Abstract
The deubiquitinases, or DUBs, are associated with various human diseases, including neurological disorders, cancer, and viral infection, making them excellent candidates for pharmacological intervention. Drug discovery campaigns against DUBs require enzymatic deubiquitination assays amenable for high-throughput screening (HTS). Although several DUB substrates and assays have been developed in recent years, they are largely limited to recombinantly purified DUBs. Many DUBs are large multi-domain proteins that are difficult to obtain recombinantly in sufficient quantities for HTS. Therefore, an assay that obviates the need of recombinant protein generation and also recapitulates a physiologically relevant environment is highly desirable. Such assay will open doors for drug discovery against many therapeutically relevant, but currently inaccessible, DUBs. Here we report a cell lysate DUB assay based on AlphaLISA technology for high throughput screening. This assay platform uses a biotin-tagged ubiquitin probe and a HA-tagged DUB expressed in human cells. The assay was validated and adapted to a 1536-well format, which enabled a screening against UCHL1 as proof of principle using a library of fifteen thousand compounds. We expect that the new platform can be readily adapted to other DUBs to allow the identification of more potent and selective small molecule inhibitors and chemical probes.
Keywords: Deubiquitinase, deubiquitination, inhibitor, AlphaScreen, AlphaLISA, ubiquitin-based probes, high-throughput screening, Ubiquitin C-terminal Hydrolases, activity-based probes, post translational modifications
Ubiquitin, a small 76 amino acid protein, plays a key role in cellular signaling and various eukaryotic cellular processes. The addition of ubiquitin to the lysine side chain in a target protein, namely ubiquitination, is a major post-translational modification (PTM) that rivals phosphorylation in both scale and complexity. Like other PTMs, ubiquitination is a reversible process that is regulated by a large class of enzymes called deubiquitinating enzymes or deubiquitinases (DUBs). DUBs remove the ubiquitin moiety from its monoubiquitinated and polyubiquitinated protein substrates, allowing tight control of various cellular pathways. Dysregulation of these pathways is a common cause of cancer, neurodegenerative, and inflammatory diseases.1
The human genome encodes close to 100 DUBs, which are divided into six families based on their structural characteristics: ubiquitin-specific proteases (USP), ubiquitin C-terminal hydrolases (UCH), JAB1/MPN/Mov34 metalloenzymes (JAMMs), Machado-Joseph domain proteases (MJD), ovarian tumor domain-containing proteases (OTUs), and the recently identified MINDY (motif interacting with ubiquitin-containing novel DUB family). Five out of the six families are cysteine proteases (USP, MJD, OTU, UCH, MINDY), while members of the JAMM family are metalloproteases.1, 2
Assaying DUB activity is central to both the investigation of DUB function, and the development of small molecule inhibitors for therapeutic intervention. To date, the most commonly used DUB assay utilizes an ubiquitin fluorophore conjugate through an isopeptide bond. Two of the most frequently used substrates are ubiquitin-7-amino-4-methylcoumarin (Ub-AMC) and ubiquitin-rhodamine 110 (Ub-Rho110). These molecules become fluorescent upon DUB-mediated cleavage of the C-terminal ubiquitin amide bond and the subsequent release of the fluorophore.3, 4 This type of substrate has been instrumental in kinetic assessment and high-throughput screening5–12 of DUBs. Among the above-mentioned substrates, Ub-Rho110 is regarded as a better substrate for HTS against DUBs. Rhodamine 110, in place of AMC, provides a red shift in excitation and emission, thus reducing the interference by fluorescent compounds in the library.13
In addition to the Ub-fluorophore assay, efforts were made to develop newer DUB assays like DUB-Glo and Ub/Ubl-CHOP-reporter assays. DUB-Glo is a bioluminescent assay with low background signal.4 Ub/Ubl-CHOP-reporter assay utilizes the reporter enzyme, phospholipase A2 (PLA2), which is conjugated to ubiquitin in tandem. Cleavage of the ubiquitin C-terminal amide bond results in an active PLA2 that cleaves a fluorescent substrate.14 This technology was later expanded to include the fusion of Ub/Ubl to the N-terminus of enterokinase light chain (EK1) and granzyme B (GZMB), improving the sensitivity and affording a multiplex assay format against DUBs and Ubl-proteases.15
Diubiquitin substrates, which represent a more physiologically relevant substrate, have been utilized in gel-, FRET-16–18 and MALDI-TOF mass spectrometry-based19 DUB assays. Among them, the gel-based assay is usually tedious and low throughput, thus not suitable for HTS. The diubiquitin FRET- and MALDI-TOF mass spectrometry-based assays enable fast and quantitative analysis of DUB-catalyzed diubiquitin cleavage. However, HTS campaigns using the FRET- and MALDI-TOF mass spectrometry-based assays have yet to be reported, likely due to the high cost associated with these assay formats.
Many human DUBs are large, multi-domain proteins. Therefore, it remains a challenge to obtain high quality DUB samples in quantities sufficient for HTS campaign. In addition, numerous DUBs are known to bind partner proteins that are essential for the DUB activity and target protein specificity.20 Cell lysate-based DUB assays represent an attractive solution to the challenges facing the conventional DUB assays described above. They circumvent the requirement of recombinant DUB protein generation, and better recapitulate the cellular milieu.
Here we report the first cell lysate-based AlphaLISA DUB assay platform amenable to HTS. It uses a biotinylated ubiquitin probe in conjunction with a HA-tagged DUB expressed in human cells. As a test case, ubiquitin C-terminal hydrolase-1 (UCHL1) was selected for the current study and this assay was adapted to a 1536-well plate format. As proof of principle, we performed a quantitative high throughput screening (qHTS) using five compound libraries totaling close to 15,000 compounds. The cell lysate AlphaLISA DUB assay proved to be robust with a Z′ factor of 0.85. Additionally, we confirmed several top hits as true inhibitors against UCHL1 through orthogonal assays. Although our work centers on DUBs, this platform may be adapted to other targets, benefiting from the many existing activity-based chemical probes against different classes of enzymes.
