −1.46 |
| ATCG00480.1 | ATP synthase subunit beta | PB | iVGEEHYETAQQVkQ | 379 | 393 | −1.04 |
| ATCG00490.1 | Ribulose‐bisphosphate carboxylases | RBCL | dDYVEkDR | 351 | 358 | −1.50 |
| Other plastid | ||||||
| AT1G12230.2 | Aldolase superfamily protein | nEIDVPHDR | 211 | 219 | −1.30 | |
| AT1G34000.1 | One‐helix protein 2 | OHP2 | sQTEGPLR | 44 | 51 | −1.47 |
| AT2G21530.1 | SMAD/FHA domain‐containing protein | lDENQSPTSGGER | 74 | 86 | −1.09 | |
| AT2G23670.1 | Homologue of Synechocystis YCF37 | YCF37 | eNIPLFGIR | 72 | 80 | −1.11 |
| AT2G44920.2 | Tetratricopeptide repeat (TPR)‐like superfamily protein | aSFFDADLTGADLSEADLR | 131 | 149 | −2.47 | |
| AT3G56910.1 | Plastid‐specific 50S ribosomal protein 5 | PSRP5 | kAAASGVDGAEPE | Ac‐64 | 76 | −1.89 |
| aAASGVDGAEPE | 65 | 76 | −1.82 | |||
| sGVDGAEPE | 68 | 76 | −1.73 | |||
| AT4G32915.1 | Glu‐tRNA Gln amidotransferase, C subunit | sSDSDSSVLQPPDVAR | 53 | 68 | −1.83 | |
| AT4G34290.1 | SWIB/MDM2 domain superfamily protein | aASSDPTTTTkTR | Ac‐51 | 63 | −1.17 | |
| AT5G02710.1 | Unknown protein | sTSGFSGGTTkE | Ac‐43 | 54 | −1.09 | |
| Apoplast/cell wall | ||||||
| AT1G68560.1 | Alpha‐xylosidase 1 | XYL1 | dEEENkSVMVEVR | 884 | 896 | −1.08 |
| AT2G05380.1 | Glycine‐rich protein 3 short isoform | GRP3S | gGGFGDNGGGR | 41 | 51 | −1.03 |
| AT2G17720.1 | 2‐Oxoglutarate (2OG) and Fe(II)‐dependent oxygenase superfamily protein | P4H5 | dVDDGGETVFPAAR | 207 | 220 | −2.10 |
| AT2G39770.1 | Glucose‐1‐phosphate adenylyltransferase family protein | VTC1, CYT1 | sTVGQWAR | 313 | 320 | −1.48 |
| AT3G13790.1 | Glycosyl hydrolases family 32 protein | ATBFRUCT1 | sPSVNQPYR | 44 | 52 | −1.78 |
| AT3G44990.1 | Xyloglucan endo‐transglycosylase‐related 8 | XTH31 | fFVDDVPIR | 162 | 170 | −1.60 |
| AT4G14130.1 | Xyloglucan endotransglucosylase/hydrolase 15 | XTH15 | yLSSQGATHDE | 92 | 102 | −1.48 |
| AT4G32460.1 | Protein of unknown function, DUF642 | gPLIDGVAmR | 172 | 181 | −1.09 | |
| AT5G20630.1 | Germin 3 | GER3 | kNPDQVTE | 42 | 49 | −1.45 |
| AT5G20710.1 | Beta‐galactosidase 7 | BGAL7 | tIVSHDER | 26 | 33 | −1.02 |
| AT5G44380.1 | FAD‐binding Berberine family protein | sASIQDQFINcVkR | 31 | 44 | −1.26 | |
| AT5G46960.1 | Plant invertase/pectin methylesterase inhibitor superfamily protein | vNSLTQDPQSkAATTLE | 44 | 60 | −2.13 | |
| AT5G56870.1 | Beta‐galactosidase 4 | BGAL4 | dITIGSGE | 475 | 482 | −1.04 |
| AT5G64100.1 | Peroxidase superfamily protein | sIPANAPGILR | 63 | 73 | −1.04 | |
| Cytoskeleton | ||||||
| AT1G71440.1 | Tubulin folding cofactor E/Pfifferling (PFI) | PFI | mkAESSNESFIIGQR | Ac‐1 | 15 | −1.14 |
| AT3G60830.1 | Actin‐related protein 7 | ARP7 | nVSGFYASE | 116 | 124 | −1.15 |
| AT5G55230.2 | Microtubule‐associated proteins 65‐1 | MAP65‐1 | aVTDTESPHLGE | 2 | 13 | −1.42 |
| Vesicle traffic and organelle biogenesis | ||||||
| AT1G35720.1 | Annexin 1 | ANNAT1 | dSVPAPSDDAE | 8 | 18 | −1.07 |
| AT1G71820.2 | SEC6 | SEC6 | mMVEDLGVEAkEAAVR | Ac1 | 16 | −1.23 |
| AT4G11380.2 | Adaptin family protein | aLFGEDGR | 802 | 809 | −1.16 | |
| Chaperones | ||||||
| AT1G24510.1 | TCP‐1/cpn60 chaperonin family protein | nDVGTNDmR | 490 | 498 | −1.81 | |
| AT2G33210.1 | Heat shock protein 60‐2 | HSP60‐2 | sVSSLLTTTE | 541 | 550 | −1.09 |
| AT3G12050.1 | Aha1 domain‐containing protein | gLVDMPYISDE | 108 | 118 | −1.20 | |
| AT3G44110.1 | DNAJ homologue 3 | J3 | eETTLHDVNIEDEmR | 375 | 389 | −1.22 |
| AT4G24190.1 | Chaperone protein htpG family protein | SHD | iSPDAVADEE | 772 | 781 | −1.33 |
| tDSDVVHR | 55 | 62 | −1.02 | |||
| AT5G53400.1 | HSP20‐like chaperones superfamily protein | BOB1 | aSSAEPIE | 111 | 118 | −1.05 |
| AT5G56030.2 | Heat shock protein 81‐2 | HSP81‐2 | gLSIDDDDAVE | 695 | 705 | −2.63 |
| iDDDDAVE | 698 | 705 | −2.26 | |||
| lSIDDDDAVE | 696 | 705 | −1.95 | |||
| Translation | ||||||
| AT2G20450.1 | Ribosomal protein L14 | sLTDIVIDINR | 54 | 64 | −1.17 | |
| dVVDQNR | 29 | 35 | −1.14 | |||
| AT2G27710.1 | 60S acidic ribosomal protein family | vASATSGGGGGGGASAAE | 75 | 92 | −1.02 | |
| AT5G47880.1 | Eukaryotic release factor 1‐1 | gLVLYTGTIVNE | 91 | 102 | −1.67 | |
| Nucleic acid binding | ||||||
| AT1G22300.1 | General regulatory factor 10 | GCRF10 | dLNEEGDER | 235 | 243 | −2.80 |
| gLAPTHPVR | 163 | 171 | −2.09 | |||
| AT2G14285.1 | Small nuclear ribonucleoprotein family protein | nVLYVRGVPE | 42 | 51 | −2.78 | |
| AT2G35410.1 | RNA‐binding (RRM/RBD/RNP motifs) family protein | aADFNPVSAR | 216 | 225 | −1.16 | |
| AT3G59980.1 | Nucleic acid‐binding, OB‐fold‐like protein | aAPDAGTTVSADE | 76 | 88 | −1.78 | |
| AT5G47210.1 | Hyaluronan/mRNA binding family | dDAEDPSQLAVALSQkVE | 12 | 29 | −1.58 | |
| Redox/stress | ||||||
| AT3G01520.1 | Adenine nucleotide alpha hydrolases‐like superfamily protein | vVDEDGFDDVDSIYASPEDFR | 55 | 75 | −1.29 | |
