Abstract
The past decade has been characterized by tremendous progress in the field of the gut microbiota and its impact on host metabolism. Although numerous studies show a strong relationship between the composition of gut microbiota and specific metabolic disorders associated with obesity, the key mechanisms are still being studied. The present review focuses on specific complex pathways as well as key interactions. For instance, the nervous routes are explored by examining the enteric nervous system, the vagus nerve, and the brain, as well as the endocrine routes (i.e., glucagon‐like peptide‐1, peptide YY, endocannabinoids) by which gut microbes communicate with the host. Moreover, the key metabolites involved in such specific interactions (e.g., short chain fatty acids, bile acids, neurotransmitters) as well as their targets (i.e., receptors, cell types, and organs) are briefly discussed. Finally, the review highlights the role of metabolic endotoxemia in the onset of metabolic disorders and the implications for alterations in gut microbiota‐host interactions and ultimately the onset of diseases.
Introduction
Over the past four decades, overweight, obesity, and related metabolic disorders have reached epidemic proportions 1. This phenomenon is the result of biological, behavioral, and environmental complex processes that still must be better deciphered. However, evidence suggests that human evolution, which has taken billions of years of continual interaction with our environment, played a major role in the way we have evolved. Among the environmental factors, intestinal microbes have conferred numerous metabolic and biological functions that we are unable to perform by our own cells. Recent data estimate that humans are colonized by trillions of microbes, and the vast majority of them reside in our gut. This tremendous number of microbial cells represents a ratio of approximately 1:1 between human and microbial cells, or even 1:10 if we take into account only the number of human nucleated cells (i.e., excluding red blood cells) 2. In addition, the gut microbiota harbors a vast number of genes that clearly outnumbers our own genome by at least 100‐fold 3. This vast catalog of genes encodes for specific metabolic activities, allowing microbes to adapt to their environment and eventually the energy sources available. Hence, the gut microbiota is considered a massive “organ” able to perform complex functions and thereby produce a myriad of different metabolites (Figures 1, 2). The current level of knowledge is progressing, with more than 10,000 papers published in 3 years (PubMed search: “gut microbiota”). Indeed, numerous publications have found an association between the microbiota and many diseases (e.g., obesity, diabetes, liver diseases, altered immunity, digestive diseases, cancer, neurodegenerative disorders), but the exact role of the gut microbiota in the onset of diseases remains a matter of debate 4. The microbial diversity (i.e., species richness of the microbiota) is another concept that has been linked with the metabolic functions of the gut bacteria. Indeed, low bacterial richness is consistently appearing in the literature as a risk factor for different diseases (e.g., obesity, low‐grade inflammation, intestinal inflammation) 5 (for review 6). Aside from the microbial diversity, evidence also suggests that we can classify subjects on the basis of the number of bacterial genes that they harbor in their gut (i.e., microbial gene richness). More precisely, Le Chatelier et al. identified a bimodal distribution of microbial genes leading to the clustering of subjects as either low gene count or high gene count according to the number of genes present in the microbiota 5. This also seems to be important for the susceptibility to respond to dietary intervention devoted to improving metabolic parameters, since dietary restriction in patients with overweight or obesity is less efficient in low gene count than in high gene count individuals in terms of improving insulin sensitivity and lowering cholesterol and inflammation 7.
Figure 1.
