This review focuses on the luminal A subtype in breast cancer: how it is defined by immunohistochemical‐based and gene‐based assays, the prognostic and predictive value of these assays and treatment implications. With population studies projecting that half of all new breast cancer diagnoses will be of the luminal A subtype, the question of how to best determine whether a tumor is luminal A and how to best approach treatment is clinically important.
Keywords: Breast neoplasm, Genotype, Immunohistochemistry, Therapeutics
Abstract
Purpose.
Chemotherapy has been the historical mainstay of treatment for patients with breast cancer, with immunohistochemical markers and tumor characteristics driving treatment decisions. The discovery of different intrinsic subtypes of breast cancer has advanced the understanding of breast cancer, with gene‐based assays shedding further light on tumor behavior and response to treatment.
Design.
This review focuses on the landscape of the luminal A subtype, its definition based on immunohistochemistry (IHC) and gene assays, the prognostic and predictive value of these assays, guideline recommendations, and treatment implications.
Results.
Clinical studies of the prognostic value of gene‐based and IHC‐based assays in patients with luminal A‐subtype breast cancers suggest a better prognosis for these patients compared with those with breast cancers of other subtypes.
Conclusion.
In today's era of precision medicine, the best treatment regimen for patients with luminal A‐subtype tumors is still undetermined, but available data raise the question whether chemotherapy can be omitted and endocrine therapy alone is sufficient for this patient population.
Implications for Practice.
Immunohistochemical markers have traditionally guided treatment decisions in breast cancer. However, advances in gene‐expression profiling and availability of gene‐based assays have launched these newer tests into everyday clinical practice. Luminal A‐subtype tumors are a unique subset that may have favorable tumor biology. Properly defining this tumor subtype is important and may identify a subset of patients for whom endocrine therapy alone is sufficient.
摘要
目的.化疗一直是乳腺癌患者治疗历史中的主要疗法, 其免疫组化标志物和肿瘤特征决定了治疗决策。对不同亚型乳腺癌的探索促进了对乳腺癌的了解, 基于基因的检测方法进一步阐明了肿瘤行为和治疗反应。
设计.本综述侧重于Luminal A亚型、基于免疫组织化学(IHC)和基因检测得出的定义、这些检测的预后和预测价值、指南建议以及治疗影响等领域。
结果.基于基因和基于IHC的检测在Luminal A亚型乳腺癌患者中的预后价值的临床研究表明, 与其他亚型的乳腺癌患者相比, 这些患者的预后更好。
结论.在当今的精准医疗时代, 仍未确定Luminal A亚型肿瘤患者的最佳治疗方案, 但现有的数据可以提出的问题是, 化疗是否可以省略, 仅采用内分泌治疗是否足以满足该患者人群的需求。
对临床实践的提示:传统上, 免疫组织化学标志物指导乳腺癌治疗决策。然而, 基因表达谱的进展和基于基因的检测的可用性已将这些较新的检测投入到日常临床实践中。Luminal A亚型肿瘤是可能具有良好肿瘤生物学特征的独特亚型。正确定义这种肿瘤亚型很重要, 并且可能确定仅采用内分泌疗法即可满足其需求的患者亚组。
Introduction
Breast cancer is a heterogeneous disease, with treatment decisions and prognosis traditionally guided by immunohistochemistry (IHC) markers such as estrogen receptor (ER), progesterone receptor (PR), human epidermal growth receptor 2 (HER2), and Ki67 (a proliferation index marker), along with tumor size, tumor grade, and nodal status. More recently, advances in the development and validation of genomic tests and a deeper understanding of intrinsic subtypes (luminal A, luminal B, HER2‐enriched, basal, and normal breast‐like) have shed further light on tumor biology and how to best provide individualized treatment [1], [2], [3], [4]. This review focuses on the landscape of the luminal A subtype, how it is defined by IHC‐based and gene‐based assays, the prognostic and predictive value of these assays, genetic mutations and novel pathways in the luminal A subtype, and treatment implications. With population studies projecting that luminal A‐subtype breast cancers compose at least half of all new breast cancer diagnoses, the question of how best to determine whether a tumor is luminal A and how best to approach treatment becomes clinically very important [5], [6], [7], [8].
Defining Luminal A Subtype
Currently, two methods can determine subtype: gene‐based assays and IHC‐based markers. In 2011, the St. Gallen expert consensus panel adopted a subtype‐based approach for treating early breast cancer in the adjuvant setting using levels of ER, PR, Ki67 and HER2 expression [9]. Based on work by Prat et al., who determined that patients with IHC‐based luminal A tumors had better disease‐free survival (DFS) if PR was >20%, the 2013 St. Gallen update defined luminal A as ER positive (ER+), PR ≥20%, HER2 negative, Ki67 <14%, and, if available, “low” recurrence risk based on gene‐based assays [10]. Luminal B‐like (HER2‐negative) tumors are ER+, HER2 negative, and at least one of the following: Ki67 ≥20%, PR negative or <20%, and, if available, “high” recurrence risk based on multi‐gene expression assay. Luminal B‐like (HER2‐positive [HER2+]) tumors are ER+, HER2+, any Ki67 level, and any PR level. HER2+ (non‐luminal) tumors are defined as HER2+ and ER and PR negative. Triple‐negative (ductal) tumors are defined as ER, PR, and HER2 negative. These definitions are frequently used in clinical practice today. However, these IHC‐based markers are only a surrogate and cannot establish the intrinsic subtype of any given cancer, with discordance rates between IHC‐based markers and gene‐based assays as high as 30% [11], [12].
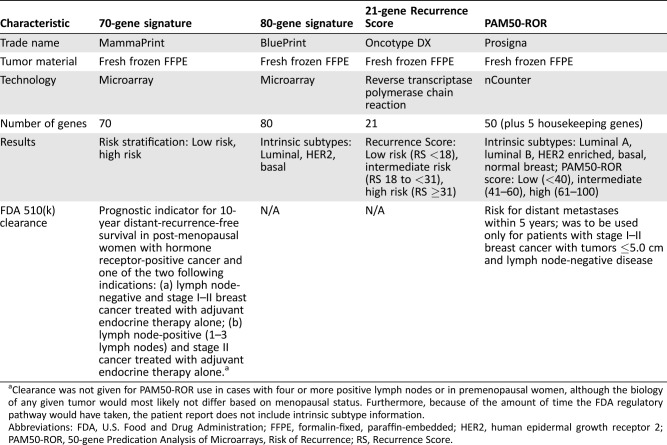
Ideally, an effective biomarker should be prognostic (determine the risk of recurrence or metastases) and predictive (determine the benefit from a given treatment). There are currently five gene‐based assays available: the 21‐gene Recurrence Score (RS; Oncotype DX; Genomic Health, Inc., Redwood City, CA), the 50‐gene Prediction Analysis of Microarrays (PAM50) and PAM50 Risk of Recurrence (PAM50‐ROR; Prosigna; NanoString Technologies, Seattle, WA), the 70‐gene and 80‐gene signatures (MammaPrint and BluePrint, respectively; Agendia, Irvine, CA), EndoPredict (Myriad Genetics, Inc., Salt Lake City, UT), and the Breast Cancer Index (BCI; Biotheranostics, Inc., San Diego, CA). This review will focus on RS, PAM50‐ROR, and the 70‐ and 80‐gene signatures (Table 1).
