The crystal structure of a supramolecular lithium complex of p-tert-butylcalix[4]arene has been determined and analyzed. Different from the majority of calixarene–alkali metal complexes, which are formed by direct coordination of the metal cation to the calixarene hydroxy groups, this complex is stabilized by an interplay of weak interactions involving the methanol molecules surrounding the metal, giving rise to a second-sphere coordination supramolecular assembly.
Keywords: crystal structure, calix[4]arene, supramolecular lithium complex, inclusion compound, hydrogen bonding, C—H⋯π interactions
Abstract
Crystals of a supramolecular lithium complex with a calix[4]arene derivative, namely tetramethanollithium 5,11,17,23-tetra-tert-butyl-25,26,27-trihydroxy-28-oxidocalix[4]arene methanol monosolvate, [Li(CH3OH)4](C44H55O4)·CH3OH or [Li(CH3OH)4]+·(calix[4]arene−)]·CH3OH (where calix[4]arene− represents a mono-anion species because of deprotonation of one H atom of the calixarene hydroxy groups), were obtained from p-tert-butylcalix[4]arene reacted with LiH in tetrahydrofuran, followed by recrystallization from methanol. The asymmetric unit comprises one mono-anionic calixarene molecule, one Li+ cation coordinated to four methanol molecules, and one methanol molecule included in the calixarene cavity. The calixarene molecule maintains a cone conformation by intramolecular hydrogen bonding between one phenoxide (–O−) and three pendent calixarene hydroxy groups (–OH). The coordinated methanol molecules around the metal cation play a significant role in forming the supramolecular assembly. The crystal structure of this assembly is stabilized by three sets of intermolecular interactions: (i) hydrogen bonds involving the –OH and –O− moieties of the calixarene molecules, the –OH groups of the coordinated methanol molecules, and the –OH group of the methanol molecule included in the calixarene cavity; (ii) C—H⋯π interactions between the calixarene molecules and/or the coordinated methanol molecules; (iii) O—H⋯π interactions between the calixarene molecule and the included methanol molecule.
Chemical context
Calixarenes are synthetic macrocyclic compounds that are composed of phenol rings, linked with methylene groups at linking positions (Gutsche, 1998 ▸). They are versatile molecules for the inclusion of organic and/or inorganic compounds into their flexible cavities and for the coordination of organic/metal ions in molecular recognition phenomena and host–guest chemistry (Vicens & Böhmer, 1991 ▸). The coordination chemistry of alkali metal cations, involving conventional calixarenes (and their corresponding functionalized derivatives) as ligands, has been intensively investigated in the past years, as a possible method of selective extraction of this class of cations using calixarenes as extractant. At the same time, the X-ray analysis of alkali metal complexes with p-tert-butylcalix[4]arene in the crystalline state has been reported (Bock et al., 1995 ▸; Davidson et al., 1997 ▸; Dürr et al., 2006 ▸; Gueneau et al., 2003 ▸; Guillemot et al., 2002 ▸; Hamada et al., 1993 ▸; Hanna et al., 2002 ▸, 2003 ▸; Harrowfield et al., 1991 ▸; Lee et al., 2009 ▸). In the majority of cases, the alkali metal complexes of p-tert-butylcalix[4]arene in the solid state show direct coordination of the metal ions to the oxygen atoms belonging to the calixarene hydroxy groups at the lower rim, with the resulting crystal structures stabilized by weak interactions with the lattice solvent molecules.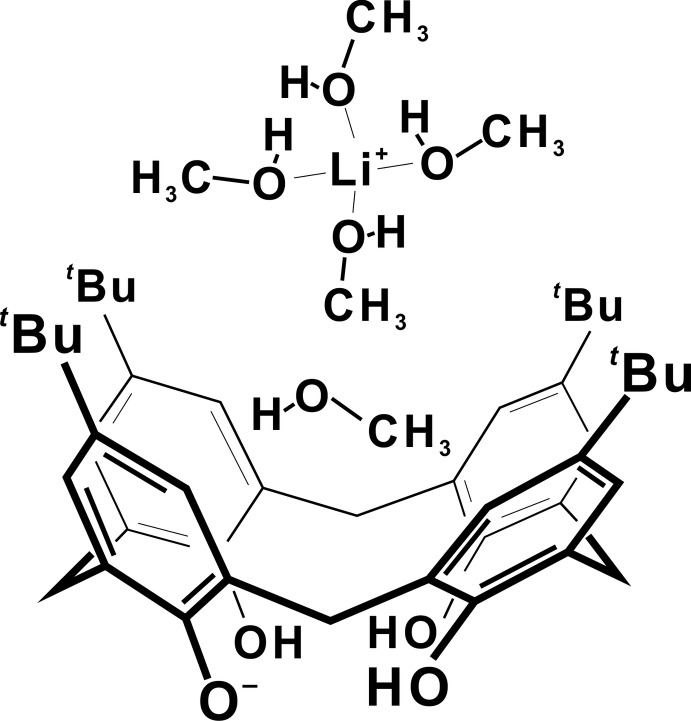
In the present paper, we report a different type of Li complex with p-tert-butylcalix[4]arene, in which no direct coordination of the metal to the oxygen atoms of the calixarene hydroxy groups takes place. The lithium cation is instead surrounded by four methanol solvent molecules, which are in turn connected to the host molecule via a series of hydrogen bonds, playing a significant role in the formation of the supramolecular assembly.
Structural commentary
Fig. 1 ▸ shows the molecular structure of the complex [Li(CH3OH)4]+·(calix[4]arene−)]·CH3OH, consisting of one mono-deprotonated calix[4]arene unit in a cone conformation, one methanol molecule included in the cavity, and one Li cation coordinated to four methanol molecules. The positive charge of the methanol–lithium complex naturally dictates that the calixarene is in a mono-anionic form. The conformation of the macrocycle is stabilized by intramolecular hydrogen bonding involving one deprotonated –O− and three –OH groups at the lower rim, as shown in Table 1 ▸. The geometrical parameters of the cone conformer are given in Table 2 ▸, which reports the angle between the mean plane passing through the oxygen atoms O1, O2, O3 and O4, and the four mean planes passing through the aromatic walls (plane A: C1–C6/O1; plane B: C7–C12/O2; plane C: C13–C18/O4; plane D: C19–C24/O3). From these values, it is possible to notice that the two neighboring aromatic rings (C1–C6 and C7–C12) are slightly outward with respect to the other two adjacent aromatic moieties. Selected bond distances and angles for the tetrakis(methanol)–lithium complex are reported in Table 3 ▸.
Figure 1.
ORTEP diagram of the Li complex of p-tert-butylcalix[4]arene with displacement ellipsoids at the 20% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O8—H74⋯O9 | 0.67 (3) | 2.01 (8) | 2.673 (3) | 167 (3) |
| O2—H68⋯O1 | 0.83 (3) | 1.66 (4) | 2.490 (2) | 172 (3) |
| O3—H69⋯O1 | 0.89 (3) | 1.64 (3) | 2.520 (2) | 169 (3) |
| O4—H70⋯O2 | 0.90 (3) | 1.77 (3) | 2.650 (2) | 166 (3) |
| O5—H71⋯O1i | 0.88 (4) | 1.87 (4) | 2.714 (3) | 160 (4) |
| O6—H72⋯O4ii | 0.94 (5) | 1.81 (5) | 2.732 (3) | 165 (4) |
| O7—H73⋯O3i | 0.79 (6) | 1.91 (6) | 2.676 (3) | 163 (6) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Table 2. Conformation of the four aromatic walls of the calix[4]arene host (°).
A–D are the mean planes passing through the four phenyl moieties of the host. The values reported are the angles formed with the mean plane passing through atoms O1–O4.
| Plane | Angle |
|---|---|
| A | 136.01 (6) |
| B | 136.80 (6) |
| C | 108.21 (6) |
| D | 119.02 (6) |
Table 3. Selected geometric parameters (Å, °).
| Li1—O5 | 1.922 (6) | Li1—O7 | 1.903 (6) |
| Li1—O6 | 1.917 (6) | Li1—O8 | 1.922 (6) |
| O5—Li1—O6 | 107.2 (3) | O6—Li1—O7 | 112.3 (3) |
| O5—Li1—O7 | 111.3 (3) | O6—Li1—O8 | 109.9 (3) |
| O5—Li1—O8 | 111.0 (3) | O7—Li1—O8 | 105.3 (3) |
As shown in Fig. 2 ▸, one methanol molecule is included in the cavity, displaying a short O—H⋯π interaction involving the hydroxy moiety and π-electrons of the calixarene aromatic ring C1–C6. The O9⋯Cg1 and the H75⋯Cg1 distances are 3.360 (6) and 2.538 (5) Å, respectively, while the angle O9—H79⋯Cg1 is of 166.34 (6)° (Cg1 is the centroid of the C1–C6 ring). On the other hand, there are no C—H⋯π interactions between the embedded methanol and the aromatic-π electrons of the calixarene, hence the included solvent is stabilized inside the calixarene cavity only by the O—H⋯π interaction.