| AT4G11600.1 | Glutathione peroxidase 6 | GPX6 | vASQcGLTNSNYTE | 101 | 114 | −1.42 |
| AT5G54430.1 | Adenine nucleotide alpha hydrolases‐like superfamily protein | PHOS32 | tQIEDPNAQPQPSQE | 101 | 115 | −1.34 |
| Other | ||||||
| AT1G77540.1 | Acyl‐CoA N‐acyltransferases (NAT) superfamily protein | tNTAATTEAkMATEPPkIVW | Ac‐2 | 21 | −2.06 | |
| AT2G01530.1 | MLP‐like protein 329 | MLP329 | aTSGTYVTEVPLkGSADkH | Ac‐2 | 20 | −1.10 |
| AT2G26210.1 | Ankyrin repeat family protein | aGLDTPQR | 90 | 97 | −1.16 | |
| AT2G38710.1 | AMMECR1 family | tVSVLTDYE | 96 | 104 | −1.24 | |
| AT2G39310.1 | Jacalin‐related lectin 22 | JAL22 | gGEGGQEWDDDVYEGVR | 12 | 28 | −1.88 |
| AT2G44060.1 | Late embryogenesis abundant protein, group 2 | kEDDDDDDEE | 316 | 325 | −1.07 | |
| AT3G02090.2 | Insulinase (Peptidase family M16) protein | MPPBETA | gTSPIAEDIGR | 456 | 466 | −1.41 |
| AT3G43810.1 | Calmodulin 7 | CAM7 | aDQLTDDQISEFkEAF | Ac‐2 | 17 | −1.61 |
| AT4G23400.1 | Plasma membrane intrinsic protein 1;5 | PIP1;5 | mEGkEEDVNVGAN | Ac‐1 | 13 | −1.03 |
| AT4G24520.1 | P450 reductase 1 | ATR1 | vATYGDGEPTDNAAR | 145 | 159 | −1.56 |
| AT5G11950.1 | Putative lysine decarboxylase family protein | LOG8 | dTGVEEGFIkPGAR | 155 | 168 | −1.66 |
| AT5G16280.1 | Tetratricopeptide repeat (TPR)‐like superfamily protein | aLTGDDIVE | 258 | 266 | −1.38 | |
| AT5G44020.1 | HAD superfamily, subfamily IIIB acid phosphatase | sSQYEDDVER | 88 | 97 | −1.26 | |
| Unknown | ||||||
| AT2G23370.1 | Unknown protein | sLEGTWDESLER | 308 | 319 | −1.04 | |
| AT2G32240.1 | FUNCTIONS IN: molecular function unknown; INVOLVED IN: response to cadmium ion | dIDLSFSSPTkR | 1267 | 1278 | −1.62 | |
| AT2G38450.1 | CONTAINS InterPro DOMAIN/s: Sel1‐like (InterPro:IPR006597) | mDSSDkDSSSTTTTSETTR | Ac‐34 | 52 | −1.87 | |
| AT3G03150.1 | Unknown protein | gHSSAYDkNVE | 40 | 50 | −1.09 | |
| AT5G40450.1 | Unknown protein | eSSDEALVSm | 1897 | 1906 | −1.14 | |
| AT5G67490.1 | Unknown protein | sSGTPPPPQAPSPNQDLNR | 29 | 47 | −1.29 | |
Peptides listed are more than two‐fold decreased in abundance in prt6, compared to Col‐0, at P < 0.05. The start and finish amino acid positions are defined with respect to TAIR10 gene models. Residues with modifications Nt‐TMT, side‐chain Lys TMT or other (e.g. oxidised Met) are indicated in lower case; full details are given in Supporting Information Table S3. Ac, N‐terminal acetylation.
Proteases are differentially regulated in prt6
The reduced abundance of protease N‐termini in prt6 was of interest in the context of delayed seed storage protein mobilisation. To gain insight into their potential regulation by the Arg/N‐end rule, transcript levels of selected proteases were quantified by RT‐qPCR. RD21A and SLP2 transcripts were less abundant in prt6‐1 seedlings than Col‐0; RD19A was unchanged and CP1 was more abundant in prt6‐1, indicative of distinct modes of control (Fig. 6). As it is generally not possible to predict protease activity from transcript or even protein abundance (van der Hoorn, 2008), we took advantage of the availability of fluorescent probes for ABPP of cysteine proteases to examine a potential role of the Arg/N‐end rule in protease regulation (Richau et al., 2012; Lu et al., 2015). Specificity of the probes has been established previously by analysis of Arabidopsis protease knock‐out lines and transient expression of proteases in Nicotiana benthamiana (Gu et al., 2012; Lu et al., 2015); labelling specificity was confirmed here by pre‐incubation with the inhibitor, E64 (Fig. S6). Figure 7 shows ABPP results for FY01 and JODGA1 probes; images of the whole gels are shown in Fig. S7. Dependent on the labelling conditions, FY01 detects aleurain‐like proteases (ALPs) and RD21A (Lu et al., 2015). In extracts of 4‐d‐old etiolated seedlings, FY01 labelled bands of 30 and 34 kDa, probably corresponding to ALPs, AALP and ALP2, respectively, and a band of c. 40 kDa, corresponding to iRD21A, the active intermediate form of RD21A (Fig. 7a; note that labelling alters the apparent relative molecular mass of the proteases). Intensity of the RD21A signal was reduced in prt6‐1 and the prt6‐1 hre1 hre2 triple mutant, but not significantly different from WT in the prt6‐1 rap2.12 rap2.2 rap2.3 quadruple mutant and the prt6 erf VII sextuple mutant, indicating that repression of RD21A activity is dependent on RAP transcription factors (Fig. 7b). Consistent with the implication of RAPs, RD21A was also downregulated in ate1 ate2, which lacks arginyl transferase function (Fig. S6). MV201, which also labels RD21A and other papain‐like cysteine proteases (PLCPs) (Richau et al., 2012), gave a similar result (Figs S6c, S7).
Figure 6.
