 Specific long- and short-range electrostatic interactions and not redox potentials determine the substrate specificity of Trx family proteins.
Specific long- and short-range electrostatic interactions and not redox potentials determine the substrate specificity of Trx family proteins.
Abstract
Thiol–disulfide oxidoreductases from the thioredoxin (Trx) family of proteins have a broad range of well documented functions and possess distinct substrate specificities. The mechanisms and characteristics that control these specificities are key to the understanding of both the reduction of catalytic disulfides as well as allosteric disulfides (thiol switches). Here, we have used the catalytic disulfide of E. coli 3′-phosphoadenosine 5′-phosphosulfate (PAPS) reductase (PR) that forms between the single active site thiols of two monomers during the reaction cycle as a model system to investigate the mechanisms of Trx and Grx protein specificity. Enzyme kinetics, ΔE′0 determination, and structural analysis of various Trx and Grx family members suggested that the redox potential does not determine specificity nor efficiency of the redoxins as reductant for PR. Instead, the efficiency of PR with various redoxins correlated strongly to the extent of a negative electric field of the redoxins reaching into the solvent outside the active site, and electrostatic and geometric complementary contact surfaces. These data suggest that, in contrast to common assumption, the composition of the active site motif is less important for substrate specificity than other amino acids in or even outside the immediate contact area.
Introduction
During the past decade the concept of cellular redox homeostasis shifted more and more from the idea of a redox balance between oxidants and antioxidants towards the concept of spatio-temporally controlled redox signalling events. These events are specific with respect to the cysteinyl residues modified and the redox compounds and enzymes involved. Thioredoxins (Trxs) and glutaredoxins (Grxs) are the master regulators of the redox state of the thiol groups of the proteome with numerous well documented functions in essentially all cellular processes, including metabolism and cell signalling, see for instance refs. 1–4. In most cases, Trxs and Grxs reduce catalytic and allosteric disulfides, a classification introduced by Hogg and coworkers.5 Although Trxs and Grxs have a broad range of functions, each member of the family has distinct substrate specificities. The mechanisms and characteristics that control these specificities are unclear and have hardly been addressed before.
Both Trxs and Grxs are part of the Trx family of proteins, characterized by a common structure, the Trx fold.6,7 This fold is defined by a central four to five-stranded β-sheet, surrounded by three to four α-helices. Trxs and Grxs catalyze thiol–disulfide exchange reactions. Two thiols in the characteristic Cys–X–X–Cys active site reduced a target disulfide in a reversible two-step reaction. The more N-terminal thiolate cysteinyl residue, located at the surface of the protein in a loop connecting β1 and α1 (in Grxs) or β2 and α2 (in Trxs), attacks the target disulfide resulting in an intermediate mixed disulfide between the redoxin and the target protein. This is directly attacked by the more C-terminal active site thiol, normally buried in the protein at the beginning of the α-helix (1 or 2), yielding a disulfide in the active site and a reduced target protein. Oxidized Trx is reduced by a Trx reductase and Grx by two molecules of glutathione. This reaction sequence was named the dithiol mechanism to distinguish it from the monothiol mechanism. The latter is used by Grxs to reduce protein–glutathione mixed disulfides and requires only one, the more N-terminal, active site cysteinyl residue.
Both Trxs and Grxs were originally identified as electron donors for ribonucleotide reductase from E. coli.8,9 The requirement for Trx in sulfate assimilation was originally described by Gonzalez-Porqué et al.10 for yeast. Reduction of sulfate to sulfite requires two electrons with a ΔE′0 of –517 mV. Adenylation and phosphorylation to 3′-phosphoadenonsine-5′-phosphosulfate (PAPS) lowers this redox potential to –60 mV. In E. coli, Trx1 and Grx1 were identified as alternative electron donors for the catalytic disulfide in PAPS reductase (PR).11,12 Enzymatically active PR (EC 1.8.99.4) forms a homo-dimer (2 × 28 kDa). It is devoid of redox-active chromophores but contains a single cysteine in a strictly conserved ECGLH motif that is located at the C-terminus.13 Steady state analysis of the reaction and mutagenesis analyses demonstrated that PR follows a ping-pong mechanism in the reduction of PAPS to sulfite and 3′-5′-adenosine diphosphate (PAP).13–17 In the first step, the PR dimer reduces PAPS directly to sulfite without any detectable sulfate or sulfite-bound intermediates. Oxidation of PR yields an intermolecular disulfide between the ECGLH cysteinyl residues of the two monomers. In the second step, this disulfide is the substrate for Trx or Grx and requires the dithiol reaction mechanism of the redoxins for reduction. Although E. coli PR is a rather promiscuous enzyme regarding its choice of electron donors, it cannot be reduced by any Trx or Grx. The high cross-reactivity of E. coli PR was, for instance, useful in the identification and purification of heterologous Trxs from spinach, Synechococcus, and yeast.18 However, the two additional dithiol Grxs of E. coli itself, Grx2 and Grx3,19 cannot reduce PR, neither in vivo20 nor in vitro.16
In this study, we have used E. coli PR as a model for the analysis of the specificity of Trx family proteins addressing two hypotheses. First, is the functionality of the redoxins determined by the redox potential of their active site dithiol–disulfide redox pair? Or, second, is their functionality determined by specific molecular interactions next to the thiol–disulfide exchange reaction?
Results
Homologous and heterologous thioredoxins and glutaredoxins as electron donors for PAPS reductase
The functionality of various thioredoxins and glutaredoxins from different species with E. coli PR was analyzed in kinetic assays. In addition to the well established E. coli Trx1 and Grx1, which are regarded as physiological reductants,12,16 and the non-active E. coli Trx2, Grx2, and Grx3,16 we analyzed E. coli Grx4 (ref. 21) and NrdH,22 T4 Grx,23Arabidopsis thaliana TrxH1, 2, 3, and 4,24 poplar Grx,25 and human Trx1 (ref. 26) and Grx2.27,28 The results are summarized in Table 1.
