Abstract
A significant aspect of the control of cellular zinc in eukarya is its subcellular re‐distribution. One of the four human vesicular zinc transporters, ZnT8, supplies the millimolar zinc concentrations of insulin granules in pancreatic β‐cells, affecting insulin processing, crystallisation and secretion. ZnT8 has a transmembrane and a C‐terminal cytosolic domain; the latter has important functions and purportedly mediates protein–protein interactions, senses cytosolic zinc and/or channels zinc to the transport site in the transmembrane domain (TMD). A common variant W325R in the C‐terminal domain (CTD) increases the risk to develop type 2 diabetes and affects autoantibody specificity in type 1 diabetes. To investigate the differences between the two protein variants, we purified and biophysically characterised both variants of the ZnT8 CTD [R325 variant of ZnT8 CTD (aa267–369) (ZnT8cR) and W325 variant of ZnT8 CTD (aa267–369) (ZnT8cW)]. The domains fold independently of the TMD. Remarkably, the ZnT8cW variant (diabetes protection in the full‐length protein) is less thermostable than the ZnT8cR variant (diabetes risk in the full‐length protein). The ZnT8cW monomers associate with higher affinity. Both CTD variants bind zinc with a stoichiometry that differs from bacterial homologues, emphasising the limitation of the latter as models for the structure and function of the human proteins. The relatively small but reproducible differences between the two ZnT8 CTD variants begin to provide a molecular basis for the different diabetes susceptibility caused by the full‐length ZnT8 proteins.
Keywords: cation diffusion facilitator, C‐terminal domain, diabetes, zinc, ZnT8
Abbreviations
- Ch. Int.
charge interlock
- CTD
C‐terminal domain
- ICP‐MS
inductively coupled plasma mass spectrometry
- MST
microscale thermophoresis
- nDSF
nano differential scanning fluorimetry
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- TMD
transmembrane domain
- ZnT8cR
R325 variant of ZnT8 CTD (aa267‐369)
- ZnT8cW
W325 variant of ZnT8 CTD (aa267–369)
- ZnT
zinc transporter
Introduction
Perturbations of cellular zinc metabolism through mutations in dozens of proteins are implicated in a growing number of human diseases 1. Among these proteins are transporters that regulate the passage of Zn2+ across biological membranes. Belonging to two families, ZIPs (Zrt/Irt‐like Protein, SLC39A) and ZnTs (zinc transporter, SLC30A), they transport zinc into and out of the cytosol, respectively 2. Humans have 14 ZIPs and 10 ZnTs, which are expressed differentially in tissues and have different cellular localisations 2.
Cellular zinc compartmentalisation is controlled by these two families of zinc transporters and by metallothioneins 3. A subgroup of four transporters (ZnT2, 3, 4 and 8 in mammals) of the cation diffusion facilitator (CDF) superfamily exports zinc ions from the cytosol into intracellular vesicles. One of them, ZnT8 (SLC30A8), is highly expressed in the membrane of the dense insulin secretory granules of pancreatic β‐cells 4. It supplies zinc for the storage of insulin as a crystalline hexamer and for other aspects of the biochemistry occurring in the granules 5. ZnT8 also has a yet to be defined role in glucagon secretion in pancreatic α‐cells and is present in some other tissues 6.
A structural issue of ZnT8 with significant functional implications is an SNP resulting in two major variants with either Arg (R) or Trp (W) at position 325 in the C‐terminal domain (CTD). Both variants are common in human populations (R: 60%–95%; W: 5%–40%) 7. A third variant encoding Gln is far rarer in populations and will not be discussed here. Remarkably, the major R325 variant increases the risk of developing type 2 diabetes (T2D) 5. These observations have led to the postulate that higher zinc transport activity is related to β‐cell health and that activation of ZnT8 might be a way of improving β‐cell health 8. However, this interpretation has now come into question, as the R325 T2D‐risk allele apparently has the higher transport activity 9, providing a plausible explanation why humans heterozygous for one of several loss‐of‐function mutations in SLC30A8 are protected against diabetes 10. Based on 3D homology models, the R/W325 replacement in ZnT8 is not directly involved in the CTD zinc‐binding site(s). However, it has functional consequences and a relatively large impact on human health, as the differences in transport activity of the two variants confer a 15% increased risk of developing T2D for each allele of the risk R325 form 9, 11.
There is no 3D structure of a eukaryotic CDF protein; therefore, concepts of how these vesicular ZnTs work are based entirely on the 3D structures of prokaryotic CDF homologues. The crystal structure of the Escherichia coli protein YiiP revealed a dimer with a transmembrane domain (TMD) and a CTD 12. The dimer binds eight zinc ions: two in sites A, which are the primary TMD transport sites, two in sites B between the domains with an unknown function and four in two binuclear sites C at the dimer interface of the CTD. The CTDs are believed to be metal sensors, working by an allosteric mechanism that links the occupancy of sites C by zinc (K d = 24 μm, measured in proteoliposomes) to induction of a conformational change of the CTD, which is then transmitted to the TMD to trigger zinc/proton antiport 13. This allosteric regulation of the TMD by the CTD is thought to be important for full transporter functionality, as though CTD‐truncated versions of the CDF proteins ZitB from E. coli and CzcD from Cupriavidus metallidurans were capable of transporting Zn2+ in vivo, their transport activity was decreased 14. However, mutations at site C in the CTD of MntE from Streptococcus pneumoniae did not affect manganese transport 15. Additionally, a cryo‐electron microscopy structure of a YiiP homologue from Shewanella oneidensis does not seem to confirm the allosteric mechanism of zinc binding; rather this study suggests that the zinc‐binding sites in the CTD are of such high affinity that zinc is always bound; therefore, any conformational changes during transport occur solely in the TMD 16. These data suggest that different CDF CTDs have varying effects on transport function even among bacterial homologues.
Aside from the structure of the full‐length E. coli protein YiiP, there are three crystal structures of bacterial CTDs: Thermus thermophilus CzrB 17, Thermotoga maritima TM0876 18 and Magnetospirillum gryphiswaldense MamM 19. The models of the bacterial proteins fail to explain the function(s) of the CTD in the family of mammalian vesicular transporters for the following reason. While the bacterial proteins sense and export an excess of zinc, there is no evidence for an excess of zinc in the cytosol of eukaryotic cells for export into granules of the secretory pathway. Unless zinc is made available by some yet unknown mechanism, the cytosolic free zinc ion concentrations are only hundreds of pm to maximally 1.5 nm, a very small fraction of the ~ 250 μm total cellular zinc concentration 20, 21. In insulin granules, estimates of free zinc ion concentrations are 120 nm (pH 6) and total zinc concentrations are tens to perhaps even hundreds of millimolar 22, 23. Thus, for both total and free zinc, ZnT8 has to work against a concentration gradient of about three orders of magnitude.
The R325W replacement in ZnT8 generates a different epitope for autoantibodies in type 1 diabetes (T1D) 24 (an issue of protein conformation in the CTD) in addition to affecting insulin biology in T2D (an issue thought to relate to zinc transport) 9. The underlying question for the basic biological chemistry addressed here is how these two amino acids affect subunit interactions, dimer dynamics and zinc binding. Thus, biophysical investigations of the CTDs of ZnT8 would solve a key issue in β‐cell granule biology central to control of energy metabolism, provide important information regarding the biology of other zinc‐containing vesicles served by ZnT2–4, and make a significant contribution to CDF biology in general. Towards this goal, we expressed both ZnT8cW (ZnT8 CTD, aa267–369, expressing Trp at position 325) and ZnT8cR (ZnT8 CTD expressing Arg at position 325). The two proteins adopt their predicted fold independent of the presence of the TMD and have different zinc‐binding characteristics compared to their bacterial homologues. Structural and stability differences between the two CTD variants affect their dimerisation. Previous deductions made from the 3D structures of bacterial homologues are therefore insufficient to explain the properties of the human proteins in health and disease.
Results
Bioinformatics structure predictions
Based on a comparison of the primary sequences of several human and two bacterial CDF CTDs, for which 3D structures are known, the two ZnT8 CTD variants (ZnT8cR and ZnT8cW) are predicted to adopt an αββαβ fold (Fig. 1A) observed in at least four bacterial CTDs of homologous zinc transporters. This fold is characteristic of the ‘heavy metal‐associated domain’, also called the ferredoxin fold βαββαβ in different metalloproteins interacting with iron, copper or zinc 25. Indeed, we predict such a structure for all mammalian ZnT CTDs with the possible exception of ZnT9 (Fig. 1A). A 3D model of the ZnT8cR homodimer based on the structure of T. thermophilus CzrB was constructed (Fig. 1B). The model predicts that residue 325 is located in a loop which is in close proximity to the second protomer in the dimer in the zinc‐bound state.
Figure 1.