Results and Discussion
Design and Generation of AlphaLISA DUB Assay Components
To enable HTS against DUBs in cell lysates, we utilized the Alpha (amplified luminescent proximity homogenous assay) technology that detects the interactions, covalent or non-covalent, of two assay components. Alpha technology is a no wash, bead-based proximity assay. It is sensitive in detection of low concentration (sub-pM) analytes and quantitative with a wide dynamic range.21–24 Our AlphaLISA DUB assay relies on an activity-based DUB probe with a biotinylated Avi-Tag introduced to the N-terminus and a vinyl-methyl ester introduced to the C-terminus of ubiquitin (biotin-UbVMe). The biotin on ubiquitin affords binding to the streptavidin coated Alpha donor beads. The DUB of interest (UCHL1) contains a C-terminal human influenza hemagglutinin (HA) tag, which allows its association with the AlphaLISA acceptor beads coated with anti-HA antibody. When the biotin-UbVMe labels the DUB active site cysteine, the covalent interaction brings the two beads close enough for singlet oxygen (generated from illumination of the donor beads at 680 nm) transfer from the donor to the acceptor beads. This results in the emission of light at 615 nm (Figure 1). Small molecules that inhibit the labeling of DUBs by the UbVMe probe reduce the Alpha signal. The combination of the Alpha technology and the activity-based DUB probe provides an excellent platform for the discovery of small molecule DUB inhibitors in cell lysates (Figure 1).
Figure 1.
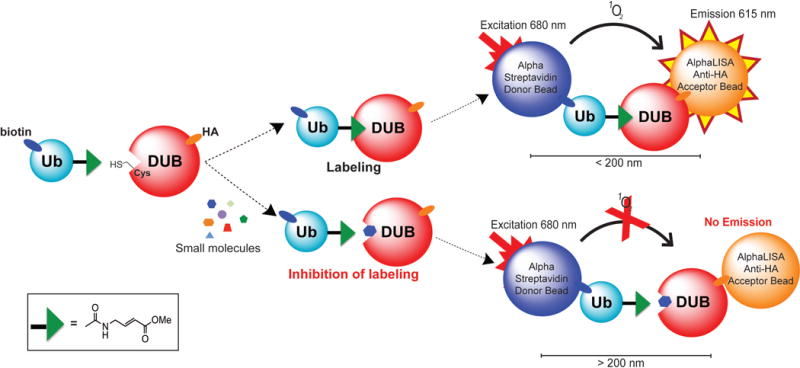
Illustration of the AlphaLISA cell lysate DUB assay platform. The ubiquitin-probe, UbVMe, containing an N-terminal biotinylated Avi-Tag reacts with a HA-tagged deubiquitinase (DUB) expressed in HEK293T cells. The epitope tags allow for their association with either the Alpha Streptavidin donor beads or the AlphaLISA Anti-HA acceptor beads. The labeling of the DUB with UbVMe brings the AlphaLISA donor and acceptor beads within 200 nm, enabling singlet oxygen transfer from the donor bead to the acceptor bead upon illumination at 680 nm. This transfer results in emission of the acceptor bead at 615 nm, which is quantified in an Alpha plate reader. Small molecules that inhibit the labeling of DUB by the UbVMe probe cause a measurable reduction in the Alpha signal.
To generate biotin-UbVMe, the ubiquitin protein was modified to contain an Avi-Tag at the N-terminus to enable efficient biotinylation (Supporting Information Figure 1a). Avi-Ub(1-75) was expressed and purified as an intein fusion and cleaved with sodium 2-mercaptoethane sulfonate (MESNa) (Supporting Information Figure 1b). The Avi-Ub(1-75)-MESNa molecular weight was confirmed by mass spectrometry analysis (10,998 Da) (Supporting Information Figure 1c). Avi-Ub(1-75)-MESNa was then converted to Avi-UbVMe following a previously reported procedure with minor modifications.25 Biotinylation of Avi-UbVMe was carried out in vitro using E. coli biotin ligase, BirA, and the modification was confirmed by mass spectrometry (MW 11,199 Da) (Supporting Information Figure 2a). Note that we also observed biotin-UbVMe with N-terminal acetylation (11,241 Da) and a low level of biotin-Ub(1-75)-COOH (11,102 Da). Biotin-Ub(1-75)-COOH is the hydrolysis product of Avi-Ub(1-75)-MESNa and does not contain the reactive Michael acceptor. N-terminal acetylation of a serine residue in recombinant protein is known to occur in E. coli when the first methionine of the expressed protein is removed post-translationally.26 The removal of the first methionine in biotin-UbVMe was confirmed by the LC-MS/MS analysis (Supporting Information Figure 3). Furthermore, the biotinylation on Lys12 and acetylation on Ser2 in the Avi-Ub fusion were also confirmed (Supporting Information Figure 3). The acetylated biotin-UbVMe and the small amount of biotin-Ub(1-75)-COOH are not expected to interfere the labeling of DUBs by the biotin-UbVMe probe. Additionally, an SDS-PAGE analysis of the final biotin-UbVMe sample showed that it was free of other contaminating proteins (Supporting Information Figure 2b).
The activity of biotin-UbVMe was validated through in vitro and cellular labeling. Upon incubation of biotin-UbVMe with purified UCHL1, UCHL3 or UCHL5, adduct formation was detected on a denaturing SDS-PAGE gel visualized by Coomassie blue staining (Supporting Information Figure 4). Biotin-UbVMe labeled endogenous DUBs in the HEK293T cell lysates with a profile similar to that obtained by the commonly used HA-UbVMe probe (Supporting Information Figure 5a). The labeling of biotin-UbVMe by endogenous UCHL1 was assessed by immunoblotting with anti-UCHL1 antibody and was comparable to HA-UbVMe probe (Supporting Information Figure 5b). Expression of HA-UCHL1 in HEK293T cells and its labeling by biotin-UbVMe probe was confirmed by Western blotting using an anti-HA antibody (Figure 2a). To access if labeling by biotin-UbVMe was dependent on UCHL1 activity, we generated two catalytically inactive mutants C90A and C90S HA-UCHL1. Expression and activity of the HA-UCHL1 mutants in HEK293T cell lysates were assessed by Western blotting using anti-HA antibody (Figure 2a). Unlike WT HA-UCHL1, we were unable to detect labeling by biotin-UbVMe of either HA-UCHL1 mutant by Western blotting using an anti-HA antibody (Figure 2a). This demonstrates that labeling of UCHL1 by biotin-UbVMe is dependent on UCHL1’s catalytic activity.
Figure 2.