Table 1. Redox potentials and kinetic parameters of various thioredoxins and glutaredoxins a .
| Redoxin |
Redoxpotential |
Kinetic parameters with PR |
|||||
| Abbr. | Source | ΔE′0, mV | Ref. | K m, μM | V max, U mg–1 | k cat K m –1, % | Ref. |
| Trx1 | E. c. | –270 | 40 | 13.7 | 6.7 | 100 | 16 |
| Trx2 | E. c. | –221 | 60 | 34.2 | 6.3 | 38 | 16 |
| Grx1 | E. c. | –233 | 30 | 14.9 | 5.1 | 70 | 16 |
| Grx2 | E. c. | (n.a.) | No activity | 16 | |||
| Grx3 | E. c. | –198 | 30 | No activity | 16 | ||
| Grx4 | E. c. | (n.a.) | No activity | p.s. | |||
| NrdH | E. c. | –248.5 | 22 | No activity | p.s. | ||
| Grx | T4 | –240 | 23 | No activity | p.s. | ||
| TrxH1 | A. t. | (n.a.) | 59.0 | 5.3 | 18 | p.s. | |
| TrxH2 | A. t. | (n.a.) | 43.1 | 4.7 | 23 | p.s. | |
| TrxH3 | A. t. | (n.a.) | 17.8 | 13.2 | 154 | p.s. | |
| TrxH4 | A. t. | (n.a.) | 26.1 | 5.7 | 45 | p.s. | |
| Grx | Poplar | (n.a.) | 63.7 | 10.4 | 33 | 25 | |
| hTrx1 | H. s. | –230 | 61 | 68.1 | 19.6 | 59 | p.s. |
| hGrx2 | H. s. | –221 | 62 | No activity | p.s. | ||
a A. t.: Arabidopsis thaliana, E. c.: Escherichia coli, H. s.: Homo sapiens, n.a.: not available; p.s.: present study.
Not surprisingly, E. coli Grx4 could not reduce PR. Grx4 is a monothiol-type Grx and the reduction of PR requires the dithiol mechanisms.16 NrdH, a protein with a Grx-like structure but Trx-like activity profile, is a specific electron donor for the alternative ribonucleotide reductase NrdEF,22 but NrdH is not functional with PR. Grx (formerly Trx) from the bacteriophage T4 was also not able to catalytically reduce PR. In contrast, the four plant Trxs H1–H4 and the plant Grx25 were able to supply PR with electrons yielding catalytic efficiencies between 18 and 154% compared to E. coli Trx1. With human Trx1, PR yielded a catalytic efficiency of 59%. Human Grx2, on the other hand, was not able to reduce the enzyme's catalytic disulfide.
Redox potential of PR
One possible explanation that was suggested for why some redoxins are functional in this assay while others are not are the different standard redox potentials of the dithiol–disulfide pairs of the redoxins. From the literature, the E′0 values of some of the functional redoxins are in the range of –233 to –270 mV, and the E′0 values of some of the non-functional redoxins are in the range of –198 to –248.5 mV, see Table 1. Although these values did not clearly discriminate the two groups, we decided to determine the redox potential of the active site ECGLH cysteinyl/intermolecular disulfide pair of dimeric PR.
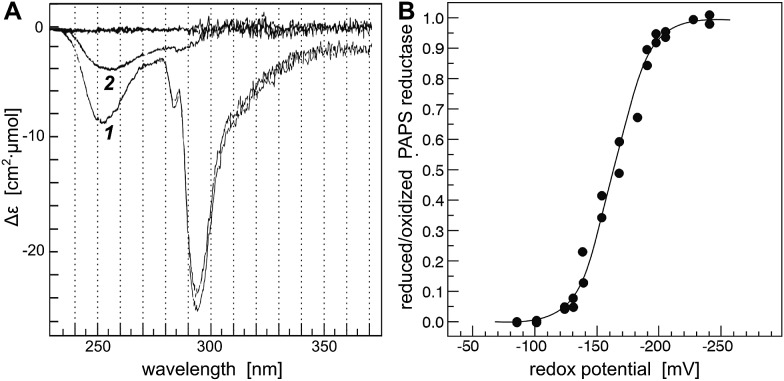
The ping-pong uni–bi–bi reaction sequence of dimeric PR requires a considerable conformational change of the protein during each reaction cycle.15–17 This conformational change causes small changes in absorbance of the protein in the UV spectrum.15 These changes were analyzed by difference absorption spectroscopy using a two beam setup with in series tandem cuvettes in each beam. PR was oxidized by incubation with the substrate PAPS and subsequent removal of the reaction products SO4– and PAP by gel filtration chromatography. This oxidation yielded exclusively intermolecular disulfides between two PR monomers (not shown). The reaction mechanism proposed in ref. 29 involving a stable sulfate-bound intermediate can be excluded since incubation of reduced PAPS reductase with [35S]–PAPS (in the absence of reductants) yielded no detectable radioactivity associated with the enzyme, confirming earlier conclusions from kinetic experiments.15–18
In the photometer, the reference beam contained PR (fully oxidized) and a glutathione redox buffer in separate cuvettes; in the sample beam the protein was in the same cuvette as the redox buffer. The glutathione redox buffer also contained a catalytic amount of Grx1 in a ratio of 1 : 100 to PR. Reduction of PR led to a decrease in absorbance at 253 and 294 nm (Fig. 1A, spectrum 1), likely caused by disulfide reduction and changes in the interactions of a tryptophanyl residue with the solvent. The differences in absorption coefficient at 294 nm were recorded over time after incubation in defined glutathione redox buffers until equilibrium was reached, i.e. up to seven hours (not shown). The ratio of reduced versus oxidized PR was plotted against the redox potential defined by the redox buffer (Fig. 1B). Non linear regression of 25 independent measurements against the Nernst equation yielded a redox potential of –162.2 mV. This potential is still 36 mV above the potential of Grx3, the non-functional redoxin with the highest potential (–198 mV)30 and does not correlate to the different activities of the redoxins towards PR, see Table 1.
Fig. 1. Determination of the redox potential of PAPS reductase. (A) Difference spectrum of oxidized (reference) versus reduced PR, (A-1) the fully reduced enzyme, (A-2) at a redox potential – defined by glutathione redox buffer – of –100 mV. (B). The differences in absorption at 294 nm were used to calculate the reduced/oxidized ratio of PR after incubation of the enzyme in various redox buffers until equilibrium was reached, i.e. up to seven hours. The 25 individual measurements were fitted to the Nernst equation (solid line) yielding a standard redox potential of PR of –162 mV. For experimental details, see Experimental procedures.