Figure 2.
Hydrogen bonds (blue dotted lines) involving the p-tert-butylcalix[4]arene anion, the methanol molecule included in the cavity, and the [Li(CH3OH)4]+ complex belonging to the asymmetric unit. The centroid of aromatic the ring, Cg1, is represented as a blue sphere. The H atoms of the calixarene host have been omitted for clarity.
Supramolecular features
The relevant feature of the title complex is that the lithium cation is not directly coordinated to the hydroxy groups of the lower rim of the calix[4]arene host. On the contrary, the interaction of the [Li(CH3OH)4]+ complex with the macrocycle in the asymmetric unit is mediated by the methanol molecule embedded in the cavity, which acts as hydrogen-bond acceptor for a methanol molecule (C48–O8) coordinated to the lithium cation (Fig. 2 ▸ and Table 1 ▸).
Moreover, the coordinated methanol molecules of [Li(CH3OH)4]+ further contribute to the stabilization of the complex in the structure, interacting with two other adjacent calixarene molecules through hydrogen bonds and C—H⋯π interactions, as illustrated in Fig. 3 ▸ and Table 1 ▸. In particular, three of the coordinated methanol molecules (C45–O5, C47–O7 and C46–O6), act as hydrogen-bond donors towards the hydroxy groups at the lower rim of the macrocycle, namely O1i, O3i and O4ii, respectively [symmetry codes: (i) −x +  , y +
, y +  , −z +
, −z +  ; (ii) x +
; (ii) x +  , −y +
, −y +  , z +
, z +  ]. In addition, the fourth coordinated methanol molecule C48–O8 interacts with the aromatic-π electrons of a calixareneii
via a C—H⋯π interaction. The C48⋯C17ii and C48—H64⋯C17ii distances are 3.603 (4) and 2.628 Å, respectively, with a C48—H64⋯C17ii angle of 173.3 (8)°.
]. In addition, the fourth coordinated methanol molecule C48–O8 interacts with the aromatic-π electrons of a calixareneii
via a C—H⋯π interaction. The C48⋯C17ii and C48—H64⋯C17ii distances are 3.603 (4) and 2.628 Å, respectively, with a C48—H64⋯C17ii angle of 173.3 (8)°.
Figure 3.
Hydrogen bonding (blue and green dotted lines) involving the [Li(CH3OH)4]+ complex and two adjacent calix[4]arene molecules in the crystal structure. [Symmetry codes: (i)  − x,
− x,  + y,
+ y,  − z; (ii)
− z; (ii)  + x,
+ x,  − y,
− y,  + z.]
+ z.]
Similarly, C—H⋯π interactions are also present between tert-butyl groups at the upper rim of the macrocycle and π-electrons of the aromatic walls of adjacent calix[4]arenes. In particular, Fig. 4 ▸ shows the spatial arrangement of four symmetry-related host molecules [the C40⋯C4i and C40—H41⋯C4i distances are 3.498 (4) and 2.770 Å, respectively and the C40—H41⋯C4i angle is 131.6 (5)° while the C42⋯C10iii and C42—H46⋯C10iii distances are 3.770 (5) and 2.828 Å, and the C42—H46⋯C10iii angle is 161.7 (8)°; symmetry code: (iii) 1 + x, y, z].
Figure 4.
C—H⋯π interactions involving four adjacent calix[4]arene anions in the crystal structure. [Symmetry codes: (i)  − x,
− x,  + y,
+ y,  − z; (iii) 1 + x, y, z.]
− z; (iii) 1 + x, y, z.]
Database survey
A search in the Cambridge Structural Database (Version 5.38, update May 2017; Groom et al., 2016 ▸) based on a fragment comprising alkali metals and unsubstituted p-tert-butylcalix[4]arenes, yielded the structures of several compounds.
In particular, inclusion complexes were found with: (i) lithium (ZESGIN, Bock et al., 1995 ▸; RILNOP and RILNUV, Davidson et al., 1997 ▸; YEMQIR, Dürr et al., 2006 ▸; RUWVIO and RUWVOU, Gueneau et al., 2003 ▸; NASWEJ, Hamada et al., 1993 ▸; QUBJIH, Lee et al., 2009 ▸; BASWEY, Hanna et al., 2003 ▸); (ii) sodium (MODYIN, Guillemot et al., 2002 ▸; NASSEF, Hamada et al., 1993 ▸); (iii) potassium (MODYOT, Guillemot et al., 2002 ▸; NASXUA, Hamada et al., 1993 ▸; RUWVUA, Gueneau et al., 2003 ▸; WUHVUQ and WUHWAX, Hanna et al., 2002 ▸); (iv) rubidium (BASTUL, Hanna et al., 2003 ▸); (v) cesium (JIVKEE, Harrowfield et al., 1991 ▸).
In all the cases reported, the alkali metals interact with the calix[4]arene molecules through the hydroxy groups at the lower rim. The only exception is the complex with cesium, JIVKEE, in which the bare cation is placed well inside the cavity, on the quaternary axis passing through the macrocycle. The metal is involved in a polyhapto coordination with the four phenolate rings of the calix[4]arene, on which the negative charge is delocalized (Harrowfield et al., 1991 ▸). This coordination mode is probably possible due to the dimensions of Cs+, which matches the cavity in size. In the case of lithium, the cationic radius is much smaller, hence a direct cavity–cation interaction is less favoured, and the metal is either coordinating the hydroxy oxygen atoms, or forming a second-sphere coordination supramolecular complex, like in the title compound.
Synthesis and crystallization
To a white suspension of p-tert-butylcalix[4]arene (2.00 g, 3.08 mmol) in THF (50 mL) was added LiH (0.245 g, 30.8 mmol), and a yellow suspension was obtained. The suspended mixture was stirred at room temperature for 5 h under a nitrogen atmosphere, after which time, the mixture became a yellow clear solution. After quenching the excess of LiH with methanol, the solvent was removed in vacuo. The resulting yellow solid material was dissolved in methanol (80 mL) and the remaining insoluble matter was filtered off. The clear solution thus obtained was allowed to stand for several weeks to get colorless, thin plate-shaped crystals of the molecular adduct of the title compound. IR (ATR): ν 2952.40 (m), 1478.65 (s), 1360.61 (m) cm−1; 1H NMR (300 MHz, CDCl3, TMS): δ 7.04 (s, 8H, Ar–H), 4.25 (s, 4H, –CH2–), 3.46 (s, 4H, –CH2–), 3.46 (s, 15H, –CH–, five methanol molecules), 1.21 (m, 36H, tert-butyl).
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. The C-bound H atoms were placed in calculated positions and refined using a riding model: C—H = 0.95–0.98 Å with U iso(H) = 1.5U eq(C-methyl) and 1.2U eq(C) for other H atoms. H atoms on O atoms were located in the difference-Fourier map and refined with U iso(H) = 1.5U eq(O).
