Abstract
The mouse vomeronasal organ is specialized in the detection of pheromones. Vomeronasal sensory neurons (VSNs) express chemosensory receptors of two large gene repertoires, V1R and V2R, which encode G‐protein‐coupled receptors. Phylogenetically, four families of V2R genes can be discerned as follows: A, B, C, and D. VSNs located in the basal layer of the vomeronasal epithelium coordinately coexpress V2R genes from two families: Approximately half of basal VSNs coexpress Vmn2r1 of family C with a single V2R gene of family A8‐10, B, or D (‘C1 type of V2Rs’), and the other half coexpress Vmn2r2 through Vmn2r7 of family C with a single V2R gene of family A1‐6 (‘C2 type V2Rs’). The regulatory mechanisms of the coordinated coexpression of V2Rs from two families remain poorly understood. Here, we have generated two mouse strains carrying a knockout mutation in Vmn2r1 by gene targeting in embryonic stem cells. These mutations cause a differential decrease in the numbers of VSNs expressing a given C1 type of V2R. There is no compensatory expression of Vmn2r2 through Vmn2r7. VSN axons coalesce into glomeruli in the appropriate region of the accessory olfactory bulb in the absence of Vmn2r1. Gene expression profiling by NanoString reveals a differential and graded decrease in the expression levels across C1 type of V2Rs. There is no change in the expression levels of C2 type of V2Rs, with two exceptions that we reclassified as C1 type. Thus, there appears to be a fixed probability of gene choice for a given C2 type of V2R.
Keywords: accessory olfactory system, gene expression, gene regulation, pheromone, V2R
Introduction
The mouse vomeronasal organ (VNO) is a chemosensory organ residing at the base of the nasal septum that is specialized in the detection of pheromones. Most VSNs express genes of two repertoires that encode chemosensory receptors with a putative seven transmembrane domain structure: vomeronasal receptor genes V1R (Dulac & Axel, 1995) and V2R (Herrada & Dulac, 1997; Matsunami & Buck, 1997; Ryba & Tirindelli, 1997). The cell bodies of V1R+ and V2R+ VSNs reside in the apical and basal layers of the VNO, respectively. The basal layer of VSNs can be subdivided into two sublayers based on the expression of nine non‐classical class I major histocompatibility complex genes termed H2‐Mv (Ishii et al., 2003; Loconto et al., 2003; Ishii & Mombaerts, 2008; Leinders‐Zufall et al., 2014). Axons of VSNs from the apical and basal layers coalesce into multiple glomeruli in the anterior and posterior halves of the accessory olfactory bulb (AOB), respectively (Belluscio et al., 1999; Rodriguez et al., 1999; Del Punta et al., 2002; Ishii & Mombaerts, 2008, 2011).
V2R+ VSNs respond physiologically to peptides and proteins (Leinders‐Zufall et al., 2004, 2009, 2014; Kimoto et al., 2005; Chamero et al., 2007, 2011, 2012; Haga et al., 2007, 2010; Sturm et al., 2013). The deduced amino acid sequences of V2Rs contain a long N‐terminal extracellular region, which may form the ligand‐binding site. The mouse genome contains 120 intact V2R genes, which can be grouped in four families (A, B, D, and C) based on amino acid sequence homology (Yang et al., 2005; Shi & Zhang, 2007; Young & Trask, 2007; Francia et al., 2014). The largest V2R gene family, family A, can be further grouped in nine subfamilies A1 to A10; family A7 exists in rat but not in mouse. Family C, which was originally referred to as mV2R 2 (Ryba & Tirindelli, 1997) and then mouse V2R2 (Martini et al., 2001), comprises seven genes in mouse, Vmn2r1 through Vmn2r7 (Silvotti et al., 2007, 2011; Francia et al., 2014).
The consensus of numerous in situ hybridization (ISH) studies is that mouse family‐ABD V2R genes are expressed in a punctate and mutually exclusive manner, most likely as a single gene per basal VSN. When we knocked out the family‐A9 gene V2rf2/Vmn2r81 and replaced it by the β‐galactosidase marker using gene targeting in embryonic stem (ES) cells, we found that ~25% of VSNs that express the marker express another family‐ABD V2R gene (Ishii & Mombaerts, 2011). This second choice of another family‐ABD V2R gene can be interpreted in terms of a hypothetical negative feedback mechanism that helps restrict expression of family‐ABD V2R genes to a single gene per VSN.
In sharp contrast to observations with family‐ABD V2R genes, ISH probes or antibodies for family‐C V2Rs label large populations of basal VSNs (Martini et al., 2001; Silvotti et al., 2007, 2011; Ishii & Mombaerts, 2011). Immunoreactive signals with antibodies against family‐C members Vmn2r1 and Vmn2r2/Vmn2r5 are detected in a mutually exclusive manner, each in approximately half of basal VSNs (Silvotti et al., 2007, 2011). VSNs expressing V2Rs of families A8‐10, B, and D are preferentially colabeled with anti‐Vmn2r1 antibody, and VSNs expressing V2Rs of family A1‐6 are preferentially colabeled with Vmn2r2/Vmn2r5 antibody (Silvotti et al., 2007, 2011). Vmn2r2 through Vmn2r7 are coexpressed in various combinations, and many VSNs appear to coexpress all six receptors (Silvotti et al., 2011). Surprisingly, more than twenty years after the discovery of mouse V2R genes, there are no reports about mouse strains with a knockout mutation in a family‐C gene.
Here, we have generated two novel mouse strains carrying a knockout mutation in Vmn2r1. We found that various subpopulations of V2R+ VSNs are differentially affected by the absence of Vmn2r1; that there is no compensatory expression of Vmn2r2 through Vmn2r7; and that glomeruli form normally if there are sufficient VSNs left. We discovered that Vmn2r65 (family A5) and Vmn2r120 (family A6) are C1 type of V2Rs, in discordance with their phylogenetic classification as C2 type of V2Rs.
Materials and methods
Animal ethics
Mouse studies were carried out in accordance with the German Animal Welfare Act, the European Communities Council Directive 2010/63/EU, and the institutional ethical and animal welfare guidelines of The Rockefeller University, the Max Planck Institute of Biophysics, and the Max Planck Research Unit for Neurogenetics. Approval came from the IACUC of The Rockefeller University, the Regierungspräsidium Darmstadt and the Veterinäramt of the City of Frankfurt.
Generation of gene‐targeted mouse strains
We have previously described the gene‐targeted strains V2r1b‐IRES‐tauGFP (Del Punta et al., 2002) and V2rf2‐IRES‐tauGFP (Ishii & Mombaerts, 2011). The internal ribosome entry site sequence (IRES) affords bicistronic translation of transcripts. We have here generated four novel mouse strains by gene targeting in the parental ES cell line E14. For the ∆C1 targeting vector, a 2.6 kilobase (kb) upstream fragment and a 4.8 kb downstream fragment flanking exon 1 of the Vmn2r1 gene were used as the 5′ and 3′ homologous arms, respectively. The Hprt (5′)‐loxP‐neo cassette (Ramírez‐Solis et al., 1995) was ligated to replace a 1065 basepair (bp) sequence from the SpeI site at the 28th nucleotide (nt) before the initiation codon to the HindIII site at the 801st nt after the end of exon 1. For the ∆C1‐GFP targeting vector, a 2.6 kb upstream fragment and a 5.6 kb downstream fragment of exon 1 of the Vmn2r1 gene were used as the 5′ and 3′ homologous arms, respectively. The tauGFP‐pA cassette and the self‐excising neo selectable marker cassette (ACNF) were ligated to replace a 230 bp sequence from the initiation codon in‐frame to the 4th nt before the end of exon 1. For the V2rf4‐IRES‐tauVenus targeting vector, a 8.2 kb HpaI‐NheI fragment containing exon 6 of the Vmn2r83 gene was used to construct the homologous arms. The IRES‐tauVenus‐ACNF cassette was inserted into an AscI site that was created one nt after the stop codon of Vmn2r83. For the V2rf1‐IRES‐taumCherry targeting vector, a 7.8 kb SpeI fragment containing exon 6 of the Vmn2r82 gene was used to construct the homologous arms. The IRES‐taumCherry‐ACNF cassette was inserted into an artificial AscI site one nt after the stop codon of Vmn2r82. All DNA sequences for homologous arms were from mouse BAC clones of 129/SvEv genomic origin. The targeting vectors were linearized and electroporated into ES cells as described (Mombaerts et al., 1996). Homologous recombination events were screened and confirmed by Southern blot hybridization with external probes. Cells from gene‐targeted ES clones were injected into C57BL/6J blastocysts, and germline transmission was obtained by mating male chimeras with C57BL/6J females. The ACNF cassette got removed during transmission through the male germline, leaving a loxP sequence behind in the targeted mutation. The strains are publicly available from The Jackson Laboratory in a mixed (129P2/OlaHsd x C57BL/6J) background, as follows: ∆C1 as stock number JR#24643 and strain name B6;129P2‐Vmn2r1<tm1Mom>/MomJ, ∆C1‐GFP as JR#26765 and B6;129P2‐Vmn2r1<tm2Mom>/MomJ, V2rf1‐mCherry as JR#7885 and B6;129P2‐Vmn2r82<tm1Mom>/MomJ, and V2rf4‐Venus as JR#7886 and B6;129P2‐Vmn2r83<tm1Mom>/MomJ.
Tissue preparation
Mice were anesthetized by intraperitoneal injection of ketamine and xylazine (210 mg/kg and 10 mg/kg body weight, respectively), and perfused transcardially with phosphate‐buffered saline (PBS) at room temperature, followed by ice‐cold 4% paraformaldehyde/PBS. The VNO and the brain including the AOB were dissected separately, and post‐fixed for 3 h at 4 °C. The samples were then decalcified in 0.45 m EDTA/PBS at 4 °C overnight (for 4‐week‐old mice) or two nights (for 10‐week‐old mice), cryoprotected in 15 and 30% sucrose/PBS successively, and frozen in OCT compound (Sakura Finetek #4583). The decalcification step was omitted for brain samples and 0‐day‐old samples.