Table 1. Overview of molecular assays.
Clearance was not given for PAM50‐ROR use in cases with four or more positive lymph nodes or in premenopausal women, although the biology of any given tumor would most likely not differ based on menopausal status. Furthermore, because of the amount of time the FDA regulatory pathway would have taken, the patient report does not include intrinsic subtype information.
Abbreviations: FDA, U.S. Food and Drug Administration; FFPE, formalin‐fixed, paraffin‐embedded; HER2, human epidermal growth receptor 2; PAM50‐ROR, 50‐gene Predication Analysis of Microarrays, Risk of Recurrence; RS, Recurrence Score.
Paik et al. used a reverse‐transcriptase polymerase‐chain‐reaction assay on prospectively selected genes to validate the 21‐gene RS and quantify the likelihood of distant recurrence in patients with node‐negative, ER+ breast cancer treated with tamoxifen into low‐, intermediate‐, and high‐risk categories [13]. Parker et al. used a 50‐gene set, PAM50, to standardize intrinsic subtype classifications and establish a risk of relapse score that correlates with the likelihood of 10‐year recurrence [14]. Using the PAM50 gene signature, Prosigna developed a risk of recurrence (ROR) score (PAM50‐ROR) that provides a numerical score estimating the probability of distant recurrence over 10 years [15]. The 70‐gene signature, which divides patients into low‐ versus high‐risk groups corresponding to 10‐year distant‐metastasis‐free survival (DMFS), is frequently used with the 80‐gene signature (BluePrint), which distinguishes between basal, luminal, and HER2 intrinsic subtypes [16], [17], [18], [19]. The 70‐ and 80‐gene signatures combined stratify patients into luminal A‐like (luminal subtype and low‐risk), luminal B‐like (luminal subtype and high‐risk), HER2, and basal subtypes [18], [19]. Other gene‐based assays such as EndoPredict and BCI have also been studied prospectively in randomized trials [20], [21]. Of these, PAM50‐ROR and the 70‐gene signature have received U.S. Food and Drug Administration 510(k) clearance [22], [23]. The prognostic and/or predictive value of these assays have been studied in clinical trials, and some of them will be discussed below.
Prognostic and Predictive Value of Gene‐Based Assays and IHC‐Based Markers
21‐Gene Recurrence Score
The National Comprehensive Cancer Network (NCCN) guidelines emphasize the 21‐gene RS as one of the best‐validated prognostic assays [24]. Studies have shown that the 21‐gene RS predicts both response to adjuvant chemotherapy and locoregional and distant recurrence for postmenopausal patients treated with tamoxifen or an aromatase inhibitor [24]. Wolmark et al. looked at the utility of the RS in predicting late (>5 year) distant recurrence in patients with stage I and II breast cancer with high and low ESR1‐expressing groups from the National Surgical Adjuvant Breast and Bowel Project B‐14 (node‐negative, tamoxifen only) and B‐28 (node‐positive, chemotherapy and tamoxifen) trials [25]. An ESR1 cutoff of 9.1 cycle threshold units (first tertile) was found to quantify the likelihood of recurrence in patients from the B‐28 and B‐14 trials. RS was found to be prognostic for early (0–5 years, p < .001) and late (>5 years, p = .02) recurrence for B‐28 patients and early recurrence for B‐14 patients (p < .001) but not for late recurrence for B‐14 patients (p = .06). RS was associated with early distant recurrence risk for ESR1‐low and ‐high expression (p < .001), but for late recurrence for >5 years, only ESR1 >9.1 was significant (p = .003, B‐28; p = .04, B‐14).
Initial results of the ongoing TAILORx (NCT00310180) trial suggest that RS is prognostic. Of note, TAILORx defined low recurrence score as ≤10, intermediate as 11–25, and high as ≥26, compared with the original definitions in Table 1. Patients with RS <10 who received only endocrine therapy had a 5‐year invasive‐DFS rate of 93.8%, freedom from distant breast cancer recurrence rate of 99.3%, freedom from distant or local recurrence rate of 98.7%, and an overall survival (OS) of 98.0%, suggesting their tumors were most likely luminal A subtype [26]. Although these results are extremely thought provoking, more research is needed to definitively identify a specific subset of patients who would not benefit from chemotherapy. Kim et al. conducted a retrospective review of RS over a 9‐year period at five medical institutions in the United States, with the goal of developing and validating a model for predicting risk categories using clinicopathologic parameters (ER, PR, Ki67, HER2, and tumor grade), and found that histopathologic markers alone determined high‐ versus low‐risk RS categories (≤25 or >25), with greater than 95% confidence in more than 55% of cases and with the validation set predicting the risk category correctly in 52.5% of cases [27]. Klein et al. used cases with known 21‐gene RS results from a single hospital to build models (i.e., new Magee equations 1, 2, and 3) to predict an estimated RS based on IHC data such as ER, PR, HER2, and Ki67 [28]. Overall, there was 54.4%–59.4% concordance between a new Magee equation and the actual 21‐gene RS; this increased to more than 95% when the intermediate‐risk categories for the actual 21‐gene RS and Magee equation‐estimated RS were excluded. When the estimated RS fell in the intermediate category based any of the Magee equations, the actual 21‐gene RS was low or intermediate in more than 80% of cases. These results suggest that histopathologic markers may serve as surrogates in certain cases and certainly warrant further research.