Structural analysis
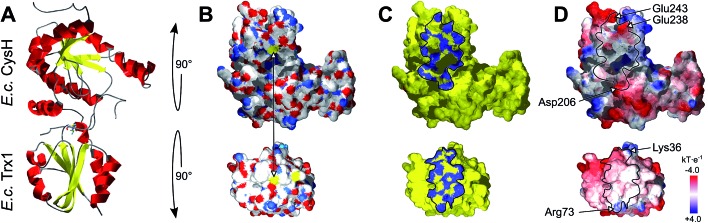
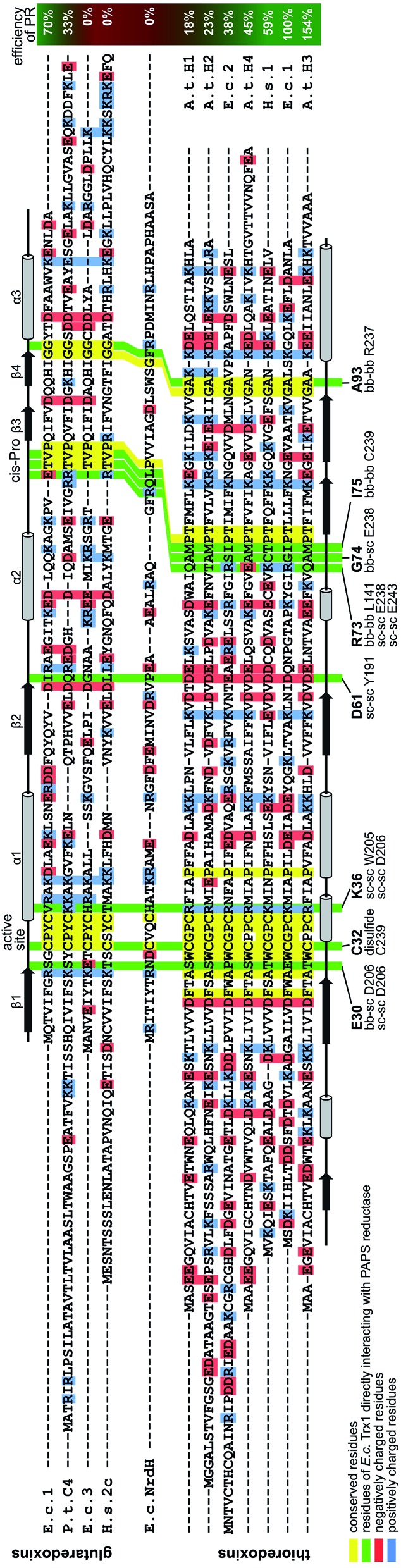
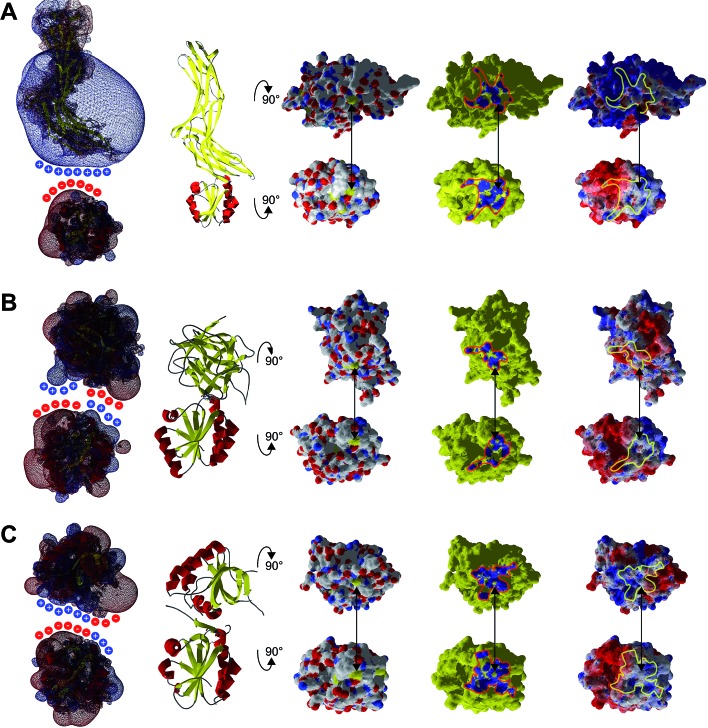
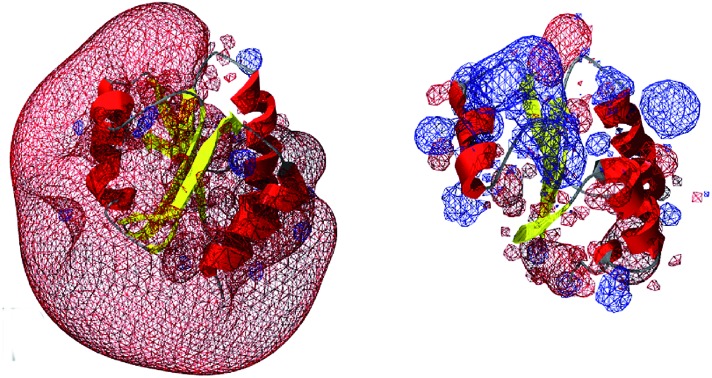
What else, if not the redox potential, could determine the different specificities of the Trx family proteins for a promiscuous protein like PR? Chartron et al. presented the structure of a complex between a monomer of PR and an active site mutant of Trx1 that represents a catalytic mixed disulfide intermediate in the reduction of PR by the redoxins in the dithiol reaction mechanism.29 In this complex, both proteins interact specifically at multiple points (see Fig. 2C and 3). Clearly, the degree of conservation of these interacting residues cannot explain the different functionalities and efficiencies of the redoxins with PR (Fig. 3). The interactions include many backbone–backbone interactions. Specific side chain interactions are rare and may be taken over by other amino acids, for instance in the case of aspartyl 61 of E. c. Trx1 by the aspartyl residue 37 of E. c. Grx1, located one amino acid N-terminally of aspartyl 61 in the structural alignment (Fig. 3). The molecular interactions between Trx and PR also include specific electrostatic interactions at the short sites/corners of the contact surface (see Fig. 2D). The ε-amino group of lysyl residue 36 of Trx1 specifically interacts with the β-carboxyl group of aspartyl residue 206 of PR and the guanidino group of arginyl residue 73 of Trx1 with the γ-carboxyl groups of glutamyl residues 238 and 243 of PR. In between these two small positive surface patches of E. coli Trx1 is a surface area with a neutral to slightly negative electrostatic surface potential (see Fig. 2D, lower row).
Fig. 2. Structure of the E. coli PAPS reductase–mutant Trx1 mixed disulfide complex. (A) Secondary structure representation of the complex, top: PR (CysH), bottom: Trx1. (B) Surfaces in atomic type coloring after rotation of PR by 90° backwards and Trx1 by 90° to the front. The arrow points to the two cysteinyl sulfur atoms that form the intermediate mixed disulfide. (C) The contact surfaces marked in blue. (D) The electrostatic surface potentials (from red = –4 to blue = +4 kT per e, using atomic partial charges) mapped to the surface and highlighting the contact surfaces and some specifically interacting amino acid site chains. All pictures and data were computed using the DeepView/Swiss-PDB Viewer 4.1 with PDB accession number ; 2o8v.29.