Immunohistochemistry and cell counts in the VNO
Coronal 14 μm cryosections were cut through the entire VNO and collected on glass slides. Sections were treated with 0.5% SDS/PBS for 30 min at room temperature. This step was omitted for V2Rp5 or GFP IHC alone. After washing with PBS, sections were blocked in 5% normal goat serum (NGS) and 0.3% Triton X‐100/PBS, and incubated with primary antibodies in the same blocking solution for 1–2 h at room temperature and then overnight at 4 °C. After washing in 0.1% Triton X‐100/PBS, sections were incubated with secondary antibodies and counterstained with DAPI (1:10 000, Invitrogen #D1306) for 2–4 h at room temperature. Primary antibodies and working dilution of each antibody were as follows: rabbit anti‐C1 (anti‐Vmn2r1) antibody (1:500; Silvotti et al., 2007), rabbit anti‐C2 (anti‐Vmn2r2/5) antibody (1:4000; Silvotti et al., 2007), rabbit anti‐panC antibody (also referred to as V2R2; 1:4000; Martini et al., 2001), rabbit anti‐V2Rp5 antibody (1:1500; Haga et al., 2010), and chicken anti‐GFP antibody (1:1500; Aves Labs #GFP‐1020). To improve the signal‐to‐noise ratio of anti‐C1, anti‐C2, and anti‐GFP IHC, the antibodies were pre‐absorbed with acetone powder prepared from brain homogenates of mice with a deletion of the family‐C V2R gene cluster generated by genome engineering (∆V2RC∆, unpublished). The antibody solution was incubated with 1% acetone powder overnight at 4 °C, and cleared by centrifugation and filtration before use. The following secondary antibodies were used at 1:1000 dilution: Alexa 488 goat anti‐rabbit IgG, Alexa 546 goat anti‐rabbit IgG, Alexa 647 goat anti‐rabbit IgG, and Alexa 488 goat anti‐chicken IgG (Invitrogen #A11034, A11035, A21245, A11039). Sections were examined under a Zeiss LSM 710 confocal microscope without knowing the genotype. Labeled cells were counted on every tenth section (for 4‐week and 10‐week‐old mice) or on every fifth section (for 0‐day‐old mice). The total cell number per mouse was estimated by multiplying the count by ten or five, respectively. The numbers given are numbers of VSN cell profiles, but for the sake of simplicity, they are referred to as numbers of VSNs.
IHC of the AOB
Serial sagittal 25 μm sections of the AOB were cut and collected in PBS. To visualize the glomerular layer, free‐floating sections were blocked in 5% NGS and 0.2% Tween 20/PBS, and incubated with rabbit anti‐VGLUT2 antibody (1:3000; Synaptic Systems #135403) in the same blocking solution for 1–2 h at room temperature, and then overnight at 4 °C. After washing in 0.1% Tween 20/PBS, sections were incubated with Alexa 546 goat anti‐rabbit IgG (1:1000) and counterstained with DAPI (1:10 000) for 2–4 h at room temperature. Sections were mounted on glass slides and examined under a Zeiss LSM 710 confocal microscope. For visualization of GFP+ axons, images of the intrinsic fluorescence were taken from serial AOB sections. Multiple sections were aligned using Photoshop, then combined in a z‐stacked file using image j. The 2D images were created by z‐projection with maximum intensity method.
NanoString multiplex gene expression analysis
Whole VNO mucosa was dissected in RNA Stabilization Reagent RNAlater (Qiagen #76106). Tissue from a single mouse was placed in a tube and homogenized by TissueLyser LT (Qiagen #85600). Total RNA was extracted using the RNeasy Micro kit with DNase I treatment on the column (Qiagen #74004), and stored at −80 °C until use. A 0.75 μg aliquot of RNA from each mouse was tested with the custom CodeSet Pao (NanoString Technologies, Seattle, WA, USA). Samples from six wild‐type mice and six homozygous mice were analyzed in one cartridge using the nCounter SPRINT Profiler system (NanoString Technologies) according to the manufacturer's instructions. Data processing and statistical analyses were performed using nsolver data analysis Software 3.0 (NanoString Technologies). We confirmed that samples passed quality control (QC) criteria based on imaging QC, binding density QC, and positive and negative control probes. We then processed the raw count data into normalized count values in three steps: normalization using a scaling factor calculated from the geometric mean of the six positive control counts, second normalization using a scaling factor calculated from the geometric mean of the counts of four reference genes (Ano2, Cnga4, Omp, Trpc2), and background correction by subtracting the mean + 2 standard deviations of eight negative control counts for each sample. Values < 1 after background subtraction were set as 1. Genes with a median count < 100 in wild‐type mice were eliminated from further analysis. Changes in gene expression were estimated by calculating the fold change (FC) of the geometric mean of the counts in mutant mice over the geometric mean of the counts in wild‐type mice, and the log2 FC values were plotted.
In situ hybridization
RNA probes were prepared for the coding sequences of Vmn2r65 (nt 948‐1603 of RefSeq NM_001105180.1), Vmn2r76 (mix of nt 508‐1028 and nt 1219‐1590 of RefSeq NM_001102580.1), Vmn2r118 (nt 822‐1306 of RefSeq NM_001104582.1), and Vmn2r120 (nt 1044‐1739 of RefSeq NM_001104591.1; Silvotti et al., 2007). Digoxigenin (DIG) labeling of RNA probes, tissue preparation, and ISH were performed as described previously (Ishii et al., 2004) with some modifications. Coronal 12 μm sections were cut through the entire VNO, and every 12th section was collected onto a slide for each probe. The incubation time of Proteinase K (Roche #03115828001) was 12 min. Hybridized probes were detected with an alkaline phosphatase‐conjugated anti‐DIG antibody (1:1000; Roche #11093274910) and BM purple substrate solution (Roche #11442074001). Labeled cells were counted on digital images of sections taken by a Pannoramic MIDI slide scanner (3D HISTECH) without knowing the genotype. The total cell number per mouse was determined by multiplying the count by twelve.
ISH combined with IHC
Tissue preparation and ISH were carried out as described above, except that the incubation time of Proteinase K was 5 min. After hybridization and washing, sections were blocked in 5% NGS and 0.3% Triton X‐100/PBS, and incubated with alkaline phosphatase‐conjugated anti‐DIG antibody (1:1000) and anti‐C1 antibody (1:500) or anti‐C2 antibody (1:4000) in the same blocking solution for two nights at 4 °C. After washing in 0.1% Triton X‐100/PBS, sections were incubated with Alexa 488 goat anti‐rabbit IgG (1:1000) and DAPI (1:10 000) for 8 h at room temperature. Sections were washed with 0.1% Triton X‐100/PBS, and DIG‐labeled probes were detected by incubation with HNPP/Fast Red substrate solution (Roche #11758888001) for 30 min. After washing with TN buffer (100 mm Tris pH 7.5, 150 mm NaCl), sections were examined under a Zeiss LSM 710 confocal microscope, directly in TN buffer without mounting.
Experimental design and statistical analysis
For statistical evaluations of cell numbers, the Mann–Whitney test was performed using graphpad prism 5. For NanoString analysis, statistical analysis was performed by nsolver data analysis Software 3.0. The false discovery rates (FDR; Benjamini & Yekutieli, 2001) from conducting multiple testing were computed based on the P‐values of the t‐test. Genes were considered differentially expressed if FDR < 0.05.
Results
Generation of gene‐targeted strains with a knockout mutation in Vmn2r1
The mouse V2R gene family C comprises seven intact genes, which are located in a single cluster extending over a 640 kb region on chromosome 3 (Fig. 1A). We generated two mouse strains with a gene‐targeted mutation in the Vmn2r1 gene (referred to as C1), which is composed of six coding exons (Fig. 1B).
Figure 1.
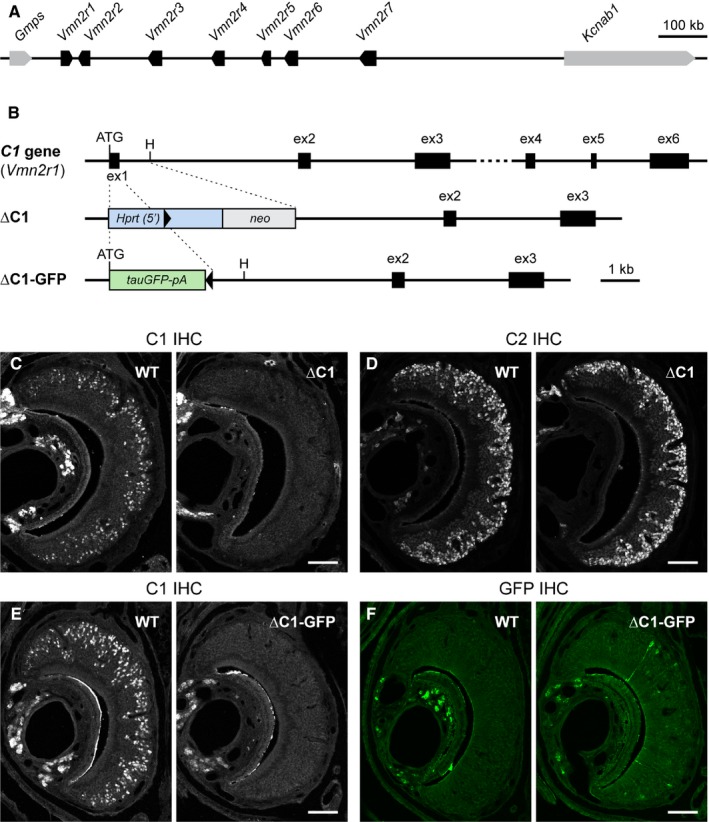
Generation of mouse strains with a knockout mutation in Vmn2r1 by gene targeting in ES cells. (A) Genomic organization of the family‐C V2R genes in mouse. The seven genes Vmn2r1 through Vmn2r7 are clustered in a 640 kb region on chromosome 3. The Vmn2r1 gene is transcribed from the (+) strand, and the other six genes are transcribed from the (−) strand. Gmps and Kcnab1 are the closest genes to the gene cluster, centromerically and telomerically, respectively. (B) Genomic structure and targeted mutagenesis of the family‐C gene Vmn2r1, abbreviated here as C1. This gene is composed of six coding exons. In the ∆C1 strain, a 1065 bp fragment that includes coding exon 1 (ex1) is replaced with a Hprt(5′)‐loxP‐neo cassette. This cassette is left behind in the targeted mutation in the mouse strain. In the ∆C1‐GFP strain, a 230 bp fragment from the initiation ATG codon to the fourth nt before the end of coding exon 1 is replaced in frame with a tauGFP‐pA cassette, followed by a self‐excising neo selectable marker cassette (ACNF). Following the removal of ACNF, one loxP site is left behind in the targeted mutation in the mouse strain, after tauGFP‐pA. The black triangles indicate a loxP site. H, HindIII. (C,D) IHC with anti‐C1 antibody (C) and anti‐C2 antibody (D) on coronal sections of the VNO from wild‐type (WT) and homozygous (∆C1) mice of the ∆C1 strain. Mice were male and 10 weeks old. There is no C1 immunoreactivity in ∆C1 mice (C), and no obvious difference in C2 immunoreactivity between WT and ∆C1 mice (D). Scale bar, 100 μm. (E,F) IHC with anti‐C1 antibody (E) and anti‐GFP antibody (F) on coronal sections of the VNO from WT and homozygous (∆C1‐GFP) mice of the ∆C1‐GFP strain. Mice were male and 4 weeks old. There is no C1 immunoreactivity in ∆C1‐GFP mice (E). Sparse GFP+ cells are detected in ∆C1‐GFP mice (F). Scale bar, 100 μm.