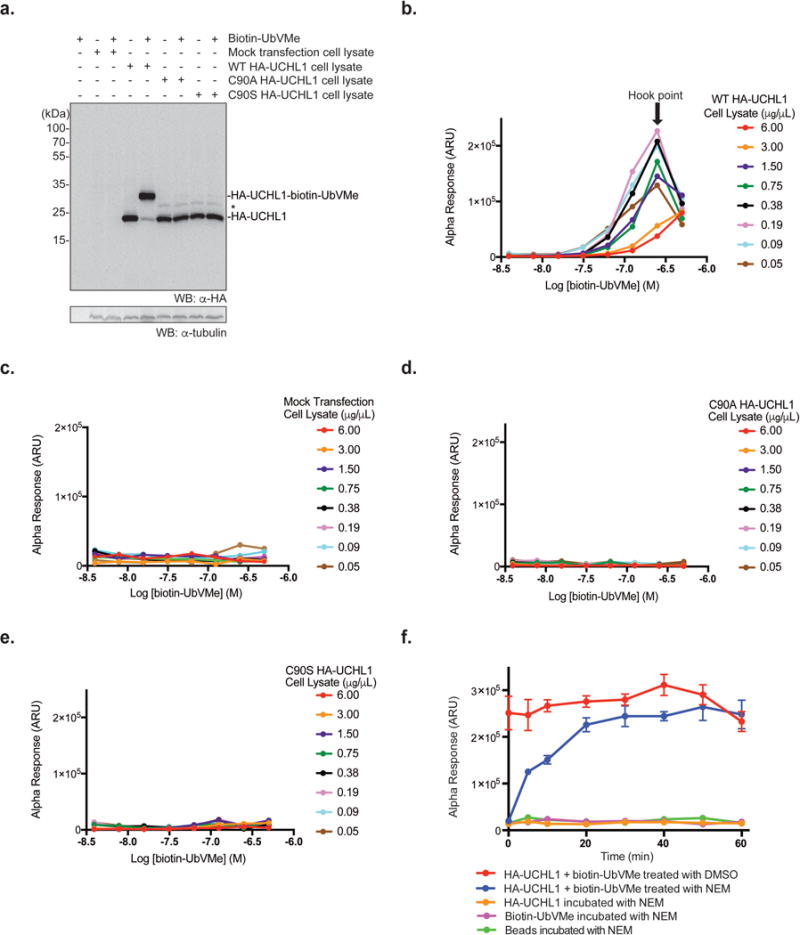
Detection of HA-UCHL1 labeling by biotin-UbVMe in the AlphaLISA cell lysate DUB assay. (a) In vitro labeling of WT HA-UCHL1 or catalytically inactive HA-UCHL1mutants (C90A, C90S) overexpressed in HEK293T cell lysates using biotin-UbVMe. The adduct was detected using anti-HA antibody. α-tubulin was utilized as a loading control detected with anti-α-tubulin antibody. Asterisk (*) indicates non-specific band. (b-e) AlphaLISA cross titration experiments using biotin-UbVMe and HEK293T cell lysates ectopically expressing WT HA-UCHL1 (b), mock transfected (c), or catalytically inactive HA-UCHL1 mutants C90A (d), C90S (e). (f) Time-dependent quenching of the biotin-UbVMe labeling reaction of HA-UCHL1 in HEK293T cell lysates using 50 mM N-ethylmaleimide (NEM). Parallel reactions (including controls) were carried out as indicated in the legend.
Development of the AlphaLISA DUB Assay
After demonstrating that the biotin-UbVMe probe can properly label recombinant and cellular DUBs including UCHL1, we used the probe in developing the AlphaLISA DUB assay in cell lysates. The optimal condition for the strongest AlphaLISA signal was determined through a cross titration experiment by varying the concentration of biotin-UbVMe and the total protein concentration of the HEK293T cell lysates expressing HA-UCHL1 (Figure 2b). We observed a hook point at 188 ng/μL of HEK293T cell lysates and 250 nM biotin-UbVMe. The hook point is the concentration of the binding partners where maximum AlphaLISA signal is achieved. The hook effect is commonly observed in bimolecular detection systems that involve saturable binding reagents (the donor and acceptor beads in AlphaLISA) that are used to capture specific binding partners. To rule out signal due to non-specific binding from the cell lysates, several control experiments were run in parallel using the mock-transfected HEK293T cell lysates and HEK293T cell lysates expressing the catalytically inactive UCHL1 mutants (C90A and C90S UCHL1). We observed little to no signal in the absence of HA-UCHL1 in the cell lysates (Figure 2c). Low signal was also observed when C90A HA-UCHL1 (Figure 2d) or C90S HA-UCHL1 (Figure 2e) HEK293T cell lysates were used. These results indicate that the observed AlphaLISA signal was due to the labeling of the UCHL1 catalytic cysteine by the biotin-UbVMe probe.
The AlphaLISA DUB assay requires an incubation period following the labeling reaction to allow efficient association of biotin-UbVMe and HA-UCHL1 with the donor and acceptor beads, respectively. Thus, it is necessary to quench the labeling reaction prior to incubation with the beads to stabilize the AlphaLISA signal and achieve a consistent result. Denaturants (SDS, urea) and reducing agents (DTT, 2-mercaptoethanol) were evaluated as quenchers to terminate the labeling reaction by biotin-UbVMe. However, no satisfactory quenching effect was observed with the above-mentioned reagents (data not shown). Cysteine modifying compounds N-ethylmaleimide (NEM), bromomaleimide, and methyl methane thiosulfonate (MMTS) were also tested given their documented reactivity with the cysteine thiol group.27, 28 Among the three reagents, NEM showed to be the most effective in quenching the labeling reaction. In a reaction mixture containing biotin-UbVMe and HA-UCHL1 cell lysates, 50 mM NEM was added at varied time points to stop the labeling reaction (Figure 2f). At time point zero, 50 mM NEM efficiently stopped the reaction. A time-dependent increase of the AlphaLISA signal within the first 20 minutes was observed and a plateau was reached after 20 minutes. In the control reaction, addition of vehicle DMSO did not stop the reaction. Based on these results, 30 minutes was chosen for the UCHL1 labeling reaction by the biotin-UbVMe probe before being quenched with NEM.