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | [Li(CH3OH)4](C44H55O4)·CH3OH |
| M r | 815.03 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 200 |
| a, b, c (Å) | 12.8434 (4), 20.0919 (6), 19.3168 (6) |
| β (°) | 92.561 (2) |
| V (Å3) | 4979.7 (3) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.58 |
| Crystal size (mm) | 0.20 × 0.20 × 0.10 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Bruker 2006 ▸) |
| T min, T max | 0.893, 0.945 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 41849, 8251, 6715 |
| R int | 0.021 |
| (sin θ/λ)max (Å−1) | 0.588 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.065, 0.203, 1.06 |
| No. of reflections | 8251 |
| No. of parameters | 557 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 1.46, −0.39 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989018001834/xi2008sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018001834/xi2008Isup2.hkl
CCDC reference: 1563055
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [Li(CH3OH)4](C44H55O4)·CH3OH | F(000) = 1776 |
| Mr = 815.03 | Dx = 1.087 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54178 Å |
| a = 12.8434 (4) Å | Cell parameters from 9823 reflections |
| b = 20.0919 (6) Å | θ = 3.2–63.8° |
| c = 19.3168 (6) Å | µ = 0.58 mm−1 |
| β = 92.561 (2)° | T = 200 K |
| V = 4979.7 (3) Å3 | Plane, colorless |
| Z = 4 | 0.20 × 0.20 × 0.10 mm |
Data collection
| Bruker APEXII CCD diffractometer | 8251 independent reflections |
| Radiation source: fine-focus sealed tube | 6715 reflections with I > 2σ(I) |
| Detector resolution: 8.333 pixels mm-1 | Rint = 0.021 |
| φ and ω scans | θmax = 65.0°, θmin = 3.2° |
| Absorption correction: multi-scan (SADABS; Bruker 2006) | h = −14→14 |
| Tmin = 0.893, Tmax = 0.945 | k = −22→23 |
| 41849 measured reflections | l = −22→22 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.065 | Hydrogen site location: mixed |
| wR(F2) = 0.203 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.1063P)2 + 3.6084P] where P = (Fo2 + 2Fc2)/3 |
| 8251 reflections | (Δ/σ)max < 0.001 |
| 557 parameters | Δρmax = 1.46 e Å−3 |
| 0 restraints | Δρmin = −0.39 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement was performed using all reflections. The weighted R-factor (wR) and goodness of fit (S) are based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 sigma(F2) is used only for calculating R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.76784 (19) | 0.08132 (12) | 0.95506 (12) | 0.0392 (5) | |
| C2 | 0.66560 (18) | 0.09379 (12) | 0.93080 (12) | 0.0373 (5) | |
| H1 | 0.621091 | 0.118191 | 0.959486 | 0.045* | |
| C3 | 0.62623 (17) | 0.07212 (11) | 0.86666 (11) | 0.0337 (5) | |
| C4 | 0.69117 (17) | 0.03573 (11) | 0.82392 (11) | 0.0326 (5) | |
| C5 | 0.79496 (17) | 0.02383 (11) | 0.84604 (12) | 0.0345 (5) | |
| C6 | 0.83058 (18) | 0.04662 (12) | 0.91089 (12) | 0.0380 (5) | |
| H2 | 0.900898 | 0.038046 | 0.925538 | 0.046* | |
| C7 | 0.45550 (17) | 0.26708 (12) | 0.78857 (12) | 0.0385 (5) | |
| C8 | 0.46863 (17) | 0.26210 (13) | 0.71766 (12) | 0.0387 (5) | |
| H3 | 0.457513 | 0.300562 | 0.689690 | 0.046* | |
| C9 | 0.49727 (16) | 0.20330 (12) | 0.68618 (12) | 0.0364 (5) | |
| C10 | 0.51265 (16) | 0.14659 (12) | 0.72660 (12) | 0.0349 (5) | |
| C11 | 0.50132 (16) | 0.14914 (12) | 0.79826 (11) | 0.0344 (5) | |
| C12 | 0.47390 (17) | 0.20927 (12) | 0.82756 (12) | 0.0369 (5) | |
| H4 | 0.467306 | 0.211298 | 0.876300 | 0.044* | |
| C13 | 0.78349 (19) | 0.26506 (12) | 0.57083 (12) | 0.0395 (5) | |
| C14 | 0.83391 (19) | 0.20539 (12) | 0.55992 (12) | 0.0390 (6) | |
| H5 | 0.905410 | 0.206537 | 0.549319 | 0.047* | |
| C15 | 0.78502 (18) | 0.14371 (12) | 0.56372 (11) | 0.0354 (5) | |
| C16 | 0.67899 (18) | 0.14313 (12) | 0.57703 (11) | 0.0354 (5) | |
| C17 | 0.62574 (18) | 0.20153 (12) | 0.59032 (11) | 0.0371 (5) | |
| C18 | 0.67867 (19) | 0.26118 (13) | 0.58751 (12) | 0.0410 (6) | |
| H6 | 0.642613 | 0.301143 | 0.597261 | 0.049* | |
| C19 | 1.05823 (18) | 0.09337 (12) | 0.70485 (13) | 0.0418 (6) | |
| C20 | 1.01017 (17) | 0.05755 (12) | 0.75598 (13) | 0.0395 (5) | |
| H7 | 1.043514 | 0.054747 | 0.800787 | 0.047* | |
| C21 | 0.91492 (17) | 0.02561 (11) | 0.74393 (12) | 0.0354 (5) | |
| C22 | 0.86546 (17) | 0.03069 (11) | 0.67883 (12) | 0.0337 (5) | |
| C23 | 0.90887 (17) | 0.06798 (12) | 0.62654 (12) | 0.0357 (5) | |
| C24 | 1.00575 (18) | 0.09739 (12) | 0.64050 (13) | 0.0401 (6) | |
| H8 | 1.037292 | 0.121225 | 0.604494 | 0.048* | |
| C25 | 0.51496 (17) | 0.08751 (12) | 0.84342 (12) | 0.0372 (5) | |
| H9 | 0.473547 | 0.093272 | 0.885011 | 0.045* | |
| H10 | 0.486068 | 0.048761 | 0.817385 | 0.045* | |
| C26 | 0.51156 (18) | 0.20106 (13) | 0.60839 (12) | 0.0406 (6) | |
| H11 | 0.477998 | 0.160327 | 0.589089 | 0.049* | |
| H12 | 0.476077 | 0.239910 | 0.586407 | 0.049* | |
| C27 | 0.84858 (18) | 0.08067 (12) | 0.55860 (12) | 0.0378 (5) | |
| H13 | 0.897651 | 0.085075 | 0.520772 | 0.045* | |
| H14 | 0.801769 | 0.042596 | 0.547653 | 0.045* | |