PAM50‐ROR
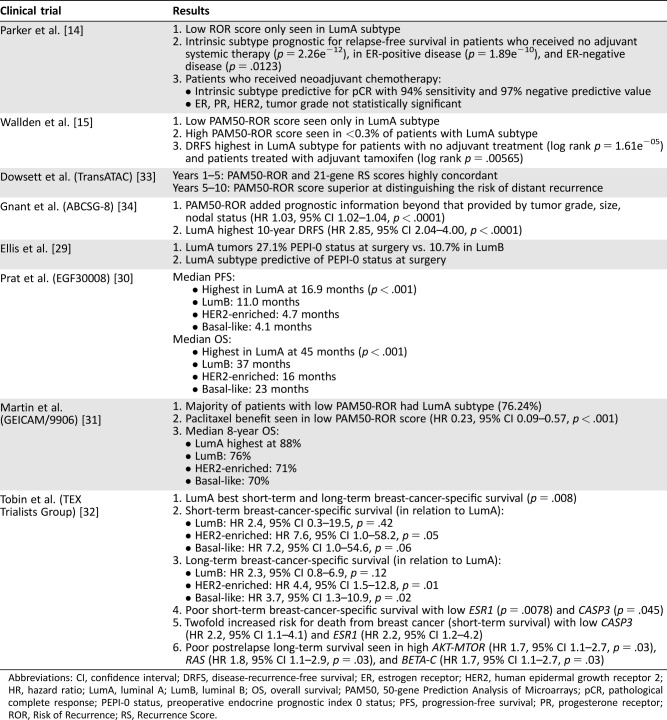
Parker et al. found the intrinsic subtype determined by PAM50 to be both predictive of pathological complete response (pCR) rate in patients who received neoadjuvant treatment and prognostic for recurrence‐free survival (RFS) [3], [14]. Subsequent studies have looked at the prognostic value of PAM50 (Table 2). Ellis et al. looked at postmenopausal patients with stage II–III, ER+, grade 2–3 breast cancer randomized to neoadjuvant aromatase inhibitors and found that preoperative endocrine prognostic index 0 (PEPI‐0) status at surgery, which predicts lower likelihood of recurrence, was higher in luminal A compared with luminal B tumors. Furthermore, luminal A subtype was the primary factor in predicting likelihood of recurrence as measured by PAM50 [29]. Prat et al. looked at EGF30008 trial tumor samples in which postmenopausal patients with hormone receptor‐positive (HR+) cancer received letrozole ± lapatinib [30]. PAM50‐based intrinsic subtype was the strongest prognostic factor associated with progression‐free survival and OS and highest in the luminal A subgroup. Martin et al. looked at GEICAM/9906 trial patients randomized to adjuvant 5‐fluorouracil, epirubicin, cyclophosphamide (FEC) ± weekly paclitaxel and showed that patients with luminal A subtype had the best 8‐year OS at 88% [31]. Tobin et al. looked at TEX Trialists Group patients with confirmed locoregional or distant relapse and showed that patients with HER2‐enriched and basal‐like subtypes had worse survival than those with luminal A [32]. These results suggest that patients with PAM50‐based luminal A subtype may have a better prognosis than those with other subtypes.
Table 2. Key clinical trials of PAM50 and PAM50‐ROR.
Abbreviations: CI, confidence interval; DRFS, disease‐recurrence‐free survival; ER, estrogen receptor; HER2, human epidermal growth receptor 2; HR, hazard ratio; LumA, luminal A; LumB, luminal B; OS, overall survival; PAM50, 50‐gene Prediction Analysis of Microarrays; pCR, pathological complete response; PEPI‐0 status, preoperative endocrine prognostic index 0 status; PFS, progression‐free survival; PR, progesterone receptor; ROR, Risk of Recurrence; RS, Recurrence Score.
Wallden et al. used the PAM50 platform to generate and validate the clinical accuracy of PAM50‐ROR [15]. PAM50‐ROR was found to be prognostic for disease‐recurrence‐free survival (DRFS), and only luminal A subtype had a low PAM50‐ROR score (Table 2) [33], [34]. The TransATAC study looked at patients who received tamoxifen or anastrozole to compare PAM50‐ROR and 21‐gene RS. Beyond 5 years, PAM50‐ROR was a better prognostic indicator [33]. The ABCSG‐8 study looked at the prognostic value of PAM50‐ROR in postmenopausal patients treated with adjuvant tamoxifen or tamoxifen followed by anastrozole and found that patients with luminal A subtype had the highest 10‐year DRFS [34]. Liu et al. looked at CALGB 9741 trial patients who received adjuvant doxorubicin, cyclophosphamide, and paclitaxel in 2‐week dose‐dense and 3‐week schedules [35]. PAM50‐based intrinsic subtype was found to be prognostic of RFS, regardless of treatment schedule, and highest in the luminal A cohort, suggesting that the better prognosis of luminal A subtype outweighed the benefit of a dose‐dense schedule. Sestak et al. compared disease recurrence (DR) of the clinical treatment score (CTS, using nodal status, grade, tumor size, age, and treatment), four IHC markers (IHC4, using IHC‐based ER, PR, Ki67, and HER2), 21‐gene RS, PAM50‐ROR, BCI, and EndoPredict in postmenopausal patients with HR+/HER2‐negative breast cancer from the TransATAC study [36]. In years 0–10, all signatures were prognostic for DR in patients with node‐negative disease. In those with node‐positive disease, PAM50‐ROR and EndoPredict identified low‐risk patients with good DR risk who would likely not need chemotherapy. Considering only years 5–10, BCI, PAM50‐ROR, and EndoPredict were prognostic for late DR. In patients with node‐positive disease, PAM50‐ROR and EndoPredict identified those at low risk of late DR. Dowsett et al. looked at the prognostic ability of PAM50‐ROR when added to the CTS (nodal status, tumor size, histopathologic grade, age, anastrozole or tamoxifen treatment), using tumor samples from the ATAC trial, and compared its performance with that of the 21‐gene RS and IHC4 in predicting the risk of DR after endocrine therapy [37]. PAM50‐ROR added more prognostic information compared with 21‐gene RS and CTS (p < .001). Compared with IHC4, PAM50‐ROR added more information in the HER2‐negative/node‐negative subgroup, but the two were similar for all patients together. Although the NCCN has acknowledged PAM50 as clinically validated for prognosis in its 2015 guidelines, ongoing studies such as the Optimal Personalised Treatment of Early Breast Cancer Using Multi‐Parameter Analysis trial (International Standard Randomised Controlled Trial Number 42400492) will hopefully shed more light on whether PAM50‐ROR guided treatment is better than standard of care [24].
The predictive value of PAM50 and PAM50‐ROR has shown mixed results. Stover et al. looked at the neoadjuvant response to an anthracycline‐taxane chemotherapy and found a high PAM50‐ROR score predictive of pCR rate [38]. Liu et al. looked at the MA.21 trial patients randomized to adjuvant anthracycline‐based chemotherapy. Patients with PAM50‐ROR‐based luminal A had the best RFS, but intrinsic subtype did not predict benefit from chemotherapy [39]. Bayraktar et al. looked at the correlation of pCR and near‐complete pCR rates of patients treated with neoadjuvant capecitabine and docetaxel ± trastuzumab using the 70‐ and 80‐gene signatures versus PAM50. Patients with 70‐ and 80‐gene signature‐based luminal A had a 7% total pCR and near‐complete pCR rate, and patients with PAM50‐based luminal A had a 10% rate. Of the total 122 tumor samples, PAM50 found 41 luminal A, whereas the 70‐ and 80‐gene signatures only found 14, suggesting a high level of discordance between the two assays [40]. Prat et al. looked at PAM50‐based intrinsic subtype of clinically HER2+ cancers from The Cancer Genome Atlas (TCGA) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) data sets [41]. Although the majority of tumors were of the HER2‐enriched subtype (47.0%), proportions were luminal B (28.2%) and luminal A (10.7%). The authors further looked at HER2 status and survival using the METABRIC data set of patients with primary resected breast cancer who did not receive adjuvant trastuzumab. Although HER2+ disease was prognostic for poorer breast cancer‐specific survival compared with HER2‐negative disease, the prognostic value of HER2 positivity disappeared when intrinsic subtype was taken into account. Patients with luminal A/HER2+ tumors had survival similar to those with luminal A/HER2‐negative tumors (hazard ratio [HR] 1.34, 95% confidence interval 0.62–2.90, p = .46), and both luminal A subgroups had better outcomes compared with other subtypes (p < .001). This suggests that HER2 amplification status was not prognostic for survival when intrinsic subtype was also taken into account.