Validation of the AlphaLISA DUB Assay
Based on the above-described experiments, we developed an assay protocol as illustrated in Figure 3a. The assay performance was tested with a known UCHL1 inhibitor, LDN-57444, in a 384-well plate format. LDN-57444 is a reversible, competitive inhibitor of UCHL1.8 LDN-57444 inhibited the labeling of HA-UCHL1 by biotin-UbVMe in our AlphaLISA assay with a well-defined dose–response curve (Figure 3b). In comparison control compound, ML323, previously reported to be inactive against UCHL1,7 showed no obvious inhibition in the AlphaLISA assay. We also carried out a parallel experiment using ubiquitin-bromide (Ub-Br), an activity-based DUB inhibitor, as a competitive UCHL1 inhibitor. The dose-response curves (Figure 3b) of the two inhibitors were fit to a four-parameter dose-response curve equation with a variable Hill slope using GraphPad Prism. We obtained IC50 of 12.5 μM for LDN-57444 and 7.5 μM for Ub-Br, respectively. These results demonstrated the ability of the cell lysate-based AlphaLISA DUB assay to reliably report the DUB inhibition in a complex sample. Moreover, this assay format is quantitative in nature and allows the determination of IC50 of small molecule and ubiquitin-based inhibitors targeting the cellular DUBs.
Figure 3.
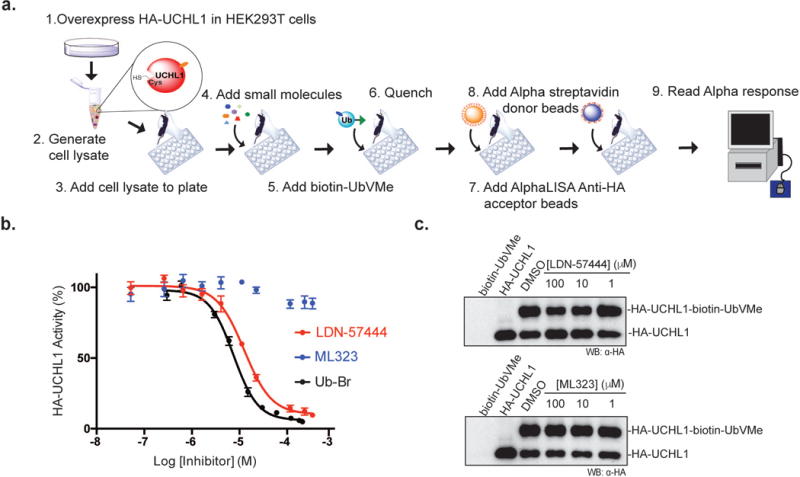
Validation of AlphaLISA DUB assay. (a) Scheme showing the AlphaLISA cell lysate DUB assay procedure. (b) Dose–response curve showing the inhibition of UCHL1 by LDN-57444, ML323, and Ubiquitin-Br using AlphaLISA cell lysate DUB assay. (c) Inhibition of HA-UCHL1 by LDN-57444 and ML323 visualized by Western blotting using anti-HA antibody.
We further used a gel-based DUB labeling assay to confirm the inhibition of biotin-UbVMe labeling of UCHL1 by LDN-57444 in cell lysates. The labeling of HA-UCHL1 was detected by Western blotting using an anti-HA antibody. LDN-57444 exhibited a dose-dependent inhibition of UCHL1 labeling by biotin-UbVMe (Figure 3c top), while ML323 did not inhibit the labeling (Figure 3c bottom). We found that although the trend of UCHL1 inhibition by LDN-57444 was consistent in the repeats, obtaining an IC50 based on the quantified band intensity proved to be challenging. Western blotting is known to be non-ideal for rigorous quantitative analysis29, 30, which may be one contributing factor of the difficulty met in our attempts. In comparison, we found that the AlphaLISA DUB assay is amenable for quantitative analysis and can be used to obtain good quality dose-response curves and allow the determination of inhibitor IC50 relatively quickly.
To show that AlphaLISA DUB assay is not limited to UCHL1 and can be adapted to other members of the UCH family, we tested the potency of LDN-57444 and ML323 against HA-UCHL3 and HA-UCHL5 overexpressed in HEK293T cells respectively. LDN-57444 potency was determined to be 19.9 μM for HA-UCHL3 (Supporting Information Figure 6a) and 42.7 μM for HA-UCHL5 (Supporting Information Figure 6b). Meanwhile, ML323 showed low potency against both HA-UCHL3 and HA-UCHL5 (>100 μM and >200 μM, respectively) (Supporting Information Figure 6a, b). Because the expression levels of the various UCHs in cells are possibly different, a direct comparison of the IC50 values obtained for the various UCHs by the same inhibitor may not be meaningful. However, for the same UCH cell lysate, the IC50 obtained in the AlphaLISA assay allows the ranking of the potency of different inhibitors against the same target.
Implementation of the AlphaLISA DUB Assay in qHTS
To miniaturize the assay into 1536-well format, a cross titration was performed and an optimal condition of 31.25 ng/μL HA-UCHL1 cell lysates and 6 nM biotin-UbVMe was determined (Figure 4a). Further optimization of the assay indicated that 10 μg/mL bead concentration yielded the best Z′ factor, with a satisfactory fold change in signal (data not shown). Under the established qHTS condition, summarizing in Supporting Information Table 1, the known UCHL1 inhibitor LDN-57444 elicited a dose-dependent decrease of AlphaLISA signal with an IC50 of 0.73 μM (Figure 4b).
Figure 4.
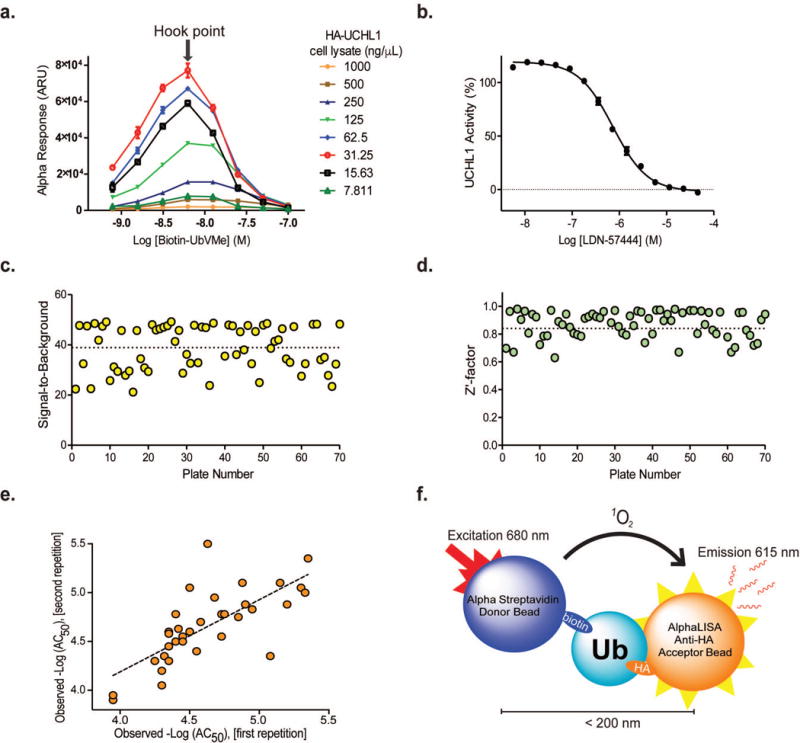
Miniaturization and adaptation of AlphaLISA DUB Assay for qHTS in 1536-well format. (a) Cross titration to determine optimal conditions for qHTS. (b)Dose–response curve for LDN-57444 inhibition during initial assay optimization. (c, d) Screening statistics of individual plates: signal-to-background ratio (c) and Z′-factor (d). (e) Correlation plot of observed –Log(AC50) values for repeated compounds (R = 0.734, MSR = 3.4 and RMSE = 0.265). (f) Scheme of biotin-Ub-HA counter-screen assay.