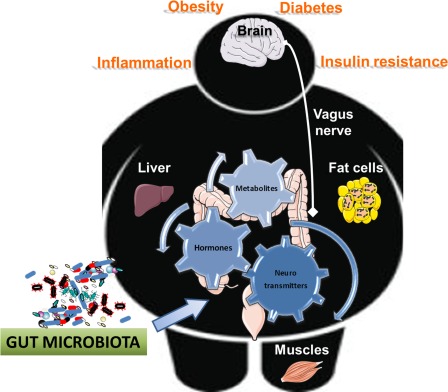
Gut microbiota is involved in a complex interaction with host metabolism. The gut microbiota is involved in complex interaction between food (i.e., dietary ingredients changing the microbiota) and consequently the metabolite produced. Gut bacteria also contribute to the regulation of the production of neurotransmitters, different hormones, and finally host metabolism. Numerous data suggest that the composition and the activity of the gut microbes are responsible for the protection or the onset of diseases associated with obesity, such as insulin resistance, low‐grade inflammation, fatty liver, and diabetes. Thus, the gut and microbes are communicating with all the organs via specific metabolites, hormones, and neurotransmitters, acting through direct or indirect pathways (i.e., the vagus nerve).
Figure 2.
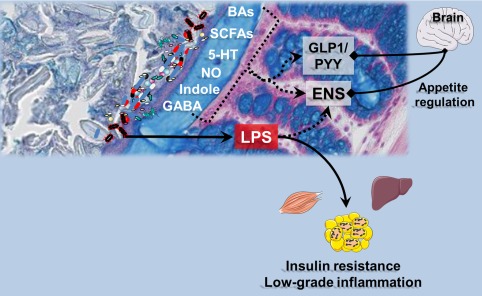
Mechanisms of interaction between bacterial products and host organs: the role of the gut lining. Numerous metabolites are produced upon the metabolic activity of the gut microbes. Most of them are chemically similar to those produced by the host cells (i.e., nitric oxide [NO]; gamma‐aminobutyric acid [GABA]; serotonin [5‐hydroxytriptamine, (5‐HT)]; short chain fatty acids [SCFAs], and indoles), whereas others result from the chemical transformations of host molecules by microbes, namely the bile acids (BAs). All these molecules are recognized by the host cells and may act on specific receptors (both nuclear and membrane receptors) or eliciting the secretion of other hormonal signals such as the gut peptides glucagon‐like peptide‐1 (GLP‐1) or peptide YY (PYY) that both act on energy metabolism by acting through nervous routes or blood relay.
Translocation of lipopolysaccharides (LPS) through the gut lining is a hallmark of obesity, diabetes, and related disorders. Leakage of LPS into the blood triggers low‐grade inflammation and thereby affects liver, adipose tissue, and muscle metabolism. In addition, those endotoxins can alter the activity of the enteric nervous system (ENS) as well as the gut‐brain axis via the vagus nerve, hence affecting appetite regulation.
Although a direct causality is not completely proven, various data support the concept that strong relationships exist among the gut microbiota composition, its metabolic activity (e.g., metabolite production), and its host metabolism (Figure 1). In other words, the activity of the gut bacteria may influence not only our health but also the risk of developing diseases 8, 9.
In this review, we will focus our attention on what is known thus far regarding key microbial metabolites and the specific mechanisms by which they interact with host cells in the context of obesity and metabolic disorders. We will also describe specific pathways by which gut microbes regulate physiological processes, including the nervous, immune, and endocrine routes.
Brief Overview of Data Suggesting Link Between Microbes and Obesity
The link between the gut microbiota and body weight has been extensively studied. Over the past 15 years, numerous studies have shown that changing the gut microbiota of both rats and mice by using, for example, prebiotics 10 reduces body weight gain and fat mass development and improves glucose metabolism 11, 12, 13. However, it was only in 2006 that Turnbaugh et al. provided the first proof of concept using germ‐free (GF) mice (i.e., mice that are devoid of any microorganisms), demonstrating that transferring the microbiota from mice with genetic obesity (i.e., ob/ob) to GF mice induced a higher body weight and fat mass gain 14. This link has been further observed with the gut microbiota from rodents with diet‐induced obesity 15, 16 and even with the microbiota from humans with obesity 17. It is important to highlight that although both body weight and fat mass were increased in all these experiments, the magnitude of the effect observed in the GF‐recipient mice receiving the microbiota from donors with obesity remains moderate as compared to diet‐induced or genetic‐induced obesity (i.e., ranging from 10%‐20% of difference between lean mice vs. mice with obesity) 15, 16, 17. For example, in the seminal paper by Turnbaugh et al., the recipient mice gained 47% of fat mass when receiving the microbiota from ob/ob mice, whereas they gained 27% with the microbiota from lean mice (i.e., a mean of 20% difference between groups). Conversely, in the study from Ridaura et al., 17 the same kind of experiment was associated with a 7% to 14% fat mass gain (ob/ob donors). A similar difference of around 10% increased fat mass was observed when comparing the impact of the microbiota from a human twin with obesity versus the lean twin 17.