| C28 | 0.86781 (18) | −0.01488 (12) | 0.80093 (12) | 0.0375 (5) | |
| H15 | 0.925062 | −0.033610 | 0.830930 | 0.045* | |
| H16 | 0.828803 | −0.052668 | 0.779515 | 0.045* | |
| C29 | 0.8047 (2) | 0.10237 (14) | 1.02850 (13) | 0.0491 (6) | |
| C30 | 0.7435 (4) | 0.0613 (2) | 1.08047 (17) | 0.0999 (15) | |
| H17 | 0.668580 | 0.067370 | 1.070758 | 0.150* | |
| H18 | 0.761205 | 0.014078 | 1.075957 | 0.150* | |
| H19 | 0.761885 | 0.076280 | 1.127723 | 0.150* | |
| C31 | 0.7776 (3) | 0.17521 (19) | 1.0412 (2) | 0.0817 (11) | |
| H20 | 0.702438 | 0.181772 | 1.033078 | 0.123* | |
| H21 | 0.797621 | 0.187096 | 1.089159 | 0.123* | |
| H22 | 0.815358 | 0.203520 | 1.009480 | 0.123* | |
| C32 | 0.9206 (3) | 0.0945 (2) | 1.04151 (19) | 0.0893 (13) | |
| H23 | 0.940253 | 0.048049 | 1.033720 | 0.134* | |
| H24 | 0.957366 | 0.123369 | 1.009797 | 0.134* | |
| H25 | 0.939630 | 0.106944 | 1.089476 | 0.134* | |
| C33 | 0.4217 (2) | 0.33149 (13) | 0.82393 (14) | 0.0483 (6) | |
| C34 | 0.3259 (3) | 0.31832 (16) | 0.86617 (17) | 0.0646 (8) | |
| H26 | 0.268945 | 0.301671 | 0.835477 | 0.097* | |
| H27 | 0.343143 | 0.285070 | 0.901997 | 0.097* | |
| H28 | 0.304356 | 0.359772 | 0.888088 | 0.097* | |
| C35 | 0.3903 (3) | 0.38581 (16) | 0.77120 (18) | 0.0744 (10) | |
| H29 | 0.333490 | 0.369450 | 0.740210 | 0.112* | |
| H30 | 0.367093 | 0.425437 | 0.795745 | 0.112* | |
| H31 | 0.450375 | 0.397248 | 0.743962 | 0.112* | |
| C36 | 0.5101 (3) | 0.3574 (2) | 0.8719 (2) | 0.0909 (12) | |
| H32 | 0.571635 | 0.365840 | 0.844952 | 0.136* | |
| H33 | 0.488380 | 0.398774 | 0.893783 | 0.136* | |
| H34 | 0.527167 | 0.324072 | 0.907692 | 0.136* | |
| C37 | 0.8372 (2) | 0.33242 (13) | 0.56622 (14) | 0.0502 (7) | |
| C38 | 0.7819 (4) | 0.3747 (2) | 0.5104 (3) | 0.1056 (16) | |
| H35 | 0.816671 | 0.417941 | 0.507589 | 0.158* | |
| H36 | 0.784179 | 0.351932 | 0.465588 | 0.158* | |
| H37 | 0.709105 | 0.381352 | 0.522014 | 0.158* | |
| C39 | 0.9517 (3) | 0.32673 (16) | 0.54956 (18) | 0.0671 (9) | |
| H38 | 0.982298 | 0.371323 | 0.547234 | 0.101* | |
| H39 | 0.988990 | 0.300901 | 0.585886 | 0.101* | |
| H40 | 0.957595 | 0.304301 | 0.504852 | 0.101* | |
| C40 | 0.8356 (3) | 0.36674 (17) | 0.6372 (2) | 0.0764 (10) | |
| H41 | 0.869993 | 0.410165 | 0.634851 | 0.115* | |
| H42 | 0.763233 | 0.372949 | 0.650028 | 0.115* | |
| H43 | 0.872446 | 0.339017 | 0.672132 | 0.115* | |
| C41 | 1.1651 (2) | 0.12642 (17) | 0.71726 (16) | 0.0569 (7) | |
| C42 | 1.2390 (3) | 0.0983 (4) | 0.6681 (3) | 0.152 (3) | |
| H44 | 1.245849 | 0.050269 | 0.675822 | 0.228* | |
| H45 | 1.212602 | 0.106484 | 0.620456 | 0.228* | |
| H46 | 1.307207 | 0.119585 | 0.675508 | 0.228* | |
| C43 | 1.2088 (3) | 0.1184 (3) | 0.7914 (2) | 0.0982 (14) | |
| H47 | 1.159602 | 0.137049 | 0.823511 | 0.147* | |
| H48 | 1.219424 | 0.070991 | 0.801543 | 0.147* | |
| H49 | 1.275553 | 0.141857 | 0.796839 | 0.147* | |
| C44 | 1.1537 (4) | 0.2011 (2) | 0.7058 (3) | 0.1234 (19) | |
| H50 | 1.104797 | 0.219191 | 0.738495 | 0.185* | |
| H51 | 1.221716 | 0.222572 | 0.713330 | 0.185* | |
| H52 | 1.127111 | 0.209471 | 0.658278 | 0.185* | |
| C45 | 1.0638 (3) | 0.4028 (2) | 0.7522 (2) | 0.0873 (12) | |
| H53 | 1.055124 | 0.421695 | 0.705518 | 0.131* | |
| H54 | 1.121242 | 0.425519 | 0.777618 | 0.131* | |
| H55 | 1.079539 | 0.355241 | 0.748939 | 0.131* | |
| C46 | 1.1121 (3) | 0.48291 (18) | 0.92075 (18) | 0.0740 (9) | |
| H56 | 1.158468 | 0.496325 | 0.959936 | 0.111* | |
| H57 | 1.153669 | 0.471653 | 0.881086 | 0.111* | |
| H58 | 1.064767 | 0.519665 | 0.908156 | 0.111* | |
| C47 | 0.7645 (3) | 0.3692 (2) | 0.9612 (2) | 0.0964 (13) | |
| H59 | 0.693686 | 0.387179 | 0.962822 | 0.145* | |
| H60 | 0.761040 | 0.320979 | 0.953826 | 0.145* | |
| H61 | 0.802949 | 0.378616 | 1.005018 | 0.145* | |
| C48 | 1.0529 (3) | 0.2521 (2) | 0.9146 (2) | 0.0881 (12) | |
| H62 | 1.036956 | 0.204507 | 0.910682 | 0.132* | |
| H63 | 1.114548 | 0.262203 | 0.888369 | 0.132* | |
| H64 | 1.066664 | 0.263650 | 0.963430 | 0.132* | |
| C49 | 0.7898 (4) | 0.2062 (3) | 0.7671 (2) | 0.1036 (15) | |
| H65 | 0.742500 | 0.244404 | 0.763110 | 0.18 (3)* | |
| H66 | 0.751149 | 0.165274 | 0.755859 | 0.28 (5)* | |
| H67 | 0.846004 | 0.211673 | 0.734910 | 0.18 (3)* | |
| Li1 | 0.9522 (4) | 0.3821 (3) | 0.8811 (3) | 0.0634 (13) | |
| O1 | 0.65262 (12) | 0.01180 (8) | 0.76275 (8) | 0.0357 (4) | |
| O2 | 0.53575 (13) | 0.08851 (9) | 0.69430 (8) | 0.0407 (4) | |
| H68 | 0.572 (3) | 0.0638 (16) | 0.7202 (17) | 0.061* | |
| O3 | 0.77306 (13) | −0.00130 (9) | 0.66416 (9) | 0.0417 (4) | |
| H69 | 0.736 (3) | 0.0003 (15) | 0.7021 (17) | 0.063* | |
| O4 | 0.62688 (13) | 0.08283 (9) | 0.57403 (9) | 0.0423 (4) | |
| H70 | 0.589 (3) | 0.0795 (16) | 0.6120 (18) | 0.063* | |
| O5 | 0.97040 (17) | 0.41161 (11) | 0.78777 (11) | 0.0619 (6) | |
| H71 | 0.933 (3) | 0.440 (2) | 0.762 (2) | 0.093* | |
| O6 | 1.05402 (19) | 0.42743 (12) | 0.93941 (12) | 0.0702 (6) | |
| H72 | 1.069 (3) | 0.420 (2) | 0.987 (3) | 0.105* | |
| O7 | 0.8138 (2) | 0.39801 (16) | 0.90840 (17) | 0.0986 (10) | |
| H73 | 0.783 (5) | 0.430 (3) | 0.894 (3) | 0.148* | |