Prognostically, CES was associated with distant relapse‐free survival in patients with node‐negative disease who were not treated with adjuvant therapy and in patients with node‐positive or ‐negative disease treated with tamoxifen.
Finally, Prat et al. retrospectively analyzed HR+/HER2‐negative tumor samples using PAM50 subtyping to derive a PAM50‐based chemoendocrine score (CES), which was calculated by looking at the tumor sample's correlation coefficients to PAM50‐based luminal A and basal‐like status [42]. CES was categorized into more chemotherapy sensitive (CES‐C) or more endocrine sensitive (CES‐E). When CES was compared with ROR score, of the samples that were ROR‐low, 94.9% were found to be CES‐E and 100% were luminal A subtype. Of the ROR‐intermediate and CES‐E samples, 77.3% were luminal A subtype. The rate of pCR was lower in the CES‐E group than in the CES‐C group across all validation data sets (p < .05 for all). CES was predictive beyond intrinsic subtype and, depending on the validation data set used, also beyond Ki67 and PAM50‐ROR score. Prognostically, CES was associated with distant relapse‐free survival in patients with node‐negative disease who were not treated with adjuvant therapy and in patients with node‐positive or ‐negative disease treated with tamoxifen (p < .0001 for both). Their results confirmed an inverse relationship between endocrine sensitivity and chemotherapy sensitivity in ER+ breast cancer and that the main driver of endocrine sensitivity within ER+/HER2‐negative tumors is whether the tumor's intrinsic biology is luminal A or basal‐like. These results suggest that the role of CES and how it could be used in clinical practice warrant further investigation.
70‐ and 80‐Gene Signature Assays
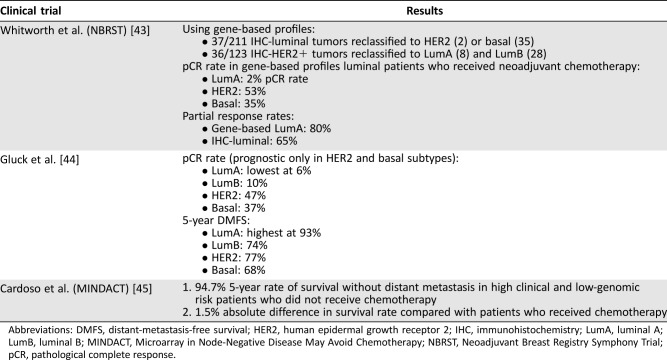
The 70‐gene and 80‐gene signatures are frequently used together to determine which subset of tumors are luminal A‐like (luminal and low‐risk). Two studies have looked at the predictive value of the 70‐ and 80‐gene signatures: the Neoadjuvant Breast Registry Symphony Trial (NBRST) and a study by Glück et al. (Table 3). The NBRST looked at IHC‐based markers versus gene‐based assays in predicting pCR or partial response in patients receiving neoadjuvant chemotherapy or endocrine therapy and showed that gene‐based luminal A subtype had the lowest pCR rate compared with other subtypes [43]. Glück et al. analyzed data from four neoadjuvant chemotherapy trials and found that although patients with luminal A tumors had the lowest pCR rate, they also had the highest 5‐year DMFS rate [44]. Taken together, these two studies suggest patients with 70‐ and 80‐gene signatures‐determined luminal A‐like tumors may have a better prognosis than other tumor subsets.
Table 3. Key clinical trials of the 70‐gene and 80‐gene signatures.
Abbreviations: DMFS, distant‐metastasis‐free survival; HER2, human epidermal growth receptor 2; IHC, immunohistochemistry; LumA, luminal A; LumB, luminal B; MINDACT, Microarray in Node‐Negative Disease May Avoid Chemotherapy; NBRST, Neoadjuvant Breast Registry Symphony Trial; pCR, pathological complete response.
MINDACT (NCT00433589) is an ongoing randomized phase III noninferiority trial looking at patients with early‐stage breast cancer to determine genomic risk and the predictive value of the 70‐gene signature using fresh tissue [45]. Patients were classified based on genomic risk (determined using 70‐gene signature) and clinical risk (using Adjuvant! Online; Adjuvant, Inc., San Antonio, TX). Of the patients at high clinical risk and low genomic risk, the absolute difference in 5‐year survival without distant metastases was 1.5% higher among those who received chemotherapy compared with those who did not (95.9% vs. 94.4%, HR 0.78, p = .27). Even among patients at low clinical risk and high genomic risk, the absolute difference in 5‐year survival was only 0.8% higher in those who received chemotherapy (95.8% vs. 95.0%, HR 1.17, p = .66). Furthermore, there were no differences between disease‐free survival and overall survival in the high clinical/low genomic or low clinical/high genomic risk groups. Assuming luminal A‐like tumors fall within the low genomic risk category, these results suggest that the benefit of adjuvant chemotherapy in this patient population is small.