Similarly, reducing the abundance of most intestinal microbes by using broad‐spectrum antibiotics also partially protects mice against diet‐induced obesity and related metabolic disorders (Figure 1) 18, 19, 20. These findings suggest a causal link between microbes and the onset of body weight gain.
Despite this series of animal experiments showing proof of concept of a clear association between microbes and host metabolism, the vast majority of the results currently published in the literature have been obtained by comparing the composition of the microbiota and/or microbial metabolites between lean subjects and subjects with obesity and/or diabetes. Nevertheless, these types of observations are consistent among cohorts and even between very different geographic situations. However, there are still numerous discussions regarding the exact composition of a healthy microbiota 5, 14, 21, 22, 23. Should we focus on the general composition only at the phyla levels (i.e., the so‐called Firmicutes/Bacteroidetes ratio) or at deeper levels (i.e., the genus and species level)? Is it more relevant to explore the metabolic capacities of the intestinal microbiota and, eventually, the metabolites produced? Aside from these considerations, another major issue is the large technical variations observed in metagenomic analysis. Therefore, it is urgent to standardize the sampling of specimens, the DNA extraction protocols, and the bioinformatics tools, not only in order to achieve sufficient reproducibility between studies but also to generate comparable data between cohorts for further meta‐analysis and an eventually interesting outcome for health 24, 25
At the current state of knowledge, there is no clear answer to these questions, but the overall scientific community has recommended obtaining a more comprehensive view of the situation by assessing not only the composition but also the metabolites (Figure 2). Thus, for this part of the review, there are still numerous factors and mechanisms that merit consideration and will be further discussed in proceeding sections.
Key Mechanisms by Which Microbes Communicate with Host Organs
Nervous routes and endocrine routes
Enteric nervous system
The gastrointestinal tract is densely innervated by intrinsic and extrinsic neurons: the differentiation relies on the localization of the soma of the neurons. The enteric nervous system (ENS) is composed of various types of neurons, including intrinsic primary afferent neurons and inter‐ and motor neurons 26. These neurons are in close proximity and in contact with spinal and vagal afferent nerves that send intestinal information to the brain. In addition to the well‐known nerve alteration observed in type 2 diabetes, the alteration specifically in the ENS observed during obesity and diabetes has an impact on the control of food intake and metabolism. In fact, the gut is considered a major partner that influences feeding behavior via the ENS. Numerous papers have described the actual cross talk among gut hormones, the ENS, and microbial factors to control digestive motility and food intake 27, and evidence suggests that alterations in the gut‐brain axis are associated with eating disorders 28 (Figure 2); this association will be further detailed in this review.
Regarding the regulation of glucose metabolism, the alteration in the ENS in the duodenum of mice with obesity participates in the development of a type 2 diabetic state 29. The mechano‐detection of duodenal hyper‐contractility then sends an aberrant message to the brain that favors insulin resistance in tissues and participates in the hyperglycemic phenotype observed in this pathology. Therefore, targeting the ENS to improve glucose tolerance, insulin resistance, and hyperglycemia is now considered a new therapeutic perspective 30.