We employed quantitative high-throughput screening (qHTS) to the optimized AlphaLISA DUB assay. First, a library of 1,912 compounds, comprising approved and investigational drugs with known mechanism of action, was screened as 11-point dilution series, with final compound concentrations ranging from 0.78 nM to 46.1 μM. LDN-57444 was used as an intraplate control and exhibited an average IC50 of 2.56 ± 0.7 μM. Minimum significant ratio (MSR) of the control compound was 1.93, showing a good reproducibility of the assay. Slight variation was observed for LDN-57444 IC50 values obtained during initial assay development, assay miniaturization, and qHTS. We attribute the differences to the assay conditions and the different target DUB concentrations in the cell lysates. The Z′-factor for the assay remained nearly constant throughout the experiment, with an average value of 0.93 ± 0.04, and a signal-to-background ratio of 45.5 ± 4.0, indicating a robust assay performance. Next, we performed a screen against 12,682 compounds from libraries of annotated small molecule tool compounds, as well as approved and investigational drugs. Each compound was tested in 5-point dilution series with final compound concentrations ranging from 5.7 nM to 91.6 μM. Overall, the assay performed well with an average signal-to-background ratio of 38.87 ± 8.91 and average Z′ of 0.85 ± 0.13 (Figure 4c, d).
In addition to assay quality assessment, we have analyzed the assay reproducibility using potency estimates expressed as AC50 values scaled in logarithmic units (-Log(AC50)). The total number of compounds measured multiple times was 623 from 14,594 (1,912 and 12,682) compounds screened across all libraries in this study. From the 623, only 36 duplicates yielded AC50 values and were used for reproducibility analysis. To estimate the assay reproducibility, we calculated the following statistical parameters: Pearson correlation coefficient (Rpearson), Minimum significant ratio (MSR) and Root mean square error (RMSE).31 The obtained results are: R = 0.734, MSR = 3.4 and RMSE = 0.265. The plot of the observed AC50 values for duplicates in logarithmic units is presented in Figure 4e. All statistical parameters reflect a good reproducibility of the developed assay.
Selection of Potential UCHL1 Inhibitors
Of the 14,594 small molecules screened, 973 unique compounds were selected for follow-up experiments (Supporting Information Figure 7 Figure 8). The selection was made based on the potency and efficacy of screened compounds as previously described.32 Selected compounds were retested to confirm inhibition using the AlphaLISA DUB assay at 11-point dilution series, with final concentrations ranging from 0.78 nM to 92.2 μM. We then conducted two counter-screens against these compounds. First, we used Perkin Elmer’s TruHits assay to detect false positive hits caused by compounds that can act as singlet oxygen or color quenchers, light scatterers, and biotin mimetics. The TruHits assay consists of a streptavidin-coated Alpha donor bead and a biotinylated AlphaLISA acceptor bead. The biotinylated AlphaLISA acceptor bead will bind to the streptavidin donor bead and emit light at 615 nm upon laser excitation at 680 nm. We identified 250 compounds as false positives based on the TruHits assay.
Potential false positive hits could also arise from disruption of the HA-tag and the bead-conjugated anti-HA antibody interaction. To uncover these compounds, we devised a second counter-screen based on a dual epitope- tagged Ub containing a biotinylated Avi-Tag at the N-terminus and a HA-tag at the C-terminus (Figure 4f). Biotin-Ub(1-75)-HA was generated in a similar way as biotin-UbVMe. Briefly, Avi-Ub(1-75)-HA was expressed as an intein fusion and cleaved using DTT. The biotinylation reaction of Avi-Ub(1-75)-HA was carried out in vitro using BirA with an excess of biotin. The ESI mass spectrometry analysis of biotin-Ub(1-75)-HA showed a molecular weight of 12,311 Da, identical to the theoretical molecular weight (Supporting Information Figure 8). Using the biotin-Ub(1-75)-HA counter-screen, we identified 446 compounds as false positives, encompassing 224 out of the 250 hits (90%) identified by the TruHits assay (Supporting Information Figure 7). Our two assay-specific counter-screens were able to collectively uncover total 472 false positive hits. After applying filters to eliminate redox cyclers, promiscuous, and other problematic compounds33, a total of 18 hits were chosen for further testing.
Validation of Hits as UCHL1 Inhibitors
Redox cyclers are common false hits against enzymes that utilize a catalytic cysteine for activity. Although our cheminformatics approach helps to filter out some common redox cyclers, we elected to run a redox cycling assay of the selected 18 hits. We employed a colorimetric assay reported by Johnston and coworkers to monitor hydrogen peroxide generation.34 Using the assay we found that one out of eighteen cherry-picked hits, DA-3003-1, was a strong redox cycling compound (Supporting Information Figure 9). We then determined the IC50 of the seventeen compounds against recombinant UCHL1 using Ub-AMC as a substrate. Two compounds, prednicarbate and lapatinib could not be assayed for an accurate IC50. We observed prednicarbate to be a strong fluorescent quencher of the AMC fluorophore, thus obstructing IC50 determination. Meanwhile, high concentrations of lapatinib in the assay caused visible precipitation, also preventing an accurate IC50 determination. The top inhibitors include acetyl isogambogic acid (IC50 of 5.4 μM), celastrol (IC50 of 6.8 μM), mangiferin (IC50 of 8.9 μM), rifampicin (IC50 of 19.5 μM), and (-)-aricine (IC50 of 47.7 μM) (Figure 5a, Supporting Information Table 2).