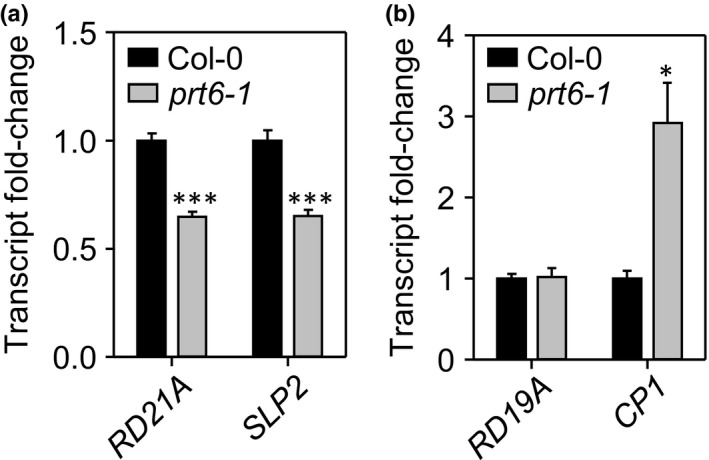
Quantification of protease transcripts in Col‐0 and prt6 seedlings. RT‐qPCR analysis of (a) RD21A and SLP2, and (b) RD19A and CP1 in 4‐d‐old etiolated Arabidopsis thaliana seedlings of Col‐0 and prt6‐1. Values are means ± SE (n = 4); *, P < 0.05; ***, P < 0.001.
Figure 7.
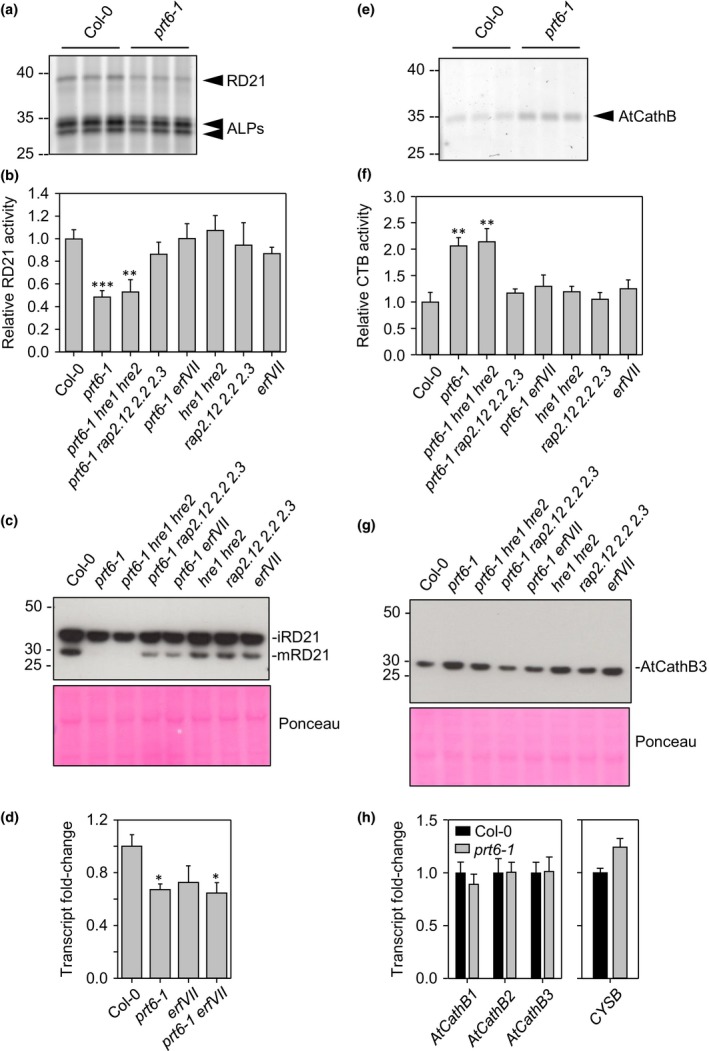
Differential regulation of proteases by the Arg/N‐end rule pathway. Activities, protein and RT‐qPCR analysis of selected Arabidopsis thaliana proteases. (a, e) Activity‐based protein profiling of 4‐d‐old etiolated seedlings of N‐end rule and erfVII combination mutants. (a) Probe FY01 labels RD21A and aleurin‐like proteases (ALPs); (e) probe JOGDA1 labels cathepsin B (AtCathB). Each lane represents a biological replicate; positions of molecular weight markers (kDa) are shown to the left of each panel. (b, f) Quantification of (b) RD21A signal and (f) AtCathB signal; values are means ± SE (n = 3). (c, g) Immunoblots probed with antisera raised to (c) RD21A and (g) AtCathB3; protein extracts from equal numbers of 4‐d‐old etiolated seedlings were loaded in each lane; positions of molecular weight markers (kDa) are shown to the left of each panel. (d, h) RT‐qPCR analysis of (d) RD21A, and (h) AtCathB1‐3 and CYSB in etiolated seedlings of Col‐0 and prt6‐1. Values are means ± SE (n = 4); *, P < 0.05; **, P < 0.01; ***, P < 0.001.
RD21A undergoes several processing steps: the signal peptide is cleaved co‐translationally and removal of the inhibitory pro‐domain generates an active, intermediate form (iRD21). The protein is further matured by removal of the C‐terminal granulin domain to produce two additional activated forms (mRD21) (Yamada et al., 2001; Gu et al., 2012). Two overlapping Nt peptides (DELPESIDWR; ELPESIDWR) corresponding to the N‐terminus of the activated form (iRD21) and indicative of ‘ragged’ processing were of significantly lower abundance in prt6‐5 (two‐ and 1.75‐fold lower than Col‐0, respectively; P < 0.05). RD21A protein abundance and processing were investigated further using an antiserum raised to the N‐terminus of the active, processed form (Kaschani et al., 2009). Bands corresponding to the intermediate form (iRD21) and the mature forms (mRD21) were detected in Col‐0, but the iRD21 band was less intense in prt6‐1 and mRD21 was undetectable (Figs 7c, S8). Combining hre1 and hre2 alleles with prt6‐1 did not recapitulate the WT phenotype, indicating that HRE1 and HRE2 do not play a role in downregulation of RD21 in the prt6 background. By contrast, removal of ERFVII function in either the sextuple prt6 erf VII mutant or the quadruple prt6 rap2.12 rap 2.2 rap 2.3 increased the abundance of iRD21 and mRD21 (Figs 7c, S8). Given the RAP‐dependence of RD21A activity and protein, we quantified transcripts in different genetic backgrounds. RD21A transcript levels were reduced not only in prt6‐1 seedlings (as in Fig. 6) but also in erf VII and prt6 erf VII mutants (Fig. 7d).
JODGA1 labelled a band of c. 34 kDa, which corresponds to the cathepsin B (AtCathB)‐specific signal detected with this probe (Lu et al., 2015). Intriguingly, this signal was significantly increased in prt6‐1, prt6‐5 and ate1 ate2, indicating an enhancement of cathepsin activity (Figs 7e, S6d). Probing extracts from combination mutants impaired in function of different ERFVII transcription factors demonstrated that this effect was dependent on RAP transcription factors but independent of HRE1 and HRE2 (Figs 7f, S7). Although no cathepsin‐derived peptides were identified in the N‐terminome dataset, immunoblotting with a specific antiserum confirmed that AtCathB3 exhibited increased abundance in prt6‐1 and the prt6‐1 hre1 hre2 triple mutant (Fig. 7g). Removing RAP function in the quadruple prt6‐1 rap2.12 rap 2.2 rap 2.3 or the sextuple prt6 erf VII mutants restored AtCathB3 protein to WT levels but the interpretation of this result was complicated by higher levels of AtCathB3 in the erf VII pentuple and lower‐order mutants (which are wild type for PRT6). Despite the apparent RAP‐dependence of increased AtCathB3 protein and activity in prt6 seedlings, cathepsin B transcripts were not increased in the mutant, pointing to post‐transcriptional or post‐translational regulation (Fig. 7h). Therefore, we tested whether altered expression of the seed‐expressed cystatin, AtCYS6/CYSB (At3g12490; Hwang et al., 2009), might contribute to post‐translational regulation of AtCathB3 in prt6‐1. Cystatins are candidate regulators of cathepsin activity and a CYSB‐derived peptide was present in the prt6‐down dataset (Table 2), but other peptides from this protein were not changed in abundance in the mutant (Table S2) and CYSB transcripts were not reduced in prt6‐1 relative to WT (Fig. 7h). Taken together, the ABPP, immunoblotting and transcript data indicate regulation of proteases by the Arg/N‐end rule at both transcriptional and post‐transcriptional levels, which is largely, but not completely, RAP‐dependent.