In the first strain (∆C1), we replaced a 1065 bp sequence including coding exon 1 with a selectable marker cassette. Immunohistochemistry (IHC) with anti‐C1 antibody revealed immunoreactivity in the basal layer of the VNO in wild‐type mice (WT) but not in homozygous (hereafter referred to as ∆C1) mice (Fig. 1C). The anti‐C1 antibody is called ‘anti‐Vmn2r1 antibody’ in Silvotti et al. (2007), and recognizes the antigen peptide region encoded by exons 3 and 4. The absence of C1 immunoreactivity in ∆C1 mice confirms that the C1 mutation is a null mutation, and that no truncated C1 protein is produced. The anti‐C2 antibody is called ‘anti‐Vmn2r2 antibody’ in Silvotti et al. (2007), and recognizes both Vmn2r2 and Vmn2r5 (Silvotti et al., 2007, 2011). We found no obvious difference in C2 immunoreactivity between WT and ∆C1 mice (Fig. 1D), indicating that the selection marker that is left in the targeted mutation does not disturb the expression of the neighboring genes.
In the second strain (∆C1‐GFP), we replaced a 230 bp sequence of exon 1 with a tauGFP‐pA cassette. Here too, there is no C1 immunoreactivity in homozygous mice (∆C1‐GFP), consistent with the null design of the targeted mutation (Fig. 1E). The tauGFP marker fused to the initiation codon should enable us to detect VSNs in which the mutant C1 locus is transcribed, but no functional C1 receptor is expressed. However, we observed only sparse GFP+ VSNs in ∆C1‐GFP mice. The intrinsic GFP fluorescence in VSNs of ∆C1‐GFP mice was weak and needed to be enhanced by IHC with anti‐GFP antibody (Fig. 1F). The number of GFP+ VSNs in ∆C1‐GFP mice and in mice heterozygous for this mutation (data not shown) is much less than the number of VSNs detected by anti‐C1 antibody in WT mice. The mutant allele may yield unstable transcripts; alternatively, transcription of the mutant allele may extinguish with time.
We established the ∆C1 and ∆C1‐GFP strains in a mixed (129P2/OlaHsd × C57BL/6J) background. We maintained the strains by mating heterozygous mice, such that WT and homozygous mice were obtained from the same litters. ∆C1 mice and ∆C1‐GFP mice are viable and fertile and show no obvious differences in appearance compared to their WT littermates. These strains are publicly available from The Jackson Laboratory.
Differential and graded decrease in the numbers of VSNs that express C1 type of V2R genes
We examined the effects of the ∆C1 mutation on the numbers of various subpopulations of V2R+ VSNs in the VNO.
First, we crossed the ∆C1 strain with the gene‐targeted strains V2r1b‐IRES‐tauGFP (abbreviated as V2r1b‐GFP; Del Punta et al., 2002) or V2rf2‐IRES‐tauGFP (abbreviated as V2rf2‐GFP; Ishii & Mombaerts, 2011). In these strains, VSNs that express the V2r1b/Vmn2r26 gene (family B, C1 type) or the V2rf2/Vmn2r81 gene (family A9, C1 type), respectively, coexpress tauGFP by virtue of IRES‐mediated bicistronic translation. We have previously shown that > 90% of V2r1b‐GFP+ VSNs and V2rf2‐GFP+ VSNs are colabeled with anti‐C1 antibody, and rarely with anti‐C2 antibody (Ishii & Mombaerts, 2011). We counted GFP+ VSNs on VNO sections of ∆C1 mice and their WT littermates in crosses of ∆C1 with V2r1b‐GFP or V2rf2‐GFP. We observed a marked decrease in the number of V2r1b‐GFP+ VSNs in ∆C1 mice (Fig. 2A). At 4 week, the number of V2r1b‐GFP+ VSNs was decreased to 21.5% of WT (mean ± SEM in ∆C1, 185 ± 16 compared to WT, 860 ± 76; n = 4 per genotype; Mann–Whitney test, P = 0.0286) (Fig. 2E). At 10 week, this number was decreased further to 9.5% of WT (∆C1, 85 ± 16 compared to WT, 898 ± 113; n = 4 per genotype; P = 0.0286) (Fig. 2E). There was no significant difference at 0 day (∆C1, 490 ± 35 compared to WT, 391 ± 24; n = 4 per genotype; P = 0.0571); we counted V2r1b‐GFP+ VSNs stained by anti‐GFP antibody, as the intrinsic GFP fluorescence was weak in WT and ∆C1 at 0 day (Fig. 2B,E). The subpopulation of V2rf2‐GFP+ VSNs was also affected in ∆C1 mice, but to a different extent and with a different time course than the subpopulation of V2r1b‐GFP+ VSNs: at 4 weeks, the number was not significantly different (∆C1, 2033 ± 184 compared to WT, 2093 ± 154; n = 4 per genotype; P = 0.8857), and at 10 weeks, it was decreased to 63.4% of WT (∆C1, 585 ± 50 compared to WT, 923 ± 43; n = 4 per genotype; P = 0.0294) (Fig. 2C,E). The anti‐V2Rp5 antibody labels VSNs that express the Vmn2r116 gene, which belongs to family A3 (Haga et al., 2010). Consistent with the preferential colabeling of family‐A3 V2Rs with anti‐C2 antibody (Silvotti et al., 2007, 2011), we found that the majority of V2Rp5+ VSNs was colabeled with anti‐C2 antibody (190 of 261, 72.8%) and rarely with anti‐C1 antibody (2 of 287, 0.7%). Accordingly, we found no significant difference in the number of V2Rp5+ VSNs between WT mice and ∆C1 mice at 10 weeks (∆C1, 2375 ± 429 compared to WT, 2450 ± 387; n = 4 per genotype; P = 0.8857) (Fig. 2D,E). Figure 2E shows a summary of VSN counts, and the differential and graded effects of the ∆C1 mutation on subpopulations of V2R+ VSNs. The numbers of V2r1b‐GFP+ and V2rf2‐GFP+ VSNs, which were colabeled with anti‐C1 antibody but not with anti‐C2 antibody, were decreased in ∆C1 mice with age and to different extents, and the numbers of V2Rp5+ VSNs, which were colabeled with anti‐C2 antibody but not with anti‐C1 antibody, were not affected in ∆C1 mice.
Figure 2.
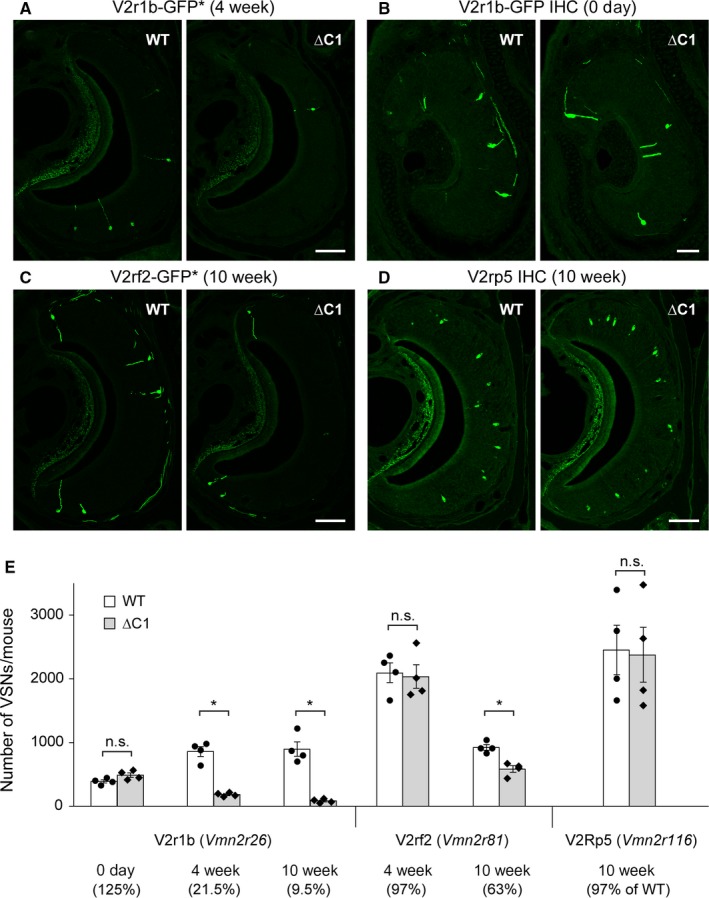
Decrease in the number of VSNs expressing V2r1b‐GFP or V2rf2‐GFP but not Vmn2r116 in ∆C1 mice. (A–C) The ∆C1 strain was crossed with the gene‐targeted V2r1b‐GFP and V2rf2‐GFP strains to generate double‐homozygous mice. Intrinsic GFP fluorescence (indicated with an asterisk) of coronal VNO sections is shown in A,C. IHC with anti‐GFP antibody is shown in B. Scale bar, 100 μm in A,C, and 50 μm in B. (D) IHC with anti‐V2Rp5 antibody on coronal VNO sections from WT and ∆C1 mice. Scale bar, 100 μm. (E) Summary of the numbers of labeled VSNs per mouse. Four mice per genotype were analyzed for each cross at the indicated ages. Mice were male, except for 0 day, when both males and females were used. Error bars represent mean ± SEM, with data points superimposed on bar charts. Mann–Whitney test was performed: n.s., not significant; *P value < 0.05.