The relationship between the gut microbiota and ENS neurons is relatively complex. First, the microbiota can influence the development of the ENS 31, and this has consequences on ENS activity and neurochemistry (such as neuronal subpopulations). Second, gut bacteria can use different modes of communication to talk with ENS neurons, including a direct “sensing” with intrinsic primary afferent neurons or the release of numerous bacterial messengers (e.g., neurotransmitters, bioactive lipids, gaseous factors) 26. Along those lines, it is worth noting that the immune cells infiltrating the gut epithelium may also communicate with the microbiota 32. For example, Monteiro‐Sepulveda et al. showed that the abundance of a specific T cell (i.e., CD8αβ) in the jejunum correlated with BMI, steatosis, or an altered lipid metabolism. The authors suggest that this may also contribute to inflammation and insulin resistance in the jejunum, with eventually systemic effect 32.
Is there a link between the gut microbiota, the ENS, and obesity? Although the link seems obvious, a direct relationship between the gut microbiota, the ENS, and obesity has never been clearly demonstrated. In addition to gut dysbiosis, mice with obesity present an alteration in myenteric neuron plasticity 33 and sensitivity 34, which could contribute to the gut dysfunctions (i.e., alternating phases of diarrhea and constipation) and hyperglycemia observed in patients with obesity and diabetes. Furthermore, evidence shows that these phenotypic characteristics (e.g., dysbiosis, alteration of gut motility, hyperglycemia) are exacerbated during aging. As demonstrated by Stenkamp‐Strahm et al. 35, aging is associated with an increase in excitatory neuronal markers, which could explain intestinal hyper‐contractility. Dysmotility of the colon during aging could also be explained by the development of fat deposition in the tunica muscularis of intestinal smooth muscle cells, which decreases the number of myenteric neurons that express the neuronal nitric oxide (NO) synthase enzyme 36. The link between obesity and gut microbiota is well established, but researchers have to focus on the capacity of the gut microbiota and its releasing factors to target the ENS in order to propose novel approaches to treat obesity and its associated phenotypes: namely, increase in food intake, intestinal dysmotility, and type 2 diabetes. However, although the link between colonic gut microbiota and the ENS is easily plausible, one may not fully explain the impact of the ENS on glucose absorption or the arrival of nutrients in the duodenal part. For instance, in humans, numerous factors, such as the nutrient composition of the diet and the hormonal response, strongly influence the gastric emptying, which in turn can affect the overall glycemic profile as well as the appetite sensation. In addition to ENS neurons, the cellular link between gut microbiota and obesity could be the enteric glial cells (EGCs). In fact, papers suggest that EGCs exert pleiotropic effects throughout the whole body, which could imply various roles in numerous pathologies, such as inflammatory bowel diseases, Parkinson disease, and obesity 37. Mice with obesity fed a high‐fat diet presented a significantly reduced number of S100 beta glial cells in the mucosal and submucosal plexuses of the duodenum 35. During development, the appearance of an EGCs network correlates with the maturation of gut microbiota 38. Similar to that observed in enteric neurons, there are multiple forms of communication between gut microbiota and EGCs. In a very interesting review, Kabouridis et al. 38 explained these different possibilities. In summary, EGC activity could be modified by bacterial metabolites and by epithelial or immune factors (which are released in response to bacterial recognition by epithelial cells and immune cells, respectively). Deciphering the cross talk among gut microbiota, EGCs, and obesity is thus of major importance.
Glucagon‐like peptide‐1, peptide YY, and endocannabinoids
In this part of the review, we briefly discuss the role of key hormones and bioactive lipids that might be involved in the microbiota and the peripheral axis. However, several recent reviews have covered the modulation of these peptides and are mentioned here for review 39, 40, 41.