Discussion
Impact of the Arg/N‐end rule on the proteome
In recent years, the N‐end rule pathway of targeted protein degradation has emerged as an important regulator of diverse processes in plants (Gibbs et al., 2014b, 2015, 2016). Whilst analysis of mutants impaired in different pathway components has provided insight into the physiological functions of the N‐end rule, knowledge regarding the identity of substrates and the impact of this pathway on the proteome is limited. Affinity purification and quantitative proteomics provide unbiased strategies to probe the N‐end rule in plants. Previously, we used TMT labelling and tandem MS to identify proteins with altered abundance in roots of prt6 and ate1 ate2 mutants, and employed dimethyl‐TAILS to isolate Nt peptides, which achieved high enrichment of protein N‐termini but did not allow reliable quantification (Zhang et al., 2015). Here, we combined the TAILS technique with TMT labelling for enrichment of Nt peptides in etiolated prt6 seedlings. The TMT‐TAILS protocol enabled robust quantification, with quantitative data obtained for 3937 peptides (Table S2; Fig. S3). Moreover, the use of two proteases increased coverage and increased the proportion of neo‐Nt peptides identified (Fig. 2b). This dataset provides a useful resource for proteogenomics: although not a focus of our study, the data can be used, for example, to identify proteolytic processing events, such as those involved in protein in/activation and signal peptide removal and to support analysis of alternative translation start sites (Hartmann & Armengaud, 2014).
Regarding the Arg/N‐end rule, neo‐Nt peptides with destabilising residues were under‐represented in the complete dataset (Fig. 2g), but were not markedly upregulated in prt6 compared to wild type (Table S3). Consistent with our results from global TMT labelling (Zhang et al., 2015), relatively few proteins exhibited altered abundance in prt6, implying that the PRT6 does not function in bulk protein turnover under normal conditions, but probably plays a role in the controlled degradation of a few regulatory proteins. In agreement with this, it is clear from global protein lifetime measurements and several N‐terminome studies that by no means all proteins with destabilising N‐termini are degraded via the N‐end rule in wild type plants (Bienvenut et al., 2012; Tsiatsiani et al., 2013; Linster et al., 2015; Venne et al., 2015; Zhang et al., 2015; Li et al., 2017). Degradation depends on the structural and subcellular context of the destabilising residue (amongst other factors) as part of a functional N‐degron (Varshavsky, 2011).
Upregulation of proteins in prt6 seedlings requires ERFVII transcription factors
Of the 45 protein groups upregulated in prt6‐5 relative to Col‐0, there were no obvious candidate Arg/N‐end rule substrates nor were the known ERFVII substrates identified, suggesting that further enrichment is required to detect low abundance, regulatory proteins. The abundance of cruciferin was dependent on activity of RAP‐type ERFVIIs, indicating that cruciferin is controlled by the known Arg/N‐end rule substrates, rather than being a substrate itself (Fig. 4). Immunoblotting confirmed the ERFVII‐dependent up‐regulation of ADH and PDC, in agreement with their known roles in the low oxygen response (Licausi et al., 2013; Gibbs et al., 2015), and demonstrated that Oleosin1 is regulated by RAP‐type ERFVIIs (Fig. 4). Finally, although not identified previously as prt6‐regulated in published transcriptome datasets, we demonstrated a RAP‐dependent increase of PYL2 transcripts in prt6 seedlings which was reflected in protein abundance; upregulation of this ABA receptor component may contribute to the ABA hypersensitivity of prt6 (Holman et al., 2009).
Protein groups associated with diverse functions are downregulated in prt6
Nt peptides representing a diverse collection of proteins were downregulated in prt6 seedlings (Table 2). Whilst these data require confirmation at the protein level with immunoblotting or quantitative shotgun proteomics, they nevertheless support the notion that stabilisation of Arg/N‐end rule substrates can impact negatively on abundance of other proteins, either directly or indirectly. Numerous transcripts are downregulated in published microarray data from different tissues of Arg/N‐end rule mutants (Choy et al., 2008; Gibbs et al., 2011; de Marchi et al., 2016). This suggests that transcriptional repression by stabilised ERFVIIs or unknown transcription factor substrates probably underpins the down‐regulation of protein abundance in prt6, although other mechanisms are also possible. N‐termini of several plastid proteins, including enzymes of Chl biosynthesis, were downregulated in prt6 seedlings, consistent with the known role for the Arg/N‐end rule pathway in co‐ordination of photomorphogenesis and oxygen sensing (Abbas et al., 2015). Transcripts of nuclear photosynthesis‐related genes are markedly lower in dark‐grown seedlings of the prt6 allele, ged1, than in wild type (Choy et al., 2008) and ERF‐dependent repression of protochlorophyllide reductase A, B and C and other Chl biosynthetic genes may serve to prevent accumulation of toxic metabolites under low oxygen, which is needed for several steps of Chl biosynthesis (Abbas et al., 2015). Our data demonstrate that these transcriptional changes are reflected at the protein level. Enzymes associated with SAM synthesis and recycling were also downregulated in the prt6 N‐terminome. These included the three SAM cycle enzymes, Met synthase, SAM synthase and S‐adenosyl‐L‐homocysteine hydrolase (Table 2). Also downregulated were methylenetetrahydrofolate reductase 1 which can serve as a methyl donor for Met synthesis, and adenosine kinase, involved in salvage of adenylates and methyl recycling. SAM is an abundant cofactor required for ethylene and polyamine biosynthesis and is an important methyl donor for numerous methyltransferase reactions (Sauter et al., 2013), so modulation of the SAM cycle by the Arg/N‐end rule could potentially have several important metabolic and developmental consequences. Whilst the ethylene biosynthetic protein ACC oxidase4 was downregulated in the prt6 N‐terminome, ACC oxidase1 was significantly upregulated (Table 1) and is a core hypoxia‐responsive gene constitutively expressed in prt6 alleles (Mustroph et al., 2009; Gibbs et al., 2011). In future studies, it will be interesting to measure SAM, polyamines and ethylene in prt6 seedlings and to determine whether ERFVII transcription factors are involved in their regulation.