Fig. 3. Structural alignment of the thioredoxin family proteins investigated in this study. The structures of the various redoxins were pairwise aligned to the structure of E. coli Trx 1 (PDB code ; 1xob) using PyMOL and thereafter manually arranged in this alignment. The residues directly interacting in the E. c. Trx1–PAPS reductase complex were highlighted with a green background. The residues were also specified below the sequence, as well as the interacting residues in PAPS reductase and the parts of the amino acids involved in these interactions (bb: backbone, sc: side chain). The interactions were calculated with ‘contact’ of the CCP4 suite.58 Conserved residues were highlighted with a yellow, positively charged residues with a blue, and negatively charged residues with a red background. The positions of the active site as well as the Trx-fold specific cis-Pro residues were also marked above the sequences. The secondary structures of E. c. Grx1 was also included above the sequences, the secondary structure of E. c. Trx1 below the sequences.
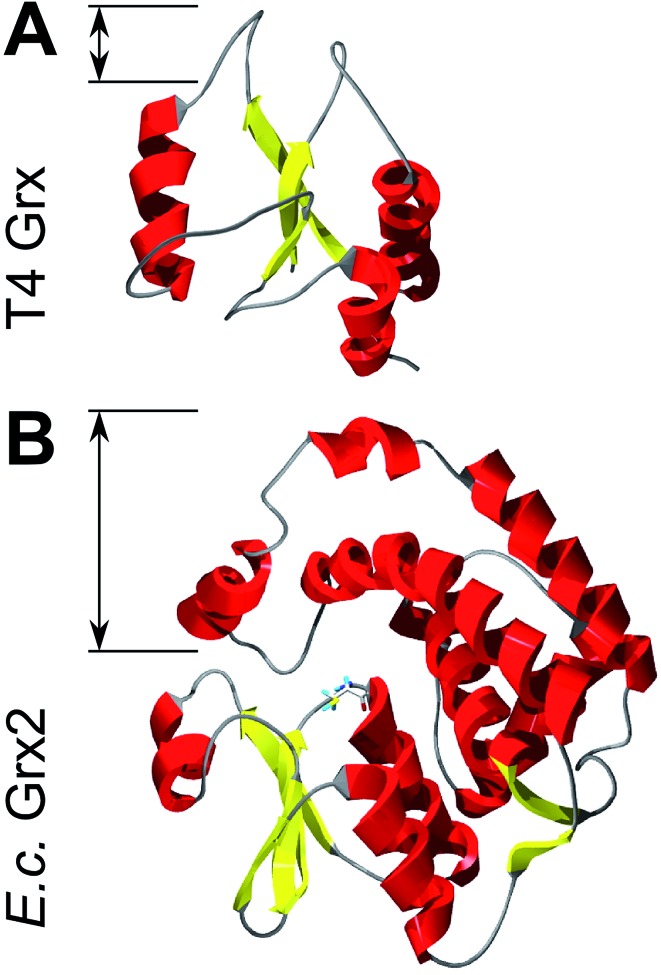
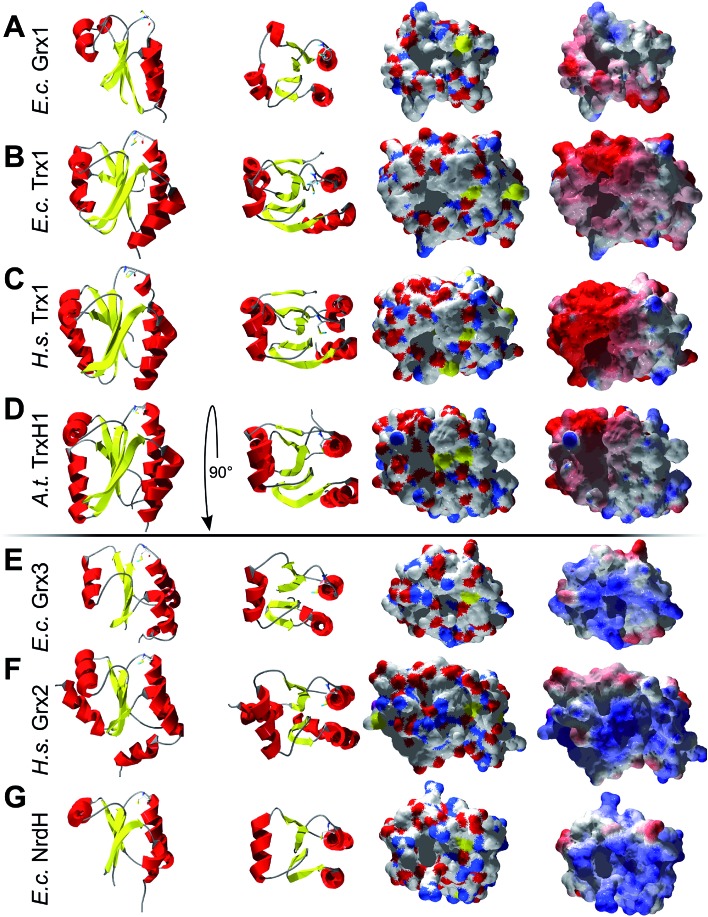
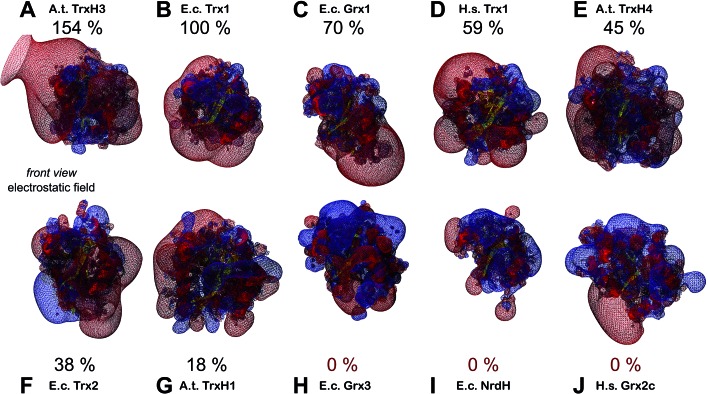
Could these specific complementary electrostatic surface potential patches be a common feature of redoxins that react kinetically with PR and be different in those that don't? To address this hypothesis, we analyzed the available structures of the redoxins analyzed kinetically, i.e. E. coli Grx1 (pdb code: ; 1egr),31 Grx2 (; 1g7o),32 Grx3 (; 3grx),33 Trx1 (; 1xob),34 and NrdH (; 1h75),35 bacteriophage T4 Grx (; 1de2),36A. thaliana TrxH1 (; 1xfl),37 as well as human Grx2 (; 2fls) and Trx1 (; 1ert).38 From these structures, T4 Grx and E. coli Grx2 differ the most, since these redoxins contain elongated loops on the interaction surface (T4 Grx, Fig. 4A) or an additional domain that partly covers the contact area (E. coli Grx2, Fig. 4B). These features obviously explain their lack of activity with PR. The remaining structures fall into two groups with respect to their electrostatic characteristics (Fig. 5). The first group consists of E. coli Grx1 (Fig. 5A) and Trx1 (Fig. 5B), human Trx1 (Fig. 5C), and A. thaliana TrxH1 (Fig. 5D). This group is characterized by a neutral to slightly negative contact area, marked by a few small positive patches (Fig. 5A–D, 5th column). And, moreover, a rather prominent negative electric field protruding into the surrounding solvent outside the contact surface characterized above (Fig. 6A–G). The second group includes E. coli Grx3 (Fig. 5E and 6I), human Grx2 (Fig. 5F and 6H), and E. coli NrdH (Fig. 5G and 6J). This group lacks prominent electric fields reaching into the solvent and displays a pronounced positive surface potential in the area corresponding to the contact surface between E. coli Trx1 and PR (Fig. 2C and D). Astonishingly, these two groups exactly correspond to the redoxins active with PR (first group) and the redoxins inactive with PR (second group). In fact, the strength and extent of the negative electric field of the redoxins outside the contact area directly correlates to the catalytic efficiency of PR with these redoxins as electron donors (Fig. 6).