Glucagon‐like peptide‐1 (GLP‐1) is a key endocrine factor that could participate in the control of the gut‐brain axis by gut microbiota because of its location (i.e., released by intestinal L cells). In the middle of the 2000s, Cani et al. showed in rodents that modulation of gut microbiota by using specific prebiotics such as oligofructose improved glucose metabolism via a mechanism dependent on GLP‐1 12. The impact of prebiotics on GLP‐1 in humans will not be discussed in the present review but is discussed elsewhere 39, 42. In addition to this discovery, whether intestinal GLP‐1 can reach the brain to affect food intake and metabolism is still a matter of debate, but it is clear that GLP‐1 has a potential anorexigenic effect in humans with obesity after bariatric surgery 43. However, whether the appetite and the glycemic impact observed after bariatric surgery are mediated by only the hormone GLP‐1 remains a matter of discussion 44. Furthermore, GLP‐1 receptors are expressed on ENS neurons 45. In addition to its direct central effect, GLP‐1 could act on ENS neurons to modify the gut‐brain axis to control food intake 46 and glucose metabolism 47 (Figure 2).
Similar to GLP‐1, the peptide YY (PYY) is released in response to nutrients by enteroendocrine cells and is known to reduce food intake and to increase energy expenditure by its actions in the brain. It is worth noting that the modulation of gut microbiota also has an impact on PYY release, mainly via the activation of mechanisms dependent on specific G protein–coupled receptors (GPRs), such as GPR43 (also known as FFAR2) or GPR41 (FFAR3) 48. In turn, PYY can also exert its effects on the brain via an endocrine route or via the ENS. Y1 receptors are located in myenteric and submucosal neurons 49, whereas Y2 receptors are located only in myenteric neurons (Figure 2) 50. The importance of PYY in obesity is reinforced by the suggestion to use GLP‐1 analogues in combination with PYY to reduce food intake in humans with obesity.
In the context of energy homeostasis, the endocannabinoid system (ECS) plays a major role. Endocannabinoids (eCBs) are bioactive lipids that are synthesized in and exert their action on several organs involved in metabolism and appetite regulation 41. Depending on the action exerted by eCBs on the intestinal mucosa, they can be clustered as a “gate opener” (anandamide) and “gate keeper” (palmitoylethanolamine, 2‐oleoylglycerol) 41. A pioneer study from Muccioli et al. demonstrated for the first time that gut microbiota can modulate intestinal eCB tone. An “obesity microbiota” is associated with an increased intestinal level of anandamide, thus increasing gut permeability 20. The daily administration of a key bacterium, Akkermansia muciniphila, was found to reverse diet‐induced obesity by a mechanism associated with increased intestinal levels of eCBs that control inflammation, the gut barrier, and gut peptide secretion 51. On the other hand, recent evidence suggests that ECS from adipose tissue influence the composition of gut microbiota. For example, selective deletion of the eCB‐synthesizing enzyme N‐acylphosphatidylethanolamine in adipose tissue induces obesity, glucose intolerance, altered lipid metabolism, and adipose tissue inflammation. These metabolic alterations were associated with a shift in gut microbiota composition that was reproduced in GF mice after the transfer of microbiota 52. To better unravel bidirectional ECS‐gut microbiota cross talk, further studies evaluating tissue‐specific modulation of the ECS would be of major interest. Notably, the gut microbiota itself is also able to produce bioactive compounds mimicking human eCBs that are able to modulate metabolic hormones and glucose homeostasis 53.
Metabolites produced by gut microbiota and acting as signaling molecules
Short chain fatty acids
Short chain fatty acids (SCFAs) are organic fatty acids containing two to six atoms of carbon and are produced in the cecum and in the colon of the host by the microbiota following the fermentation of nondigestible dietary fibers, proteins, and glycoproteins. Acetate, propionate, and butyrate represent 95% of SCFAs, and they are the most studied bacterial metabolites 54.
Bacterial SCFAs locally modulate the physiology of the large intestine, but they can also be absorbed (only 5%‐10% are excreted in feces) 55 and control the metabolism of other organs (such as adipose, liver, muscle, and brain tissue), thus influencing the energetic homeostasis of the host, including appetite regulation 56. Of note, aside from the activation of GPR43 or GPR41, one of the primary roles of SCFAs is the modulation of the activity of histone deacetylase, and SCFAs are used by the colonic cells as an energy source 56, 57 (Figure 2) (for review 56, 58).