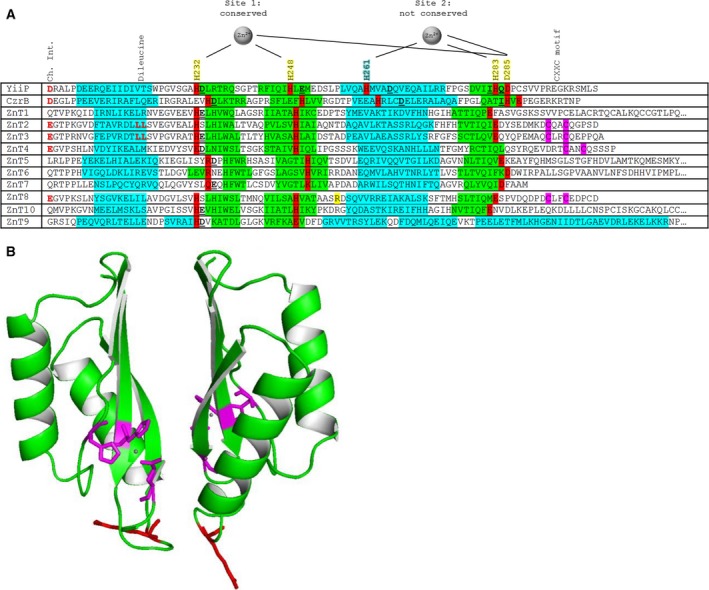
Metal‐binding and structural motifs in the CTDs of ZnTs/CDFs. (A) Primary sequence comparison between the CTDs of selected bacterial and human CDFs, indicating both conserved and non‐conserved motifs. Protein secondary structure was predicted using jpred 4 (Materials and methods); α‐helices in blue and β‐sheets in green. Metal‐binding residues are highlighted in red with black text; site 2 of the binuclear zinc site described in the 3D structure of Escherichia coli YiiP (shown on top) is not conserved in mammalian ZnTs. Metal‐binding residues annotated in the alignment are contributed from one protomer (yellow) or the other protomer (blue) in the dimer. Both metal‐binding sites in E. coli YiiP utilise a water molecule as the fourth ligand in the tetrahedral coordination of each Zn2+ ion. Specific residue numbering is based on the sequence of the E. coli YiiP protein. The arginine at position 325 in ZnT8 is highlighted in yellow. Residues involved in the charge interlock (Ch. Int.) are indicated in red text; notably, these residues are only partially conserved (Glu replacing Asp) between the bacterial and the vesicular ZnT subfamily (ZnT2, 3, 4 and 8). The CXXC motif, which is also specific to vesicular ZnTs, is highlighted in purple. Dileucine motifs in ZnT2 and 3 are purportedly involved in protein localisation 34. The ligands forming the purported third weaker zinc‐binding site in CzrB 17 are not indicated. (B) 3D homology model of human ZnT8cR based on Thermus thermophilus CzrB using swiss‐model (Materials and methods), highlighting the conserved metal‐binding ligands in magenta with bound zinc ions in grey and the T2D‐risk variant residue R325 in red. The triple β‐sheet face is predicted to form the dimer interface, while residue 325 is located in a loop at the apex of the dimer.
Our analysis further shows that in the CTDs of human ZnTs, the ligands for a second metal ion in the binuclear site C are not strictly conserved and, importantly, the ligand stemming from the other subunit, His261 in E. coli YiiP, is not conserved (Fig. 1A). Therefore, an important issue is if the CTDs of these human transporters bind fewer or even no metal ions at all. The conservation of only three predicted metal ligands, that is, two histidines and one glutamate/aspartate in the vesicular ZnTs (Fig. 1A), raises the questions of whether the CTDs in these human transporters sense zinc ion concentrations and how the protomers forming the dimer interact. The metal ligands that are conserved do not form a bridge between the two protomer CTDs in the dimer; thus, the CTD dimerisation‐induced conformational change seen upon zinc binding to the CTD in E. coli YiiP 13 may not occur and may not have the same consequences in human ZnTs.
Remarkably, there is a high density of potential metal binding residues in the C‐terminal tail of ZnT8, including a CXXC motif, which is present only in the vesicular subfamily of human ZnTs (ZnT2, 3, 4 and 8). This motif is conserved in all verified vesicular ZnT sequences available from the UniProt database, including mouse, rat, cow and frog. The significance of this motif is not known although CXXC motifs have redox functions or a metal‐binding role in metalloproteins, such as in some copper chaperones where they can mediate metal transfer to client proteins 26. However, in copper chaperones, this motif is typically in a different position in the primary sequence.
A ‘charge interlock’ (Ch. Int.) comprised of Asp207 in the CTD and Lys77 in the TMD is thought to be important for dimer formation in the full‐length E. coli YiiP protein 13. However, these residues are not conserved in non‐vesicular human ZnTs (i.e. not ZnT2–4 or 8). The charge of these residues is conserved in vesicular ZnTs, but Asp207 in the E. coli YiiP CTD is replaced by Glu in the vesicular ZnT subfamily (Fig. 1A), while the TMD Lys77 is replaced by Arg.
Protein yield
A typical 2 L bacterial culture (of either variant, aa267–369 in addition to an N‐terminal hexahistidine tag and a TEV protease cleavage site) yielded ~ 1 mg of > 95% pure ZnT8 CTD protein (Fig. 2A). Protein samples were concentrated to ~ 100–300 μm. There is a tendency for the proteins to aggregate and ultimately precipitate completely after a period of ~ 2 weeks. To alleviate the aggregation issues, many buffer constituents and several different E. coli expression strains were screened; the most effective conditions for expression of a folded protein were used herein (Materials and methods). Addition of fresh Tris(2‐carboxyethyl)phosphine hydrochloride (TCEP) during the size exclusion purification step is necessary to obtain a high yield of pure protein.
Figure 2.
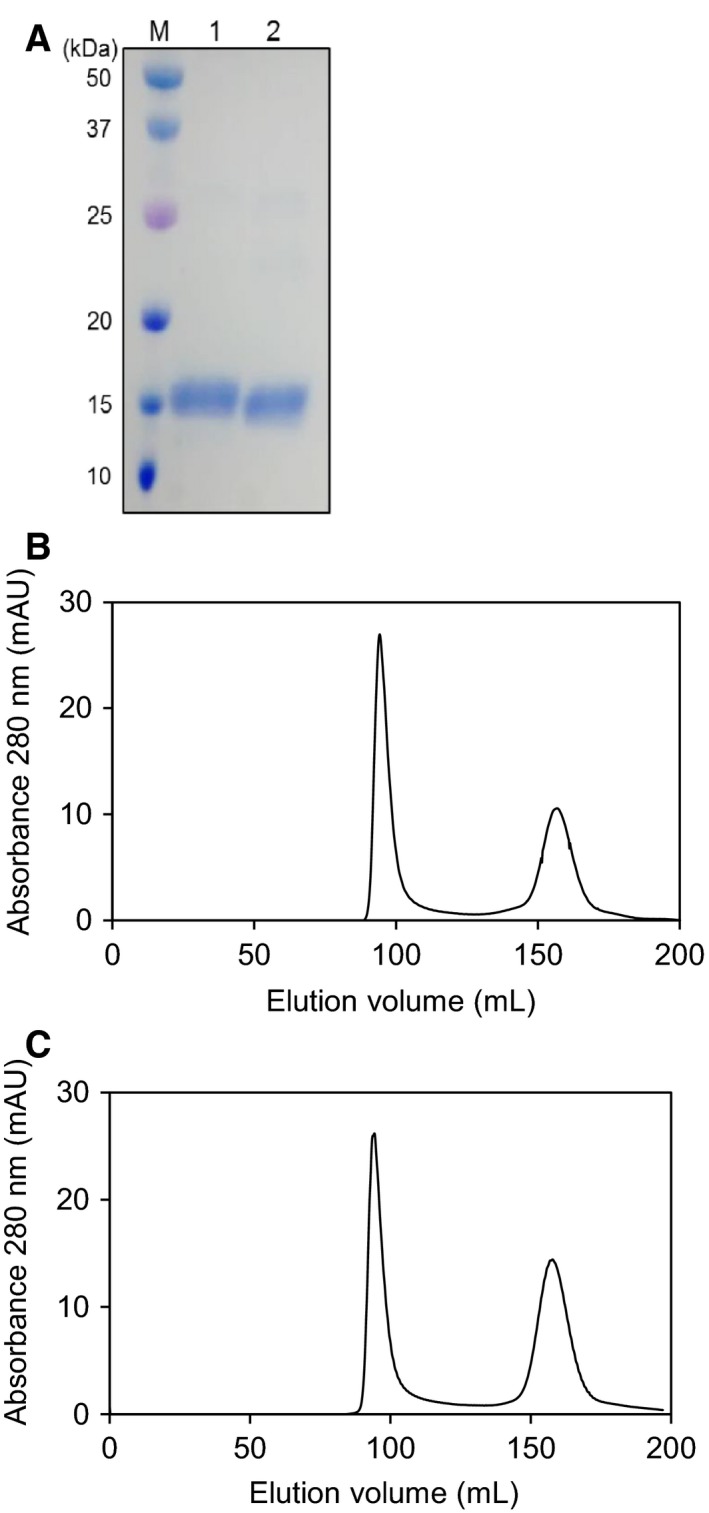
Purity and elution profiles of human ZnT8 CTD proteins. (A) Protein in the minor elution peaks at ~ 160 mL was analysed by SDS/PAGE and is > 95% pure ZnT8 CTD. Lane ‘M’ contains molecular weight markers; lane ‘1’ contains purified apo‐ZnT8cR; and lane ‘2’ contains purified apo‐ZnT8cW. The protein in the major elution peaks at ~ 95 mL was also analysed by SDS/PAGE (not shown) and is aggregated ZnT8. (B) Size exclusion chromatogram using a Superdex S75 26/60 column for ZnT8cR protein and, (C) ZnT8cW protein. Following calibration of the column (Materials and methods), the proteins in the fractions eluting at ~ 160 mL have a molecular mass of 34.9 kDa (calculated ZnT8 CTD monomer mass is 13.3 kDa).