Using the same design as for the V2r1b‐GFP and V2rf2‐GFP strains, we next generated two gene‐targeted strains called V2rf4‐Venus and V2rf1‐mCherry. We inserted IRES‐tauVenus and IRES‐taumCherry after the stop codon of the V2rf4/Vmn2r83 and V2rf1/Vmn2r82 genes, respectively (Fig. 3A). These two genes belong to family A9, a family of four C1 type of V2Rs: V2rf1/Vmn2r82, V2rf2/Vmn2r81, V2rf3/Vmn2r80, V2rf4/Vmn2r83. We found that, indeed, V2rf4‐Venus+ VSNs and V2rf1‐mCherry+ VSNs were colabeled preferentially with anti‐C1 antibody (93 of 123, 76% and 22 of 38, 58%, respectively), but not with anti‐C2 antibody (0 of 140 and 0 of 40, respectively). In ∆C1 mice at 10 weeks, the number of V2rf4‐Venus+ VSNs was decreased to 15.5% of WT (∆C1, 208 ± 28 compared to WT, 1345 ± 82; n = 4 per genotype; P = 0.0286) (Fig. 3B), and the number of V2rf1‐mCherry+ VSNs was decreased to 35.5% of WT (∆C1, 285 ± 32 compared to WT, 803 ± 67; n = 4 per genotype; P = 0.0286) (Fig. 3C). In ∆C1‐GFP mice at 10 weeks, the number of V2rf1‐mCherry+ VSNs was decreased to 35.8% of WT (∆C1‐GFP, 263 ± 17 compared to WT, 733 ± 117; n = 4 per genotype; P = 0.0286) (Fig. 3D). Thus, the two mutations in Vmn2r1 had the same effect on the subpopulation of V2rf1‐mCherry+ VSNs (Fig. 3E).
Figure 3.
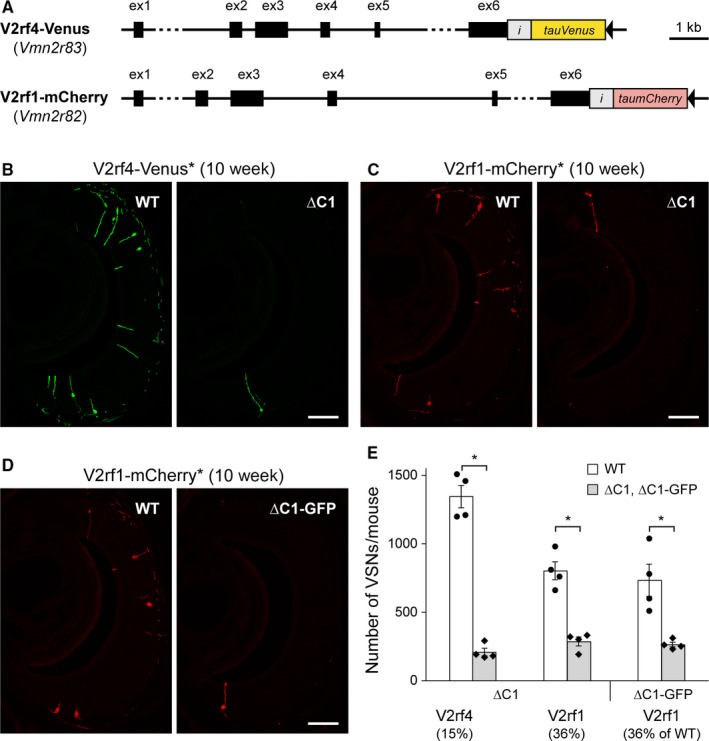
Decrease in the number of VSNs expressing V2rf4‐Venus or V2rf1‐mCherry in ∆C1 mice. (A) The V2rf4‐Venus and V2rf1‐mCherry gene‐targeted strains have an insertion of IRES‐tauVenus or IRES‐taumCherry after the stop codon of the Vmn2r83 or Vmn2r82 genes, respectively. The coding sequences remain intact. The black triangles indicate a loxP site. i, IRES. (B,C) The ∆C1 strain was crossed with the V2rf4‐Venus and V2rf1‐mCherry strains to generate double‐homozygous mice. Intrinsic fluorescence (indicated with an asterisk) of coronal VNO sections is shown in green (B) or red (C). Scale bar, 100 μm. (D) The ∆C1‐GFP strain was crossed with the V2rf1‐mCherry strain to generate double‐homozygous mice. Intrinsic fluorescence of coronal VNO sections is shown in red. Scale bar, 100 μm. (E) Summary of the numbers of labeled VSNs per mouse. Four 10‐week‐old male mice per genotype were analyzed for each cross. Error bars represent mean ± SEM, with data points superimposed on bar charts. Mann–Whitney test was performed: *P value < 0.05.
In summary, the Vmn2r1 mutations affect differentially the four populations of C1 type of V2R+ VSNs that we examined with gene‐targeted strains of the V2R‐IRES‐marker design. At 10 weeks, V2r1b‐GFP+ VSNs and V2rf4‐Venus+ VSNs were strongly affected (decreased to 9.5 and 15.5% of WT, respectively), and V2rf2‐GFP+ VSNs and V2rf1‐mCherry+ VSNs were less affected (decreased to 63.4 and 35.5% of WT, respectively).
No compensatory expression of other family‐C V2R genes
Conceivably, other members of family‐C V2Rs may become expressed in the C1 type of V2R+ VSNs that remain in ∆C1 and ∆C1‐GFP mice. Such expression might occur to compensate for the loss of C1 receptor function and as a result of the absence of a hypothetical negative feedback, analogous to what we observed in V2rf2 knockout mice (Ishii & Mombaerts, 2011).
To test this possibility, we examined coexpression of other family‐C V2Rs in C1 type of V2R+ VSNs by immunohistochemistry with anti‐C2 antibody. Representative images of VNO sections from 10‐week‐old mice with intrinsic signal of the fluorescent marker and C2 immunoreactivity are shown for the crosses ∆C1 × V2rf2‐GFP (Fig. 4A) and ∆C1 × V2rf1‐mCherry (Fig. 4B). Only a few V2rf2‐GFP+ VSNs (4 of 369, 1%) and V2rf1‐mCherry+ VSNs (5 of 321, 2%) were colabeled with anti‐C2 antibody in WT mice, consistent with the preferential colabeling with anti‐C1 antibody. In ∆C1 mice, the colabeling percentages with anti‐C2 antibody in V2rf2‐GFP+ VSNs (2 of 234, 1%) and V2rf1‐mCherry+ VSNs (3 of 114, 3%) were equally low. Likewise, in the cross ∆C1‐GFP × V2rf1‐mCherry (Fig. 4C), V2rf1‐mCherry+ VSNs were rarely colabeled with the anti‐C2 antibody in WT mice (2 of 293, 1%) and in ∆C1‐GFP mice (4 of 105, 4%). Figure 4D shows the summary of colabeling analysis in 10‐week‐old mice (data from 4 mice per genotype for each cross) including the results for V2r1b‐GFP+ and V2rf4‐Venus+ VSNs. We found no compensatory expression of Vmn2r2 and Vmn2r5 (as detected with anti‐C2 antibody) in these VSNs in the absence of C1 expression.
Figure 4.
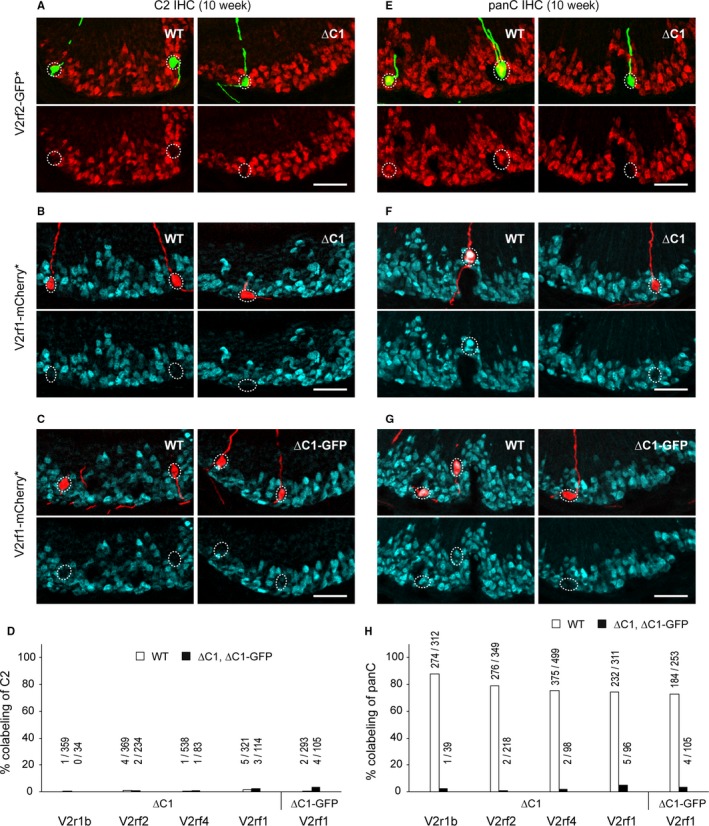
No compensatory expression of Vmn2r2 through Vmn2r7 in the absence of Vmn2r1. (A,B) IHC with anti‐C2 antibody on coronal VNO sections of WT and ∆C1 mice from the crosses ∆C1 × V2rf2‐GFP (A) and ∆C1 × V2rf1‐mCherry (B). In (A) intrinsic GFP fluorescence (indicated with an asterisk) is shown in green, and C2 immunoreactivity in red. In (B) intrinsic mCherry fluorescence is shown in red, and C2 immunoreactivity in blue. Mice were male and 10 weeks old. Scale bar, 50 μm. (C) IHC with anti‐C2 antibody (blue) on coronal VNO sections of WT and ∆C1‐GFP mice from the cross ∆C1‐GFP × V2rf1‐mCherry at 10 weeks. Intrinsic mCherry fluorescence is shown in red. Scale bar, 50 μm. (D) Summary of the percentage of colabeling with anti‐C2 antibody in VSNs that express a fluorescence marker from a gene‐targeted locus. Four mice per genotype were analyzed for each cross. Samples were from the 10‐week‐old male mice that were used for VSN counting in Figs 2 and 3. Above each bar, the number of double‐labeled VSNs/number of fluorescence marker‐positive VSNs is given. (E,F) IHC with anti‐panC antibody on coronal VNO sections of WT and ∆C1 mice from the crosses ∆C1 × V2rf2‐GFP (E) and ∆C1 × V2rf1‐mCherry (F) at 10 weeks. In (E) intrinsic GFP fluorescence is shown in green, and panC immunoreactivity in red. In (F) intrinsic mCherry fluorescence is shown in red, and panC immunoreactivity in blue. Scale bar, 50 μm. (G) IHC with anti‐panC antibody (blue) on coronal VNO sections of WT and ∆C1‐GFP mice from the cross ∆C1‐GFP × V2rf1‐mCherry at 10 weeks. Intrinsic mCherry fluorescence is shown in red. Scale bar, 50 μm. (H) Summary of the percentage of colabeling with panC antibody in fluorescence marker‐expressing VSNs. Four 10‐week‐old male mice per genotype were analyzed for each cross.