Figure 5.
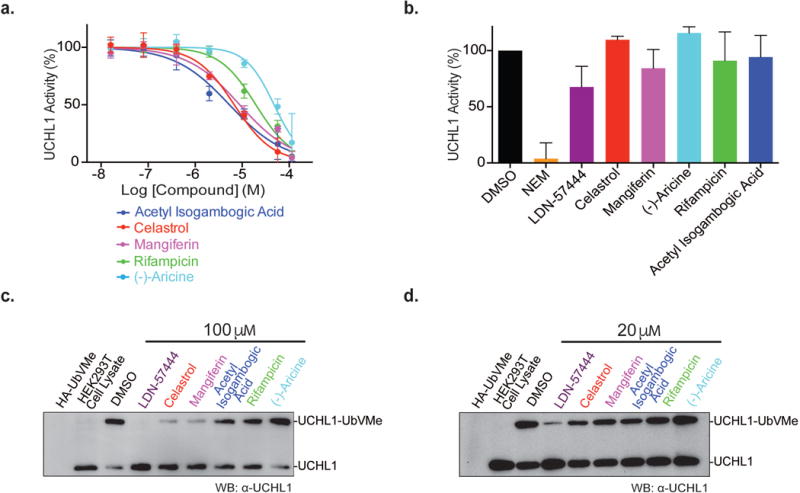
Assessment of small molecule inhibitors identified from the AlphaLISA qHTS against UCHL1. (a) Dose–response curve of recombinant UCHL1 inhibition by selected inhibitors using Ub-AMC as substrate. (b) Reversibility of inhibition by the compounds graphed as percent of remaining activity of UCHL1 in a rapid dilution assay. (c, d) Inhibition of endogenous UCHL1 labeling by HA-UbVMe in the presence of 100 μM (c) and 20 μM (d) of each compound. The blot shown is representative of three repeats.
Next we assessed the reversibility of the inhibition of UCHL1 by the top inhibitors following a protocol by Copeland and coworkers.35 Briefly, UCHL1 was incubated with the compound at a concentration ten times its IC50 then, rapidly diluted (100-fold) into the assay buffer. The incubated UCHL1-inhibitor mixture was then assayed for enzymatic activity using Ub-AMC. Recovered activity was normalized to UCHL1 samples treated with DMSO. LDN-57444 has previously been reported as a reversible inhibitor of UCHL1, and was thus utilized as a comparison.8 After treatment with LDN-57444 and rapid dilution, approximately 68% of UCHL1 activity was recovered compared to DMSO (Figure 5b). NEM irreversibly inhibits UCHL1 and resulted in low level of UCHL1 activity (3.9%) after treatment (Figure 5b). Our results indicate that celastrol, mangiferin, (-)-aricine, rifampicin, acetyl isogambogic acid were largely reversible inhibitors of UCHL1 with >80% recovered activity in the rapid dilution assay (Figure 5b).
We then tested the selectivity of the five compounds on inhibiting other human DUBs, UCHL3, UCHL5, USP2 catalytic core and full length USP15 using Ub-AMC as substrate. We found that celastrol and mangiferin were selective for UCHL1 among the UCHs and USPs tested with at least a 4-fold difference in IC50 value. (-)-Aricine displays weaker potency (47.7 μM) towards UCHL1. However when other members of the UCH, or USP families were tested no significant inhibition was observed at the highest inhibitor concentration of 114 μM. Acetyl isogambogic acid and rifampicin were found to be more promiscuous inhibitors, with lower IC50 values for UCHL1 (Supporting Information Table 2).
The effect of the small molecules on endogenous UCHL1 activity in HEK293T cell lysates was also investigated by assessing the inhibition of UCHL1 labeling by HA-UbVMe in the presence of the compound. Cell lysates were incubated with either 100 μM (Figure 5c), 20 μM (Figure 5d) of each compound, or equal volume of DMSO as control. We examined the inhibition of endogenous UCHL1 labeling by HA-UbVMe by immunoblotting against UCHL1. At both inhibitor concentrations, LDN-57444, celastrol, mangiferin, and acetyl isogambogic acid showed stronger inhibition among the inhibitors tested (Figure 5). The ranking of the inhibitor potency based on the inhibition of the labeling of endogenous UCHL1 agreed well with the IC50 values obtained using the recombinant UCHL1 in the fluorogenic assay. Additionally, the ranking of inhibitor potency agreed with the IC50 values determined in the qHTS: celastrol (IC50 of 4.46 μM), mangiferin (IC50 of 1.12 μM), acetyl isogambogic acid (IC50 of 30.6 μM), rifampicin (IC50 of 38.5 μM), and (-)-aricine (IC50 of 38.5 μM) (data not shown). The general agreement between the different assays in assessing the inhibition of UCHL1 by small molecules suggests that reliable evaluation and ranking of inhibitors is possible without purified DUB enzyme or complexes.
CONCLUSION
In recent years, diverse biological roles have emerged for DUBs. The connection of DUBs with many human diseases makes them promising therapeutic targets for drug development. The current DUB assay methods for HTS largely rely on recombinantly purified enzymes. Our cell lysate-based AlphaLISA assay represents a new platform for DUB inhibitor discovery and adds to the growing toolbox for studying DUBs. Because our assay affords screening in cell lysates, it better recapitulates the physiological environment of the target protein. Additionally, our assay enables screening of disease-associated DUBs that are inaccessible in recombinant form.
A challenge of using Alpha technology for HTS is the potential high hit rate due to interference by the small molecules on the AlphaLISA technology. However, this can be managed with counter-screens, including one developed in this study using biotin-Ub-HA. Through the use of counter-screens and applying filters33, we were able to identify most of the false positives, allowing us to focus on the true inhibitors for further characterization.
In addition to the current monoubiquitin–based DUB probe, other DUB probes, such as the recently developed diubiquitin-VMe probes36–39 can be implemented in this new assay to afford more physiologically relevant HTS assay. The AlphaLISA assay can also be adapted to the AlphaPlex format to allow quick screening against multiple DUBs for identification of selective DUB inhibitors. Furthermore, ubiquitin-like protein based probes (i.e. ISG15, NEDD8, SUMO) coupled with the AlphaLISA cell lysate assay will allow HTS against deISGylases, deNeddylases and deSUMOylases in whole cell lysates. This platform may also be adapted to a number of enzyme classes, including hydrolases40–43, proteases44–49, kinases50–53, phosphatases54, histone deacetylases55–57, of which activity based-probes have already been developed.