The two CTD variants share structural similarities
ZnT8cR and ZnT8cW elute at the same volume in size exclusion chromatography (160 mL, Fig. 2B,C). Calibration of the Superdex S75 26/60 column with protein standards (Materials and methods) indicates that both variant ZnT8 CTD proteins have an apparent molecular mass of 34.9 kDa. The expected mass of the monomer is 13.3 kDa including the His‐tag and TEV protease site. Native PAGE analyses of the purified proteins indicate that both variants are dimeric. SDS/PAGE analysis of the largest peak at 95 mL indicates that it is aggregated but soluble ZnT8 CTD protein. The secondary structure of both apo‐ZnT8 CTD variants was investigated using CD spectroscopy; the two variants yield similar far‐UV CD spectra (Fig. 3A). The spectra did not change significantly upon addition of two molar equivalents of ZnCl2 or NiSO4, or replacement of NaCl with KCl. Inserting individual CD spectra into BeStSel 27, a fold recognition algorithm, showed that the two variants contain similar helix and sheet content to both each other and the CTD of the 3D‐characterised E. coli homologue YiiP (Fig. 3B). Thus, as predicted, the secondary structure and fold are highly conserved.
Figure 3.
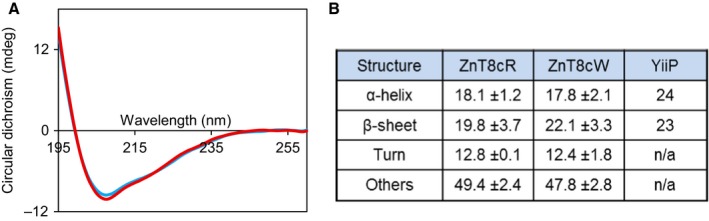
CD spectroscopy of the two human ZnT8 CTD variants. (A) Representative (n = 3) far‐UV CD spectra of 0.2 mg·mL−1 apo‐ZnT8cR(blue) and apo‐ZnT8cW (red) variants in 10 mm K2 HPO 4, 60 mm NaCl, 20 mm sucrose, pH 8. Separate far‐UV CD spectra measured in the presence of two molar equivalents of Zn2+, two molar equivalents of Ni2+ or with KCl replacing NaCl, were not significantly different from those of the apo‐proteins. (B) Protein secondary structure, expressed as % ± standard deviation (n = 3), was determined using individual CD spectra for both apo‐ZnT8cR and ZnT8cW variants with the BeStSel algorithm (Materials and methods). The difference in secondary structure between the two variants is not statistically significant. Helix and sheet content of Escherichia coli YiiP CTD were calculated from the 3D structure (PDB ID: 2qfi), while turns and other structures could not be readily differentiated.
ZnT8cR is more thermostable than ZnT8cW; Zn2+ stabilises both variants
The thermal stability of both CTD variants in the presence and absence of ZnCl2 was investigated using melting analysis by both CD spectroscopy between 6 and 92 °C (Fig. 4A,B) and nano differential scanning fluorimetry (nDSF) between 20 and 85 °C. This type of DSF utilises intrinsic protein fluorescence; the ratio of the emission at 350 nm to that at 330 nm as a function of temperature reveals the point(s) at which the protein structure changes significantly. There is a significant difference between the stability of the CTD variants in the apo (Zn2+‐free) form as measured with both CD and nDSF; the ZnT8cR T m is 42.8 ± 0.5 °C, whereas the ZnT8cW T m is 41.4 ± 0.4 °C (n = 3, P = 0.013). Remarkably, apo‐ZnT8cR (T2D‐risk in the full‐length protein) has higher thermostability than apo‐ZnT8cW (T2D‐protective in the full‐length protein). Both CTD variants are significantly more stable in the presence of two molar equivalents of Zn2+; ZnT8cR‐2Zn T m is 54.5 ± 2.1 °C and ZnT8cW‐2Zn T m is 51.0 ± 1.8 °C (in each comparison n = 3, P < 0.001), but not in the presence of two molar equivalents of Ni2+. The numerical difference in stability between the two CTD variants in the presence of Zn2+ is not statistically significant (P = 0.093).
Figure 4.
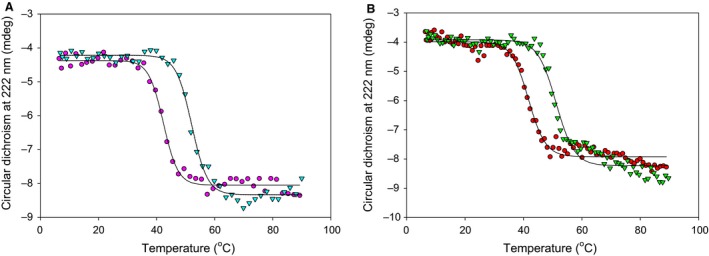
Thermostability of the two human ZnT8 CTD variants. (A) Representative (n ≥ 3) melting curves for apo‐ZnT8cR (magenta circles, T m = 42.8 ± 0.5 °C) and ZnT8cR with two molar equivalents of Zn2+ (teal triangles, T m = 54.5 ± 2.1 °C) measuring the change in CD at 222 nm from 6 to 92 °C with a heating rate of 1 °C·min−1. (B) Representative (n = 3) CD melting curves of apo‐ZnT8cW (red circles, T m = 41.4 ± 0.4 °C) and ZnT8cW in the presence of two molar equivalents of Zn2+ (green triangles, T m = 51.0 ± 1.8 °C). There are significant differences between thermal stability of apo‐ZnT8cR and apo‐ZnT8cW (n = 3, P = 0.013) and between both apo‐variants and the variant in the presence of Zn2+ (for each comparison n = 3, P < 0.001). The difference in stability between the two variants in the presence of Zn2+ is not statistically significant (P = 0.093).
The two Trp residues in ZnT8cW are in different local environments
ZnT8cW contains two tryptophan residues (W306 and W325), whereas ZnT8cR contains only one (W306). The emission spectrum (λEx = 295 nm) of ZnT8cR provides information on the tryptophan residue shared by both variants (i.e. W306). Therefore, by subtracting the ZnT8cR emission spectrum from that of ZnT8cW, information about W325 in ZnT8cW can be obtained (Fig. 5). The emission maximum of ZnT8cR was 340 nm, corresponding to W306, while that of ZnT8cW was 345 nm. The emission maximum of W325, calculated by subtracting the ZnT8cR spectrum from that of ZnT8cW, is 350 nm. For comparison, a pure N‐acetyl‐dl‐tryptophan solution measured in the same buffer has an emission maximum at 363 nm. The degree of blue shift of a tryptophan residue's emission from that of pure tryptophan in solution depends on how hydrophobic the local environment is. In the ZnT8cW protein, W325 is therefore in a less hydrophobic environment than W306, and therefore more solvent accessible.
Figure 5.
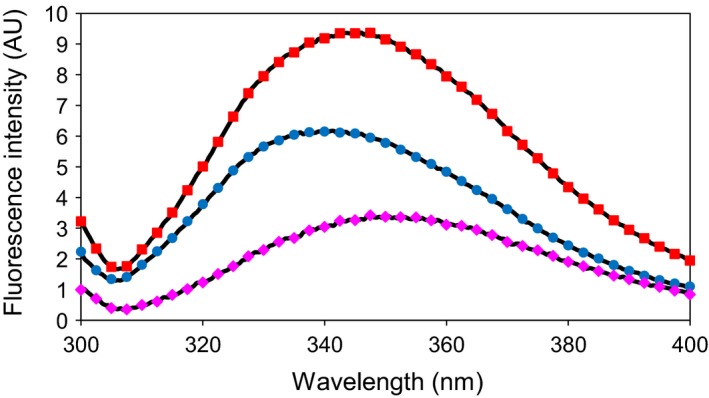
Fluorescence spectroscopy of the two human ZnT8 CTD variants. Representative (n = 3) fluorescence spectra of ZnT8cW (red squares) and ZnT8cR (blue circles) protein, both 2.8 μm, in 50 mm Tris/HCl, pH 8, 300 mm NaCl (λEx = 295 nm). The ZnT8cR variant contains one tryptophan residue (W306), while the ZnT8cW variant contains two (W306 and W325). Therefore, by subtracting the ZnT8cR signal from that of ZnT8cW the fluorescence spectrum of W325 was obtained (magenta diamonds).