The Arg/N‐end rule differentially regulates protease activities
The N‐termini of several proteases were downregulated in the prt6 N‐terminome, two of which, RD21A and SLP2, were also downregulated at the transcript level (Fig. 6). However, as proteases are subject to post‐translational regulation in planta to avoid deleterious consequences of uncontrolled proteolysis, transcript and protein levels often do not predict activity (van der Hoorn, 2008). Therefore, we analysed protease activity in prt6 seedlings using well‐characterised, subfamily‐specific cysteine protease activity probes (Richau et al., 2012; Lu et al., 2015). Two ABPP probes, FY01 and MV201, provided evidence for reduced RD21A activity in prt6‐1, prt6‐5 and ate1 ate2 seedlings (Figs 7, S6). RD21A is responsible for the dominant PLCP activity in Arabidopsis extracts (Gu et al., 2012) and has been associated with functions in immunity, herbivore defence, senescence, cell death and response to stresses (Shindo et al., 2012; Lampl et al., 2013; Rustgi et al., 2017; and references therein). Following activation via a proteolytic cascade, RD21A activity is tightly regulated at different developmental stages by a Kunitz‐type protease inhibitor, water‐soluble Chl binding protein (reversible inhibition) and by AtSerpin1 (irreversible inhibition) (Lampl et al., 2013; Boex‐Fontvieille et al., 2015; Rustgi et al., 2017). RD21A protein is also subject to ubiquitin‐dependent degradation mediated by the E3 ligase AtAIRP3/LOG2 (Kim & Kim, 2013). In this study, we provide evidence for another layer of regulation via the Arg/N‐end rule pathway. RD21A activity (as quantified by ABPP) was reduced in prt6 alleles and correlated well with protein levels assessed by MS and immunoblotting. Whilst the reduction in activity and protein was largely dependent on RAP‐type ERFVIIs, surprisingly, the transcriptional repression/downregulation of RD21A in prt6 could not be clearly attributed to ERFVII function (Fig. 7).
ABPP also revealed increased Cathepsin B activity in prt6 and ate1 ate2 (Figs 7, S6d). Arabidopsis has three Cathepsin B genes, AtCathB1 (At1g02300), AtCathB2 (At1g02305) and AtCathB3 (At4g01610), which are ubiquitously expressed (Iglesias‐Fernández et al., 2014) and functionally redundant in the hypersensitive response and programmed cell death (McLellan et al., 2009; Ge et al., 2016). However, AtCathB3 exhibits the highest level of transcript, is very strongly induced in germination and accounts for the strong ABPP signal in young seedlings (Iglesias‐Fernández et al., 2014; Lu et al., 2015). Although AtCathB3 protein and activity were RAP‐dependent, surprisingly, transcript abundance was unaltered in prt6‐1 seedlings (Fig. 7h); moreover, none of the Arabidopsis cathepsins exhibits significant differential regulation in published microarray studies of Arg/N‐end rule mutants (Choy et al., 2008; Gibbs et al., 2011). Taken together, these lines of evidence point to post‐translational regulation of AtCathB3 by the Arg/N‐end rule. Although cystatins are known regulators of Cathepsin B activity in plants, we did not find evidence for altered expression of a major seed cystatin, CYSB in prt6 seedlings (Fig. 7h), so the RAP‐type ERFVIIs may influence AtCathB3 protein and activity via more than one cystatin or a different mechanism.
RD21A and Cathepsin B represent only a subset of the large number of proteases encoded by the Arabidopsis genome (van der Hoorn, 2008), but we have discovered that they are regulated in an opposing and complex manner by the Arg/N‐end rule. Aside from roles in seed protein mobilisation, regulation of protease activity may be important to prevent potentially deleterious accumulation of neo‐peptides in prt6 mutants. An interesting challenge for future studies will be to determine to what extent the N‐end rule pathway regulates other proteases and to investigate the potential homeostatic interplay between different protease activities.
The Arg/N‐end rule regulates seed storage protein mobilisation through RAP‐type ERFVII transcription factors
Identification of PRT6 in a genetic screen for seeds with reduced germination potential provided the first link between the Arg/N‐end rule and germination completion and established a role for this pathway in storage oil mobilisation (Holman et al., 2009). Subsequently, we demonstrated that RAP‐type ERFVII transcription factors underpin the germination phenotype of prt6 seeds (Gibbs et al., 2014a). Our quantitative proteomics data set show that the PRT6 branch of the Arg/N‐end rule pathway also plays a role in regulating breakdown of endosperm storage protein reserves (Fig. 4). In wild type plants, cruciferins are laid down during seed development and mobilised upon germination, but can also be neosynthesised following germination (Galland et al., 2014). Dry prt6 and Col‐0 seeds contained similar amounts of seed storage proteins; in agreement with this, developing seeds of the prt6 allele, ged1 (Riber et al., 2015), contain wild‐type amounts of transcripts encoding seed proteins including At12S4/CRU1 (Choy et al., 2008). Cruciferin is almost completely depleted in 4‐d‐old wild type seedlings grown in culture (Heath et al., 1986). The presence of cruciferin in the endosperm of prt6 seedlings at 4 d post‐germination is suggestive of delayed mobilisation (Fig. 4). Multiple neo‐Nt peptides derived from the cruciferin α‐subunits were identified as upregulated in prt6 seedlings, consistent with aberrant degradation and suggesting that different enzymes degrade SSPs when the protease complement of seeds is disrupted. In support of this notion, genetic removal of vacuolar processing enzymes has been shown to result in compensatory, aberrant SSP processing by alternative proteases (Gruis et al., 2002).
Although the reduced activity of RD21A correlates with delayed α‐cruciferin degradation in the endosperm (Fig. 5c), a causative link has not been established. Surprisingly, proteases responsible for the mobilisation of Arabidopsis SSPs have remained poorly defined until recently: whilst the activity of several proteases parallels the disappearance of cruciferins post‐imbibition, storage protein profiles were unaffected in single and multiple protease mutants (inclusive of rd21a alleles), indicative of substantial redundancy (Lu et al., 2015). Cathepsins have been associated with storage protein mobilisation in Arabidopsis (Iglesias‐Fernández et al., 2014), yet AtCathB3 activity and protein were increased in prt6 seedlings, which would have been predicted to promote, not retard, SSP mobilisation. Moreover, AtCathB3 is absent from endosperm, as judged by immunoblotting (Fig. 5c) and fluorescence in situ hybridisation (Iglesias‐Fernández et al., 2014), and therefore AtCathB3 regulation by PRT6 does not affect mobilisation of SSPs in the endosperm. During the course of this study, CP1/RDL1 was shown to play a quantitatively important role in endosperm cruciferin degradation (Piskurewicz et al., 2016). The decay of cruciferin levels was delayed by 12 h in endosperm of cp1 mutants, independently of germination, but embryo cruciferin was degraded normally. The SSP mobilisation phenotype of prt6 seedlings, in which endosperm α‐cruciferin degradation is specifically retarded, is consistent with the potential regulation of CP1 by the Arg/N‐end rule. Neither an activity probe nor a specific antibody is available for CP1, so we were unable to test this hypothesis but as CP1 was represented in our prt6 downregulated dataset (Table 2) it is plausible that it contributes to the delayed SSP mobilisation phenotype of prt6.