| O8 | 0.9716 (3) | 0.28755 (14) | 0.8890 (2) | 0.1247 (15) | |
| H74 | 0.937 (6) | 0.269 (4) | 0.870 (4) | 0.187* | |
| O9 | 0.8315 (5) | 0.2023 (2) | 0.8336 (3) | 0.196 (3) | |
| H75 | 0.804567 | 0.170190 | 0.854065 | 0.295* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0394 (13) | 0.0415 (13) | 0.0365 (12) | 0.0012 (10) | −0.0019 (10) | 0.0006 (10) |
| C2 | 0.0352 (12) | 0.0438 (13) | 0.0331 (12) | 0.0029 (10) | 0.0024 (10) | 0.0002 (10) |
| C3 | 0.0304 (11) | 0.0381 (12) | 0.0330 (11) | −0.0005 (9) | 0.0043 (9) | 0.0051 (9) |
| C4 | 0.0317 (12) | 0.0350 (12) | 0.0310 (11) | −0.0035 (9) | 0.0007 (9) | 0.0032 (9) |
| C5 | 0.0328 (12) | 0.0364 (12) | 0.0346 (12) | 0.0008 (9) | 0.0029 (9) | 0.0034 (9) |
| C6 | 0.0309 (12) | 0.0435 (13) | 0.0392 (13) | −0.0001 (10) | −0.0021 (10) | 0.0043 (10) |
| C7 | 0.0274 (11) | 0.0481 (14) | 0.0400 (13) | −0.0041 (10) | 0.0021 (9) | 0.0018 (10) |
| C8 | 0.0270 (11) | 0.0491 (14) | 0.0398 (13) | 0.0001 (10) | 0.0013 (9) | 0.0074 (11) |
| C9 | 0.0224 (11) | 0.0543 (14) | 0.0325 (12) | 0.0006 (10) | −0.0007 (9) | 0.0037 (10) |
| C10 | 0.0209 (10) | 0.0475 (14) | 0.0363 (12) | −0.0020 (9) | −0.0003 (9) | −0.0004 (10) |
| C11 | 0.0198 (10) | 0.0486 (14) | 0.0348 (12) | −0.0016 (9) | 0.0011 (9) | 0.0038 (10) |
| C12 | 0.0266 (11) | 0.0516 (14) | 0.0326 (12) | −0.0015 (10) | 0.0019 (9) | 0.0025 (10) |
| C13 | 0.0439 (13) | 0.0453 (14) | 0.0296 (11) | 0.0006 (11) | 0.0045 (10) | −0.0008 (10) |
| C14 | 0.0356 (12) | 0.0508 (15) | 0.0310 (12) | −0.0005 (11) | 0.0063 (9) | −0.0014 (10) |
| C15 | 0.0376 (12) | 0.0441 (13) | 0.0245 (10) | −0.0011 (10) | 0.0031 (9) | −0.0027 (9) |
| C16 | 0.0358 (12) | 0.0468 (14) | 0.0235 (10) | −0.0020 (10) | −0.0003 (9) | −0.0011 (9) |
| C17 | 0.0351 (12) | 0.0513 (14) | 0.0249 (11) | 0.0026 (10) | 0.0000 (9) | 0.0042 (10) |
| C18 | 0.0431 (14) | 0.0479 (14) | 0.0325 (12) | 0.0077 (11) | 0.0059 (10) | 0.0022 (10) |
| C19 | 0.0285 (12) | 0.0463 (14) | 0.0510 (14) | −0.0005 (10) | 0.0053 (10) | −0.0066 (11) |
| C20 | 0.0293 (12) | 0.0465 (14) | 0.0425 (13) | 0.0035 (10) | 0.0002 (10) | −0.0060 (11) |
| C21 | 0.0304 (12) | 0.0363 (12) | 0.0397 (12) | 0.0032 (9) | 0.0042 (9) | −0.0043 (10) |
| C22 | 0.0285 (11) | 0.0342 (12) | 0.0387 (12) | −0.0005 (9) | 0.0038 (9) | −0.0062 (9) |
| C23 | 0.0313 (12) | 0.0395 (13) | 0.0370 (12) | 0.0020 (10) | 0.0074 (9) | −0.0058 (10) |
| C24 | 0.0321 (12) | 0.0433 (13) | 0.0458 (14) | −0.0014 (10) | 0.0102 (10) | −0.0015 (10) |
| C25 | 0.0289 (12) | 0.0491 (14) | 0.0341 (12) | −0.0010 (10) | 0.0059 (9) | 0.0044 (10) |
| C26 | 0.0321 (12) | 0.0570 (15) | 0.0326 (12) | 0.0068 (11) | −0.0011 (9) | 0.0064 (11) |
| C27 | 0.0365 (12) | 0.0457 (13) | 0.0319 (12) | −0.0014 (10) | 0.0083 (10) | −0.0067 (10) |
| C28 | 0.0332 (12) | 0.0403 (13) | 0.0390 (12) | 0.0046 (10) | 0.0020 (10) | 0.0017 (10) |
| C29 | 0.0502 (15) | 0.0572 (16) | 0.0391 (14) | 0.0016 (13) | −0.0063 (11) | −0.0075 (12) |
| C30 | 0.142 (4) | 0.117 (3) | 0.0401 (17) | −0.036 (3) | −0.008 (2) | −0.0003 (19) |
| C31 | 0.073 (2) | 0.081 (2) | 0.088 (2) | 0.0166 (19) | −0.0279 (19) | −0.039 (2) |
| C32 | 0.066 (2) | 0.129 (3) | 0.069 (2) | 0.030 (2) | −0.0309 (18) | −0.043 (2) |
| C33 | 0.0541 (16) | 0.0450 (14) | 0.0459 (14) | −0.0036 (12) | 0.0046 (12) | −0.0010 (11) |
| C34 | 0.073 (2) | 0.0594 (18) | 0.0626 (18) | 0.0134 (16) | 0.0228 (16) | 0.0010 (15) |
| C35 | 0.105 (3) | 0.0514 (18) | 0.069 (2) | 0.0131 (18) | 0.0228 (19) | 0.0064 (15) |
| C36 | 0.085 (3) | 0.084 (3) | 0.102 (3) | −0.010 (2) | −0.015 (2) | −0.036 (2) |
| C37 | 0.0540 (16) | 0.0452 (15) | 0.0522 (15) | −0.0020 (12) | 0.0103 (12) | 0.0011 (12) |
| C38 | 0.103 (3) | 0.078 (3) | 0.134 (4) | −0.023 (2) | −0.020 (3) | 0.058 (3) |
| C39 | 0.068 (2) | 0.0579 (18) | 0.078 (2) | −0.0197 (15) | 0.0283 (17) | −0.0135 (16) |
| C40 | 0.083 (2) | 0.0604 (19) | 0.087 (2) | −0.0145 (17) | 0.0283 (19) | −0.0277 (17) |
| C41 | 0.0343 (14) | 0.073 (2) | 0.0632 (18) | −0.0130 (13) | 0.0023 (12) | −0.0065 (15) |
| C42 | 0.043 (2) | 0.264 (7) | 0.151 (5) | −0.049 (3) | 0.038 (3) | −0.105 (5) |
| C43 | 0.054 (2) | 0.143 (4) | 0.096 (3) | −0.033 (2) | −0.0148 (19) | −0.004 (3) |
| C44 | 0.106 (4) | 0.096 (3) | 0.164 (5) | −0.055 (3) | −0.042 (3) | 0.019 (3) |
| C45 | 0.064 (2) | 0.102 (3) | 0.096 (3) | 0.032 (2) | 0.003 (2) | 0.001 (2) |
| C46 | 0.085 (2) | 0.074 (2) | 0.062 (2) | −0.0032 (19) | −0.0116 (17) | 0.0150 (17) |
| C47 | 0.083 (3) | 0.114 (3) | 0.091 (3) | −0.004 (2) | −0.010 (2) | 0.039 (3) |
| C48 | 0.104 (3) | 0.086 (3) | 0.074 (2) | 0.034 (2) | 0.000 (2) | 0.003 (2) |
| C49 | 0.115 (4) | 0.113 (4) | 0.080 (3) | −0.023 (3) | −0.024 (3) | 0.025 (2) |
| Li1 | 0.057 (3) | 0.058 (3) | 0.073 (3) | 0.007 (2) | −0.015 (2) | 0.014 (2) |
| O1 | 0.0329 (8) | 0.0413 (9) | 0.0329 (8) | −0.0036 (7) | 0.0006 (6) | −0.0022 (6) |
| O2 | 0.0393 (9) | 0.0495 (10) | 0.0330 (9) | 0.0039 (8) | −0.0017 (7) | 0.0001 (7) |
| O3 | 0.0363 (9) | 0.0506 (10) | 0.0387 (9) | −0.0126 (7) | 0.0052 (7) | −0.0054 (7) |