The amino acid at position 325 affects dimer formation
The homodimerisation affinities of both ZnT8 CTD variants were measured using microscale thermophoresis (MST) in the presence of EDTA, eliminating any influence of divalent metal ions (Fig. 6). Titrating 100 nm labelled apo‐protein with 180 μm–5.5 nm (ZnT8cR) or 124 μm–3.8 nm (ZnT8cW) unlabelled apo‐protein yielded homodimerisation K d values of 4.3 ± 1.3 μm for ZnT8cR and 1.8 ± 0.1 μm for ZnT8cW. This difference is statistically significant (n = 3, P = 0.034). Thus, the dimerisation of ZnT8cR (T2D‐risk in the full‐length protein) occurs with less affinity than ZnT8cW (T2D‐protective in the full‐length protein) in the presence of EDTA. The directionality of this difference is opposite to that observed for the thermostability of the two forms.
Figure 6.
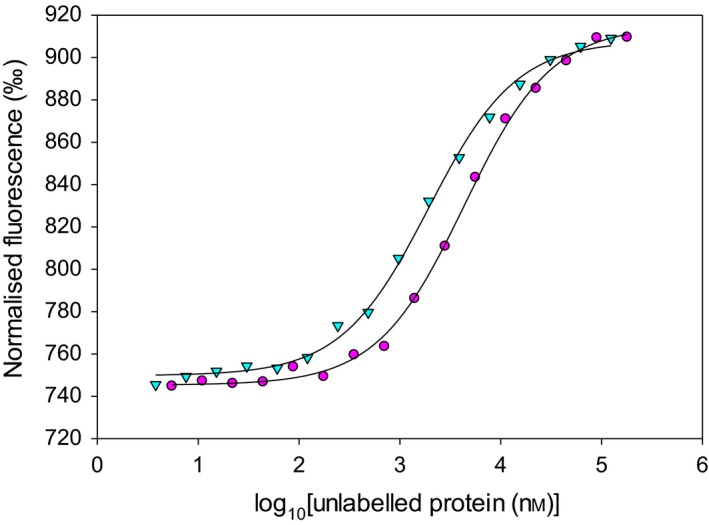
Dimerisation of the two human ZnT8 CTD variants. Representative (n = 3) MST traces for dimerisation of ZnT8 CTD protein. Fluorescently labelled apo‐ZnT8cR (100 nm, magenta circles) was titrated (in the presence of 1 mm EDTA) with unlabelled apo‐ZnT8cR protein (180 μm–5.5 nm), yielding a homodimerisation K d of 4.3 ± 1.3 μm. Fluorescently labelled apo‐ZnT8cW (100 nm, teal triangles) was titrated (in the presence of 1 mm EDTA) with unlabelled apo‐ZnT8cW protein (124 μm–3.8 nm), with a homodimerisation K d of 1.8 ± 0.1 μm. There is a significant difference between the homodimerisation K d of each variant in the presence of EDTA (n = 3, P = 0.034).
The amino acid at position 325 does not directly affect metal binding
Based on sequence analysis, the expected divalent metal ion binding capacity for ZnT8 CTDs is one ion per monomer (Fig. 1A). The two variant apo‐proteins (10 μm protein) were incubated with 0–10 molar equivalents of Zn2+ and subjected to gel filtration to remove any loosely bound zinc. Inductively coupled plasma mass spectrometry (ICP‐MS) analysis of the apo‐ZnT8 CTD proteins incubated with no additional Zn2+ showed that 0.21 ± 0.07 (n = 6) divalent metal ions (Zn2+ and Ni2+) were residually bound per monomer. The vast majority of this residual metal load (> 90%) was contributed by Ni2+. Supplementing up to 10 molar equivalents of Zn2+ indicates that both variants bind approximately three Zn2+ ions per monomer; an average of 2.6 ± 0.4 Zn2+ ions bind to ZnT8cR, whereas 3.2 ± 0.5 Zn2+ ions bind to ZnT8cW (Fig. 7). This difference between the two variants is not statistically significant (n = 3, P = 0.156). Upon addition of 4–10 molar equivalents of Zn2+, the small amount of Ni2+ residually bound to both CTD variants was displaced.
Figure 7.
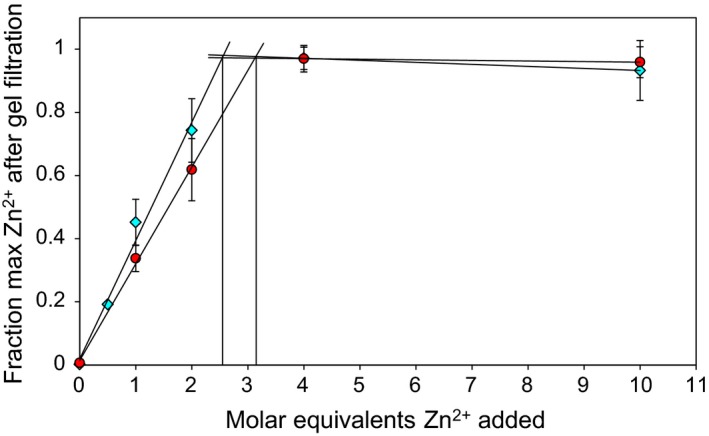
Zinc stoichiometry of the two ZnT8 CTD variants. Fraction of the maximum Zn2+ content of 10 μm ZnT8cR (teal diamonds) and ZnT8cW (red circles) following incubation with 0–10 molar equivalents of Zn2+ and subsequent gel filtration to remove unbound Zn2+. Protein concentration was determined spectrophotometrically (Materials and methods). The intersection points in the titration data indicate that ZnT8cR binds Zn2+ with a stoichiometry of 2.6 ± 0.4 per monomer, whereas ZnT8cW binds 3.2 ± 0.5 per monomer. The difference between the two variants is not statistically significant (n = 3 for both variants, P = 0.156).
A competition assay with the chromophoric chelating agent Zincon shows similar results for both ZnT8 CTD variants (Fig. 8A,B). When titrated with zinc in buffer alone, 70 μm Zincon is saturated with 70 μm ZnCl2 and an initial increase in absorbance at 620 nm is measured upon addition of 1 μm ZnCl2. Zincon has a K d of 214 nm for a 1 : 1 complex with zinc at pH 8 28. However, when competing with 5 μm apo‐ZnT8 CTD (either variant), the initial increase in absorbance is not seen until 10 μm Zn2+ is added, indicating that both ZnT8 CTD variants contain two Zn2+‐binding sites that have a tighter affinity than 214 nm and thus outcompete the zinc binding to Zincon. Following this initial 10 μm ZnCl2, an additional 75 μm ZnCl2 is required to saturate zinc binding to Zincon in the presence of 5 μm apo‐ZnT8 CTD protein (both variants). Thus, both variants have an additional zinc site with low affinity competing directly with Zincon. When both ZnT8 CTD protein variants have their cysteines blocked by alkylation with iodoacetamide, only 5 μm ZnCl2 is required to measure a change in absorbance at 620 nm. This result indicates that cysteines in the C‐terminal tail, which contains three cysteines, constitute one of the two high affinity binding sites that outcompete the binding of zinc to Zincon. With protein modified by iodoacetamide (both variants), an additional 75 μm ZnCl2 is still required to saturate the Zincon, indicating that the lower affinity site is not lost upon cysteine alkylation.
Figure 8.
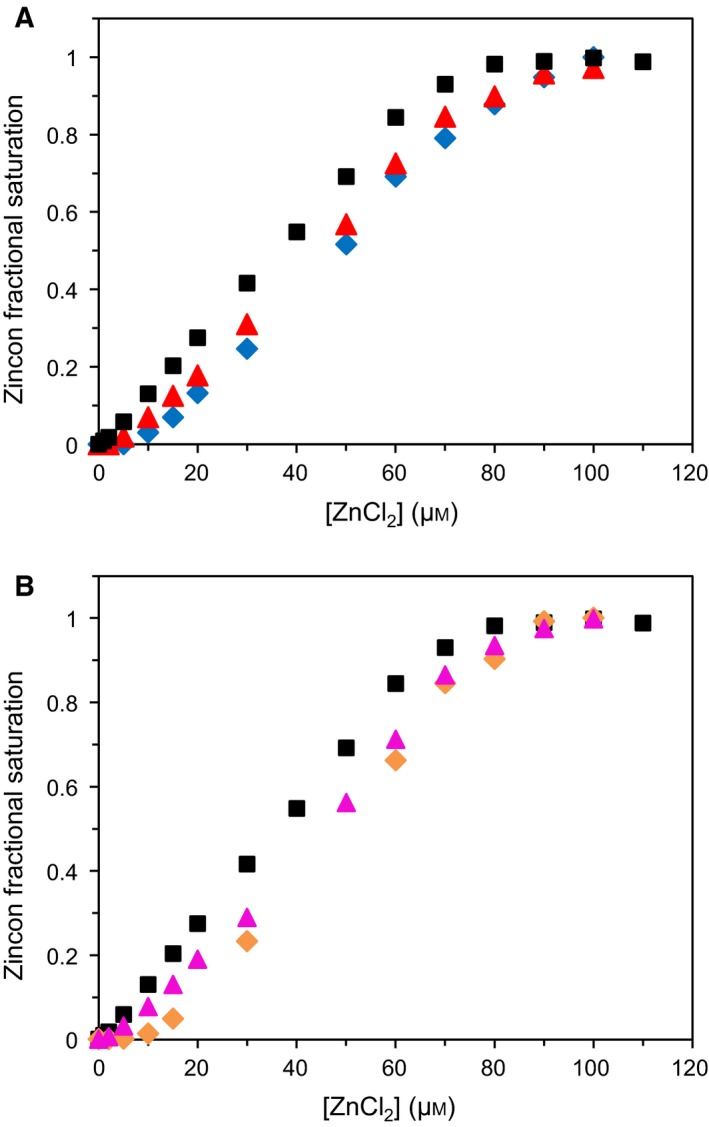
Zinc affinity of the two ZnT8 CTD variants. (A) Zinc binding to ZnT8cR in competition with Zincon. Measuring absorbance of 620 nm, it takes 70 μm ZnCl2 to saturate 70 μm Zincon in 50 mm HEPES, pH 8, 300 mm NaCl, 100 mm sucrose (black squares). In competition with 5 μm apo‐ZnT8cR (blue diamonds), no signal at 620 nm is detected until 10 μm ZnCl2 is added, revealing two high affinity zinc‐binding sites in ZnT8cR which outcompete Zincon. An additional 75 μm ZnCl2 is required to fully saturate Zincon, identifying one lower affinity (~ μm) site that directly competes with Zincon. When ZnT8cR is reduced and incubated with iodoacetamide for 1 h prior to the Zincon assay (red triangles), only 5 μm ZnCl2 is required to elicit the initial signal at 620 nm. An additional 75 μm ZnCl2 is required to saturate the Zincon. Thus, when the cysteines are blocked by alkylation, ZnT8cR retains one high affinity and one low affinity Zn2+‐binding site. B, Zinc binding to ZnT8cW in competition with Zincon. ZnCl2 titration of Zincon alone in HEPES buffer (black squares), in competition with apo‐ZnT8cW (orange diamonds), and in competition with ZnT8cW modified with iodoacetamide (magenta triangles), demonstrating that ZnT8cW also contains two high affinity and one low affinity zinc binding sites, and that one high affinity binding site is lost upon alkylation of the three cysteines in ZnT8cW.