The anti‐C2 antibody does not detect Vmn2r3, Vmn2r6, and Vmn2r7 (Silvotti et al., 2011). We further examined the possibility of compensatory expression of other members of family‐C V2Rs using an anti‐panC antibody (Fig. 4E–H), which recognizes all seven family‐C V2Rs (Martini et al., 2001; Silvotti et al., 2007). In WT mice, large fractions of V2r1b‐GFP+, V2rf2‐GFP+, V2rf4‐Venus+, and V2rf1‐mCherry+ VSNs were colabeled with panC antibody at 10 weeks, reflecting the preferential coexpression of Vmn2r1 in these VSNs, respectively, 87.8, 79.1, 75.2, and 74.6 or 72.7%. In sharp contrast, these subpopulations of VSNs were rarely colabeled with panC antibody in ∆C1 and ∆C1‐GFP mice, respectively, 3, 1, 2, 5, or 4% (data from 4 mice per genotype for each cross).
Taken together, we found no evidence for compensatory expression of other family‐C V2Rs in the absence of Vmn2r1. The subpopulations of C1 type of V2R+ VSNs that we examined do not appear to express any functional family‐C receptor.
Axonal projections of subpopulations of VSNs in ∆C1 mice
V2R+ VSNs project their axons to the pAOB, where they coalesce into multiple glomeruli. V2r1b‐GFP+ glomeruli are located in the anterior part of the pAOB (Del Punta et al., 2002; Ishii & Mombaerts, 2008), and V2rf2‐GFP+ glomeruli in the middle part of the pAOB (Ishii & Mombaerts, 2011; Leinders‐Zufall et al., 2014). Deletion of the V2rf2 gene results in a diffuse distribution of axons across the pAOB (Ishii & Mombaerts, 2011).
As a first step in elucidating possible biological functions of Vmn2r1, we asked if it is required for axonal wiring of VSNs that express C1 type of V2Rs. We examined the intrinsic GFP fluorescence in AOB sections of 4‐week‐old mice from the crosses ∆C1 × V2r1b‐GFP and ∆C1 × V2rf2‐GFP. We found that ∆C1 mice had only a few and small V2r1b‐GFP+ glomeruli (Fig. 5A), consistent with a decrease in the number of V2r1b‐GFP+ VSNs at 4 weeks to 21.5% of WT (Fig. 2E). The small glomeruli in ∆C1 mice resided in the anterior part of pAOB, where V2r1b‐GFP+ axons normally coalesce. We then visualized the glomerular layer of AOB, where VSN axons terminate and form synaptic connections, by IHC for a presynaptic marker using an anti‐VGLUT2 antibody. We found that the few and small V2r1b‐GFP+ glomeruli in ∆C1 mice were located in the VGLUT2+ glomerular layer, as was the case in WT mice (Fig. 5B). But for V2rf2‐GFP+ VSNs, which were not affected in ∆C1 mice at 4 weeks (97% of WT, Fig. 2E), we observed multiple glomeruli with strong GFP intensity in the middle part of the pAOB in WT and ∆C1 mice, and without obvious difference in the pattern (Fig. 5C).
Figure 5.
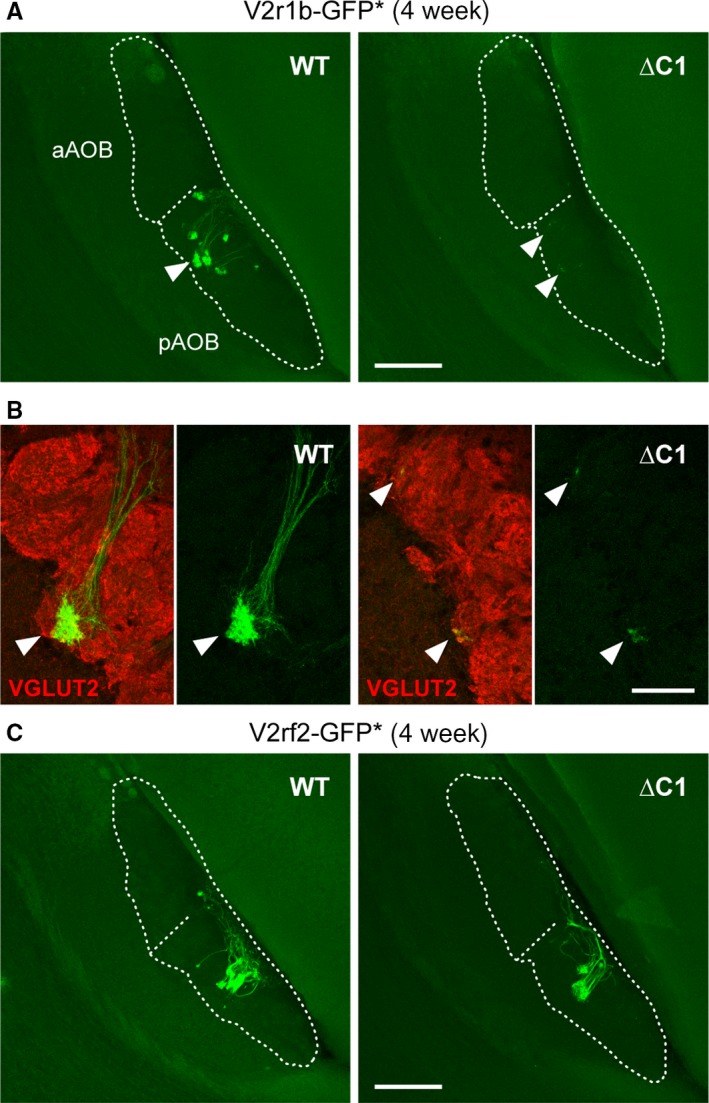
Axonal projections of subpopulations of VSNs to the AOB. (A) Axonal projections of V2r1b‐GFP+ VSNs to the AOB of WT and ∆C1 mice from the cross ∆C1 × V2r1b‐GFP. Samples were from 4‐week‐old male and female mice. Multiple images of intrinsic GFP fluorescence from serial sagittal sections were z‐stacked and projected into a 2D image. Dashed lines indicate the contours of the nerve layer and the glomerular layer of the AOB, and the border between the anterior AOB (aAOB) and the posterior AOB (pAOB). Arrowheads indicate GFP+ glomeruli (WT) and GFP+ axons (∆C1) that are shown in B. Scale bar, 200 μm. (B) IHC with anti‐VGLUT2 antibody (red) on sagittal AOB sections of WT and ∆C1 mice from the cross ∆C1 × V2r1b‐GFP, combined with intrinsic GFP fluorescence. Arrowheads V2r1b‐GFP+ glomeruli in WT, and GFP+ axons terminating in the VGLUT2+ glomerular layer in ∆C1. Scale bar, 50 μm. (C) Axonal projections of V2rf2‐GFP+ VSNs to the AOB in WT and ∆C1 mice from the cross ∆C1 × V2rf2‐GFP. Samples were from 4‐week‐old female mice. Axons with intrinsic GFP fluorescence form multiple glomeruli of various sizes in the pAOB both in WT and ∆C1 mice. Scale bar, 200 μm.
Thus, Vmn2r1 appear not to be required for the guidance of axons of V2rf2‐GFP+ VSNs from the VNO to the appropriate region of the AOB, and for the formation of V2rf2 glomeruli. This finding excludes a major biological function of Vmn2r1 in axonal wiring and coalescence.
Gene expression profiling by NanoString multiplex analysis
The NanoString platform enables multiplex analysis of RNA levels of large gene repertoires such as ORs and VRs with high specificity and sensitivity (Khan et al., 2011, 2013; Leinders‐Zufall et al., 2014). We designed a custom CodeSet Pao consisting of probes to analyze V2R gene expression in ∆C1 and ∆C1‐GFP mice. Specific probes against the coding sequences were designed for 45 V2R genes, which is one‐third of the repertoire of 120 V2R genes. To obtain higher coverage, we designed additional probes against 3′ UTR sequences for 24 V2R genes, based on the sequence data from a transcriptome study by RNA‐seq in the mouse VNO (Ibarra‐Soria et al., 2014). We were unable to design specific NanoString probes for family‐A4 V2Rs due to the high sequence homology among the family members. Of 69 V2R probes in CodeSet Pao, 10 probes gave very low counts in 10‐week‐old WT mice. Genes with a median count < 100 in WT samples were considered as not expressed in whole VNO mucosa at the age of analysis, and deleted from data. Of the probes designed against 3′UTR sequences, two gave extraordinary high counts; they were considered as non‐specific, and these data were also deleted from the analysis. CodeSet Pao thus enabled us to examine expression of 57 V2R genes, covering 48% of the V2R gene repertoire and all V2R families except for family A4.
We examined total RNA samples from whole VNO mucosae of individual 10‐week‐old mice from the ∆C1 and ∆C1‐GFP strains. Six WT mice and six homozygous (MUT) mice were analyzed for each strain. Differential expression between WT and MUT mice was evaluated by the false discovery rates (FDR) based on the P‐value of the Student's t‐test for each gene. If FDR is < 0.05, a gene was considered differentially expressed (DE). Counts for Vmn2r1 were at background level in MUT mice of the ∆C1 and ∆C1‐GFP strains, confirming that these gene‐targeted mutations are null mutations.
Figure 6A plots the log2 of the fold change (FC) of the mean count in MUT mice over the mean count in WT mice for the 57 V2R genes, arranged in order of gene families from C, A1–10, B, to D. Comparison of the results from ∆C1 and ∆C1‐GFP strains revealed that there were no significant differences between the two strains: Gene expression profiles of V2R genes were very similar between the strains, as shown by close log2 FC values and overlap of the 99% confidence intervals (CI) for each gene. Only a few genes showed a discrepancy: Vmn2r94 and Vmn2r104 are non‐DE genes in the data from the ∆C1 strain, whereas these genes were markedly decreased in the data from the ∆C1‐GFP strain. An explanation is that for both Vmn2r94 and Vmn2r104, counts in the corresponding WT samples were substantially lower in the ∆C1 strain than in the ∆C1‐GFP strain, resulting in differences of log2 FC values between the strains. The differences in counts in WT samples for Vmn2r94 and Vmn2r104 may reflect the mixed genetic background of the ∆C1 and ∆C1‐GFP strains.
Figure 6.