| O4 | 0.0384 (9) | 0.0508 (10) | 0.0376 (9) | −0.0085 (8) | 0.0012 (7) | −0.0047 (7) |
| O5 | 0.0633 (13) | 0.0662 (13) | 0.0553 (12) | 0.0252 (10) | −0.0069 (10) | 0.0018 (10) |
| O6 | 0.0821 (16) | 0.0774 (15) | 0.0493 (12) | −0.0152 (12) | −0.0174 (11) | 0.0159 (11) |
| O7 | 0.0702 (16) | 0.105 (2) | 0.122 (2) | 0.0284 (15) | 0.0196 (15) | 0.0684 (19) |
| O8 | 0.109 (2) | 0.0535 (15) | 0.203 (4) | 0.0076 (15) | −0.082 (2) | 0.0104 (19) |
| O9 | 0.273 (6) | 0.114 (3) | 0.191 (4) | −0.084 (3) | −0.111 (4) | 0.046 (3) |
Geometric parameters (Å, º)
| C1—C6 | 1.387 (3) | C33—C34 | 1.529 (4) |
| C1—C2 | 1.397 (3) | C33—C35 | 1.535 (4) |
| C1—C29 | 1.535 (3) | C34—H26 | 0.9800 |
| C2—C3 | 1.387 (3) | C34—H27 | 0.9800 |
| C2—H1 | 0.9500 | C34—H28 | 0.9800 |
| C3—C4 | 1.405 (3) | C35—H29 | 0.9800 |
| C3—C25 | 1.511 (3) | C35—H30 | 0.9800 |
| C4—O1 | 1.349 (3) | C35—H31 | 0.9800 |
| C4—C5 | 1.402 (3) | C36—H32 | 0.9800 |
| C5—C6 | 1.392 (3) | C36—H33 | 0.9800 |
| C5—C28 | 1.521 (3) | C36—H34 | 0.9800 |
| C6—H2 | 0.9500 | C37—C39 | 1.524 (4) |
| C7—C8 | 1.391 (3) | C37—C38 | 1.524 (5) |
| C7—C12 | 1.399 (3) | C37—C40 | 1.536 (4) |
| C7—C33 | 1.535 (4) | C38—H35 | 0.9800 |
| C8—C9 | 1.386 (3) | C38—H36 | 0.9800 |
| C8—H3 | 0.9500 | C38—H37 | 0.9800 |
| C9—C10 | 1.390 (3) | C39—H38 | 0.9800 |
| C9—C26 | 1.523 (3) | C39—H39 | 0.9800 |
| C10—O2 | 1.362 (3) | C39—H40 | 0.9800 |
| C10—C11 | 1.399 (3) | C40—H41 | 0.9800 |
| C11—C12 | 1.386 (3) | C40—H42 | 0.9800 |
| C11—C25 | 1.520 (3) | C40—H43 | 0.9800 |
| C12—H4 | 0.9500 | C41—C42 | 1.484 (5) |
| C13—C14 | 1.383 (3) | C41—C44 | 1.522 (6) |
| C13—C18 | 1.400 (3) | C41—C43 | 1.523 (5) |
| C13—C37 | 1.524 (4) | C42—H44 | 0.9800 |
| C14—C15 | 1.393 (3) | C42—H45 | 0.9800 |
| C14—H5 | 0.9500 | C42—H46 | 0.9800 |
| C15—C16 | 1.397 (3) | C43—H47 | 0.9800 |
| C15—C27 | 1.512 (3) | C43—H48 | 0.9800 |
| C16—O4 | 1.384 (3) | C43—H49 | 0.9800 |
| C16—C17 | 1.388 (3) | C44—H50 | 0.9800 |
| C17—C18 | 1.380 (4) | C44—H51 | 0.9800 |
| C17—C26 | 1.522 (3) | C44—H52 | 0.9800 |
| C18—H6 | 0.9500 | C45—O5 | 1.420 (4) |
| C19—C20 | 1.389 (4) | C45—H53 | 0.9800 |
| C19—C24 | 1.390 (4) | C45—H54 | 0.9800 |
| C19—C41 | 1.534 (4) | C45—H55 | 0.9800 |
| C20—C21 | 1.392 (3) | C46—O6 | 1.397 (4) |
| C20—H7 | 0.9500 | C46—H56 | 0.9800 |
| C21—C22 | 1.387 (3) | C46—H57 | 0.9800 |
| C21—C28 | 1.516 (3) | C46—H58 | 0.9800 |
| C22—O3 | 1.368 (3) | C47—O7 | 1.354 (5) |
| C22—C23 | 1.394 (3) | C47—H59 | 0.9800 |
| C23—C24 | 1.393 (3) | C47—H60 | 0.9800 |
| C23—C27 | 1.515 (3) | C47—H61 | 0.9800 |
| C24—H8 | 0.9500 | C48—O8 | 1.340 (5) |
| C25—H9 | 0.9900 | C48—H62 | 0.9800 |
| C25—H10 | 0.9900 | C48—H63 | 0.9800 |
| C26—H11 | 0.9900 | C48—H64 | 0.9800 |
| C26—H12 | 0.9900 | C49—O9 | 1.370 (6) |
| C27—H13 | 0.9900 | C49—H65 | 0.9800 |
| C27—H14 | 0.9900 | C49—H66 | 0.9800 |
| C28—H15 | 0.9900 | C49—H67 | 0.9800 |
| C28—H16 | 0.9900 | Li1—O5 | 1.922 (6) |
| C29—C32 | 1.507 (4) | Li1—O6 | 1.917 (6) |
| C29—C31 | 1.527 (4) | Li1—O7 | 1.903 (6) |
| C29—C30 | 1.542 (5) | Li1—O8 | 1.922 (6) |
| C30—H17 | 0.9800 | Li1—H74 | 2.29 (8) |
| C30—H18 | 0.9800 | O2—H68 | 0.83 (3) |
| C30—H19 | 0.9800 | O3—H69 | 0.89 (3) |
| C31—H20 | 0.9800 | O4—H70 | 0.90 (3) |
| C31—H21 | 0.9800 | O5—H71 | 0.88 (4) |
| C31—H22 | 0.9800 | O6—H72 | 0.94 (5) |
| C32—H23 | 0.9800 | O7—H73 | 0.79 (6) |
| C32—H24 | 0.9800 | O8—H74 | 0.68 (8) |
| C32—H25 | 0.9800 | O9—H75 | 0.8400 |
| C33—C36 | 1.524 (5) | ||
| C6—C1—C2 | 116.5 (2) | C34—C33—C7 | 110.0 (2) |
| C6—C1—C29 | 122.9 (2) | C35—C33—C7 | 112.0 (2) |
| C2—C1—C29 | 120.5 (2) | C33—C34—H26 | 109.5 |
| C3—C2—C1 | 122.9 (2) | C33—C34—H27 | 109.5 |
| C3—C2—H1 | 118.6 | H26—C34—H27 | 109.5 |
| C1—C2—H1 | 118.6 | C33—C34—H28 | 109.5 |
| C2—C3—C4 | 119.1 (2) | H26—C34—H28 | 109.5 |
| C2—C3—C25 | 120.2 (2) | H27—C34—H28 | 109.5 |
| C4—C3—C25 | 120.7 (2) | C33—C35—H29 | 109.5 |
| O1—C4—C5 | 120.9 (2) | C33—C35—H30 | 109.5 |
| O1—C4—C3 | 119.7 (2) | H29—C35—H30 | 109.5 |
| C5—C4—C3 | 119.4 (2) | C33—C35—H31 | 109.5 |
| C6—C5—C4 | 119.2 (2) | H29—C35—H31 | 109.5 |
| C6—C5—C28 | 119.9 (2) | H30—C35—H31 | 109.5 |
| C4—C5—C28 | 120.9 (2) | C33—C36—H32 | 109.5 |
| C1—C6—C5 | 122.8 (2) | C33—C36—H33 | 109.5 |
| C1—C6—H2 | 118.6 | H32—C36—H33 | 109.5 |
| C5—C6—H2 | 118.6 | C33—C36—H34 | 109.5 |
| C8—C7—C12 | 116.5 (2) | H32—C36—H34 | 109.5 |
| C8—C7—C33 | 123.2 (2) | H33—C36—H34 | 109.5 |
| C12—C7—C33 | 120.3 (2) | C13—C37—C39 | 112.9 (2) |
| C9—C8—C7 | 122.6 (2) | C13—C37—C38 | 109.8 (3) |
| C9—C8—H3 | 118.7 | C39—C37—C38 | 108.5 (3) |
| C7—C8—H3 | 118.7 | C13—C37—C40 | 108.8 (2) |
| C8—C9—C10 | 119.1 (2) | C39—C37—C40 | 105.9 (3) |
| C8—C9—C26 | 120.3 (2) | C38—C37—C40 | 110.9 (3) |
| C10—C9—C26 | 120.6 (2) | C37—C38—H35 | 109.5 |
| O2—C10—C9 | 118.2 (2) | C37—C38—H36 | 109.5 |
| O2—C10—C11 | 121.3 (2) | H35—C38—H36 | 109.5 |
| C9—C10—C11 | 120.5 (2) | C37—C38—H37 | 109.5 |
| C12—C11—C10 | 118.4 (2) | H35—C38—H37 | 109.5 |
| C12—C11—C25 | 120.0 (2) | H36—C38—H37 | 109.5 |
| C10—C11—C25 | 121.6 (2) | C37—C39—H38 | 109.5 |