IHC‐Based Markers
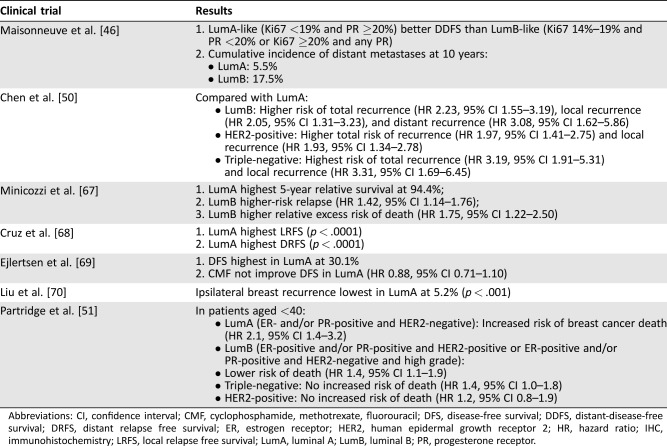
Although the data on the prognostic and predictive value of gene‐based assays raise the question of whether they should be incorporated into everyday treatment decision‐making to help individualize therapy, especially for patients with luminal A subtypes, surrogate IHC‐based markers are frequently still the preferred testing modality for establishing subtype. Studies have looked at the prognostic and predictive value of IHC‐based markers, and some are discussed below (Table 4). Maisonneuve et al. looked at distant‐disease‐free survival (DDFS) in patients treated for a first primary nonmetastatic breast cancer with varying levels of Ki67 and PR and found those with luminal A tumors had higher DDFS and lower incidence of distant metastases at 10 years compared with patients with luminal B tumors [46]. Rocca et al. looked at the benefit of first‐line endocrine therapy in patients with advanced breast cancer and found PR >20% predictive of longer time to tumor progression (p = .012) [47]. Bonnefoi et al. looked at the prognostic implications of pCR and taxane‐ versus nontaxane‐based chemotherapy and found that although patients with luminal A tumors had the lowest pCR rate at 7.5%, they had the best event‐free survival, DMFS, and OS [48]. Nielsen et al. looked at high‐risk premenopausal patients and found that patients with luminal A tumors had no benefit from chemotherapy, whereas those with other subtypes did [49]. In a meta‐analysis of the role of molecular subtypes and recurrence risk after breast‐conserving therapy, Chen et al. found luminal B, HER2+, and triple‐negative tumors had higher risks of recurrence compared with luminal A tumors [50]. Various other studies have confirmed that patients with IHC‐based luminal A tumors have a much better prognosis than those with other subtypes. Currently, the ongoing Trial of Perioperative Endocrine Therapy ‐ Indivisualising Care (NCT02338310) is looking at more than 4,000 postmenopausal patients with ER+ disease randomized to 2 weeks of neoadjuvant aromatase inhibitor versus no treatment, focusing on the clinical utility of Ki67 as a predictor of long‐term outcomes.
Table 4. Key clinical trials of IHC‐based subtypes.
Abbreviations: CI, confidence interval; CMF, cyclophosphamide, methotrexate, fluorouracil; DFS, disease‐free survival; DDFS, distant‐disease‐free survival; DRFS, distant relapse free survival; ER, estrogen receptor; HER2, human epidermal growth receptor 2; HR, hazard ratio; IHC, immunohistochemistry; LRFS, local relapse free survival; LumA, luminal A; LumB, luminal B; PR, progesterone receptor.
IHC‐Based Markers versus Gene‐Based Assay Concordance
These previous studies suggest that luminal A subtype, when determined by IHC‐based markers, has better survival even with lower pCR rates. However, Partridge et al. recently looked at IHC‐based subtypes in patients aged <40 with breast cancer and found that those with luminal A subtype had an increased risk of breast cancer death, whereas those with luminal B, HER2+, and triple‐negative subtypes did not (Table 4) [51]. This raises the question of whether these patients would also have luminal A‐subtype tumors when analyzed by gene‐based assays, whether there are different types of luminal A tumors, and whether IHC‐based markers serve as a reliable surrogate. Prat et al. reviewed the concordance between surrogate IHC‐based and PAM50‐based intrinsic subtype and found a discordance rate of 30.72% between the two classification systems [11]. Of the 637 samples thought to be IHC‐based luminal A (HR+/HER2‐negative), only 396 were PAM50‐based luminal A. Of the 317 IHC‐based luminal B samples, 108 were PAM50‐based luminal A. Chia et al. evaluated the prognostic and predictive value of PAM50‐based versus IHC‐based intrinsic subtype from patients in the National Cancer Institute of Canada Clinical Trials Group MA.12 trial [52]. Patients with PAM50‐based luminal A had the best 5‐year DFS at 84.2% and OS at 95.7%, and PAM50 was prognostic for DFS (p = .0003) and OS (p = .0002). When IHC was used to determine subtype, patients with luminal A tumors had better prognosis for both DFS and OS, but the findings were not statistically significant. PAM50 luminal subtype predicted tamoxifen benefit compared with nonluminal subtypes when looking at DFS, but neither subtyping by IHC, ER or PR, was predictive. Whitworth et al. and Cristofanilli et al. looked at the concordance of the 70‐ and 80‐gene signatures versus IHC in identifying the intrinsic subtype and found a 22%–25% discordance rate between the two [12], [43]. These studies suggest that perhaps the IHC‐based and gene‐based methods of identifying a tumor's subtype are not the same and cannot be used interchangeably.
Just as important as determining IHC‐based markers versus gene‐based assay concordance is determining concordance between gene‐based assays. Esserman et al. performed a secondary analysis of a randomized trial of tamoxifen versus no systemic therapy in postmenopausal women with node‐negative tumors smaller than 3 cm and more than 20 years’ follow‐up [53]. The 70‐gene signature was used to determine an ultralow‐risk threshold (≥0.355) above which no breast cancer deaths occurred after 15 years without systemic therapy. Patients in the ultralow‐risk category had significantly better breast‐cancer‐specific survival overall (p < .001), regardless of whether they received tamoxifen (p = .003) or were untreated (p = .004). The tamoxifen‐treated patients had a 20‐year disease‐specific survival rate of 97% versus 94% for those who did not receive tamoxifen. All ultralow‐risk tumors were HR+, HER2 negative, and luminal subtype using 80‐gene signature assay, and of these, 89% were also PAM50‐based luminal A intrinsic subtype. Conversely, however, only 25% of tumors characterized as PAM50‐based luminal A and 26% of those characterized as 80‐gene signature‐determined luminal A were found to be ultralow‐risk. Fan et al. looked at the 70‐gene signature, RS, and PAM50 to compare the prognostic value of each gene‐expression‐based model [54]. All were found to be prognostic for relapse‐free survival and overall survival, with statistically significant p values. Of the intrinsic‐subtype luminal A tumors, low RS identified 62 out of 70 samples, whereas the other models were more heterogeneous. In a direct comparison of ER+ samples, the 70‐gene assay and RS were found to be concordance in 76.9% of cases and highly correlated (p < .001). These studies question not only whether IHC‐based markers are interchangeable with gene‐based assays, but also whether gene‐based assays are interchangeable with each other.
Genetic Mutations and Novel Pathways for Luminal A
Currently, studies targeting luminal A cancers using novel pathways are underway. Santarpia et al. found PIK3CA to be the most frequently mutated gene in IHC‐based ER+/luminal cancers, with data from TCGA suggesting that the PIK3CA E545K mutation is found almost exclusively in the luminal A subtype [55]. Ciriello et al. looked at the molecular diversity of luminal A tumors and found that they had fewer mutations per sample but that the mutations tended to recur and affect similar genes. Patients with luminal A had not only the longest survival but also the most variability, with the risk of late mortality, after 10 years from diagnosis, higher than patients with other subtypes [56]. Tobin et al. found ESR1, CASP3, AKT‐MTOR, RAS, and BETA‐C genes to affect long‐term and short‐term survival (Table 2) [32]. Ross et al. looked at comprehensive genomic profiling to help identify targetable genomic alterations and reclassify the intrinsic subtypes based on sensitivity to treatment [57]. Kroemer et al. looked at the role of immunotherapy in gene‐based luminal breast cancer and found that the frequency of CD47+ circulating tumor cells correlates with metastatic spread and that their presence in ER+ tumors is a negative predictor of OS [58]. Tumor‐infiltrating lymphocytes (TILs) are also currently under investigation, and although studies suggest that TILs are higher in ER‐negative/HER2‐negative and HER2+ tumor subgroups compared with ER+/HER2‐negative subgroups, more information is needed on their prognostic and predictive value in breast cancer [59]. Results such as these suggest that not all luminal A tumors are the same and that further stratification based on mutational analyses may be needed.