However, additional findings suggest that SCFAs are also involved in the modulation of colonic physiology. For example, SCFAs induce colon motility. Fukumoto et al. demonstrated that intracolonic administration of SCFAs significantly accelerated colonic transit and that this effect was reduced after administration of a 5‐hydroxytryptamine (5‐HT) 3 receptor antagonist 59. In this same work, it was also shown ex vivo that SCFA administration increased the luminal release of serotonin (5‐HT) (Figure 2). (The role of serotonin will be further discussed under “Neurotransmitters”). It has also been suggested that butyrate, but not acetate or propionate, has a colonic prokinetic effect by increasing the proportion of cholinergic (excitatory) myenteric neurons; it seems that the change in neuronal phenotype is associated with increased acetylation of histone 3 60. In addition to modulating motility, SCFAs modulate colonic secretion in response to 5HT: the gut microbiota downregulates 5‐HT3 expression via acetate production, thus lowering the host secretory response 61.
Recently, many studies have focused on the role played by SCFAs in the gut‐brain axis. The finding that the GPR41 receptor is expressed in sensory ganglia (afferent fibers) and in autonomic ganglia (efferent fibers) strongly supports the role played by SCFAs in the gut‐brain axis (Figure 1) 62. De Vadder et al. 63 also showed that butyrate and propionate activated intestinal gluconeogenesis in the colon via complementary mechanisms. Butyrate increased the expression of intestinal gluconeogenesis enzymes through a cAMP‐dependent mechanism, while the same genes were activated by propionate via a gut‐brain axis involving GPR41 expressed on periportal neural afferents 63.
It is worth noting that acetate can also influence metabolism via a gut‐brain axis. Indeed, Frost et al. demonstrated that fermentable carbohydrates such as inulin altered hypothalamic neuronal activity specifically in the arcuate nucleus (ARC). They demonstrated that intraperitoneal administration of acetate or acetate directly produced by the gut microbiota through fermentation entered the hypothalamus and reduced appetite by increasing the expression of anorectic pro‐opiomelanocortin and suppressing agouti‐related peptide 64. In contrast with those findings, Perry et al. 65 showed that acetate production from an altered gut microbiota increased glucose‐stimulated insulin secretion, ghrelin secretion, hyperphagia, and other alterations in the metabolism associated with obesity by activating parasympathetic neurons. One of the potential discrepancies between these two papers is that Perry et al. did not explore the fact that the microbiota rapidly transform acetate into other SCFAs. In addition, the protocol used may warrant discussion, since using 10 days of intragastric infusion of SCFAs probably changed both the microbiota and the overall SCFA profile in the gut, but this was not investigated. Indeed, the ratios of SCFAs are also important to consider: if the ratio of acetate to propionate is changed, for example, the real abundance of acetate can be increased. In other words, it remains unclear whether the observed effects are attributable to the acetate itself or to other products of the cross feeding. Another key factor is the route of administration; comparing the SCFAs produced by fermentation versus oral administration can strongly influence the outcome. Thus, acetate administration for 10 days by using the intragastric route suggests that acetate will be rapidly absorbed in the upper gut, and this may also change hormonal routes (via the GPRs). Thus, the question of whether changes in secretion of specific hormones are due to luminal mechanisms (in the intestine/stomach) or blood‐driven mechanisms is not yet resolved.
In any case, these two observations shed light on the important role played by the location of action of acetate in modulating the different effects on host metabolism 66.