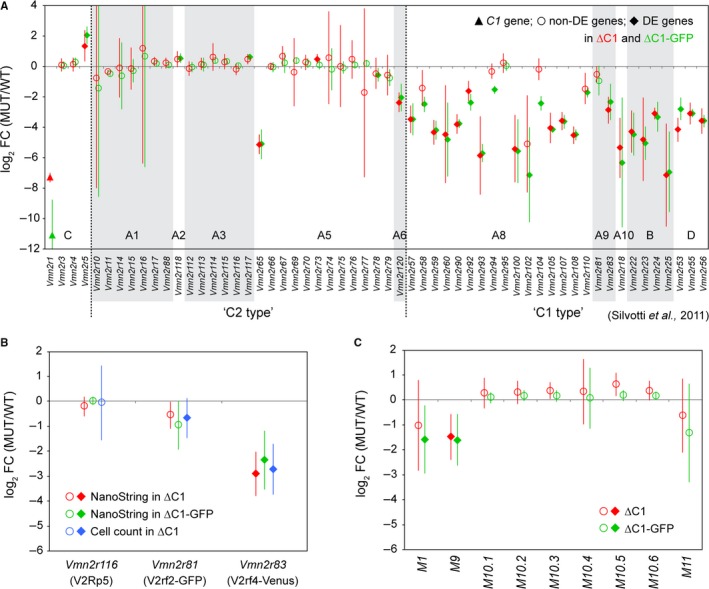
NanoString multiplex gene expression analysis of total RNA from whole VNO mucosae. (A) Gene expression profiling of V2R genes by NanoString analysis with the custom CodeSet Pao for strains ∆C1 (red) and ∆C1‐GFP (green). Six WT and six homozygous (MUT) mice were analyzed for each strain. Mice were 10‐week‐old males. The log2 values of the fold change (FC) of MUT over WT are plotted. Filled triangles indicate the Vmn2r1/C1 gene: Its NanoString count in MUT mice is near background level. Filled diamonds indicate differentially expressed (DE) genes, with a false discovery rate (FDR) < 0.05. Open circles indicate non‐DE genes. Columns are alternatively gray‐shaded and not shaded per gene family. Expression profiles are very similar between two strains, as shown by similar log2 FC values and overlap of error bars (±99% CI). (B) NanoString results in strains ∆C1 (red) and ∆C1‐GFP (green) are consistent with differences in cell counts in the ∆C1 strain (blue). Filled diamonds indicate DE genes in NanoString analysis or a significant decrease in cell count. Open circles indicate non‐DE genes. Error bars represent log2 FC ±99% CI. (C) Gene expression profiling of H2‐Mv genes by NanoString analysis with CodeSet Pao in strains ∆C1 (red) and ∆C1‐GFP (green). Filled diamonds indicate DE genes. Open circles indicate non‐DE genes. Error bars represent log2 FC ±99% CI. Expression profiles are very similar between the two strains.
Family‐ABD V2Rs are classified into two groups according to the colabeling patterns with antibodies against family‐C V2Rs (Silvotti et al., 2011). V2Rs of family‐A1 to A6 are preferentially colabeled with anti‐C2 antibody, and appear to coexpress Vmn2r2 through Vmn2r7 (C2 type of V2Rs). V2Rs of family‐A8, A9, A10, B, and D are preferentially colabeled with anti‐C1 antibody and coexpress Vmn2r1 (C1 type of V2Rs). Of the 26 C1 type of V2Rs analyzed with CodeSet Pao (the right side of the diagram in Fig. 6A), 24 showed a significant decrease in MUT mice of the ∆C1 and ∆C1‐GFP strains; of these 24, 19 were DE in both strains, and the remaining 5 were DE in one but not the other strain (Fig. 6A). In sharp contrast, there was no difference in expression of 25 of the 27 C2 type of V2R genes analyzed with CodeSet Pao (the left side of the diagram in Fig. 6A). This analysis is consistent with the differential effects of the Vmn2r1 mutations on the various subpopulations of VSNs that express C1 type of V2Rs. We identified two exceptions: Vmn2r65 of family‐A5 and Vmn2r120 of family‐A6 were significantly decreased in MUT mice. Vmn2r5 of family‐C is the only gene that was increased in MUT mice in both strains.
The NanoString results are highly consistent with the cell counts on VNO sections (Fig. 6B, data from 10‐week‐old mice were used for the comparison). Vmn2r81/V2rf2 gave lower NanoString counts in ∆C1 and ∆C1‐GFP mice, but the FC values were not significantly different; interestingly, the log2 FC values in NanoString were close to the log2 values of the ratio of the number of V2rf2‐GFP+ VSNs in MUT over WT mice. Vmn2r83/V2rf4 showed a significant decrease in NanoString analysis, and to the same extent as the decrease of the number of V2rf4‐Venus+ VSNs in cell counts. There was no difference in Vmn2r116 expression or in the number of V2Rp5+ VSNs.
CodeSet Pao includes probes for the nine non‐classical major histocompatibility H2‐Mv genes that are expressed in V2R+ VSNs (Ishii et al., 2003; Loconto et al., 2003; Ishii & Mombaerts, 2008; Leinders‐Zufall et al., 2014). We found that H2‐M1 and H2‐M9 counts were decreased significantly in ∆C1 and ∆C1‐GFP mice; H2‐M9 was DE in both strains, H2‐M1 was DE in one strain (Fig. 6C). The other H2‐Mv genes were not affected. The decrease in H2‐Mv gene expression level is consistent with the preferential expression of H2‐M10 genes in VSNs colabeled with anti‐C2 antibody (Silvotti et al., 2007), the coexpression of H2‐M9 with V2rf1‐3 genes (Ishii et al., 2003; Ishii & Mombaerts, 2008), and the decreased number of V2rf2‐GFP+ VSNs and V2rf1‐mCherry+ VSNs.
Reclassification of Vmn2r65 and Vmn2r120 as C1 type of V2R genes
Studies with family‐specific antibodies have classified families A5 and A6 as C2 type of V2Rs (Silvotti et al., 2007, 2011). Our NanoString results, however, indicate that counts for Vmn2r65 (family A5) and Vmn2r120 (family A6) were decreased significantly in ∆C1 and ∆C1‐GFP mice. To follow up on these unexpected NanoString results, we performed ISH analyses using gene‐specific probes for Vmn2r65 and Vmn2r120, and counted the numbers of labeled VSNs on VNO sections from 10‐week‐old mice of the ∆C1‐GFP strain (Fig. 7A,B). In ∆C1‐GFP mice, the number of Vmn2r65+ VSNs was decreased to 5.7% of WT (∆C1‐GFP, 87 ± 8 compared to WT, 1515 ± 108; n = 4 per genotype; Mann–Whitney test, P = 0.0294), and the number of Vmn2r120+ VSNs was decreased to 37.0% of WT (∆C1‐GFP, 615 ± 82 compared to WT, 1662 ± 153; n = 4 per genotype; P = 0.0284). As a control experiment, we tested Vmn2r118 (family A2) and Vmn2r76 (family A5). These are non‐DE genes in NanoString analysis, and they also showed no changes in ISH cell counts in ∆C1‐GFP mice: Vmn2r118 in ∆C1‐GFP, 816 ± 47 compared to WT, 744 ± 27 (n = 4 per genotype; P = 0.2454), and Vmn2r76 in ∆C1‐GFP, 1545 ± 129 compared to WT, 1650 ± 108 (n = 4 per genotype; P = 0.6857). A comparison of NanoString and ISH results validates the concordance in the differences for these four V2R genes (Fig. 7C).
Figure 7.
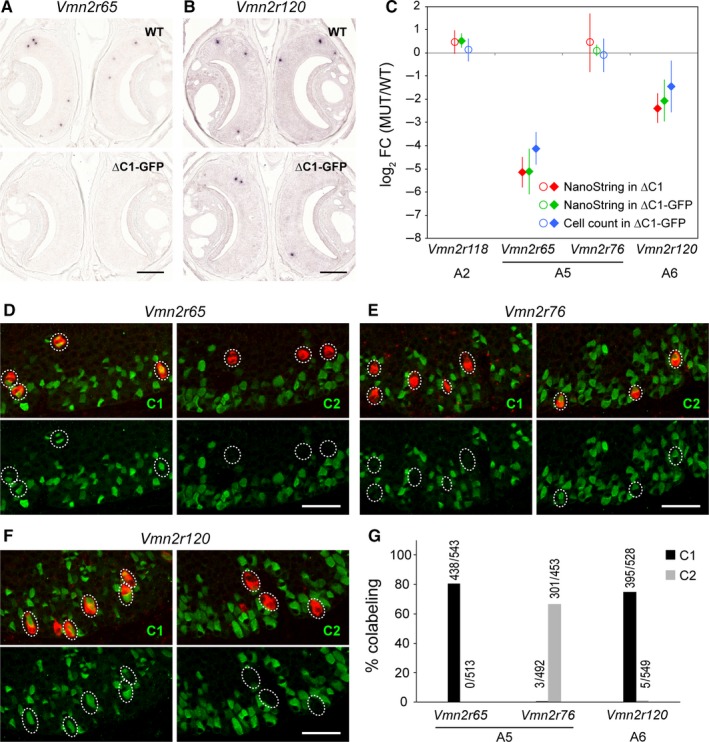
Reclassification of Vmn2r65 and Vmn2r120 as C1 type of V2R genes. (A,B) ISH with gene‐specific probes for Vmn2r65 (A) and Vmn2r120 (B) on coronal VNO sections of WT and ∆C1‐GFP mice. Labeled cells were visualized chromogenically. Four 10‐week‐old male mice per genotype were used. Scale bar, 200 μm. (C) Comparison of NanoString counts with ISH cell counts. Filled diamonds indicate DE genes in NanoString analysis or a significant decrease in cell count by ISH. Open circles indicate non‐DE genes. Vmn2r65 and Vmn2r120 were decreased in MUT mice consistently in NanoString and ISH analyses, and to the same extent for each gene: The log2 FC values are close, and there is overlap of the error bars (±99% CI). By contrast, there was no difference in ISH counts using gene‐specific probes for Vmn2r118 and Vmn2r76 genes, consistent with NanoString results. (D–F) Colabeling of Vmn2r65 (D), Vmn2r76 (E), and Vmn2r120 (F) by fluorescence ISH (red) in combination with IHC using anti‐C1 or anti‐C2 antibodies (green) on coronal VNO sections of C57BL/6J mice at 10 weeks. Scale bar, 50 μm. (G) Summary of the combined ISH/IHC analysis. VNO sections from three 10‐week‐old C57BL/6J male mice were analyzed. Above each bar, the number of double‐labeled VSNs/number of VSNs labeled by ISH with a gene‐specific probe is shown. The majority of VSNs expressing Vmn2r65 or Vmn2r120 in ISH were colabeled with C1 antibody, thus reclassifying these genes as C1 type of V2Rs.