| C11—C12—C7 | 122.9 (2) | C37—C39—H39 | 109.5 |
| C11—C12—H4 | 118.5 | H38—C39—H39 | 109.5 |
| C7—C12—H4 | 118.5 | C37—C39—H40 | 109.5 |
| C14—C13—C18 | 116.6 (2) | H38—C39—H40 | 109.5 |
| C14—C13—C37 | 123.0 (2) | H39—C39—H40 | 109.5 |
| C18—C13—C37 | 120.4 (2) | C37—C40—H41 | 109.5 |
| C13—C14—C15 | 123.2 (2) | C37—C40—H42 | 109.5 |
| C13—C14—H5 | 118.4 | H41—C40—H42 | 109.5 |
| C15—C14—H5 | 118.4 | C37—C40—H43 | 109.5 |
| C14—C15—C16 | 117.6 (2) | H41—C40—H43 | 109.5 |
| C14—C15—C27 | 119.7 (2) | H42—C40—H43 | 109.5 |
| C16—C15—C27 | 122.6 (2) | C42—C41—C44 | 110.0 (4) |
| O4—C16—C17 | 120.4 (2) | C42—C41—C43 | 110.0 (4) |
| O4—C16—C15 | 118.2 (2) | C44—C41—C43 | 105.7 (3) |
| C17—C16—C15 | 121.3 (2) | C42—C41—C19 | 109.2 (3) |
| C18—C17—C16 | 118.6 (2) | C44—C41—C19 | 108.9 (3) |
| C18—C17—C26 | 119.7 (2) | C43—C41—C19 | 113.0 (3) |
| C16—C17—C26 | 121.7 (2) | C41—C42—H44 | 109.5 |
| C17—C18—C13 | 122.5 (2) | C41—C42—H45 | 109.5 |
| C17—C18—H6 | 118.7 | H44—C42—H45 | 109.5 |
| C13—C18—H6 | 118.7 | C41—C42—H46 | 109.5 |
| C20—C19—C24 | 116.9 (2) | H44—C42—H46 | 109.5 |
| C20—C19—C41 | 122.4 (2) | H45—C42—H46 | 109.5 |
| C24—C19—C41 | 120.6 (2) | C41—C43—H47 | 109.5 |
| C19—C20—C21 | 122.4 (2) | C41—C43—H48 | 109.5 |
| C19—C20—H7 | 118.8 | H47—C43—H48 | 109.5 |
| C21—C20—H7 | 118.8 | C41—C43—H49 | 109.5 |
| C22—C21—C20 | 118.8 (2) | H47—C43—H49 | 109.5 |
| C22—C21—C28 | 121.1 (2) | H48—C43—H49 | 109.5 |
| C20—C21—C28 | 120.1 (2) | C41—C44—H50 | 109.5 |
| O3—C22—C21 | 120.7 (2) | C41—C44—H51 | 109.5 |
| O3—C22—C23 | 118.3 (2) | H50—C44—H51 | 109.5 |
| C21—C22—C23 | 121.0 (2) | C41—C44—H52 | 109.5 |
| C24—C23—C22 | 118.1 (2) | H50—C44—H52 | 109.5 |
| C24—C23—C27 | 120.9 (2) | H51—C44—H52 | 109.5 |
| C22—C23—C27 | 120.8 (2) | O5—C45—H53 | 109.5 |
| C19—C24—C23 | 122.8 (2) | O5—C45—H54 | 109.5 |
| C19—C24—H8 | 118.6 | H53—C45—H54 | 109.5 |
| C23—C24—H8 | 118.6 | O5—C45—H55 | 109.5 |
| C3—C25—C11 | 114.87 (18) | H53—C45—H55 | 109.5 |
| C3—C25—H9 | 108.6 | H54—C45—H55 | 109.5 |
| C11—C25—H9 | 108.6 | O6—C46—H56 | 109.5 |
| C3—C25—H10 | 108.6 | O6—C46—H57 | 109.5 |
| C11—C25—H10 | 108.6 | H56—C46—H57 | 109.5 |
| H9—C25—H10 | 107.5 | O6—C46—H58 | 109.5 |
| C17—C26—C9 | 112.70 (18) | H56—C46—H58 | 109.5 |
| C17—C26—H11 | 109.1 | H57—C46—H58 | 109.5 |
| C9—C26—H11 | 109.1 | O7—C47—H59 | 109.5 |
| C17—C26—H12 | 109.1 | O7—C47—H60 | 109.5 |
| C9—C26—H12 | 109.1 | H59—C47—H60 | 109.5 |
| H11—C26—H12 | 107.8 | O7—C47—H61 | 109.5 |
| C15—C27—C23 | 109.92 (18) | H59—C47—H61 | 109.5 |
| C15—C27—H13 | 109.7 | H60—C47—H61 | 109.5 |
| C23—C27—H13 | 109.7 | O8—C48—H62 | 109.5 |
| C15—C27—H14 | 109.7 | O8—C48—H63 | 109.5 |
| C23—C27—H14 | 109.7 | H62—C48—H63 | 109.5 |
| H13—C27—H14 | 108.2 | O8—C48—H64 | 109.5 |
| C21—C28—C5 | 114.67 (19) | H62—C48—H64 | 109.5 |
| C21—C28—H15 | 108.6 | H63—C48—H64 | 109.5 |
| C5—C28—H15 | 108.6 | O9—C49—H65 | 109.5 |
| C21—C28—H16 | 108.6 | O9—C49—H66 | 109.5 |
| C5—C28—H16 | 108.6 | H65—C49—H66 | 109.5 |
| H15—C28—H16 | 107.6 | O9—C49—H67 | 109.5 |
| C32—C29—C31 | 107.7 (3) | H65—C49—H67 | 109.5 |
| C32—C29—C1 | 112.8 (2) | H66—C49—H67 | 109.5 |
| C31—C29—C1 | 110.5 (2) | O5—Li1—O6 | 107.2 (3) |
| C32—C29—C30 | 111.3 (3) | O5—Li1—O7 | 111.3 (3) |
| C31—C29—C30 | 106.4 (3) | O5—Li1—O8 | 111.0 (3) |
| C1—C29—C30 | 108.0 (2) | O6—Li1—O7 | 112.3 (3) |
| C29—C30—H17 | 109.5 | O6—Li1—O8 | 109.9 (3) |
| C29—C30—H18 | 109.5 | O7—Li1—O8 | 105.3 (3) |
| H17—C30—H18 | 109.5 | O7—Li1—H74 | 97 (2) |
| C29—C30—H19 | 109.5 | O6—Li1—H74 | 126 (2) |
| H17—C30—H19 | 109.5 | O8—Li1—H74 | 16 (2) |
| H18—C30—H19 | 109.5 | O5—Li1—H74 | 103 (2) |
| C29—C31—H20 | 109.5 | C10—O2—H68 | 111 (2) |
| C29—C31—H21 | 109.5 | C22—O3—H69 | 108 (2) |
| H20—C31—H21 | 109.5 | C16—O4—H70 | 108 (2) |
| C29—C31—H22 | 109.5 | C45—O5—Li1 | 123.8 (3) |
| H20—C31—H22 | 109.5 | C45—O5—H71 | 105 (3) |
| H21—C31—H22 | 109.5 | Li1—O5—H71 | 130 (3) |
| C29—C32—H23 | 109.5 | C46—O6—Li1 | 125.8 (2) |
| C29—C32—H24 | 109.5 | C46—O6—H72 | 107 (3) |
| H23—C32—H24 | 109.5 | Li1—O6—H72 | 127 (3) |
| C29—C32—H25 | 109.5 | C47—O7—Li1 | 127.7 (3) |
| H23—C32—H25 | 109.5 | C47—O7—H73 | 111 (4) |
| H24—C32—H25 | 109.5 | Li1—O7—H73 | 120 (4) |
| C36—C33—C34 | 109.3 (3) | C48—O8—Li1 | 130.6 (3) |
| C36—C33—C35 | 109.1 (3) | C48—O8—H74 | 113 (7) |
| C34—C33—C35 | 106.5 (3) | Li1—O8—H74 | 115 (7) |
| C36—C33—C7 | 109.9 (2) | C49—O9—H75 | 109.5 |
| C6—C1—C2—C3 | 1.2 (4) | C20—C21—C22—O3 | 178.3 (2) |
| C29—C1—C2—C3 | −175.8 (2) | C28—C21—C22—O3 | −0.6 (3) |
| C1—C2—C3—C4 | 0.3 (4) | C20—C21—C22—C23 | −1.1 (3) |
| C1—C2—C3—C25 | −179.8 (2) | C28—C21—C22—C23 | 179.9 (2) |
| C2—C3—C4—O1 | 177.1 (2) | O3—C22—C23—C24 | −176.5 (2) |
| C25—C3—C4—O1 | −2.8 (3) | C21—C22—C23—C24 | 3.0 (3) |
| C2—C3—C4—C5 | −1.9 (3) | O3—C22—C23—C27 | 8.3 (3) |
| C25—C3—C4—C5 | 178.2 (2) | C21—C22—C23—C27 | −172.2 (2) |
| O1—C4—C5—C6 | −177.0 (2) | C20—C19—C24—C23 | 0.6 (4) |
| C3—C4—C5—C6 | 1.9 (3) | C41—C19—C24—C23 | 179.7 (2) |
| O1—C4—C5—C28 | 1.6 (3) | C22—C23—C24—C19 | −2.8 (4) |