Treatment Implications for Luminal A Cancers
In 2012, the Early Breast Cancer Trialists’ Collaborative Group meta‐analysis found that adding taxanes to anthracycline‐based regimens reduced the risk of recurrence and mortality [60]. The authors pointed out that no information was available on the tumor subtype and that this study was not able to directly inform about the effects of chemotherapy on low‐risk luminal A tumors, emphasizing that patients may even be harmed by the toxicities of chemotherapy. However, more recent data suggest that for patients with ER+ cancer who receive endocrine‐based therapy for only 5 years, the annual risk of distant recurrence is 1.4%–1.8% and up to 21% at 20 years even for T1N0 disease [61]. These data also raise the question of whether these patients with ER+ disease based on IHC truly have gene‐based luminal A subtype, which would predict a better prognosis, or another subtype. Lannin et al. pointed out that although tumor size can play a role in determining prognosis, a smaller but unfavorable tumor type (defined as grade 2 or 3 and ER‐negative/PR‐negative or grade 3 and ER‐negative/PR+) may have worse prognosis than a larger but favorable tumor type (defined as grade 1 and ER‐ and/or PR‐positive) [62].
More recent data suggest that for patients with ER+ cancer who receive endocrine‐based therapy for only 5 years, the annual risk of distant recurrence is 1.4%–1.8% and up to 21% at 20 years even for T1N0 disease. These data also raise the question of whether these patients with ER+ disease based on IHC truly have gene‐based luminal A subtype.
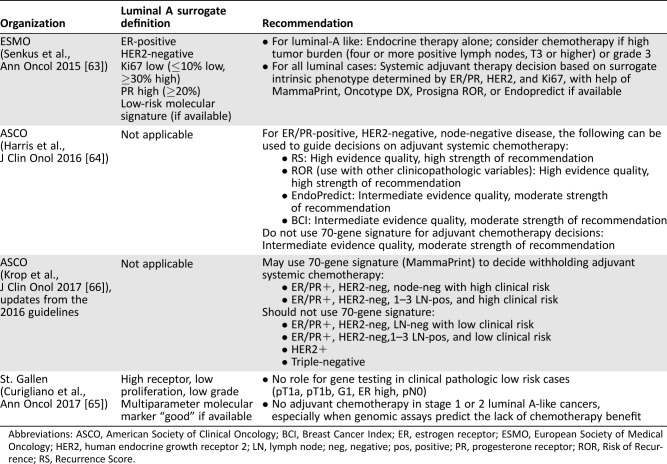
Increasingly, expert panels and societies such as the American Society of Clinical Oncology, the European Society of Medical Oncology, and the St. Gallen Expert Panel are incorporating molecular assays such as PAM50‐ROR and the 70‐gene signature into their recommendations and guidelines (Table 5) [63], [64], [65], [66]. Definitive guidelines, however, are still lacking. Currently available data suggests patients with luminal A subtype breast cancer and favorable clinical and genomic profiles may not need chemotherapy and could be treated with endocrine therapy alone. Ultimately, both tumor anatomy and tumor biology should be taken into consideration when making clinical treatment decisions.
Table 5. Guideline recommendations.
Abbreviations: ASCO, American Society of Clinical Oncology; BCI, Breast Cancer Index; ER, estrogen receptor; ESMO, European Society of Medical Oncology; HER2, human endocrine growth receptor 2; LN, lymph node; neg, negative; pos, positive; PR, progesterone receptor; ROR, Risk of Recurrence; RS, Recurrence Score.
The Future of Luminal A Breast Cancer Treatment
Our knowledge of breast cancer molecular biology and heterogeneity has significantly evolved over the past few decades. Breast cancer is no longer a single entity but rather comprises multiple subtypes, each with its own set of genomic and immunohistochemical signatures, response to treatment, and survival implications. In particular, studies suggest that patients with the luminal A subtype may have a better prognosis, raising the question of whether de‐escalating treatment and omitting chemotherapy in certain circumstances are warranted. In the current era of precision medicine, in which the goal is to neither overtreat nor undertreat patients, these individual tumor characteristics will become increasingly important in determining the right treatment for any individual patient. The authors of this review believe that given the currently available data and guideline recommendations, patients with low‐risk clinical disease and luminal A‐like tumors, whether determined by IHC or intrinsic subtyping, may not need chemotherapy. However, further definitive research is needed in this area.
Author Contributions
Conception/design: Jennifer J. Gao, Sandra M. Swain
Provision of study material or patients: Jennifer J. Gao, Sandra M. Swain
Collection and/or assembly of data: Jennifer J. Gao, Sandra M. Swain
Data analysis and interpretation: Jennifer J. Gao, Sandra M. Swain
Manuscript writing: Jennifer J. Gao, Sandra M. Swain
Final approval of manuscript: Jennifer J. Gao, Sandra M. Swain
Disclosures
Sandra M. Swain: Novartis, Genentech/Roche (H); Genentech/Roche (RF‐institutional); Inivata, IDMC, AstraZeneca (SAB); Genentech/Roche, Inivata, Caris (other‐travel). The other author indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1. Paik S, Shak S, Tang G et al. A multigene assay to predict recurrence of tamoxifen‐treated, node‐negative breast cancer. N Engl J Med 2004;351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 2. van de Vijver MJ, He YD, van't Veer LJ et al. A gene‐expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347:1999–2009. [DOI] [PubMed] [Google Scholar]
- 3. Perou CM, Sørlie T, Eisen MB et al. Molecular portraits of human breast tumours. Nature 2000;406:747–752. [DOI] [PubMed] [Google Scholar]
- 4. Sørlie T, Perou CM, Tibshirani R et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001;98:10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenberg PS, Barker KA, Anderson WF. Estrogen receptor status and the future burden of invasive and in situ breast cancers in the United States. J Natl Cancer Inst 2015;107:pii.djv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jatoi I, Anderson WF, Jeong JH et al. Breast cancer adjuvant therapy: Time to consider its time‐dependent effects. J Clin Oncol 2011;29:2301–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sweeney C, Bernard PS, Factor RE et al. Intrinsic subtypes from PAM50 gene expression assay in a population‐based breast cancer cohort: Differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev 2014;23:714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson WF, Rosenberg PS, Katki HA. Tracking and evaluating molecular tumor markers with cancer registry data: HER2 and breast cancer. J Natl Cancer Inst 2014;106:pii.dju093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldhirsch A, Wood WC, Coates AS et al. Strategies for subtypes–dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011;22:1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goldhirsch A, Winer EP, Coates AS et al. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013;24:2206–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prat A, Pineda E, Adamo B et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015;24(suppl 2):S26–S35. [DOI] [PubMed] [Google Scholar]
- 12. Cristofanilli M, Kaul K, Turk M et al. Molecular subtyping improves stratification of patients into diagnostically more meaningful risk groups. Presented at: San Antonio Breast Cancer Symposium; December 4–8, 2012; San Antonio, TX.