Methods
The details of the experimental procedures are provided in the Supporting Information.
Supplementary Material
Acknowledgments
We thank C. Apgar (Perkin Elmer) for the help with the Alpha technology during the initial development of this assay. We also thank K. Yang, A. Tencer, J. Cargill and L. Yuan for providing reagents used in this work. Additionally, we thank P. Gong for assistance with the MS/MS experiment and analysis, and H. Sun for qHTS data processing. We would like to thank W. J. Harper for the UCHL3-pDEST, UCHL5-pDEST, and USP15-pDEST plasmids and P. Loll for the pET-SUMO vector.
Funding Sources
This work was supported in part by the National Institutes of Health grants (R01GM097468 and R21NS085509) to Z. Zhuang. It was also supported by the Delaware COBRE program for instrumentation facilities, with a grant from the National Institute of General Medical Sciences (1 P30 GM110758-01), and, in part, by the Intramural Research Program of NCATS, NIH.
Footnotes
Supporting Information
Supporting Information Figures 1-9, Supporting Information Tables 1-2, experimental procedures and additional references are provided in the Supporting Information. The Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 2.Maurer T, Wertz IE. Length Matters: MINDY Is a New Deubiquitinase Family that Preferentially Cleaves Long Polyubiquitin Chains. Mol Cell. 2016;63(1):4–6. doi: 10.1016/j.molcel.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 3.Goldenberg SJ, McDermott JL, Butt TR, Mattern MR, Nicholson B. Strategies for the identification of novel inhibitors of deubiquitinating enzymes. Biochem Soc Trans. 2008;36(Pt 5):828–32. doi: 10.1042/BST0360828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orcutt SJ, Wu J, Eddins MJ, Leach CA, Strickler JE. Bioluminescence assay platform for selective and sensitive detection of Ub/Ubl proteases. Biochim Biophys Acta. 2012;1823(11):2079–86. doi: 10.1016/j.bbamcr.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A, Zhuang Z. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol. 2011;18(11):1390–400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, Trouplin V, Bianchi J, Aushev VN, Camonis J, Calabrese A, Borg-Capra C, Sippl W, Collura V, Boissy G, Rain JC, Guedat P, Delansorne R, Daviet L. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8(8):2286–95. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 7.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, Luci DK, Yuan B, Simeonov A, Jadhav A, Xiao H, Wang Y, Maloney DJ, Zhuang Z. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10(4):298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, Yeh LA, Cuny GD, Stein RL, Lansbury PT. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10(9):837–46. doi: 10.1016/j.chembiol.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee BH, Finley D, King RW. A High-Throughput Screening Method for Identification of Inhibitors of the Deubiquitinating Enzyme USP14. Curr Protoc Chem Biol. 2012;4(4):311–30. doi: 10.1002/9780470559277.ch120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mistry H, Hsieh G, Buhrlage SJ, Huang M, Park E, Cuny GD, Galinsky I, Stone RM, Gray NS, D’Andrea AD, Parmar K. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther. 2013;12(12):2651–62. doi: 10.1158/1535-7163.MCT-13-0103-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, Colland F. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19(4):467–77. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51(8):2363–71. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17(6):1035–43. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian X, Isamiddinova NS, Peroutka RJ, Goldenberg SJ, Mattern MR, Nicholson B, Leach C. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. Assay Drug Dev Technol. 2011;9(2):165–73. doi: 10.1089/adt.2010.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnst JL, Davies CW, Raja SM, Das C, Natarajan A. High-throughput compatible fluorescence resonance energy transfer-based assay to identify small molecule inhibitors of AMSH deubiquitinase activity. Anal Biochem. 2013;440(1):71–7. doi: 10.1016/j.ab.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geurink PP, van Tol BD, van Dalen D, Brundel PJ, Mevissen TE, Pruneda JN, Elliott PR, van Tilburg GB, Komander D, Ovaa H. Development of Diubiquitin-Based FRET Probes To Quantify Ubiquitin Linkage Specificity of Deubiquitinating Enzymes. Chembiochem. 2016;17(9):816–20. doi: 10.1002/cbic.201600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Y, Blaser G, Horrocks MH, Ruedas-Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492(7428):266–70. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritorto MS, Ewan R, Perez-Oliva AB, Knebel A, Buhrlage SJ, Wightman M, Kelly SM, Wood NT, Virdee S, Gray NS, Morrice NA, Alessi DR, Trost M. Screening of DUB activity specificity by MALDI-TOF mass spectrometry. Nat Commun. 2014;5:4763. doi: 10.1038/ncomms5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahtoe DD, Sixma TK. Layers of DUB regulation. Trends Biochem Sci. 2015;40(8):456–67. doi: 10.1016/j.tibs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Eglen RM, Reisine T, Roby P, Rouleau N, Illy C, Bossé R, Bielefeld M. The use of AlphaScreen technology in HTS: current status. Curr Chem Genomics. 2008;1:2–10. doi: 10.2174/1875397300801010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glickman JF, Wu X, Mercuri R, Illy C, Bowen BR, He Y, Sills M. A comparison of ALPHAScreen, TR-FRET, and TRF as assay methods for FXR nuclear receptors. J Biomol Screen. 2002;7(1):3–10. doi: 10.1177/108705710200700102. [DOI] [PubMed] [Google Scholar]
- 23.Von Leoprechting A, Kumpf R, Menzel S, Reulle D, Griebel R, Valler MJ, Buttner FH. Miniaturization and validation of a high-throughput serine kinase assay using the AlphaScreen platform. J Biomol Screen. 2004;9(8):719–25. doi: 10.1177/1087057104268805. [DOI] [PubMed] [Google Scholar]
- 24.Warner G, Illy C, Pedro L, Roby P, Bosse R. AlphaScreen kinase HTS platforms. Curr Med Chem. 2004;11(6):721–30. doi: 10.2174/0929867043455693. [DOI] [PubMed] [Google Scholar]
- 25.Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9(10):1149–59. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 26.Charbaut E, Redeker V, Rossier J, Sobel A. N-terminal acetylation of ectopic recombinant proteins in Escherichia coli. FEBS Lett. 2002;529(2–3):341–5. doi: 10.1016/s0014-5793(02)03421-x. [DOI] [PubMed] [Google Scholar]