| C3—C4—C5—C28 | −179.5 (2) | C27—C23—C24—C19 | 172.4 (2) |
| C2—C1—C6—C5 | −1.1 (4) | C2—C3—C25—C11 | 95.4 (3) |
| C29—C1—C6—C5 | 175.7 (2) | C4—C3—C25—C11 | −84.7 (3) |
| C4—C5—C6—C1 | −0.4 (4) | C12—C11—C25—C3 | −97.4 (2) |
| C28—C5—C6—C1 | −179.0 (2) | C10—C11—C25—C3 | 84.6 (3) |
| C12—C7—C8—C9 | 0.7 (3) | C18—C17—C26—C9 | −83.1 (3) |
| C33—C7—C8—C9 | −178.8 (2) | C16—C17—C26—C9 | 96.4 (3) |
| C7—C8—C9—C10 | 0.7 (3) | C8—C9—C26—C17 | 102.9 (3) |
| C7—C8—C9—C26 | −179.4 (2) | C10—C9—C26—C17 | −77.1 (3) |
| C8—C9—C10—O2 | 176.83 (19) | C14—C15—C27—C23 | 76.7 (3) |
| C26—C9—C10—O2 | −3.1 (3) | C16—C15—C27—C23 | −98.7 (2) |
| C8—C9—C10—C11 | −1.2 (3) | C24—C23—C27—C15 | −85.5 (3) |
| C26—C9—C10—C11 | 178.8 (2) | C22—C23—C27—C15 | 89.5 (3) |
| O2—C10—C11—C12 | −177.59 (19) | C22—C21—C28—C5 | −89.8 (3) |
| C9—C10—C11—C12 | 0.4 (3) | C20—C21—C28—C5 | 91.3 (3) |
| O2—C10—C11—C25 | 0.4 (3) | C6—C5—C28—C21 | −100.5 (3) |
| C9—C10—C11—C25 | 178.4 (2) | C4—C5—C28—C21 | 81.0 (3) |
| C10—C11—C12—C7 | 1.0 (3) | C6—C1—C29—C32 | 12.1 (4) |
| C25—C11—C12—C7 | −177.0 (2) | C2—C1—C29—C32 | −171.1 (3) |
| C8—C7—C12—C11 | −1.6 (3) | C6—C1—C29—C31 | 132.7 (3) |
| C33—C7—C12—C11 | 177.9 (2) | C2—C1—C29—C31 | −50.6 (4) |
| C18—C13—C14—C15 | 1.1 (3) | C6—C1—C29—C30 | −111.3 (3) |
| C37—C13—C14—C15 | −179.7 (2) | C2—C1—C29—C30 | 65.4 (4) |
| C13—C14—C15—C16 | 2.1 (3) | C8—C7—C33—C36 | −113.7 (3) |
| C13—C14—C15—C27 | −173.5 (2) | C12—C7—C33—C36 | 66.8 (3) |
| C14—C15—C16—O4 | 173.58 (19) | C8—C7—C33—C34 | 125.9 (3) |
| C27—C15—C16—O4 | −10.9 (3) | C12—C7—C33—C34 | −53.5 (3) |
| C14—C15—C16—C17 | −4.0 (3) | C8—C7—C33—C35 | 7.8 (4) |
| C27—C15—C16—C17 | 171.6 (2) | C12—C7—C33—C35 | −171.7 (3) |
| O4—C16—C17—C18 | −175.0 (2) | C14—C13—C37—C39 | −0.9 (4) |
| C15—C16—C17—C18 | 2.5 (3) | C18—C13—C37—C39 | 178.4 (2) |
| O4—C16—C17—C26 | 5.6 (3) | C14—C13—C37—C38 | 120.3 (3) |
| C15—C16—C17—C26 | −176.9 (2) | C18—C13—C37—C38 | −60.5 (4) |
| C16—C17—C18—C13 | 0.9 (3) | C14—C13—C37—C40 | −118.2 (3) |
| C26—C17—C18—C13 | −179.6 (2) | C18—C13—C37—C40 | 61.1 (3) |
| C14—C13—C18—C17 | −2.7 (3) | C20—C19—C41—C42 | 120.0 (4) |
| C37—C13—C18—C17 | 178.1 (2) | C24—C19—C41—C42 | −58.9 (5) |
| C24—C19—C20—C21 | 1.3 (4) | C20—C19—C41—C44 | −119.8 (4) |
| C41—C19—C20—C21 | −177.7 (2) | C24—C19—C41—C44 | 61.2 (4) |
| C19—C20—C21—C22 | −1.1 (3) | C20—C19—C41—C43 | −2.7 (4) |
| C19—C20—C21—C28 | 177.8 (2) | C24—C19—C41—C43 | 178.3 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O8—H74···O9 | 0.67 (3) | 2.01 (8) | 2.673 (3) | 167 (3) |
| O2—H68···O1 | 0.83 (3) | 1.66 (4) | 2.490 (2) | 172 (3) |
| O3—H69···O1 | 0.89 (3) | 1.64 (3) | 2.520 (2) | 169 (3) |
| O4—H70···O2 | 0.90 (3) | 1.77 (3) | 2.650 (2) | 166 (3) |
| O5—H71···O1i | 0.88 (4) | 1.87 (4) | 2.714 (3) | 160 (4) |
| O6—H72···O4ii | 0.94 (5) | 1.81 (5) | 2.732 (3) | 165 (4) |
| O7—H73···O3i | 0.79 (6) | 1.91 (6) | 2.676 (3) | 163 (6) |
Symmetry codes: (i) −x+3/2, y+1/2, −z+3/2; (ii) x+1/2, −y+1/2, z+1/2.
Funding Statement
This work was funded by The Mazda Foundation. grant 17KK-077. Japan Society for the Promotion of Science grant 16F16353.
References
- Bock, H., John, A., Naether, C. & Havlas, Z. (1995). J. Am. Chem. Soc. 117, 9367–9368.
- Bruker (2006). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Davidson, M. G., Howard, J. A. K., Lamb, S. & Lehmann, C. W. (1997). Chem. Commun. pp. 1607–1608.
- Dürr, S., Bechlars, B. & Radius, U. (2006). Inorg. Chim. Acta, 359, 4215–4226.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gueneau, E. D., Fromm, K. M. & Goesmann, H. (2003). Chem. Eur. J. 9, 509–514. [DOI] [PubMed]
- Guillemot, G., Solari, E., Rizzoli, C. & Floriani, C. (2002). Chem. Eur. J. 8, 2072–2080. [DOI] [PubMed]
- Gutsche, C. D. (1998). Calixarenes Revisited. Cambridge, UK: Royal Society of Chemistry.
- Hamada, F., Robinson, K. D., Orr, G. W. & Atwood, J. L. (1993). Supramol. Chem. 2, 19–24.
- Hanna, T. A., Liu, L., Angeles-Boza, A. M., Kou, X., Gutsche, C. D., Ejsmont, K., Watson, W. H., Zakharov, L. N., Incarvito, C. D. & Rheingold, A. L. (2003). J. Am. Chem. Soc. 125, 6228–6238. [DOI] [PubMed]
- Hanna, T. A., Liu, L., Zakharov, L. N., Rheingold, A. L., Watson, W. H. & Gutsche, C. D. (2002). Tetrahedron, 58, 9751–9757.
- Harrowfield, J. M., Ogden, M. I., Richmond, W. R. & White, A. H. (1991). J. Chem. Soc. Chem. Commun. pp. 1159–1161.
- Kabuto, C., Akine, S., Nemoto, T. & Kwon, E. (2009). J. Crystallogr. Soc. Jpn, 51, 218–224.
- Lee, D. S., Elsegood, M. R. J., Redshaw, C. & Zhan, S. (2009). Acta Cryst. C65, m291–m295. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Vicens, J. & Böhmer, V. (1991). Editors. Calixarenes: A Versatile Class of Macrocyclic Compounds, Kluwer Academic Publishers: Dordrecht, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989018001834/xi2008sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018001834/xi2008Isup2.hkl
CCDC reference: 1563055
Additional supporting information: crystallographic information; 3D view; checkCIF report