- 13. Paik S, Shak S, Tang G et al. A multigene assay to predict recurrence of tamoxifen‐treated, node‐negative breast cancer. N Engl J Med 2004;351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 14. Parker JS, Mullins M, Cheang MC et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wallden B, Storhoff J, Nielsen T et al. Development and verification of the PAM50‐based Prosigna breast cancer gene signature assay. BMC Med Genomics 2015;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buyse M, Loi S, van't Veer L et al. Validation and clinical utility of a 70‐gene prognostic signature for women with node‐negative breast cancer. J Natl Cancer Inst 2006;98:1183–1192. [DOI] [PubMed] [Google Scholar]
- 17. Krijgsman O, Roepman P, Zwart W et al. A diagnostic gene profile for molecular subtyping of breast cancer associated with treatment response. Breast Cancer Res Treat 2012;133:37–47. [DOI] [PubMed] [Google Scholar]
- 18. van't Veer LJ, Dai H, van de Vijver MJ et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530–536. [DOI] [PubMed] [Google Scholar]
- 19. Kittaneh M, Montero AJ, Glück S. Molecular profiling for breast cancer: A comprehensive review. Biomark Cancer 2013;5:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fitzal F, Filipits M, Rudas M et al. The genomic expression test EndoPredict is a prognostic tool for identifying risk of local recurrence in postmenopausal endocrine receptor‐positive, her2neu‐negative breast cancer patients randomised within the prospective ABCSG 8 trial. Br J Cancer 2015;112:1405–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sgroi DC, Sestak I, Cuzick J et al. Prediction of late distant recurrence in patients with oestrogen‐receptor‐positive breast cancer: A prospective comparison of the breast‐cancer index (BCI) assay, 21‐gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol 2013;14:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.U.S. Food and Drug Administration. 510(k) Decision Summary for Prosigna Breast Cancer Prognostic Gene Signature Assay. K130010. In: 510(k) Premarket Notification [database]. September 6, 2013.
- 23.U.S. Food and Drug Administration. 510(k) Decision Summary for MammaPrint FFPE. K141142. In: 510(k) Premarket Notification [database]. January 23, 2015.
- 24.National Comprehe nsive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Breast Cancer. Version 2.2017. 2017.
- 25. Wolmark N, Mamounas EP, Baehner FL et al. Prognostic impact of the combination of recurrence score and quantitative estrogen receptor expression (ESR1) on predicting late distant recurrence risk in estrogen receptor‐positive breast cancer after 5 years of tamoxifen: Results from NRG Oncology/National Surgical Adjuvant Breast and Bowel Project B‐28 and B‐14. J Clin Oncol 2016;34:2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sparano JA, Gray RJ, Makower DF et al. Prospective validation of a 21‐gene expression assay in breast cancer. N Engl J Med 2015;373:2005–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim HS, Umbricht CB, Illei PB et al. Optimizing the use of gene expression profiling in early‐stage breast cancer. J Clin Oncol 2016;34:4390–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klein ME, Dabbs DJ, Shuai Y et al. Prediction of the Oncotype DX recurrence score: Use of pathology‐generated equations derived by linear regression analysis. Mod Pathol 2013;26:658–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ellis MJ, Suman VJ, Hoog J et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor‐rich stage 2 to 3 breast cancer: Clinical and biomarker outcomes and predictive value of the baseline PAM50‐based intrinsic subtype–ACOSOG Z1031. J Clin Oncol 2011;29:2342–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prat A, Cheang MC, Galvan P et al. Prognostic value of intrinsic subtypes in hormone receptor‐positive metastatic breast cancer treated with letrozole with or without lapatinib. JAMA Oncol 2016;2:1287–1294. [DOI] [PubMed] [Google Scholar]
- 31. Martin M, Brase JC, Ruiz A et al. Prognostic ability of EndoPredict compared to research‐based versions of the PAM50 risk of recurrence (ROR) scores in node‐positive, estrogen receptor‐positive, and HER2‐negative breast cancer. A GEICAM/9906 sub‐study. Breast Cancer Res Treat 2016;156:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tobin NP, Harrell JC, Lövrot J et al. Molecular subtype and tumor characteristics of breast cancer metastases as assessed by gene expression significantly influence patient post‐relapse survival. Ann Oncol 2015;26:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dowsett M, Cuzick J, Wale C et al. Prediction of risk of distant recurrence using the 21‐gene recurrence score in node‐negative and node‐positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: A TransATAC study. J Clin Oncol 2010;28:1829–1834. [DOI] [PubMed] [Google Scholar]
- 34. Gnant M, Filipits M, Greil R et al. Predicting distant recurrence in receptor‐positive breast cancer patients with limited clinicopathological risk: Using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG‐8 trial treated with adjuvant endocrine therapy alone. Ann Oncol 2014;25:339–345. [DOI] [PubMed] [Google Scholar]
- 35. Liu MC, Pitcher BN, Mardis ER et al. PAM50 gene signature is prognostic for breast cancer patients treated with adjuvant anthracycline and taxane based chemotherapy. Cancer Res 2012;72(suppl 24):P2–10–01A. [Google Scholar]
- 36. Sestak I, Buus R, Cuzick J et al. Comprehensive comparison of prognostic signatures for breast cancer in TransATAC. Cancer Res 2017;77(suppl 4):S6–05A. [Google Scholar]
- 37. Dowsett M, Sestak I, Lopez‐Knowles E et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol 2013;31:2783–2790. [DOI] [PubMed] [Google Scholar]
- 38. Stover DG, Coloff JL, Barry WT et al. The role of proliferation in determining response to neoadjuvant chemotherapy in breast cancer: A gene expression‐based meta‐analysis. Clin Cancer Res 2016;22:6039–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu S, Chapman JA, Burnell MJ et al. Prognostic and predictive investigation of PAM50 intrinsic subtypes in the NCIC CTG MA.21 phase III chemotherapy trial. Breast Cancer Res Treat 2015;149:439–448. [DOI] [PubMed] [Google Scholar]
- 40. Bayraktar S, Royce M, Stork‐Sloots L et al. Molecular subtyping predicts pathologic tumor response in early‐stage breast cancer treated with neoadjuvant docetaxel plus capecitabine with or without trastuzumab chemotherapy. Med Oncol 2014;31:163. [DOI] [PubMed] [Google Scholar]