As we found previously (Zhang et al., 2015) many of the proteins whose abundance is influenced by the Arg/N‐end rule in this study are not bona fide substrates of the pathway, but are regulated (directly or indirectly) by the ERFVII transcription factors. ERFVIIs underpin many of the known prt6 phenotypes and are emerging as the dominant substrates of PRT6 under conditions tested to date. Their role in storage reserve mobilisation during skotomorphogenesis is interesting in the context of hypoxia signalling: RAP‐type ERFVIIs play an important role in monitoring the gaseous environment during germination, which has an adaptive value in waterlogged soils and prevents precocious photomorphogenesis (Abbas et al., 2015). Our proteomic, biochemical and genetic data expand and complement this view, suggesting that controlled degradation of ERFVII transcription factors by the PRT6 branch of the Arg/N‐end rule pathway serves to co‐ordinate germination and seedling establishment with environmental factors by optimising storage reserve mobilisation.
Author contributions
H.Z., K.S.L. and F.L.T. designed research, H.Z., M.J.D. and L.G. performed research, D.J.G., R.A.L.v.d.H. and M.J.H. contributed new analytical tools, H.Z., K.L.H. and F.L.T. analysed data, F.L.T. and H.Z. wrote the paper.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Overview of the plant N‐end rule pathway.
Fig. S2 The Arg/N‐end rule is active in etiolated seedlings.
Fig. S3 Analysis of peptide quantification in Col‐0 and prt6.
Fig. S4 Abundance of proteins in different prt6 alleles.
Fig. S5 Cruciferin peptides identified by TMT‐TAILS.
Fig. S6 Activity‐based protein profiling probe specificity.
Fig. S7 ERF‐dependence of protease activity.
Fig. S8 ERF‐dependence of RD21 abundance.
Table S1 Primers used in this study
Methods S1 TMT labelling and enrichment of N‐termini by TAILS.
Methods S2 Immunoblotting.
Table S2 Identification of protein N‐termini with TMT‐TAILS
Table S3 Quantification of peptides using TMT‐TAILS
Acknowledgements
We thank Sabine D'Andrea, Patrick Gallois, Carol Mackintosh, Dwayne Hegedus and Cathy Coutu for generous gifts of antisera. We thank Chris Overall for helpful suggestions regarding TMT‐TAILS and Kyoko Morimoto for assistance with ABPP. Research in this study was funded by BBSRC grants BB/J016276/1 and BB/J017647/1 to F.L.T. and K.S.L., respectively. Rothamsted Research receives grant‐aided support from the BBSRC of the UK.
See also the Commentary on this article by https://doi.org/10.1111/nph.15156.
References
- Abbas M, Berckhan S, Rooney DJ, Gibbs DJ, Vicente Conde J, Sousa Correia C, Bassel GW, Marín‐de la Rosa N, León J, Alabadí D et al 2015. Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Current Biology 25: 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. 1986. In vivo half‐life of a protein is a function of its amino‐terminal residue. Science 234: 179–186. [DOI] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepinie L. 2008. Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. Arabidopsis Book 6: e0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenut WV, Sumpton D, Martinez A, Lilla S, Espagne C, Meinnel T, Giglione C. 2012. Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N‐α‐acetylation features. Molecular & Cellular Proteomics: MCP 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biniossek ML, Schilling O. 2012. Enhanced identification of peptides lacking basic residues by LC‐ESI‐MS/MS analysis of singly charged peptides. Proteomics 12: 1303–1309. [DOI] [PubMed] [Google Scholar]
- Boex‐Fontvieille E, Rustgi S, von Wettstein D, Reinbothe S, Reinbothe C. 2015. Water‐soluble chlorophyll protein is involved in herbivore resistance activation during greening of Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 112: 7303–7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonissone S, Gupta N, Romine M, Bradshaw RA, Pevzner PA. 2013. N‐terminal protein processing: a comparative proteogenomic analysis. Molecular & Cellular Proteomics: MCP 12: 14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SJ, Wu X, Wadas B, Oh JH, Varshavsky A. 2017. An N‐end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355: eaal3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MK, Sullivan JA, Theobald JC, Davies WJ, Gray JC. 2008. An Arabidopsis mutant able to green after extended dark periods shows decreased transcripts of seed protein genes and altered sensitivity to abscisic acid. Journal of Experimental Botany 59: 3869–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andréa S, Jolivet P, Boulard C, Larré C, Froissard M, Chardot T. 2007. Selective one‐step extraction of Arabidopsis thaliana seed oleosins using organic solvents. Journal of Agriculture and Food Chemistry 55: 10008–10015. [DOI] [PubMed] [Google Scholar]
- Galland M, Huguet R, Arc E, Cueff G, Job D, Rajjou L. 2014. Dynamic proteomics emphasizes the importance of selective mRNA translation and protein turnover during Arabidopsis seed germination. Molecular & Cellular Proteomics: MCP 13: 252–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzón M, Eifler K, Faust A, Scheel H, Hofmann K, Koncz C, Yephremov A, Bachmair A. 2007. PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N‐end rule pathway with arginine specificity and is not the CER3 locus. FEBS Letters 581: 3189–3196. [DOI] [PubMed] [Google Scholar]
- Ge Y, Cai YM, Bonneau L, Rotari V, Danon A, McKenzie EA, McLellan H, Mach L, Gallois P. 2016. Inhibition of cathepsin B by caspase‐3 inhibitors blocks programmed cell death in Arabidopsis . Cell Death and Differentiation 23: 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ. 2014b. The eukaryotic N‐end rule pathway: conserved mechanisms and diverse functions. Trends in Cell Biology 24: 603–611. [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Bailey M, Tedds HM, Holdsworth MJ. 2016. From start to finish: amino‐terminal protein modifications as degradation signals in plants. New Phytologist 211: 1188–1194. [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey‐Serres J et al 2011. Homeostatic response of plants to hypoxia is regulated by the N‐end rule pathway. Nature 479: 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, Lozano‐Juste J, Mendiondo GM, Berckhan S, Marín‐de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP et al 2014a. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Molecular Cell 53: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Vicente Conde J, Berckhan S, Mendiondo GM, Prasad G, Holdsworth MJ. 2015. Group VII ethylene response factors co‐ordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiology 169: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Cardarelli F, Novi G, Parlanti S, Pucciariello C, Bassolino L, Banti V, Licausi F, Perata P. 2015. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nature Plants 1: 15151. [DOI] [PubMed] [Google Scholar]
- Graciet E, Mesiti F, Wellmer F. 2010. Structure and evolutionary conservation of the plant N‐end rule pathway. Plant Journal 61: 741–751. [DOI] [PubMed] [Google Scholar]