Bile acids
Bile acids (BAs) also represent an important class of metabolites modulated by the gut microbes. Primary BAs are endogenous molecules synthesized in hepatocytes starting from cholesterol and subsequently conjugated with the amino acid glycine (mainly in humans) or taurine (in mice). BAs accumulate in the gallbladder and are released in the small intestine after a meal, where they mediate the absorption of dietary fat and lipophilic vitamins. In the intestine, primary BAs are converted into secondary BAs following microbiota‐driven deconjugation and dehydroxylation. Of the total BA pool, approximately 90% is absorbed in the ileum and is redirected to the liver where they will be recycled (enterohepatic circulation), while a small part of these remaining absorbed BAs enters the systemic circulation from which they can reach the brain (Figure 2) 67.
Unconjugated and conjugated BAs can cross the blood‐brain barrier by diffusion or active transport, respectively 68; both types of metabolites were detected in the human and rodent brain in healthy and pathological conditions. Moreover, nuclear farnesoid X receptor and Takeda GPR 5 (TGR5) (also known as G protein–coupled BA receptor 1) are abundantly expressed in the brain 67. Taken together, those observations suggest that BAs could directly act in the brain.
On the other hand, several studies suggest that BAs can indirectly modulate host metabolism by signaling to the central nervous system (CNS). Intestinal activation of farnesoid X receptors by BAs induces the intestinal expression of fibroblast growth factor 19 (FGF19), which can pass through the blood‐brain barrier and enter the brain, whereas FGF19 receptor has been detected in the hypothalamus 69. In mice, ARC responds to FGF19 administered in the periphery, whereas intracerebroventricular administration suppresses agouti‐related peptide/neuropeptide Y neuronal activity in addition to improving glycemic status 70. Moreover, BAs indirectly send signals to the CNS via the TGR5–GLP‐1 pathway. The activation of TGR5 on the basolateral side of enteroendocrine L cells stimulates GLP‐1 release, which can exert a positive effect on host metabolism signaling via the vagus nerve 71 or activate the GLP‐1 receptor expressed in the brain (Figure 2) 72. From a more macroscopic perspective, it is also important to highlight the bidirectional interaction existing between BAs and gut microbiota composition 73. A study by Mertens provides a complete review of BA signaling to the CNS 67.
Neurotransmitters
It is often suggested that the levels of most neurotransmitters found in the gut are equal to or exceed those found in the brain, thus highlighting that microbial production of neurotransmitters may have a major role in bacteria‐neuron communication 74. A growing number of studies have revealed that the gut microbiota modulates the concentration of neurotransmitters in the periphery and in the brain, either directly producing/consuming the metabolites or modulating host biosynthetic pathways (Figures 1, 2) 74. However, most of the studies have been done in rodents, and therefore, although mechanistically interesting, the direct translation into humans warrants further investigation.
Serotonin (5‐HT)
In addition to what was previously described in this review regarding the role of serotonin in intestinal motility and secretion, this neurotransmitter regulates several other physiological functions.
Serotonin is synthesized in the brain, but it is also produced in enterochromaffin cells and enteric nerves 75. Moreover, it was recently confirmed in vivo that gut microbes promote colonic 5‐HT production by increasing the mRNA and protein expression of tryptophan hydroxylase (the rate‐limiting enzyme for 5‐HT synthesis) in enterochromaffin cells 76.
The precursor of serotonin is tryptophan, an essential amino acid that must be introduced by the diet. The gut microbiota directly uses tryptophan, reducing its availability to the host 77, and different bacterial species metabolize tryptophan into indole. On the other hand, bacteria can synthesize tryptophan or even produce 5‐HT from tryptophan (Figure 2) (reviewed in 77).
Regarding the exact role of the microbiota in the overall pool of serotonin, there is contrasting evidence. For example, studies show that GF mice have higher plasma levels of tryptophan and serotonin 78, 79, whereas other evidence shows that the gut microbiota promotes serotonin synthesis in enterochromaffin cells 76. Moreover, contrasting evidence related to the central level of serotonin in GF rodents has been reported 80, 81 (for review 77).