- 41. Prat A, Carey LA, Adamo B et al. Molecular features and survival outcomes of the intrinsic subtypes within HER2‐positive breast cancer. J Natl Cancer Inst 2014;106:pii.dju152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prat A, Lluch A, Turnbull AK et al. A PAM50‐based chemoendocrine score for hormone receptor‐positive breast cancer with an intermediate risk of relapse. Clin Cancer Res 2017;23:3035–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitworth P, Stork‐Sloots L, de Snoo FA et al. Chemosensitivity predicted by BluePrint 80‐gene functional subtype and MammaPrint in the Prospective Neoadjuvant Breast Registry Symphony Trial (NBRST). Ann Surg Oncol 2014;21:3261–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Glück S, de Snoo F, Peeters J et al. Molecular subtyping of early‐stage breast cancer identifies a group of patients who do not benefit from neoadjuvant chemotherapy. Breast Cancer Res Treat 2013;139:759–767. [DOI] [PubMed] [Google Scholar]
- 45. Cardoso F, van't Veer LJ, Bogaerts J et al. 70‐Gene signature as an aid to treatment decisions in early‐stage breast cancer. N Engl J Med 2016;375:717–729. [DOI] [PubMed] [Google Scholar]
- 46. Maisonneuve P, Disalvatore D, Rotmensz N et al. Proposed new clinicopathological surrogate definitions of luminal A and luminal B (HER2‐negative) intrinsic breast cancer subtypes. Breast Cancer Res 2014;16:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rocca A, Farolfi A, Maltoni R et al. Efficacy of endocrine therapy in relation to progesterone receptor and Ki67 expression in advanced breast cancer. Breast Cancer Res Treat 2015;152:57–65. [DOI] [PubMed] [Google Scholar]
- 48. Bonnefoi H, Litière S, Piccart M et al. Pathological complete response after neoadjuvant chemotherapy is an independent predictive factor irrespective of simplified breast cancer intrinsic subtypes: A landmark and two‐step approach analyses from the EORTC 10994/BIG 1‐00 phase III trial. Ann Oncol 2014;25:1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nielsen TO, Jensen MB, Burugu S et al. High‐risk premenopausal luminal A breast cancer patients derive no benefit from adjuvant cyclophosphamide‐based chemotherapy: Results from the DBCG77B clinical trial. Clin Cancer Res 2017;23:946–953. [DOI] [PubMed] [Google Scholar]
- 50. Chen J, Jiang P, Wang HJ et al. The efficacy of molecular subtyping in predicting postoperative recurrence in breast‐conserving therapy: A 15‐study meta‐analysis. World J Surg Oncol 2014;12:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Partridge AH, Hughes ME, Warner ET et al. Subtype‐dependent relationship between young age at diagnosis and breast cancer survival. J Clin Oncol 2016;34:3308–3314. [DOI] [PubMed] [Google Scholar]
- 52. Chia SK, Bramwell VH, Tu D et al. A 50‐gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res 2012;18:4465–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Esserman LJ, Yau C, Thompson CK et al. Use of molecular tools to identify patients with indolent breast cancers with ultralow risk over 2 decades. JAMA Oncol 2017;3:1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fan C, Oh DS, Wessels L et al. Concordance among gene‐expression‐based predictors for breast cancer. N Engl J Med 2006;355:560–569. [DOI] [PubMed] [Google Scholar]
- 55. Santarpia L, Bottai G, Kelly CM et al. Deciphering and targeting oncogenic mutations and pathways in breast cancer. The Oncologist 2016;21:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ciriello G, Sinha R, Hoadley KA et al. The molecular diversity of Luminal A breast tumors. Breast Cancer Res Treat 2013;141:409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ross JS, Gay LM, Elvin JA et al. Comprehensive genomic profiling of 8,654 breast carcinomas reveals therapeutically targetable molecular subtypes beyond those defined by hormone‐receptor expression: Abstract presented at the European Society for Medical Oncology 2016 Congress; October 7–11, 2016; Copenhagen, Denmark; 229PD.
- 58. Kroemer G, Senovilla L, Galluzzi L et al. Natural and therapy‐induced immunosurveillance in breast cancer. Nat Med 2015;21:1128–1138. [DOI] [PubMed] [Google Scholar]
- 59. Ingold Heppner B, Untch M, Denkert C et al. Tumor‐infiltrating lymphocytes: A predictive and prognostic biomarker in neoadjuvant treated HER2‐positive breast cancer. Clin Cancer Res 2016;22:5747–5754. [DOI] [PubMed] [Google Scholar]
- 60.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) ; Peto R, Davies C, Godwin J et al. Comparisons between different polychemotherapy regimens for early breast cancer: Meta‐analyses of long‐term outcome among 100,000 women in 123 randomised trials. Lancet 2012;379:432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pan H, Gray RG, Davies C et al. Predictors of recurrence during years 5–14 in 46,138 women with ER+ breast cancer allocated 5 years only of endocrine therapy (ET). J Clin Oncol 2016;34(suppl 15):505A. [Google Scholar]
- 62. Lannin DR, Wang S. Are small breast cancers good because they are small or small because they are good? N Engl J Med 2017;376:2286–2291. [DOI] [PubMed] [Google Scholar]
- 63. Senkus E, Kyriakides S, Ohno S et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2015;26(suppl 5):v8–v30. [DOI] [PubMed] [Google Scholar]
- 64. Harris LN, Ismaila N, McShane LM et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early‐stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 2016;34:1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Curigliano G, Burstein HJ, P Winer E et al. De‐escalating and escalating treatments for early‐stage breast cancer: The St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Ann Oncol 2017;28:1700–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Krop I, Ismaila N, Andre F et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early‐stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline focused update. J Clin Oncol 2017;35:2838–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Minicozzi P, Bella F, Toss A et al. Relative and disease-free survival for breast cancer in relation to subtype: A population-based study. J Cancer Res Clin Oncol 2013;139:1569–1577. [DOI] [PubMed] [Google Scholar]
- 68. Cruz RP, Pedrini JL, Zettler CG et al. How to identify patients with increased risk of breast cancer relapse? Appl Immunohistochem Mol Morphol 2014;22:488–497. [DOI] [PubMed] [Google Scholar]
- 69. Ejlertsen B, Jensen MB, Elversang J et al. One year of adjuvant tamoxifen compared with chemotherapy and tamoxifen in postmenopausal patients with stage II breast cancer. Eur J Cancer 2013;49:2986–2994. [DOI] [PubMed] [Google Scholar]
- 70. Liu FF, Shi W, Done SJ et al. Identification of a low-risk luminal A breast cancer cohort that may not benefit from breast radiotherapy. J Clin Oncol 2015;33:2035–2040. [DOI] [PubMed] [Google Scholar]