Finally, we examined the coexpression patterns by ISH using gene‐specific probes in combination with IHC using anti‐C1 and anti‐C2 antibodies, in three 10‐week‐old WT C57BL/6J mice (Fig. 7D–G). We found that the majority of Vmn2r65+ VSNs (family A5) were colabeled with anti‐C1 antibody (438 of 543, 81%), and none were colabeled with the anti‐C2 antibody (0 of 513, 0%). In contrast, a large fraction of Vmn2r76+ VSNs (also family A5) was colabeled with anti‐C2 antibody (301 of 453, 66%), and a few were colabeled with anti‐C1 antibody (3 of 492, 1%). The NanoString CodeSet Pao contains probes for 12 of the 15 family‐A5 V2R genes, and of these 12, only Vmn2r65 was affected by the Vmn2r1 mutation. This phenotype is readily explained by the coexpression patterns: family‐A5 V2Rs are colabeled with anti‐C2 antibody, but Vmn2r65 is an exception. As Vmn2r120 is the sole gene in family‐A6, the ISH probe is likely to be specific to Vmn2r120. We found that the majority of Vmn2r120+ VSNs were colabeled with anti‐C1 antibody (395 of 528, 74.8%), and few Vmn2r120+ VSNs were colabeled with anti‐C2 antibody (5 of 549, 1%).
Taken together, the histological analyses validate the apparent exceptions in the NanoString results, and lead us to reclassify Vmn2r65 and Vmn2r120 firmly as C1 type of V2Rs, in discordance with their phylogenetic classification as C2 type of V2Rs.
Discussion
Sequential and dependent model of coordinated coexpression of V2R genes
We have here described mouse strains with a knockout mutation in the Vmn2r1 gene, twenty years after mouse V2R genes were reported (Matsunami & Buck, 1997; Ryba & Tirindelli, 1997). The anti‐C1 and anti‐C2 antibodies (Silvotti et al., 2007, 2011) were critical reagents for our study, as gene‐specific ISH probes for family‐C genes have proved difficult to design.
A first insight into the regulatory mechanisms of the coordinated expression of a single family‐ABD gene with one or more family‐C genes came from our analysis of a mouse strain with a gene‐targeted deletion of the coding sequence of V2rf2 (Ishii & Mombaerts, 2011). We found that ~25% of VSNs that express the mutant V2rf2 allele coexpress another family‐ABD gene, consistent with the absence of a hypothetical negative feedback from the mutant V2rf2 allele. Interestingly, 9.5% of VSNs expressing the mutant V2rf2 allele were colabeled with anti‐C2 antibody, vs. 0% of V2rf2‐GFP+ VSNs. We have proposed a sequential and dependent model for the coordinated expression of two V2R genes: a family‐ABD gene is expressed first during differentiation of a VSN, and then the appropriate family‐C gene(s) is/are expressed next, in a dependent manner (Ishii & Mombaerts, 2011). The lack of compensatory expression of family‐C genes in VSNs expressing C1 type of V2Rs in the absence of Vmn2r1 is consistent with our sequential and dependent model.
Differential and graded dependence of V2R+ VSN subpopulations on Vmn2r1
We analyzed the impact of a Vmn2r1 knockout mutation on various subpopulations of VSNs that express C1 type of V2Rs in three complementary ways: by cell counts in crosses of ∆C1 and ∆C1‐GFP with gene‐targeted strains carrying V2R mutations of the IRES‐marker design, by cell counts using ISH with V2R gene‐specific probes, and by RNA counts with the NanoString multiplex gene expression platform. The four available gene‐targeted strains carrying V2R mutations of the IRES‐marker design provide the highest degree of analytical specificity for cell counting: V2r1b‐IRES‐tauGFP (Del Punta et al., 2002), V2rf2‐IRES‐tauGFP (Ishii & Mombaerts, 2011), V2rf4‐IRES‐tauVenus and V2rf1‐IRES‐taumCherry (this paper). The number of marker‐positive VSNs in the ∆C1 or ∆C1‐GFP background was reduced to 9.5, 63.4, 15.5, 35.5, or 35.8% of WT, respectively. By ISH with V2R gene‐specific probes, we found that the number of VSNs was reduced to 5.7% for Vmn2r65 and 37.0% for Vmn2r120. These decreases in cell counts correlate remarkably well with the FC values in the NanoString analysis, which reflect expression levels measured in total, non‐reverse transcribed RNA from whole VNO mucosa. Assuming that these correlations can be generalized to other family‐ABD genes, the FC values in the NanoString analysis can thus be converted into reductions in cell numbers. The number of presumptive VSNs expressing a given C1 type of V2R would then be reduced to an average of 21.83% of WT in ∆C1 and 15.13% in ∆C1‐GFP, and to a median of 7.68% of WT in ∆C1 and 8.04% in ∆C1‐GFP. Interestingly, V2r1b+ VSNs and V2rf4+ VSNs are H2‐Mv− and were affected much more than V2rf2+ VSNs and V2rf1+ VSNs, which are H2‐Mv+ (Ishii & Mombaerts, 2008). Further work is needed to determine whether subpopulations of H2‐Mv− VSNs are reduced to a greater extent than H2‐Mv+ VSNs in the absence of Vmn2r1, and what the effects would be of eliminating H2‐Mv expression completely by generating double mutants with our ∆H2Mv strain (Leinders‐Zufall et al., 2014).
Subpopulations of VSNs that express C2 type of V2R genes are not dependent on Vmn2r1, suggesting cell‐autonomous effects of the Vmn2r1 knockout mutations. By IHC analysis, we found no significant difference in the number of V2Rp5+ VSNs. By NanoString analysis, we found no significant difference in expression of 25 of the 27 C2 type of V2Rs; the two C2 type of V2Rs that were DE (Vm2nr65 and Vmn2r120) could be reclassified as C1 type of V2R genes. By ISH analysis, there was no significant difference in cell counts for C2 type of V2R genes Vmn2r118 and Vmn2r76.
Varying percentages of colabeling with anti‐C1 antibody and varying reductions in the ∆C1 or ∆C1‐GFP background
Not all VSNs expressing a given C1 type of V2R are colabeled with anti‐C1 antibody; the colabeling is preferential, not absolute. The percentage of colabeling at 10 weeks was 58% for V2rf1 (in V2rf1‐mCherry mice), 74.8% for Vmn2r120 (in C57BL6/J mice), 76% for V2rf4 (in V2rf4‐Venus mice), 80.7% for Vmn2r65 (ISH in C57BL6/J mice), 91.1% for V2rf2 (in V2rf2‐GFP mice, Ishii & Mombaerts, 2011), and 93% for V2r1b (in V2r1b‐GFP mice, Ishii & Mombaerts, 2011). There could be differences in sensitivity of the analytical techniques and fluorescent markers used, but the general conclusion is that most—but not all—VSNs of a subpopulation expressing a given C1 type of V2R coexpress Vmn2r1. At 3 weeks, this percentage of colabeling is lower (Ishii & Mombaerts, 2011): 68.2% for V2rf2 (in V2rf2‐GFP mice), and 81.3% for V2r1b (in V2r1b‐GFP mice). We speculate that with increasing age, a higher fraction of existing VSNs may start to coexpress Vmn2r1, perhaps as final maturation or induction by an activity‐dependent mechanism. An alternative, but not mutually exclusive hypothesis, is that with increasing age, Vmn2r1− VSNs get replaced by Vmn2r1+ VSNs during the continuous process of cell death and neurogenesis in the vomeronasal epithelium.
As not all VSNs of a subpopulation of expressing a C1 type of V2R are colabeled with anti‐C1 antibody in WT mice, and the percentage of colabeling varies among C1 type of V2Rs, it is not surprising that the reductions of C1 type of V2Rs in the ∆C1 or ∆C1‐GFP background were not complete, and varied. But, there is no correlation between the percentage of colabeling in WT mice and the percentage of reduction in the ∆C1 or ∆C1‐GFP background. Therefore, the simple explanation cannot be offered that there would be two subsets for each C1 type of V2R+ VSN subpopulation: a subset of VSNs that normally express Vmn2r1 and do not survive in the absence of Vmn2r1, and a subset of VSNs that never express Vmn2r1 and survive in the absence of Vmn2r1.
Possible functions of Vmn2r1
The observation of few and small glomeruli formed by axons of V2r1b+ VSNs in the absence of Vmn2r1 is consistent with the high reduction in cell number. A minimal number of olfactory sensory neurons expressing a given odorant receptor is required to maintain glomeruli in the olfactory bulb (Ebrahimi & Chess, 2000). Nonetheless, some axons of V2r1b+ VSNs reach the glomerular layer. Importantly, axons of V2rf2+ VSNs formed normal glomeruli in the appropriate region of the AOB, consistent with the lower reduction in cell number compared to V2r1b+ VSNs. These findings exclude a major biological function for Vmn2r1 in axon guidance from the VNO to the AOB, and in the coalescence of axons into glomeruli.
Results from a heterologous HEK293 expression system suggest that Vmn2r1 is a calcium‐dependent, low‐sensitivity receptor for isoleucine, leucine, and valine (DeMaria et al., 2013). If these responses can be extended in vivo or ex vivo to VSNs of WT mice and can be shown to be reduced or abolished in the ∆C1 or ∆C1‐GFP background, the reduced cell number could be interpreted in terms of reduced VSN survival due to absent or aberrant physiological responses. Vmn2r1 may respond to its own ligands, such as amino acids, independently of the family‐ABD receptor responding to, for instance, peptides (Leinders‐Zufall et al., 2009, 2014). Vmn2r1 may promote surface expression of a family‐ABD receptor in native VSNs, as suggested by studies in the heterologous HEK293 system (DeMaria et al., 2013).
Reclassification of Vmn2r65 and Vmn2r120 as C1 type of V2R genes
An unexpected observation in the NanoString analysis was that Vmn2r65 and Vmn2r120 were DE, although they were thought to be C2 type of V2Rs based on their phylogenetic classification in families A5 and A6, respectively. This analysis turned out to have predictive power. We found that the numbers of VSNs expressing Vmn2r65 and Vmn2r120 were reduced in ∆C1‐GFP mice, and to the same extent as the reduction in NanoString counts. We then showed that VSNs expressing Vmn2r65 and Vmn2r120 were colabeled preferentially with anti‐C1 antibody but rarely with anti‐C2 antibody in C57BL6/J mice. We have thus reclassified Vmn2r65 and Vmn2r120 firmly as C1 type of V2Rs. Application of RNA‐seq may result in the reclassification of more V2Rs.