- Graciet E, Walter F, Ó’Maoiléidigh DS, Pollmann S, Meyerowitz EM, Wellmer F. 2009. The N‐end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proceedings of the National Academy of Sciences, USA 106: 13618–13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravot A, Richard G, Lime T, Lemarié S, Jubault M, Lariagon C, Lemoine J, Vicente J, Robert‐Seilaniantz A, Holdsworth MJ et al 2016. Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot. BMC Plant Biology 16: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruis DF, Selinger DA, Curran JM, Jung R. 2002. Redundant proteolytic mechanisms process seed storage proteins in the absence of seed‐type members of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 14: 2863–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Shabab M, Strasser R, Wolters PJ, Shindo T, Niemer M, Kaschani F, Mach L, van der Hoorn RA. 2012. Post‐translational regulation and trafficking of the granulin‐containing protease RD21 of Arabidopsis thaliana . PLoS ONE 7: e32422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann EM, Armengaud J. 2014. N‐terminomics and protegenomics, getting off to a good start. Proteomics 14: 2637–2646. [DOI] [PubMed] [Google Scholar]
- Heath JD, Weldon R, Monnot C, Meinke D. 1986. Analysis of storage proteins in normal and aborted seeds from embryo‐lethal mutants of Arabidopsis thaliana . Planta 169: 304–312. [DOI] [PubMed] [Google Scholar]
- Herman E, Larkins B. 1999. Protein storage bodies and vacuoles. Plant Cell 11: 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Hirai MY, Fujiwara T, Naito S, Noji M, Saito S. 2006. Proteomic and transcriptomic analysis of Arabidopsis seeds: molecular evidence for successive processing of seed proteins and its implication in the stress response to sulphur nutrition. Plant Journal 48: 557–571. [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ. 2008. Molecular networks regulating Arabidopsis seed maturation, after‐ripening, dormancy and germination. New Phytologist 179: 33–54. [DOI] [PubMed] [Google Scholar]
- Holman T, Jones PD, Russell L, Medhurst A, Úbeda‐Tomás S, Talloji P, Marquez J, Schmuths H, Tung S‐A, Taylor I et al 2009. The N‐end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis . Proceedings of the National Academy of Sciences, USA 106: 4549–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA. 2008. Plant proteases: from phenotypes to molecular mechanisms. Annual Review of Plant Biology 59: 191–223. [DOI] [PubMed] [Google Scholar]
- Hu RG, Sheng J, Qi X, Xu Z, Takahashi TT, Varshavsky A. 2005. The N‐end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437: 981–986. [DOI] [PubMed] [Google Scholar]
- Huesgen PF, Overall CM. 2012. N‐ and C‐terminal degradomics: new approaches to reveal biological roles for plant proteases from substrate identification. Physiologia Plantarum 145: 5–17. [DOI] [PubMed] [Google Scholar]
- Hwang JE, Hong JK, Je JH, Lee KO, Kim DY, Lee SY, Lim CO. 2009. Regulation of seed germination and seedling growth by an Arabidopsis phytocystatin isoform, AtCYS6 . Plant Cell Reports 28: 1623–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. 2010. N‐terminal acetylation of cellular proteins creates specific degradation signals. Science 327: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias‐Fernández R, Wozny D, Iriondo‐de Hond M, Oñate‐Sánchez L, Carbonero P, Barrero‐Sicilia C. 2014. The AtCathB3 gene, encoding a cathepsin B‐like protease, is expressed during germination of Arabidopsis thaliana and transcriptionally repressed by the basic leucine zipper protein GBF1. Journal of Experimental Botany 65: 2009–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F, Verhelst SH, van Swieten PF, Verdoes M, Wong CS, Wang Z, Kaiser M, Overkleeft HS, Bogyo M, van der Hoorn RA. 2009. Minitags for small molecules: detecting targets of reactive small molecules in living plant tissues using ‘click chemistry’. Plant Journal 57: 373–385. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim WT. 2013. The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high‐salt and drought stress responses. Plant Physiology 162: 1733–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleifeld O, Doucet A, auf dem Keller U, Prudova A, Schilling O, Kainthan RK, Starr AE, Foster LJ, Kizhakkedathu JN, Overall CM. 2010. Isotopic labeling of terminal amines in complex samples identifies protein N‐termini and protease cleavage products. Nature Biotechnology 28: 281–288. [DOI] [PubMed] [Google Scholar]
- Kleifeld O, Doucet A, Prudova A, auf dem Keller U, Gioia M, Kizhakkedathu JN, Overall CM. 2011. Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nature Protocols 6: 1578–1611. [DOI] [PubMed] [Google Scholar]
- Klein T, Fung SY, Renner F, Blank MA, Dufour A, Kang S, Bolger‐Munro M, Scurll JM, Priatel JJ, Schweigler P et al 2015. The paracaspase MALT1 cleaves HOIL1 reducing linear ubiquitination by LUBAC to dampen lymphocyte NF‐κB signalling. Nature Communications 6: 8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampl N, Alkan N, Davydov O, Fluhr R. 2013. Set‐point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant Journal 74: 498–510. [DOI] [PubMed] [Google Scholar]
- Lange PF, Huesgen PF, Nguyen K, Overall CM. 2014. Annotating N termini for the human proteome project: N termini and Nα‐acetylation status differentiate stable cleaved protein species from degradation remnants in the human erythrocyte proteome. Journal of Proteome Research 13: 2028–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH. 2017. Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29: 207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang B‐C, Xu Y, Zhu Y‐X. 2007. Systematic studies of 12S seed storage protein accumulation and degradation patterns during Arabidopsis seed maturation and early seedling germination stages. Journal of Biochemistry and Molecular Biology 40: 373–381. [DOI] [PubMed] [Google Scholar]
- Licausi F, Kosmacz M, Weits DA, Giuntoli B, Giorgi FM, Voesenek LA, Perata P, van Dongen JT. 2011. Oxygen sensing in plants is mediated by an N‐end rule pathway for protein destabilization. Nature 479: 419–422. [DOI] [PubMed] [Google Scholar]
- Licausi F, Pucciariello C, Perata P. 2013. New role for an old rule: N‐end rule‐mediated degradation of ethylene responsive factor proteins governs low oxygen response in plants. Journal of Integrative Plant Biology 55: 31–39. [DOI] [PubMed] [Google Scholar]
- Linster E, Stephan I, Bienvenut WV, Maple‐Grødem J, Myklebust LM, Huber M, Reichelt M, Sticht C, Møller SG, Meinnel T et al 2015. Downregulation of N‐terminal acetylation triggers ABA‐mediated drought responses in Arabidopsis . Nature Communications 6: 7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Chandrasekar B, Oeljeklaus J, Misas‐Villamil JC, Wang Z, Shindo T, Bogyo M, Kaiser M, van der Hoorn RA. 2015. Subfamily‐specific fluorescent probes for cysteine proteases display dynamic protease activities during seed germination. Plant Physiology 168: 1462–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. 1996. The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. International Journal of Plant Sciences 157: 280–295. [Google Scholar]
- de Marchi R, Sorel M, Mooney B, Fudal I, Goslin K, Kwaśniewska K, Ryan PT, Pfalz M, Kroymann J, Pollmann S et al 2016. The N‐end rule pathway regulates pathogen responses in plants. Scientific Reports 6: 26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan H, Gilroy EM, Yun BW, Birch PR, Loake GJ. 2009. Functional redundancy in the Arabidopsis Cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytologist 183: 408–418. [DOI] [PubMed] [Google Scholar]