A dityrosine bond does not form between ZnT8 CTD protomers
Using a specific anti‐dityrosine antibody, an inter‐protomer dityrosine bond in the CTDs of ZnT3 and ZnT4 homodimers was detected 29. Dityrosine bonds have a high quantum yield at 407 nm when using an excitation wavelength of 325 nm, well above the excitation maximum of individual tyrosine residues. There is one tyrosine residue in ZnT8 CTD (Y284) although it is not at the same position as the three tyrosine residues involved in ZnT3 and ZnT4 homodimerisation. Nevertheless, using fluorescence spectroscopy, we could not detect any emission associated with a dityrosine in either ZnT8 CTD variant.
Discussion
The mammalian ZnTs are thought to function with the Zn2+/H+ antiport mechanism elucidated for ZnT1 and the bacterial homologues 30. The antiport is likely coupled to induced conformational changes that alternately open the channel inward or outward as shown for the bacterial homologues 13, 16. In contrast to the E. coli YiiP protein, which has a zinc/cadmium selectivity filter in the TMD with one histidine and three aspartates, the mammalian ZnTs utilise two histidine and two aspartate side chains to transport zinc specifically 31. Amongst mammalian ZnTs (with the exception of ZnT10, which has an asparagine instead of one of the two aspartates in the TMD and accordingly transports manganese in addition to zinc 32), the zinc transport site and the overall structure of the TMD are highly conserved 3. The CTD, however, is much more variable and is thought to be critical in the evolution of these transporters for different functions, such as the subset of four vesicular transporters, ZnT2–4 and ZnT8. This subgroup supplies exocytotic vesicles with zinc for various purposes, such as synaptic vesicles (ZnT3) involved in neurotransmission 33 and vesicles in mammary epithelial cells (ZnT2) that supply zinc in the milk of lactating women 34. How ZnTs acquire and deliver sufficient zinc to exocytotic vesicles is an unresolved biochemical issue.
Despite the lack of high sequence homology between CTDs in mammalian ZnTs, various structural features are conserved, for instance the overall fold. Based on prediction of secondary structure and CD data, both ZnT8cR and ZnT8cW form the αββαβ structure observed in the structure of E. coli YiiP, and most other ZnT CTDs are predicted to adopt this structure (Fig. 1A). Referred to as a ‘ferredoxin’ fold as it was originally found in iron proteins, it is also commonly found in copper proteins, in particular copper chaperones 25. However, the metal‐binding sites are at different positions in these proteins.
Due to the predicted location of ZnT8 residue 325 at the CTD dimer interface (Fig. 1B), the R325W replacement is likely to affect dimer formation and stability. A significant difference in dimer association between the two human ZnT8 CTD variants was detected in this investigation. The directionality of the difference was not expected, however; despite an increased thermostability of ZnT8cR, its dimerisation affinity was lower than that of ZnT8cW. Collectively, these data show that both dimer formation and stability are affected in the two CTD variants. The 2.9‐Å crystal structure of E. coli YiiP revealed that the CTD dimer interface is highly charged and that in the absence of bound Zn2+ , the repulsive charges in the dimer cause the protomers to swing apart 13. The exchange of the charged R325 residue for uncharged W325 may disrupt this charge interaction in ZnT8, and may explain the biophysical differences in the two variant CTDs observed. Neither the Arg nor the Trp at position 325 are conserved among the ZnTs, or even among species; murine and rat ZnT8 encode Gln325. The position is at a variable loop between the conserved secondary structures.
The identity of residue 325 in ZnT8 alters the specificity of ZnT8 autoantibodies in T1D 24, with sera from at least 22% of patients reacting with one of either R325 or W325 antibodies but not the other. Previous studies expressing ZnT8 CTDs to investigate antibody epitopes did not prove that the protein folds correctly 35. A properly folded protein may be crucial for presenting the correct 3D epitope to the antibody. Indeed, the ZnT8 autoantibody epitope has been confirmed to be conformational rather than linear 24. Therefore, the availability and characterisation of the folded CTDs may be important for efficient antibody production.
Another model for the dimerisation in mammalian vesicular ZnTs, namely the formation of a dityrosine, has been advanced for ZnT3 29. The ZnT8 CTD contains one tyrosine (Y284) although its location in the primary sequence is not conserved with any of the tyrosine residues implicated in ZnT3 homodimerisation. We found no evidence for dityrosine bond formation in either ZnT8 CTD variant. A charge interlock with residues from both the TMD and CTD serves as a hinge in the dimerisation of full‐length CDF proteins 13. The charge interlock CTD residues (albeit Glu replacing Asp207 and Arg replacing Lys77 in YiiP) are conserved in vesicular ZnTs (Fig. 1A) but, because of the absence of the TMD, isolated CDF CTDs do not interrogate this aspect of intersubunit linkages. Intriguingly, these charge interlock residues are not conserved in non‐vesicular ZnTs, suggesting that the intersubunit linkages differ amongst mammalian ZnTs.
A characteristic feature of CTDs in bacterial CDFs is two zinc‐binding sites per monomer, harbouring four zinc ions in the dimer 12 (although the T. thermophilus CzrB CTD contains an additional weak zinc‐binding site 17). One of these sites utilises ligands from both protomers, therefore bridging between the dimer subunits, while the other(s) are formed of ligands from only one protomer. Both metal‐binding sites utilise a water molecule as the fourth ligand in the tetrahedral coordination of the Zn2+ ions. Remarkably, the ligands for the intersubunit metal‐binding site are not conserved in the human ZnTs (Fig. 1A). Specifically, a ligand corresponding to His261 is missing. This is the only residue contributing a metal‐binding ligand from the second protomer in the dimer in E. coli YiiP, and is involved in the CTD conformational changes seen upon zinc binding, or ‘zinc sensing’, when the cytosolic zinc concentration reaches an upper threshold 13. The primary biological function of these bacterial transporters is to protect the cytosol from zinc overload, and current evidence suggests micromolar K m values for transport 13. The problem with this model for the four vesicular ZnTs (ZnT2–4 and 8) is that there is only picomolar free zinc available in the cytosol of human cells, and the total vesicular zinc concentrations are high millimolar. Therefore, either the vesicular ZnT CTDs are able to sense much lower cytosolic zinc concentrations than their bacterial homologues, for which there is no evidence at present, or the role of the CTD is different from that of the bacterial proteins and not involved in sensing zinc directly, as suggested by our findings.
Our measurements show that both apo‐ZnT8 CTD variants form stable dimers. Addition of two molar equivalents of zinc significantly increases the stability of both variant CTDs, without significantly altering their secondary structures. Following zinc addition up to saturation with 10 molar equivalents of zinc, three zinc ions were tightly bound per protein monomer. The difficulty in relating the metal binding to a particular binding site in the CTD stems from the fact that the expressed protein has a hexahistidine tag. It was possible to remove this tag, but the resulting protein was unstable and precipitated, rendering further experimentation impossible. ZnT8 has three C‐terminal cysteine residues, including a CXXC motif that has been shown to bind zinc in the metal‐binding domains of E. coli ZnTA 36 and Synechocystis PCC 6803 ZiaA 37, and copper in copper chaperones 38. Amongst human ZnTs, the CXXC motif is only conserved in the vesicular subfamily (Fig. 1A). Competition assays performed with the chromophoric zinc chelator Zincon and protein modified with iodoacetamide (Fig. 8) reveal that one of the two < 214 nm affinity zinc‐binding sites identified in both ZnT8 CTD variants is formed of the C‐terminal cysteines. The small amount of residual Ni2+ that was bound to both variant apo‐proteins was only displaced upon supplementing the protein with 4–10 molar equivalents of zinc. This agrees with published data indicating that the His‐tag has a higher affinity for Ni2+ than it does for Zn2+ 39. Protein‐bound His‐tags bind Ni2+ with an affinity of ~ 700 nm 40. These data support the hypothesis that the low affinity site (approximately micromolar), identified in both ZnT8 CTD variants with the Zincon competition assay, is contributed by the His‐tag. Therefore, our metal‐binding data can be reconciled with the prediction from the sequence alignment that indeed maximally only one metal ion binds with high affinity at the canonical interface site in the protein as isolated, the second with high affinity at the C‐terminal cysteines and the third with lower affinity to the His‐tag.