- 27.Chalker JM, Bernardes GJ, Lin YA, Davis BG. Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chem Asian J. 2009;4(5):630–40. doi: 10.1002/asia.200800427. [DOI] [PubMed] [Google Scholar]
- 28.Tedaldi LM, Smith ME, Nathani RI, Baker JR. Bromomaleimides: new reagents for the selective and reversible modification of cysteine. Chem Commun (Camb) 2009;43:6583–5. doi: 10.1039/b915136b. [DOI] [PubMed] [Google Scholar]
- 29.Degasperi A, Birtwistle MR, Volinsky N, Rauch J, Kolch W, Kholodenko BN. Evaluating strategies to normalise biological replicates of Western blot data. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0087293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: pitfalls of densitometry. Electrophoresis. 2009;30(11):1845–55. doi: 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- 31.Haas JVEBJ, Iversen PW, Weidner JR. Minimum Significant Ratio – A Statistic to Assess Assay Variability. In: Sittampalam GS, Coussens NP, Brimacombe K, Grossman A, Arkin M, Auld D, Austin C, Baell J, Bejcek B, Chung TDY, Dahlin JL, Devanaryan V, Foley TL, Glicksman M, Hall MD, Hass JV, Inglese J, Iversen PW, Lal-Nag M, Li Z, McGee J, McManus O, Riss T, Trask OJ, Weidner JR, Xia M, Xu X, editors. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences; Bethesda (MD): 2004. [PubMed] [Google Scholar]
- 32.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103(31):11473–8. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jadhav A, Ferreira RS, Klumpp C, Mott BT, Austin CP, Inglese J, Thomas CJ, Maloney DJ, Shoichet BK, Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J Med Chem. 2010;53(1):37–51. doi: 10.1021/jm901070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston PA, Soares KM, Shinde SN, Foster CA, Shun TY, Takyi HK, Wipf P, Lazo JS. Development of a 384-well colorimetric assay to quantify hydrogen peroxide generated by the redox cycling of compounds in the presence of reducing agents. Assay Drug Dev Technol. 2008;6(4):505–18. doi: 10.1089/adt.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Copeland RA. Evaluation of Enzyme Inhibitors in Drug Discovery. John Wiley & Sons, Inc; 2013. Reversible Modes of Inhibitor Interactions with Enzymes; pp. 57–121. [Google Scholar]
- 36.Haj-Yahya N, Hemantha HP, Meledin R, Bondalapati S, Seenaiah M, Brik A. Dehydroalanine-based diubiquitin activity probes. Org Lett. 2014;16(2):540–3. doi: 10.1021/ol403416w. [DOI] [PubMed] [Google Scholar]
- 37.Li G, Liang Q, Gong P, Tencer AH, Zhuang Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem Commun (Camb) 2014;50(2):216–8. doi: 10.1039/c3cc47382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGouran JF, Gaertner SR, Altun M, Kramer HB, Kessler BM. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem Biol. 2013;20(12):1447–55. doi: 10.1016/j.chembiol.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulder MP, El Oualid F, ter Beek J, Ovaa H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. Chembiochem. 2014;15(7):946–9. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kidd D, Liu Y, Cravatt BF. Profiling serine hydrolase activities in complex proteomes. Biochemistry. 2001;40(13):4005–15. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96(26):14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Direct visualization of serine hydrolase activities in complex proteomes using fluorescent active site-directed probes. Proteomics. 2001;1(9):1067–71. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 43.Schwaid AG, Ruangsiriluk W, Reyes AR, Cabral S, Rajamohan F, Tu M, Ward J, Carpino PA. Development of a selective activity-based probe for glycosylated LIPA. Bioorg Med Chem Lett. 2016;26(8):1993–6. doi: 10.1016/j.bmcl.2016.02.089. [DOI] [PubMed] [Google Scholar]
- 44.Edgington LE, van Raam BJ, Verdoes M, Wierschem C, Salvesen GS, Bogyo M. An optimized activity-based probe for the study of caspase-6 activation. Chem Biol. 2012;19(3):340–52. doi: 10.1016/j.chembiol.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes CS, Shaw G, Burden RE, Scott CJ, Gilmore BF. The application of a novel, cell permeable activity-based probe for the detection of cysteine cathepsins. Biochem Biophys Res Commun. 2016;472(3):444–50. doi: 10.1016/j.bbrc.2016.01.062. [DOI] [PubMed] [Google Scholar]
- 46.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol. 2005;1(1):33–8. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen MT, Van Kersavond T, Verhelst SH. Chemical Tools for the Study of Intramembrane Proteases. ACS Chem Biol. 2015;10(11):2423–34. doi: 10.1021/acschembio.5b00693. [DOI] [PubMed] [Google Scholar]
- 48.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci U S A. 2004;101(27):10000–5. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sieber SA, Niessen S, Hoover HS, Cravatt BF. Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat Chem Biol. 2006;2(5):274–81. doi: 10.1038/nchembio781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen MS, Hadjivassiliou H, Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat Chem Biol. 2007;3(3):156–60. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JY, Stewart PA, Borne AL, Fang B, Welsh EA, Chen YA, Eschrich SA, Koomen JM, Haura EB. Activity-Based Proteomics Reveals Heterogeneous Kinome and ATP-Binding Proteome Responses to MEK Inhibition in KRAS Mutant Lung Cancer. Proteomes. 2016;4(2) doi: 10.3390/proteomes4020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, Aban A, Chun D, Tanner S, Kozarich JW. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46(2):350–8. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 53.Yee MC, Fas SC, Stohlmeyer MM, Wandless TJ, Cimprich KA. A cell-permeable, activity-based probe for protein and lipid kinases. J Biol Chem. 2005;280(32):29053–9. doi: 10.1074/jbc.M504730200. [DOI] [PubMed] [Google Scholar]
- 54.Kumar S, Zhou B, Liang F, Wang WQ, Huang Z, Zhang ZY. Activity-based probes for protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 2004;101(21):7943–8. doi: 10.1073/pnas.0402323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci U S A. 2007;104(4):1171–6. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salisbury CM, Cravatt BF. Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes. J Am Chem Soc. 2008;130(7):2184–94. doi: 10.1021/ja074138u. [DOI] [PubMed] [Google Scholar]
- 57.Xie Y, Ge J, Lei H, Peng B, Zhang H, Wang D, Pan S, Chen G, Chen L, Wang Y, Hao Q, Yao SQ, Sun H. Fluorescent Probes for Single-Step Detection and Proteomic Profiling of Histone Deacetylases. J Am Chem Soc. 2016;138(48):15596–15604. doi: 10.1021/jacs.6b07334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