Fig. 4. Structures of Grxs non-functional with PAPS reductase with altered active site geometry. Secondary structure representations of (A) T4 Grx and (B) E. coli Grx2. The loops and domain protruding from the area corresponding to the contact surface in Trx1 are marked. The pictures were computed using the DeepView/Swiss-PDB Viewer 4.1 with PDB accession numbers ; 1de2 (T4)36 and ; 1g7o (Grx2).32.
Fig. 5. Structural analysis of some of the Trxs and Grxs tested as electron donor for PAPS reductase. First column: secondary structure representation from the front, second column: secondary structure representation from the top, i.e. rotated by 90° to the front. Third column: surface representation of the active site and potential contact areas in atomic type coloring. Fourth column: electrostatic surface potentials (from red = –4 to blue = +4 kT/e, using atomic partial charges) mapped to the surface. The pictures were computed using the DeepView/Swiss-PDB Viewer 4.1 with PDB accession numbers: (A) E. coli Grx1 (; 1egr),31, (B) E. coli Trx1 (; 1xob),34 (C) human Trx1 (; 1ert),38 (D) A. thaliana TrxH1 (; 1xfl),37 (E) E. coli Grx3 (; 3grx),33 (F) human Grx2 (; 2fls), and (G) E. coli NrdH (; 1h75).35.
Fig. 6. Electrostatic potential maps of some of the redoxins tested as electron donors for PAPS reductase. The figures show the secondary structure representations (in front view, see Fig. 4, first column) with the computed electric fields protruding into the surrounding solvent. Additionally, the efficiency of PR with the various redoxins as electron donor were included. The electrostatic potential maps (from red = –4 to blue = +4 kT per e, using atomic partial charges) and the pictures were computed using the DeepView/Swiss-PDB Viewer 4.1. The structures marked with an asterisk were not experimentally determined but computed here by molecular modelling using the Swiss-model server. (A) A. thaliana TrxH3* (modelled with template ; 1wmj), (B) E. coli Trx1 (; 1xob), (C) E. coli Grx1 (; 1egr), (D) human Trx1 (; 1ert), (E) A. thaliana TrxH4*(modelled with template ; 1wmj), (F) E. coli Trx2* (modelled with template ; 3p2a), (G) A. thaliana TrxH1 (; 1xfl), (H) human Grx2 (; 2fls), (I) E. coli Grx3 (; 3grx), and E. coli NrdH (; 1h75).
Discussion
The specificity of distinct Trx and Grx proteins for any given catalytic or allosteric disulfide, see ref. 5, is a key element for the controlled flow of metabolites and the operation of thiol switches in redox signalling.39 Using E. coli PR as model, our study suggests that geometric and electrostatic complementarity as well as electric fields and thus long-distance electrostatic interactions between the redoxin and their target proteins are the key elements for Trx family proteins' specificity and efficiency.
The differences in standard redox potentials between two redox pairs, such as a Trx (red/ox) and a metabolic enzyme such as PR (red/ox), are a measure of the free energy of the reaction and thus how thermodynamically favorable this reaction is. Thermodynamics, however, do not determine physiological reactions. Rate constants are determined by, most of all, the magnitude of the activation energy barrier. This is what enzymes facilitate – increasing reaction rates by lowering activation energies. In vivo, the flow of metabolites is controlled by metabolite concentrations, enzymes' specificities, the regulation of enzyme activity, and compartmentalization. As long as a reaction is thermodynamically favorable (or made favorable by, for instance, detracting a reaction product) reaction rates are controlled by proteins. This was nicely confirmed here. The differences in redox potentials of the redoxins and PR did not correlate to the efficiency of the enzyme with these redoxins as electron donors. Similar conclusions were drawn before. The standard redox potential of Trx1 and Grx1 are –270 mV (ref. 40) and –233 mV,30 respectively, making Trx the thermodynamically more favorable reductant. E. coli ribonucleotide reductase, however, is catalytically more efficient with Grx1 as electron donor.41 Moreover, the redoxins which are able or unable to reduce ribonucleotide reductase correspond to the same two groups as the redoxins investigated as reductants for PR here. E. coli Trxs 1, Trx2, and Grx1 as well as poplar Grx are able to reduce E. coli ribonucleotide reductase,8,9,25,42 whereas E. coli Grx2, Grx4, and NrdH display marginal or no activity.22,43,44
The importance of geometric and electrostatic complementary surfaces has been highlighted before in a study that aimed at the determination of the structure of the peroxiredoxin–glutaredoxin (Prx–Grx) hybrid protein from Haemophilus influenza.45 This study revealed two interaction sites on the surface of the Prx domain, depending on the reaction cycle of the peroxidase that involves a conformational change of the active site peroxidatic cysteinyl residue. These areas interact with essentially the same contact surface on the Grx domain of a second hybrid protein in the homo tetrameric quaternary structure. Both modes of interaction involve specific electrostatic interactions of two small positive patches on the Grx domain defined by lysyl residue 177 and arginyl residue 212, with two negative patches on the Prx domain, defined by glutamyl 59 and aspartyl–glutamyl residues 89–90 or aspartyl residues 148, 154, and 156.45 This interaction is quite similar to the interaction of Trx1 with PR, see Fig. 2D. Another example for the importance of complementary surfaces was provided by the crystal structures of the two barley TrxH isoforms 1 and 2 and a complex of TrxH2 with the α-amylase/subtilisin inhibitor BASI.46 This study also concluded that substrate specificity and reaction efficiency may be mainly based on complementary contact areas and specific molecular interactions between the Trxs and their target, and not on differences in redox potentials, that are almost identical for these two Trxs.47
Our conclusions are further supported by an analysis of three more complexes between Trx family proteins and their targets that are available in the protein data base. The complexes of human Trx1 and thioredoxin interacting protein (TXNIP),48 namely TrxH2 and BASI, and between yeast Trx1 and methionine sulfoxide reductase A (MsrA)49 are depicted in Fig. 7. The proteins do not only demonstrate surface complementarity but, when separated, also a perfect complementarity between their electrostatic field maps when the interacting surfaces face each other (Fig. 7, first column).