A recent report, in which the activity of ZnT8 reconstituted in liposomes was investigated, concluded that the transport activity is dependent on the lipid environment, and inferred that the lipid environment affects zinc loading during insulin granule biogenesis 9. This report also noted that the T2D‐risk R325 ZnT8 variant consistently showed a small increase in zinc transport activity compared to the T2D‐protective W325 variant, which was revealed only with specific lipid compositions of the liposomes. In accordance with higher transport activity, it was noted that human islets with the R325 ZnT8 variant had a higher zinc content 41. Another report on ZnT8 transport in Xenopus laevis oocytes did not detect a difference in transport kinetics between the ZnT8 R/W325 variants, supporting the conclusion that the liposome lipid composition may be crucial for revealing differences between the two variants 42.
There are two main conclusions. First, the mammalian vesicular ZnTs are significantly different from bacterial CDF ZnTs in their CTD zinc binding. The loss of the subunit‐bridging ‘sensing’ zinc binding site in the CTD, the additional high affinity zinc binding at the C‐terminal cysteines and the disparity between the very low concentration (pm) of free cytosolic zinc and the very high (mm) total zinc levels found in secretory vesicles, strongly suggest that the sensing of excess cytosolic zinc and the concomitant transport in bacteria would need to function differently in mammalian systems supplying zinc to exocytotic vesicles. Bacterial zinc exporters need only function when the cell is experiencing high and/or toxic levels of zinc, whereas loading of insulin and other secretory vesicles, for instance synaptic vesicles by ZnT3 33, must occur under conditions of normal cytosolic zinc concentrations. Second, this is the first report detailing that the W325R mutation causes significant differences in ZnT8 CTD dimer formation and stability. How or even if these effects in the CTDs lead to the altered transport function of the full‐length proteins reported elsewhere 9, and ultimately to the relatively large increased risk of developing T2D for carriers of the R325 variant, will require further investigation. That the T2D‐risk ZnT8 R325 variant is the more active form of the transporter suggests that people with the R325 variant may have an increased zinc content in their insulin granules as indicated by the data on human islets 41. This increased granular zinc uptake may deplete cytosolic zinc and affect β‐cell function. If so, it may need to be ameliorated with a higher dietary zinc intake 43.
Materials and methods
Materials
Tris(2‐carboxyethyl)phosphine hydrochloride, HEPES, iodoacetamide, Zincon sodium salt, NaCl, K2HPO4, KH2PO4, MgCl2, ZnCl2 and N‐acetyl‐dl‐tryptophan were purchased from Sigma Aldrich (St. Louis, MO, USA); Tris‐base and SDS from Severn Biotech (Kidderminster, UK); DTT and PMSF from Thermo Fisher Scientific (Waltham, MA, USA); Tween‐20 and NiSO4.6H2O from Acros Organics (Geel, Belgium); sucrose from Merck Millipore (Burlington, MA, USA); IPTG from Promega (Madison, WI, USA); l,l‐dityrosine dihydrochloride from Santa Cruz Biotechnology (Dallas, TX, USA); 5,5′‐dithiobis(2‐nitrobenzoic acid) (DTNB; Ellman's reagent) from Invitrogen (Carlsbad, CA, USA); EDTA from Cambridge Bioscience (Cambridge, UK); LB media powder from MP Biomedicals (Santa Ana, CA, USA); and imidazole from Apollo Scientific (Stockport, UK).
Protein expression and purification
The sequence encoding residues 267–369 of human R325 ZnT8 (ZnT8cR; any residue numbering refers to full‐length human ZnT8 long isoform, Ensembl transcript SLC30A8‐002) was optimised for E. coli expression and the cDNA synthesised by DNA2.0 (Menlo Park, CA, USA). It was inserted into pET6H encoding an N‐terminal hexahistidine tag and a TEV protease cleavage site. Mutagenesis to produce the W325 variant (ZnT8cW) was carried out by Mutagenex (Suwanee, GA, USA) using PCR‐based substitution, followed by sequence verification of the inserts of both plasmids. The two plasmids were transformed into E. coli strain SoluBL21™ (AMS Biotechnology, Abingdon, UK) and grown at 30 °C in LB media containing 100 μg·mL−1 ampicillin until the OD600 reached 0.60. Cells were then kept at 16 °C on an orbital shaker (G25 Incubator Shaker, New Brunswick, Edison, NJ, USA; 210 r.p.m.) for 30 min before protein expression was induced with 0.5 mm IPTG and the cells kept at 16 °C and at 210 r.p.m. for an additional 42 h. Cells were harvested by centrifugation and resuspended in 10 mL lysis buffer [50 mm Tris/HCl, pH 8, 100 mm NaCl, 100 mm sucrose, 5 mm DTT, 2 mm MgCl2, 1 mm PMSF, 5 U·mL−1 DNase (Thermo Fisher Scientific)] until a homogenous solution was obtained. The homogenate was diluted 1 : 6 with equilibration buffer [50 mm Tris/HCl, pH 8, 100 mm NaCl, 100 mm sucrose, 2 mm DTT, 20 mm imidazole, containing one tablet of Complete ULTRA mini EDTA‐free protease inhibitors (Roche, Basel, Switzerland)], and sonicated (Model 2000U, Ultrasonic Power Corp. (Freeport, IL, USA); +285 output, 0.5 s−1 pulse) in an ice‐water bath for 20 s pulse and 40 s rest settings for a total of 15 min, followed by centrifugation at 45 000 g for 40 min at 4 °C. The 60 mL supernatant was added to 2 mL of pre‐equilibrated Ni2+ affinity gel (Sigma Aldrich) and incubated end‐over‐end on a roller at 4 °C for 40 min. After centrifugation at 500 g for 1 min at 4 °C, the pellet was washed three times with equilibration buffer containing 500 mm NaCl. The bound protein was eluted in five 1 mL washes with elution buffer (50 mm Tris/HCl, pH 8, 500 mm NaCl, 100 mm sucrose, 2 mm DTT, 300 mm imidazole). The eluate was loaded onto a Superdex S75 26/60 column (GE Healthcare, Chicago, IL, USA) equilibrated with purification buffer (50 mm Tris/HCl, pH 8, 300 mm NaCl, 100 mm sucrose, 100 μm TCEP) using a flow of 2.2 mL·min−1 and a column temperature of 4 °C. The column was calibrated with yeast alcohol dehydrogenase, bovine serum albumin, bovine erythrocyte carbonic anhydrase and aprotinin. To produce ZnT8c apo‐protein, 2 mm TCEP and 1 mm EDTA were added to the 10 mL fractions from size exclusion chromatography and then concentrated to ~ 0.5 mL using a 15 mL 3 kDa MWCO centrifugal concentrator (Merck Millipore). Samples were stored in this state for up to 1 week at 4 °C. Up to 0.5 mL protein was then diluted 30× with EDTA‐ and TCEP‐free buffer (exact constituents dependent on assay to be carried out as given in the corresponding experiments) and concentrated to 0.5 mL with the same centrifugal concentrators, performing this dilution/concentration step at least three times. Reduced apo (zinc‐free) protein was then used fresh within 1 day. Concentrated samples were analysed using 16% (w/v) Tris‐glycine SDS/PAGE (Thermo Fisher Scientific) 44. Protein samples used for MST were purified as above, except that an amine‐free buffer (50 mm potassium phosphate, pH 8, 300 mm NaCl, 100 mm sucrose, 100 μm TCEP) was used for the size exclusion chromatography.
Determination of protein concentration
The concentration of the protein samples was determined spectrophotometrically using a modified Bradford assay 45 with Bio‐Rad protein assay reagent (Hercules, CA, USA). Bovine serum albumin (Thermo Fisher Scientific) dissolved in purification buffer was used as the standard. The accuracy of the determination was corroborated by spectroscopic measurements, using extinction coefficients of 13 980 m −1·cm−1 for ZnT8cW and 8480 m −1·cm−1 for ZnT8cR (calculated from the primary protein sequence).