- Mendiondo GM, Gibbs DJ, Szurman‐Zubrzycka M, Korn A, Marquez J, Szarejko I, Maluszynski M, King J, Axcell B, Smart K et al 2016. Enhanced waterlogging tolerance in barley by manipulation of expression of the N‐end rule pathway E3 ligase PROTEOLYSIS6 . Plant Biotechnology Journal 14: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mot AC, Prell E, Klecker M, Naumann C, Faden F, Westermann B, Dissmeyer N. 2018. Real‐time detection of N‐end rule‐mediated ubiquitination via fluorescently labeled substrate probes. New Phytologist 217: 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey‐Serres J. 2009. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis . Proceedings of the National Academy of Sciences, USA 106: 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Pérez‐Salamó I, Joseph MP, Giuntoli B, Bögre L, Koncz C, Szabados L. 2015. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3 . Plant Journal 82: 772–784. [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. 2006a. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Pinfield‐Wells HM, Graham IA. 2006b. Storage reserve mobilisation and seedling establishment in Arabidopsis. Arabidopsis Book 4: e0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. 2004. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1 . Plant Cell 16: 2705–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Iwasaki M, Susaki D, Megies C, Kinoshita T, Lopez‐Molina L. 2016. Dormancy‐specific imprinting underlies maternal inheritance of seed dormancy in Arabidopsis thaliana . eLife 5: e19573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Stary S, Schlögelhofer P, Becker F, Nejinskaia V, Bachmair A. 1998. PRT1 of Arabidopsis thaliana encodes a component of the plant N‐end rule pathway. Proceedings of the National Academy of Sciences, USA 95: 7904–7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudova A, Gocheva V, Auf dem Keller U, Eckhard U, Olson OC, Akkari L, Butler GS, Fortelny N, Lange PF, Mark JC et al 2016. TAILS N‐terminomics and proteomics show protein degradation dominates over proteolytic processing by cathepsins in pancreatic tumors. Cell Reports 16: 1762–1773. [DOI] [PubMed] [Google Scholar]
- Riber W, Müller JT, Visser EJ, Sasidharan R, Voesenek LA, Mustroph A. 2015. The greening after extended darkness1 is an N‐end rule pathway mutant with high tolerance to submergence and starvation. Plant Physiology 167: 1616–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stüber K, Colby T, Overkleeft HS, Bogyo M, van der Hoorn RA. 2012. Subclassification and biochemical analysis of plant papain‐like cysteine proteases displays subfamily‐specific characteristics. Plant Physiology 158: 1583–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustgi S, Boex‐Fontvieille E, Reinbothe C, von Wettstein D, Reinbothe S. 2017. Serpin1 and WSCP differentially regulate the activity of the cysteine protease RD21 during plant development in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 114: 2212–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M, Moffat B, Saechao MC, Hell R, Wirtz M. 2013. Methionine salvage and S‐adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochemical Journal 451: 145–154. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Schuessele C, Hoernstein SN, Mueller SJ, Rodriguez‐Franco M, Lorenz T, Lang D, Igloi GL, Reski R. 2016. Spatio‐temporal patterning of arginyl‐tRNA protein transferase (ATE) contributes to gametophytic development in a moss. New Phytologist 209: 1014–1027. [DOI] [PubMed] [Google Scholar]
- Shindo T, Misas‐Villamil JC, Hörger AC, Song J, van der Hoorn RA. 2012. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS ONE 7: e29317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary S, Yin XJ, Potuschak T, Schlögelhofer P, Nizhynska V, Bachmair A. 2003. PRT1 of Arabidopsis is a ubiquitin protein ligase of the plant N‐end rule pathway with specificity for aromatic amino‐terminal residues. Plant Physiology 133: 1360–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoulou FL, Eastmond PJ. 2012. Seed storage oil catabolism: a story of give and take. Current Opinion in Plant Biology 15: 322–328. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Gevaert K, Van Breusegem F. 2012. Natural substrates of plant proteases: how can protease degradomics extend our knowledge? Physiologia Plantarum 145: 28–40. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Timmerman E, De Bock PJ, Vercammen D, Stael S, van de Cotte B, Staes A, Goethals M, Beunens T, van Damme P et al 2013. The Arabidopsis METACASPASE9 degradome. Plant Cell 25: 2831–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. 2011. The N‐end rule pathway and regulation by proteolysis. Protein Science 20: 1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venne AS, Solari FA, Faden F, Paretti T, Dissmeyer N, Zahedi RP. 2015. An improved workflow for quantitative N‐terminal charge‐based fractional diagonal chromatography (ChaFRADIC) to study proteolytic events in Arabidopsis thaliana . Proteomics 15: 2458–2469. [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del‐Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez‐Riverol Y, Reisinger F, Ternent T et al 2016. 2016 update of the PRIDE database and related tools. Nucleic Acids Research 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Ross AR, Yang J, Hegedus DD, Kermode AR. 2007. Phosphorylation of the 12 S globulin cruciferin in wild‐type and abi1‐1 mutant Arabidopsis thaliana (thale cress) seeds. Biochemical Journal 404: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten HM, Riegler H, Hoefgen R, Perata P, van Dongen JT, Licausi F. 2014. Plant cysteine oxidases control the oxygen‐dependent branch of the N‐end‐rule pathway. Nature Communications 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MD, Klecker M, Hopkinson RJ, Weits DA, Mueller C, Naumann C, O'Neill R, Wickens J, Yang J, Brooks‐Bartlett JC et al 2017. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase‐catalysed arginylation of N‐end rule targets. Nature Communications 23: 14690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P, Ndah E, Jonckheere V, Stael S, Sticker A, Martens L, van Breusegem F, Gevaert K, van Damme P. 2017. N‐terminal proteomics assisted profiling of the unexplored translation initiation landscape in Arabidopsis thaliana . Molecular & Cellular Proteomics: MCP 16: 1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large‐scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit M, Galvão VC, Fankhauser C. 2016. Light‐mediated hormonal regulation of plant growth and development. Annual Review of Plant Biology 67: 513–537. [DOI] [PubMed] [Google Scholar]
- Wu SH. 2014. Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annual Review of Plant Biology 65: 311–333. [DOI] [PubMed] [Google Scholar]
- Yamada K, Matsushima R, Nishimura M, Hara‐Nishimura I. 2001. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiology 127: 1626–1634. [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Callis J, Nishida I, Watanabe A. 2002. A delayed leaf senescence mutant is defective in arginyl‐tRNA:protein arginyltransferase, a component of the N‐end rule pathway in Arabidopsis . Plant Journal 32: 129–137. [DOI] [PubMed] [Google Scholar]
- Zhang H, Deery MJ, Gannon L, Powers SJ, Lilley KS, Theodoulou FL. 2015. Quantitative proteomics analysis of the Arg/N‐end rule pathway of targeted degradation in Arabidopsis roots. Proteomics 15: 2447–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Overview of the plant N‐end rule pathway.
Fig. S2 The Arg/N‐end rule is active in etiolated seedlings.
Fig. S3 Analysis of peptide quantification in Col‐0 and prt6.
Fig. S4 Abundance of proteins in different prt6 alleles.
Fig. S5 Cruciferin peptides identified by TMT‐TAILS.
Fig. S6 Activity‐based protein profiling probe specificity.
Fig. S7 ERF‐dependence of protease activity.
Fig. S8 ERF‐dependence of RD21 abundance.
Table S1 Primers used in this study
Methods S1 TMT labelling and enrichment of N‐termini by TAILS.
Methods S2 Immunoblotting.
Table S2 Identification of protein N‐termini with TMT‐TAILS
Table S3 Quantification of peptides using TMT‐TAILS