Fig. 7. Interaction analysis of other Trx–target protein complexes from the protein data bank. A ribbon representation of the complexes is shown in the second column. The atomic, interacting and electrostatic surfaces are shown in columns three to five, arranged as in Fig. 2. The first column features the electrostatic maps of the interacting proteins separated from each other. A crude representation of the surface charges was added to highlight the complementarity of the electrostatic fields of the proteins pointing to each other with their interacting surfaces. (A) Complex between human Trx1 and the thioredoxin interacting protein (TXNIP), pdb code ; 4ll4. (B) Complex between Hordeum vulgare (barley) TrxH2 and BASI, pdb code ; 2iwt. (C) Complex between yeast Trx1 and methionine sulfoxide reductase A (MsrA), pdb code ; 3pin.
Long-range electrostatic interactions are thought to be a driving force for facilitating and enhancing the rate of specific binding of a protein to a target.50,51 Not surprisingly, the best studied mechanisms of signalling and regulation – reversible phosphorylation – primarily affect electrostatic interactions. Here, we have seen a strong correlation between the efficiency of PR with various Trx and Grx proteins as electron donor and the extent and strength of the negative electric fields of the redoxins protruding into the solvent, mostly outside the immediate contact area (Fig. 5). The only study addressing this point so far by Bunik et al.52 analyzed the activation of α-ketoglutarate dehydrogenase by various Trxs. Supporting the conclusions drawn here, this study identified that the length of the α-helix 1 (where part of the active site is located) and the surrounding charges correlate with the influence of the Trxs on the α-ketoglutarate dehydrogenase complex. The efficiency of the Trxs tested directly correlated to the strength of the, in this case positive, electric fields/polarization and the highest dipole vector. Bunik et al. concluded that the ‘selective action of a thioredoxin should stem from specific recognition upon formation of the thioredoxin–target complex […] before the highly reactive catalytic groups are brought together’.52 This hypothesis is fully supported by our study. Astonishingly, for these long range electrostatic interactions, amino acid residues outside the contact area can be as or even more important than the residues forming the complementary contact surfaces.
The importance of cellular redox potentials for efficient electron transfer, catalytic or in signalling events, is questionable.53,54 The results and analyses presented here using PR, the substrate tested for the greatest variety of redoxins, suggest that both short- and long-range electrostatic interactions are the major determinants of the specificity of Trx family proteins. Additional limits are set by the necessity for geometric complementarity. Supported by previous studies and all available structures of Trx–target complexes, this could be the major mechanism for the target specificity of Trxs and Grxs.
Experimental
General methods
Chemicals and enzymes were purchased from Sigma-Aldrich (St. Louis MO, USA), unless otherwise stated, and of analytical grade or better. Electrophoresis and Western blotting were run as described in ref. 16.
Cloning
E. coli Grx1 was cloned as a 256 bp NdeI/XhoI PCR-fragment from genomic DNA using the oligonucleotides 5′-CACACACATATGCAAACCGTTATTTTTGGTCG-3′ and 5′-CACACACTCGAGGGCGTCCAGATTTTCTTTCACCC-3′ and cloned into vector pET16b.
Protein expression and purification
PR was expressed and purified from a pET16b derivative as described in ref. 16, likewise Grx1. 40 mg of PR and 75 mg Grx1 were obtained per liter LB broth at a purity of >98% as judged by SDS-PAGE. Human Grx2 was produced as described in ref. 55. Human Trx1, E. coli NrdH, and T4 Grx were a kind gift by Arne Holmgren (Karolinska Institutet Stockholm). Recombinant plant thioredoxins were a kind gift by Yves Meyer (Perpignan, France) and Jean-Pierre Jacquot (Nancy, France).
PAPS reductase assay
Activity of PR was measured as acid labile 35[S]–SO32– formation from 35[S]–PAPS18. 35[S]–PAPS was prepared enzymatically from 35[S]-SO42– (Amersham-Buchler, Braunschweig) as described in16 using recombinant APS kinase from Arabidopsis thaliana.56 Kinetic constants were calculated from a series of measurements repeated independently at least three times. The assay mixture (100 μl) contained 100–250 ng ml–1 of purified PAPS-reductase, 100 mM Tris/HCl pH 8.0, 10 mM Na2SO3, 100 μM 35[S]–PAPS (specific radioactivity: 4.2 kBq·nmol–1), 0.5–50 μM redoxin. The redoxins were kept reduced by 10–25 mM dithiothreitol (DTT) and/or 10 mM reduced glutathione.
Spectroscopic methods
We used a double beam spectrophotometer (Sigma ZWS-II, Berlin) for UV-absorbance difference spectroscopy19 fitted with two sets of tandem cuvettes. Oxidized PR (5–50 μM) and redox buffer in separate compartments of the tandem cuvettes were measured against PR plus redox buffer in the same compartment. The reactants were allowed to equilibrate in stoppered cuvettes at ambient temperature until the reading of the absorbance difference was constant. The spectral bandwidth was adjusted to 1 nm with Grx1 as reductant and 2 nm for the PR–GrxC14S complex. The scanning speed was 2 nm s–1. The measured absorbance difference was converted into molar absorption changes Δε based on the concentration of PR used. The standard redox potential of PR was determined using glutathione (10–100 mM) redox buffers containing a catalytical amount (1 : 100 in relation to PR, i.e. 0.05–0.5 μM) of E. coli Grx1 at 25 °C in 100 mM potassium phosphate pH 7.0. Before use, PAPS–reductase was converted to the fully oxidized state (PRox) by incubating the protein with PAPS at 25 °C for 1 h in the absence of reductants. PAPS, PAP, and sulfite were removed by gel filtration using SephadexG25 (Pharmacia). The enzyme was concentrated using Centriprep concentrators YM3 (Millipore). The samples were placed in the photometer as described above and the change in absorbance was followed continuously at 294 nm versus PRox until equilibrium was reached, i.e. up to seven hours. The ratio of reduced to oxidized PAPS-reductase was obtained by normalizing Δε to Δεmax. The equilibrium constant keq for the thiol disulfide exchange reaction via glutaredoxin involves only the oxidized and reduced forms of PR and glutathione. Given the standard redox potential of GSH as –240 mV,30 the results were fitted by non linear regression using the Nernst equation with E0′ and n (the number of electrons) as variable parameters.