CD spectroscopy
CD spectra were acquired on a Chirascan Plus spectrometer (Applied Photophysics, Leatherhead, UK). Far‐UV spectra in the 195–260 nm range were recorded using a 0.5 mm path length cuvette, a bandwidth of 2 nm, a step size of 1 nm and a time‐per‐point of 1 s. Spectra were taken at 20 °C. For all spectra, the protein concentrations were 0.2 mg·mL−1 in 10 mm potassium phosphate, pH 8, 60 mm NaCl, 20 mm sucrose. Protein secondary structure content based on CD spectra was determined using BeStSel 27. Melting analyses involved monitoring the CD signal at 222 nm between 6 and 92 °C at a heating rate of 1 °C·min−1. Separate far‐UV spectra and melting analyses were performed with both variants in the presence of two molar equivalents of ZnCl2, two molar equivalents of NiSO4 and by replacing the 60 mm NaCl with 60 mm KCl to examine any possible influence of the cation.
Nano differential scanning fluorimetry (nDSF)
Samples of 5 μm ZnT8cR and ZnT8cW in 10 mm Tris/HCl, pH 8, 60 mm NaCl, 20 mm sucrose, 2 mm DTT were loaded into nDSF‐grade standard capillaries (NanoTemper Technologies, Munich, Germany). Protein unfolding was measured by detecting fluorescence emission changes at 350 and 330 nm, between 20 and 85 °C at a rate of 1 °C·min−1, using a Prometheus NT.48 instrument (NanoTemper Technologies). Protein melting temperatures were calculated by taking the first derivative of the 350/330 nm ratio, and are given as the average of at least three samples ± standard deviation.
Microscale thermophoresis (MST)
Samples of 20 μm ZnT8cR or ZnT8cW were labelled with amine‐reactive Monolith NT.115 labelling kit Red‐NHS (#MO‐L001; NanoTemper Technologies) as per the manufacturer's instructions. Labelled proteins were diluted to a final concentration of 100 nm in 10 mm potassium phosphate, pH 8, 60 mm NaCl, 20 mm sucrose, 100 μm TCEP, 1 mm EDTA, 0.05% (v/v) Tween‐20. ZnT8cR‐Red and ZnT8cW‐Red were titrated with 180 μm–5.5 nm unlabelled ZnT8cR or 124 μm–3.8 nm unlabelled ZnT8cW, respectively. All experiments were performed using a Monolith NT.115 instrument (NanoTemper Technologies) at 25 °C in standard‐coated capillaries (NanoTemper Technologies). Binding affinities were calculated using MO.Affinity Analysis software (NanoTemper Technologies) and are expressed as the average of at least three separate measurements ± standard deviation.
Inductively coupled plasma mass spectrometry (ICP‐MS)
Concentrated apo‐protein samples of both variants were prepared and diluted with ≥ 18.2 MΩ.cm water (ELGA LabWater, High Wycombe, UK). These samples were analysed for zinc and nickel with ICP‐MS (Perkin Elmer Life Science, Waltham, MA, USA, model NexION 350D) and were shown to have reduced metal content compared with protein samples that had not been incubated with additional TCEP and EDTA.
To measure zinc‐binding stoichiometry, ZnT8cR and ZnT8cW samples were prepared in 10 mm Tris/HCl, pH 8, 60 mm NaCl and 20 mm sucrose. Samples of each isoform were diluted to 10 μm. Each protein sample was then incubated with 0, 1, 2, 4 or 10 molar equivalents of Zn2+ (ZnCl2) for 10 min at 21 °C, followed by gravity‐flow gel filtration using PD MiniTrap G‐25 desalting columns (GE Healthcare) to remove excess zinc. Samples were then diluted with ≥ 18.2 MΩ.cm water and analysed for zinc and nickel using ICP‐MS.
Fluorescence
Fluorescence measurements were taken at 21 °C using a Perkin Elmer LS50 fluorimeter. Separate ZnT8cR and ZnT8cW samples were diluted to 2.8 μm in 50 mm Tris/HCl, pH 8, 300 mm NaCl in a volume of 2 mL. Emission spectra (λEx = 295 nm) were collected between 300 and 400 nm at a 100 nm·min−1 scan rate. Emission spectra for 3 μm N‐acetyl‐dl‐tryptophan solutions were collected in the same buffer. Emission spectra were recorded between 300 and 450 nm (λEx = 325 nm) for examining the presence of a dityrosine bond, using 1 μm l,l‐dityrosine dihydrochloride in 1 m Tris/HCl, pH 8 as a positive control.
Assessing redox state of the cysteines and alkylation of cysteine residues
DTNB (Ellman's reagent) was used to quantify the free sulfhydryls present in the protein samples, following the manufacturer's instructions, using 50 mm HEPES, pH 8, 300 mm NaCl and 100 mm sucrose as the reaction buffer and the molar extinction coefficient at 412 nm of TNB (14 150 m −1·cm−1). Protein was diluted to 2 μm and the production of TNB measured after 15 min. Iodoacetamide was used to alkylate the sulfhydryls on the cysteine side chains, as per the manufacturer's instructions. Alkylation was confirmed upon measuring no free sulfhydryls in the protein with DTNB.
Zincon competition assay
Zincon forms a 1 : 1 complex with Zn2+ with a reported dissociation constant of 214 nm at pH 8 28. A stock solution of Zincon was prepared in ≥ 18.2 MΩ.cm water, diluted and the concentration determined at 488 nm using ε488 = 26 900 m −1·cm−1 and a 0.5 cm quartz cuvette in a Jenway 7315 spectrophotometer. Zincon was diluted to 70 μm in 50 mm HEPES, pH 8, 300 mm NaCl, 100 mm sucrose and titrated with ZnCl2, measuring the change in absorbance at 620 nm. Competition assays between 70 μm Zincon and 5 μm ZnT8 CTD protein were performed with both apo‐protein and protein modified with iodoacetamide. The titration break points and thus the stoichiometries were analysed by extrapolating the linear portion of the titration and comparing it with the buffer‐only control.
Bioinformatics and molecular modelling
Primary protein sequences were compared using clustal omega (EMBL, Heidelberg, Germany) and secondary structure predictions were carried out using jpred 4 46. The ZnT8cR homodimer was modelled with SWISS‐MODEL 47 based on the 3D structure of the T. thermophilus homologue CzrB (PDB ID: 3byr) with two zinc ions bound, while PyMOL was used for visual representation of 3D structure. CzrB was chosen as the primary basis for the model rather than E. coli YiiP owing to the availability of the zinc‐bound conformation of CzrB in the SWISS‐MODEL template selection algorithm. Models constructed based on YiiP and other available bacterial CDF CTDs returned similar models.
Statistics
All statistical significance was assessed by unpaired Student's t‐tests using sigmaplot 14.0 (Systat Software Inc., Chicago, IL, USA).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Author contributions
DSP performed the experiments and wrote the manuscript. WM supervised the work and wrote the manuscript. CH co‐supervised the work and edited the manuscript.
Acknowledgements
We thank Drs Mitla Garcia‐Maya and Paul Brown for assistance with protein purification, Dr Mark Pfuhl for MST, Andy Cakebread for the ICP‐MS, Dr Tam Bui for the CD measurements and Prof Cláudio Gomes for insightful discussion. This work was supported by a proof of concept scheme by Johnson and Johnson and King's College London, and a Medical Research Council funded PhD studentship for DSP. Metal analyses were performed in the London Metallomics Facility funded by the Wellcome Trust (grant reference 202902/Z/16/Z).