Conclusion
Subpopulations of VSNs expressing C2 type of V2Rs are not upregulated in the absence of Vmn2r1; they do not appear to ‘fill the void’ created by the decrease in VSN subpopulations expressing C1 type of V2Rs. There thus appears to be a fixed probability of gene choice for a given C2 type of V2R, resulting in subpopulations of C2 type of V2R+ VSNs that are tightly regulated in terms of cell numbers.
Conflict of interest
The authors have no financial or other relationships to report that might lead to a conflict of interest.
Data accessibility
Original data are available upon request from the corresponding author. The NanoString probe sequences and data have been deposited in the Gene Expression Omnibus (NCBI) and are accessible by GEO accession number GSE104703.
Author contributions
S.A. and P.M. designed research, reviewed data, and wrote the manuscript. S.A., T.I., and Z.B. performed research.
Abbreviations
- AOB
accessory olfactory bulb
- CI
confidence intervals
- DE
differentially expressed
- DIG
digoxigenin
- ES
embryonic stem
- FC
fold change
- FDR
false discovery rate
- IHC
immunohistochemistry
- IRES
internal ribosome entry site
- ISH
in situ hybridization
- NGS
normal goat serum
- PBS
phosphate‐buffered saline
- VNO
vomeronasal organ
- VSNs
vomeronasal sensory neurons
Supporting information
Acknowledgements
The authors are grateful to Dr. Roberto Tirindelli and Dr. Kazushige Touhara for gifts of antibodies. P.M. thanks the Max Planck Society for generous financial support. The gene‐targeted strains V2rf1‐IRES‐taumCherry and V2rf4‐IRES‐tauVenus were generated by T.I. in the laboratory of P.M. at The Rockefeller University, New York, NY.
Edited by C. Giovanni Galizia. Reviewed by Trese Leinders‐Zufall, University of Saarland, Germany; Enrique Lanuza, University of Valencia, Spain; and Daisuke Kondoh, Obihiro University of Agriculture and Veterinary Medicine, Japan
All peer review communications can be found with the online version of the article.
References
- Belluscio, L. , Koentges, G. , Axel, R. & Dulac, C. (1999) A map of pheromone receptor activation in the mammalian brain. Cell, 97, 209–220. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. & Yekutieli, D. (2001) The control of the false discovery rate in multiple testing under dependency. Ann. Stat., 29, 1165–1188. [Google Scholar]
- Chamero, P. , Marton, T.F. , Logan, D.W. , Flanagan, K. , Cruz, J.R. , Saghatelian, A. , Cravatt, B.F. & Stowers, L. (2007) Identification of protein pheromones that promote aggressive behaviour. Nature, 450, 899–902. [DOI] [PubMed] [Google Scholar]
- Chamero, P. , Katsoulidou, V. , Hendrix, P. , Bufe, B. , Roberts, R. , Matsunami, H. , Abramowitz, J. , Birnbaumer, L. et al (2011) G protein Gαo is essential for vomeronasal function and aggressive behavior in mice. Proc. Natl. Acad. Sci. USA, 108, 12898–12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamero, P. , Leinders‐Zufall, T. & Zufall, F. (2012) From genes to social communication: molecular sensing by the vomeronasal organ. Trends Neurosci., 35, 597–606. [DOI] [PubMed] [Google Scholar]
- Del Punta, K. , Puche, A. , Adams, N.C. , Rodriguez, I. & Mombaerts, P. (2002) A divergent pattern of sensory axonal projections is rendered convergent by second‐order neurons in the accessory olfactory bulb. Neuron, 35, 1057–1066. [DOI] [PubMed] [Google Scholar]
- DeMaria, S. , Berke, A.P. , Van Name, E. , Heravian, A. , Ferreira, T. & Ngai, J. (2013) Role of a ubiquitously expressed receptor in the vertebrate olfactory system. J. Neurosci., 33, 15235–15247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac, C. & Axel, R. (1995) A novel family of genes encoding putative pheromone receptors in mammals. Cell, 83, 195–206. [DOI] [PubMed] [Google Scholar]
- Ebrahimi, F.A.W. & Chess, A. (2000) Olfactory neurons are interdependent in maintaining axonal projections. Curr. Biol., 10, 219–222. [DOI] [PubMed] [Google Scholar]
- Francia, S. , Silvotti, L. , Ghirardi, F. , Catzeflis, F. , Percudani, R. & Tirindelli, R. (2014) Evolution of spatially coexpressed families of type‐2 vomeronasal receptors in rodents. Genome Biol. Evol., 7, 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga, S. , Kimoto, H. & Touhara, K. (2007) Molecular characterization of vomeronasal sensory neurons responding to a male‐specific peptide in tear fluid: sexual communication in mice. Pure Appl. Chem., 79, 775–783. [Google Scholar]
- Haga, S. , Hattori, T. , Sato, T. , Sato, K. , Matsuda, S. , Kobayakawa, R. , Sakano, H. , Yoshihara, Y. et al (2010) The male mouse pheromone ESP1 enhances female sexual receptive behavior through a specific vomeronasal receptor. Nature, 466, 118–122. [DOI] [PubMed] [Google Scholar]
- Herrada, G. & Dulac, C. (1997) A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell, 90, 763–773. [DOI] [PubMed] [Google Scholar]
- Ibarra‐Soria, X. , Levitin, M.O. , Saraiva, L.R. & Logan, D.W. (2014) The olfactory transcriptomes of mice. PLoS Genet., 10, e1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T. & Mombaerts, P. (2008) Expression of nonclassical class I major histocompatibility genes defines a tripartite organization of the mouse vomeronasal system. J. Neurosci., 28, 2332–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, T. & Mombaerts, P. (2011) Coordinated coexpression of two vomeronasal receptor V2R genes per neuron in the mouse. Mol. Cell Neurosci., 46, 397–408. [DOI] [PubMed] [Google Scholar]
- Ishii, T. , Hirota, J. & Mombaerts, P. (2003) Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr. Biol., 13, 394–400. [DOI] [PubMed] [Google Scholar]
- Ishii, T. , Omura, M. & Mombaerts, P. (2004) Protocols for two‐ and three‐color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J. Neurocytol., 33, 657–669. [DOI] [PubMed] [Google Scholar]
- Khan, M. , Vaes, E. & Mombaerts, P. (2011) Regulation of the probability of mouse odorant receptor gene choice. Cell, 147, 907–921. [DOI] [PubMed] [Google Scholar]
- Khan, M. , Vaes, E. & Mombaerts, P. (2013) Temporal patterns of odorant receptor gene expression in adult and aged mice. Mol. Cell Neurosci., 57, 120–129. [DOI] [PubMed] [Google Scholar]
- Kimoto, H. , Haga, S. , Sato, K. & Touhara, K. (2005) Sex‐specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature, 437, 898–901. [DOI] [PubMed] [Google Scholar]
- Leinders‐Zufall, T. , Brennan, P. , Widmayer, P. , Chandramani, P.S. , Maul‐Pavicic, A. , Jager, M. , Li, X.H. , Breer, H. et al (2004) MHC class I peptides as chemosensory signals in the vomeronasal organ. Science, 306, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Leinders‐Zufall, T. , Ishii, T. , Mombaerts, P. , Zufall, F. & Boehm, T. (2009) Structural requirements for the activation of vomeronasal sensory neurons by MHC peptides. Nat. Neurosci., 12, 1551–1558. [DOI] [PubMed] [Google Scholar]
- Leinders‐Zufall, T. , Ishii, T. , Chamero, P. , Hendrix, P. , Oboti, L. , Schmid, A. , Kircher, S. , Pyrski, M. et al (2014) A family of nonclassical class I MHC genes contributes to ultrasensitive chemodetection by mouse vomeronasal sensory neurons. J. Neurosci., 34, 5121–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loconto, J. , Papes, F. , Chang, E. , Stowers, L. , Jones, E.P. , Takada, T. , Kumanovics, A. , Lindahl, K.F. et al (2003) Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell, 112, 607–618. [DOI] [PubMed] [Google Scholar]
- Martini, S. , Silvotti, L. , Shirazi, A. , Ryba, N.J. & Tirindelli, R. (2001) Co‐expression of putative pheromone receptors in the sensory neurons of the vomeronasal organ. J. Neurosci., 21, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami, H. & Buck, L.B. (1997) A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell, 90, 775–784. [DOI] [PubMed] [Google Scholar]
- Mombaerts, P. , Wang, F. , Dulac, C. , Chao, S.K. , Nemes, A. , Mendelsohn, M. , Edmondson, J. & Axel, R. (1996) Visualizing an olfactory sensory map. Cell, 87, 675–686. [DOI] [PubMed] [Google Scholar]
- Ramírez‐Solis, R. , Liu, P. & Bradley, A. (1995) Chromosome engineering in mice. Nature, 378, 720–724. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I. , Feinstein, P. & Mombaerts, P. (1999) Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell, 97, 199–208. [DOI] [PubMed] [Google Scholar]
- Ryba, N.J. & Tirindelli, R. (1997) A new multigene family of putative pheromone receptors. Neuron, 19, 371–379. [DOI] [PubMed] [Google Scholar]
- Shi, P. & Zhang, J. (2007) Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res., 17, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvotti, L. , Moiani, A. , Gatti, R. & Tirindelli, R. (2007) Combinatorial co‐expression of pheromone receptors, V2Rs. J. Neurochem., 103, 1753–1763. [DOI] [PubMed] [Google Scholar]
- Silvotti, L. , Cavalca, E. , Gatti, R. , Percudani, R. & Tirindelli, R. (2011) A recent class of chemosensory neurons developed in mouse and rat. PLoS One, 6, e24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm, T. , Leinders‐Zufall, T. , Maĉek, B. , Walzer, M. , Jung, S. , Pömmerl, B. , Stevanovíc, S. , Zufall, F. et al (2013) Mouse urinary peptides provide a molecular basis for genotype discrimination by nasal sensory neurons. Nat. Commun., 4, 1616. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Shi, P. , Zhang, Y.P. & Zhang, J. (2005) Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics, 86, 306–315. [DOI] [PubMed] [Google Scholar]
- Young, J.M. & Trask, B. (2007) V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet., 23, 212–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data are available upon request from the corresponding author. The NanoString probe sequences and data have been deposited in the Gene Expression Omnibus (NCBI) and are accessible by GEO accession number GSE104703.