Computational methods
Protein structures and secondary structure assignments were obtained from the Protein Data Bank (RCBS, http://www.rcsb.org/pdb/home/home.do). Molecular surfaces, contact surfaces and electrostatics were visualized using the Swiss PDB Viewer57 and rendered using the ‘Persistance of Vision Raytracer’ (Povray, ; http://www.povray.org/). Detailed molecular interactions were analyzed with ‘contact’ of the CCP4 suite.58 Structural alignments between all redoxins analysed and E. coli Trx1 were calculated using the PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC (; http://www.pymol.org/). Electrostatic potential maps were calculated using the Swiss PDB Viewer. The dielectric constant of the solvent and proteins were set to 78.54 and 4, respectively, for computation applying the Poisson–Boltzmann method. When mapped to the surfaces, electrostatic maps were depicted from –4 (red) to +4 kT per e (blue). The structures of A. thaliana Trxs H2 and H3 (template for both: PDB accession ; 1wmj), as well as E. coli Trx2 (template: ; 3p2a) were modelled using the Swiss-Model server.59 Grace (; http://plasma-gate.weizmann.ac.il/Grace/) was used for data plotting and regression analysis, Inkscape (; http://inkscape.org/) and the ‘GNU Image Manipulation Program’ (GIMP, ; http://www.gimp.org/) for preparing the figures.
Conclusions
The rationale for substrate specificity of the proteins from the thioredoxin family is key to the understanding of redox signalling in physiology and pathology. From this work, we conclude that the recognition of disulfide substrates by thioredoxins and glutaredoxins is not determined by redox potentials, but by specific long-distance electrostatic interactions and complementary contact surfaces.
Acknowledgments
The Authors wish to thank Arne Holmgren (Stockholm, Sweden), Jean-Pierre Jacquot (Nancy, France), and Yves Meyer (Perpignan, France) for providing proteins for analyses. We express our gratitude to Marcel Deponte (Heidelberg, Germany) for discussing the manuscript and inspiring this work. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Li 984/3-1 and Be 3259/5-1).
References
- Lillig C. H., Holmgren A. Antioxid. Redox Signaling. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- Gallogly M. M., Mieyal J. J. Curr. Opin. Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Lillig C. H., Berndt C., Holmgren A. Biochim. Biophys. Acta, Gen. Subj. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Hanschmann E.-M., Godoy J. R., Berndt C., Hudemann C., Lillig C. H. Antioxid. Redox Signaling. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B., Ho L., Hogg P. J. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- Martin J. L. Struct. Lond. Engl. 1993. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- Qi Y., Grishin N. V. Proteins. 2005;58:376–388. doi: 10.1002/prot.20329. [DOI] [PubMed] [Google Scholar]
- Laurent T. C., Moore E. C., Reichard P. J. Biol. Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- Holmgren A. J. Biol. Chem. 1979;254:3664–3671. [PubMed] [Google Scholar]
- Gonzalez-Porqué P., Baldesten A., Reichard P. J. Biol. Chem. 1970;245:2371–2374. [PubMed] [Google Scholar]
- Tsang M. L., Schiff J. A. J. Bacteriol. 1978;134:131–138. doi: 10.1128/jb.134.1.131-138.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M. L. J. Bacteriol. 1981;146:1059–1066. doi: 10.1128/jb.146.3.1059-1066.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone F. A., Westphal G., Schwenn J. D. Mol. Genet. Genomics. 1991;225:314–319. doi: 10.1007/BF00269864. [DOI] [PubMed] [Google Scholar]
- Schwenn J. D., Krone F. A., Husmann K. Arch. Microbiol. 1988;150:313–319. doi: 10.1007/BF00408300. [DOI] [PubMed] [Google Scholar]
- Berendt U., Haverkamp T., Prior A., Schwenn J. D. Eur. J. Biochem. 1995;233:347–356. doi: 10.1111/j.1432-1033.1995.347_1.x. [DOI] [PubMed] [Google Scholar]
- Lillig C. H., Prior A., Schwenn J. D., Aslund F., Ritz D., Vlamis-Gardikas A., Holmgren A. J. Biol. Chem. 1999;274:7695–7698. doi: 10.1074/jbc.274.12.7695. [DOI] [PubMed] [Google Scholar]
- Lillig C. H., Potamitou A., Schwenn J.-D., Vlamis-Gardikas A., Holmgren A. J. Biol. Chem. 2003;278:22325–22330. doi: 10.1074/jbc.M302304200. [DOI] [PubMed] [Google Scholar]
- Schwenn J. D., Schriek U. Z. für Naturforschung. 1987;42:93–102. doi: 10.1515/znc-1987-1-216. [DOI] [PubMed] [Google Scholar]
- Aslund F., Ehn B., Miranda-Vizuete A., Pueyo C., Holmgren A. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P., Holmgren A. J. Bacteriol. 1990;172:1923–1929. doi: 10.1128/jb.172.4.1923-1929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A. P., Fladvad M., Berndt C., Andresen C., Lillig C. H., Neubauer P., Sunnerhagen M., Holmgren A., Vlamis-Gardikas A. J. Biol. Chem. 2005;280:24544–24552. doi: 10.1074/jbc.M500678200. [DOI] [PubMed] [Google Scholar]
- Jordan A., Aslund F., Pontis E., Reichard P., Holmgren A. J. Biol. Chem. 1997;272:18044–18050. doi: 10.1074/jbc.272.29.18044. [DOI] [PubMed] [Google Scholar]
- Berglund O., Sjöberg B. M. J. Biol. Chem. 1970;245:6030–6035. [PubMed] [Google Scholar]
- Meyer Y., Reichheld J. P., Vignols F. Photosynth. Res. 2005;86:419–433. doi: 10.1007/s11120-005-5220-y. [DOI] [PubMed] [Google Scholar]
- Rouhier N., Vlamis-Gardikas A., Lillig C. H., Berndt C., Schwenn J.-D., Holmgren A., Jacquot J.-P. Antioxid. Redox Signaling. 2003;5:15–22. doi: 10.1089/152308603321223504. [DOI] [PubMed] [Google Scholar]