References
- 1. Hogstrand C & Maret W (2016) Genetics of human zinc deficiencies eLS, pp. 1–8. John Wiley & Sons Ltd, Chichester, UK. [Google Scholar]
- 2. Kambe T, Tsuji T, Hashimoto A & Itsumura N (2015) The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol Rev 95, 749–784. [DOI] [PubMed] [Google Scholar]
- 3. Hogstrand C & Fu D (2014) Zinc In Binding, Transport and Storage of Metal Ions in Biological Cells (Maret W. & Wedd AG, eds), pp. 666–694. The Royal Society of Chemistry, Cambridge, UK. [Google Scholar]
- 4. Chimienti F, Devergnas S, Favier A & Seve M (2004) Identification and cloning of a beta‐cell‐specific zinc transporter, ZnT‐8, localized into insulin secretory granules. Diabetes 53, 2330–2337. [DOI] [PubMed] [Google Scholar]
- 5. Davidson HW, Wenzlau JM & O'Brien RM (2014) Zinc transporter 8 (ZnT8) and β cell function. Trends Endocrinol Metab 25, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Solomou A, Philippe E, Chabosseau P, Migrenne‐Li S, Gaitan J, Lang J, Magnan C & Rutter GA (2016) Over‐expression of Slc30a8/ZnT8 selectively in the mouse α cell impairs glucagon release and responses to hypoglycemia. Nutr Metab (Lond) 13, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boesgaard TW, Žilinskait≐ J, Vänttinen M, Laakso M, Jansson P‐A, Hammarstedt A, Smith U, Stefan N, Fritsche A, Häring H et al (2008) The common SLC30A8 Arg325Trp variant is associated with reduced first‐phase insulin release in 846 non‐diabetic offspring of type 2 diabetes patients—the EUGENE2 study. Diabetologia 51, 816–820. [DOI] [PubMed] [Google Scholar]
- 8. Rutter GA & Chimienti F (2014) SLC30A8 mutations in type 2 diabetes. Diabetologia 58, 31–36. [DOI] [PubMed] [Google Scholar]
- 9. Merriman C, Huang Q, Rutter GA & Fu D (2016) Lipid‐tuned zinc transport activity of human ZnT8 protein correlates with risk for type‐2 diabetes. J Biol Chem 291, 26950–26957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flannick J, Thorleifsson G, Beer NL, Jacobs SBR, Grarup N, Burtt NP, Mahajan A, Fuchsberger C, Atzmon G, Benediktsson R et al (2014) Loss‐of‐function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet 46, 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S et al (2007) A genome‐wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885. [DOI] [PubMed] [Google Scholar]
- 12. Lu M & Fu D (2007) Structure of the zinc transporter YiiP. Science 317, 1746–1748. [DOI] [PubMed] [Google Scholar]
- 13. Lu M, Chai J & Fu D (2009) Structural basis for autoregulation of the zinc transporter YiiP. Nat Struct Mol Biol 16, 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anton A, Weltrowski A, Haney CJ, Franke S, Grass G, Rensing C & Nies DH (2004) Characteristics of zinc transport by two bacterial cation diffusion facilitators from ralstonia metallidurans CH34 and Escherichia coli . J Bacteriol 186, 7499–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin JE & Giedroc DP (2016) Functional determinants of metal ion transport and selectivity in paralogous cation diffusion facilitator transporters CzcD and MntE in Streptococcus pneumoniae . J Bacteriol 198, 1066–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coudray N, Valvo S, Hu M, Lasala R, Kim C, Vink M, Zhou M, Provasi D, Filizola M, Tao J et al (2013) Inward‐facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy. Proc Natl Acad Sci USA 110, 2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cherezov V, Höfer N, Szebenyi DME, Kolaj O, Wall JG, Gillilan R, Srinivasan V, Jaroniec CP & Caffrey M (2008) Insights into the mode of action of a putative zinc transporter CzrB in Thermus thermophilus . Structure 16, 1378–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higuchi T, Hattori M, Tanaka Y, Ishitani R & Nureki O (2009) Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein. Proteins 76, 768–771. [DOI] [PubMed] [Google Scholar]
- 19. Zeytuni N, Offer T, Davidov G & Zarivach R (2012) Crystallization and preliminary crystallographic analysis of the C‐terminal domain of MamM, a magnetosome‐associated protein from Magnetospirillum gryphiswaldense MSR‐1. Acta Crystallogr Sect F Struct Biol Cryst Commun 68, 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chabosseau P & Rutter GA (2016) Zinc and diabetes. Arch Biochem Biophys 611, 79–85. [DOI] [PubMed] [Google Scholar]
- 21. Krężel A & Maret W (2006) Zinc‐buffering capacity of a eukaryotic cell at physiological pZn. J Biol Inorg Chem 11, 1049–1062. [DOI] [PubMed] [Google Scholar]
- 22. Hessels AM, Chabosseau P, Bakker MH, Engelen W, Rutter GA, Taylor KM & Merkx M (2015) eZinCh‐2: a versatile, genetically encoded FRET sensor for cytosolic and intraorganelle Zn 2+ imaging. ACS Chem Biol 10, 2126–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maret W (2015) Analyzing free zinc(II) ion concentrations in cell biology with fluorescent chelating molecules. Metallomics 7, 202–211. [DOI] [PubMed] [Google Scholar]
- 24. Wenzlau JM, Liu Y, Yu L, Moua O, Fowler KT, Rangasamy S, Walters J, Eisenbarth GS, Davidson HW & Hutton JC (2008) A common nonsynonymous single nucleotide polymorphism in the SLC30A8 gene determines ZnT8 autoantibody specificity in type 1 diabetes. Diabetes 57, 2693–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blindauer CA (2015) Advances in the molecular understanding of biological zinc transport. Chem Commun (Camb) 51, 4544–4563. [DOI] [PubMed] [Google Scholar]
- 26. Larin D, Mekios C, Das K, Ross B, Yang AS & Gilliam TC (1999) Characterization of the interaction between the Wilson and Menkes disease proteins and the cytoplasmic copper chaperone, HAH1p. J Biol Chem 274, 28497–28504. [DOI] [PubMed] [Google Scholar]
- 27. Micsonai A, Wien F, Kernya L, Lee Y, Goto Y, Réfrégiers M & Kardos J (2015) Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci USA 112, E3095–E3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kocyła A, Pomorski A & Krężel A (2017) Molar absorption coefficients and stability constants of Zincon metal complexes for determination of metal ions and bioinorganic applications. J Inorg Biochem 176, 53–65. [DOI] [PubMed] [Google Scholar]
- 29. Salazar G, Falcon‐Perez JM, Harrison R, Faundez V & Hastrup H (2009) SLC30A3 (ZnT3) oligomerization by dityrosine bonds regulates its subcellular localization and metal transport capacity. PLoS ONE 4, e5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shusterman E, Beharier O, Shiri L, Zarivach R, Etzion Y, Campbell CR, Lee I‐H, Okabayashi K, Dinudom A, Cook DI et al (2014) ZnT‐1 extrudes zinc from mammalian cells functioning as a Zn2+/H+ exchanger. Metallomics 6, 1656–1663. [DOI] [PubMed] [Google Scholar]
- 31. Hoch E, Lin W, Chai J, Hershfinkel M, Fu D & Sekler I (2012) Histidine pairing at the metal transport site of mammalian ZnT transporters controls Zn2+ over Cd2+ selectivity. Proc Natl Acad Sci USA 109, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nishito Y, Tsuji N, Fujishiro H, Takeda TA, Yamazaki T, Teranishi F, Okazaki F, Matsunaga A, Tuschl K, Rao R et al (2016) Direct comparison of manganese detoxification/efflux proteins and molecular characterization of ZnT10 protein as a manganese transporter. J Biol Chem 291, 14773–14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palmiter RD, Cole TB, Quaife CJ & Findley SD (1996) ZnT‐3, a putative transporter of zinc into synaptic vesicles. Proc Natl Acad Sci USA 93, 14934–14939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee S, Hennigar SR, Alam S, Nishida K & Kelleher SL (2015) Essential role for zinc transporter 2 (ZnT2)‐mediated zinc transport in mammary gland development and function during lactation. J Biol Chem 290, 13064–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wenzlau JM, Juhl K, Yu L, Moua O, Sarkar SA, Gottlieb P, Rewers M, Eisenbarth GS, Jensen J, Davidson HW et al (2007) The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci USA 104, 17040–17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Banci L, Bertini I, Ciofi‐Baffoni S, Finney LA, Outten CE & O'Halloran TV (2002) A new zinc‐protein coordination site in intracellular metal trafficking: solution structure of the Apo and Zn(II) forms of ZntA(46‐118). J Mol Biol 323, 883–897. [DOI] [PubMed] [Google Scholar]
- 37. Banci L, Bertini I, Ciofi‐Baffoni S, Poggi L, Vanarotti M, Tottey S, Waldron KJ & Robinson NJ (2010) NMR structural analysis of the soluble domain of ZiaA‐ATPase and the basis of selective interactions with copper metallochaperone Atx1. J Biol Inorg Chem 15, 87–98. [DOI] [PubMed] [Google Scholar]
- 38. Pufahl RA, Singer CP, Peariso KL, Lin SJ, Schmidt PJ, Fahrni CJ, Culotta VC, Penner‐Hahn JE & O'Halloran TV (1997) Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278, 853–856. [DOI] [PubMed] [Google Scholar]
- 39. Watly J, Simonovsky E, Barbosa N, Spodzieja M, Wieczorek R, Rodziewicz‐Motowidlo S, Miller Y & Kozlowski H (2015) African viper poly‐his tag peptide fragment efficiently binds metal ions and is folded into an α‐helical structure. Inorg Chem 54, 7692–7702. [DOI] [PubMed] [Google Scholar]
- 40. Nieba L, Nieba‐Axmann SE, Persson A, Hämäläinen M, Edebratt F, Hansson A, Lidholm J, Magnusson K, Karlsson ÅF & Plückthun A (1997) BIACORE analysis of histidine‐tagged proteins using a chelating NTA sensor chip. Anal Biochem 252, 217–228. [DOI] [PubMed] [Google Scholar]
- 41. Wong WP, Allen NB, Meyers MS, Link EO, Zhang X, MacRenaris KW & El Muayed M (2017) Exploring the association between demographics, SLC30A8 genotype, and human islet content of zinc, cadmium, copper, iron, manganese and nickel. Sci Rep 7, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Carvalho S, Molina‐López J, Parsons D, Corpe C, Maret W & Hogstrand C (2017) Differential cytolocation and functional assays of the two major SLC30A8 (ZnT8) isoforms. J Trace Elem Med Biol 44, 116–124. [DOI] [PubMed] [Google Scholar]
- 43. Hosseini‐Esfahani F, Mirmiran P, Koochakpoor G, Daneshpour MS, Guity K & Azizi F (2017) Some dietary factors can modulate the effect of the zinc transporters 8 polymorphism on the risk of metabolic syndrome. Sci Rep 7, 1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 45. Zor T & Selinger Z (1996) Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem 236, 302–308. [DOI] [PubMed] [Google Scholar]
- 46. Drozdetskiy A, Cole C, Procter J & Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43, W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L et al (2014) SWISS‐MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42, W252–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]