- Tagaya Y., Maeda Y., Mitsui A., Kondo N., Matsui H., Hamuro J., Brown N., Arai K., Yokota T., Wakasugi H. EMBO J. 1989;8:757–764. doi: 10.1002/j.1460-2075.1989.tb03436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg M., Johansson C., Chandra J., Enoksson M., Jacobsson G., Ljung J., Johansson M., Holmgren A. J. Biol. Chem. 2001;276:26269–26275. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- Gladyshev V. N., Liu A., Novoselov S. V., Krysan K., Sun Q. A., Kryukov V. M., Kryukov G. V., Lou M. F. J. Biol. Chem. 2001;276:30374–30380. doi: 10.1074/jbc.M100020200. [DOI] [PubMed] [Google Scholar]
- Chartron J., Shiau C., Stout C. D., Carroll K. S. Biochemistry. 2007;46:3942–3951. doi: 10.1021/bi700130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslund F., Berndt K. D., Holmgren A. J. Biol. Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- Sodano P., Xia T. H., Bushweller J. H., Björnberg O., Holmgren A., Billeter M., Wüthrich K. J. Mol. Biol. 1991;221:1311–1324. doi: 10.1016/0022-2836(91)90935-y. [DOI] [PubMed] [Google Scholar]
- Xia B., Vlamis-Gardikas A., Holmgren A., Wright P. E., Dyson H. J. J. Mol. Biol. 2001;310:907–918. doi: 10.1006/jmbi.2001.4721. [DOI] [PubMed] [Google Scholar]
- Nordstrand K., slund F., Holmgren A., Otting G., Berndt K. D. J. Mol. Biol. 1999;286:541–552. doi: 10.1006/jmbi.1998.2444. [DOI] [PubMed] [Google Scholar]
- Jeng M. F., Campbell A. P., Begley T., Holmgren A., Case D. A., Wright P. E., Dyson H. J. Struct. Lond. Engl. 1993. 1994;2:853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- Stehr M., Schneider G., Aslund F., Holmgren A., Lindqvist Y. J. Biol. Chem. 2001;276:35836–35841. doi: 10.1074/jbc.M105094200. [DOI] [PubMed] [Google Scholar]
- Wang Y., Amegbey G., Wishart D. S. J. Biomol. NMR. 2004;29:85–90. doi: 10.1023/B:JNMR.0000019506.30351.ca. [DOI] [PubMed] [Google Scholar]
- Peterson F. C., Lytle B. L., Sampath S., Vinarov D., Tyler E., Shahan M., Markley J. L., Volkman B. F. Protein Sci. 2005;14:2195–2200. doi: 10.1110/ps.051477905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichsel A., Gasdaska J. R., Powis G., Montfort W. R. Struct. Lond. Engl. 1993. 1996;4:735–751. doi: 10.1016/s0969-2126(96)00079-2. [DOI] [PubMed] [Google Scholar]
- Deponte M., Lillig C. H. Biol. Chem. 2015 doi: 10.1515/hsz-2014-0280. [DOI] [PubMed] [Google Scholar]
- Krause G., Lundström J., Barea J. L., Pueyo de la Cuesta C., Holmgren A. J. Biol. Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
- Fernandes A. P., Holmgren A. Antioxid. Redox Signaling. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- Miranda-Vizuete A., Damdimopoulos A. E., Gustafsson J., Spyrou G. J. Biol. Chem. 1997;272:30841–30847. doi: 10.1074/jbc.272.49.30841. [DOI] [PubMed] [Google Scholar]
- Vlamis-Gardikas A., Aslund F., Spyrou G., Bergman T., Holmgren A. J. Biol. Chem. 1997;272:11236–11243. doi: 10.1074/jbc.272.17.11236. [DOI] [PubMed] [Google Scholar]
- Ortenberg R., Gon S., Porat A., Beckwith J. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7439–7444. doi: 10.1073/pnas.0401965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Woo J. R., Hwang Y. S., Jeong D. G., Shin D. H., Kim K., Ryu S. E. J. Biol. Chem. 2003;278:10790–10798. doi: 10.1074/jbc.M209553200. [DOI] [PubMed] [Google Scholar]
- Maeda K., Hägglund P., Finnie C., Svensson B., Henriksen A. Protein Sci. 2008;17:1015–1024. doi: 10.1110/ps.083460308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Hägglund P., Björnberg O., Winther J. R., Svensson B. FEBS Lett. 2010;584:3376–3380. doi: 10.1016/j.febslet.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Hwang J., Suh H.-W., Jeon Y. H., Hwang E., Nguyen L. T., Yeom J., Lee S.-G., Lee C., Kim K. J., Kang B. S., Jeong J.-O., Oh T.-K., Choi I., Lee J.-O., Kim M. H. Nat. Commun. 2014;5:2958. doi: 10.1038/ncomms3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.-X., Guo P.-C., Shi W.-W., Luo M., Tan X.-F., Chen Y., Zhou C.-Z. J. Biol. Chem. 2011;286:13430–13437. doi: 10.1074/jbc.M110.205161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M. K. Curr. Opin. Struct. Biol. 1995;5:216–223. doi: 10.1016/0959-440x(95)80079-4. [DOI] [PubMed] [Google Scholar]
- Zhou H.-X. Phys. Biol. 2005;2:R1–R25. doi: 10.1088/1478-3975/2/3/R01. [DOI] [PubMed] [Google Scholar]
- Bunik V., Raddatz G., Lemaire S., Meyer Y., Jacquot J. P., Bisswanger H. Protein Sci. 1999;8:65–74. doi: 10.1110/ps.8.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohé L. Biochim. Biophys. Acta. 2013;1830:3139–3142. doi: 10.1016/j.bbagen.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Berndt C., Lillig C. H., Flohé L. Front. Pharmacol. 2014;5 doi: 10.3389/fphar.2014.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig C. H., Berndt C., Vergnolle O., Lonn M. E., Hudemann C., Bill E., Holmgren A. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8168–8173. doi: 10.1073/pnas.0500735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig C. H., Schiffmann S., Berndt C., Berken A., Tischka R., Schwenn J. D. Arch. Biochem. Biophys. 2001;392:303–310. doi: 10.1006/abbi.2001.2453. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M. C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M. C. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hajjaji H., Dumoulin M., Matagne A., Colau D., Roos G., Messens J., Collet J.-F. J. Mol. Biol. 2009;386:60–71. doi: 10.1016/j.jmb.2008.11.046. [DOI] [PubMed] [Google Scholar]
- Watson W. H., Pohl J., Montfort W. R., Stuchlik O., Reed M. S., Powis G., Jones D. P. J. Biol. Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- Sagemark J., Elgán T. H., Bürglin T. R., Johansson C., Holmgren A., Berndt K. D. Proteins. 2007;68:879–892. doi: 10.1002/prot.21416. [DOI] [PubMed] [Google Scholar]