Catecholamines
There are three main catecholamines: adrenalin and noradrenalin are produced by postganglionic sympathetic neurons or by chromaffin cells in the adrenal gland and are responsible for the “fight or flight” autonomic response, whereas dopamine is mainly synthesized in the brain and is involved in the reward system. At the level of the gut, catecholamines modulate gastrointestinal blood flow, nutrient absorption, and gut motility 75. The gut microbiota contributes to catecholamine levels and availability (Figures 1, 2). Compared to specific pathogen‐free mice, GF mice have lower levels of catecholamines in the cecum, and > 90% of them are biologically inactive 82.
Gamma‐aminobutyric acid
Gamma‐aminobutyric acid (GABA) is the principal inhibitory neurotransmitter of the vertebrate CNS and is produced in neurons following the action of glutamate decarboxylase on glutamic acid. It has been demonstrated that certain strains of Lactobacillus produce GABA by this same biosynthetic pathway because they express glutamate decarboxylase 83, 84, 85. More recently, Lactobacillus brevis and Bifidobacterium dentium were identified as the most efficient GABA producers among all human‐derived strains cultured 85. Interestingly, in mice, chronic administration of Lactobacillus rhamnosus modulates the central expression of GABA receptors in a region‐specific manner and reduces anxiety‐ and depression‐related behavior; both effects were lost after vagotomy 84, suggesting that the intestinal microbiota could also modulate central neurotransmission in the host (Figure 2). On the other hand, in support of a bidirectional interplay, growing evidence suggests that some bacteria of the human microbiota could sense and probably respond to intestinal GABA or glutamate 86.
NO: gaseous neurotransmitters
Enteric bacteria can produce gaseous neurotransmitters such as NO 87 through the reduction of nitrite or by bacterial NO synthase 88. NO is known to have important local effects; however, a previous report suggested a microbial‐induced gut‐to‐brain mechanism for this neurotransmitter (for review 26) (Figure 2).
Caseinolytic protease B: specific bacterial component
In 2014, Fetissov's team identified a protein expressed by commensal bacteria (caseinolytic protease B [ClpB]) as an antigen mimetic of an anorexigenic protein expressed by the host α‐melanocyte‐stimulating hormone [MSH] 89. In mice, administration of Escherichia coli–expressing ClpB protein stimulated host production of α‐MSH autoantibody and affected food intake, whereas this effect was not observed in mice treated with E. coli lacking ClpB expression 89. Moreover, the same team demonstrated that ClpB directly modulated appetite‐controlling pathways. In mice, acute injection of microbial protein extract containing ClpB increased the activation of pro‐opiomelanocortin neurons in the ARC, which is known to decrease food intake. Other brain regions known to modulate appetite were also involved 90.
Aside from the markers and metabolites mentioned here above, it is clear that numerous others exist and have been correlated with metabolic parameters, such as for instance cresol, indole, hippurate, and many other molecules that may be also directly influenced by our dietary habits 8, 91, 92, 93, 94. Therefore, the need to accurately investigate the nutrient ingestion in these kinds of studies should be strongly highlighted 95, 96.
Gut Microbes and Metabolic Endotoxemia: 10 Years Later
Currently, there is no controversy regarding the statement that a low‐grade inflammatory tone is characteristic of obesity and metabolic disorders. Although this inflammation may come from different potential origins, a profound change in the gut microbiota composition was found to be strongly correlated with higher blood levels of a constituent found in Gram‐negative bacteria; that is, the lipopolysaccharide (LPS) 97. Cani et al. demonstrated that the translocation of LPS was higher in both mice with genetic obesity and mice with diet‐induced obesity 18, 97 (Figure 2). This phenomenon was called “metabolic endotoxemia,” in contrast to endotoxic shock, in which the LPS levels are far higher (i.e., 2 to 3 log higher). Using genetic tools (i.e., toll‐like receptor [TLR] knockout mice [CD14 or TLR‐4]), it was unequivocally demonstrated that LPS was a triggering factor involved in the onset of inflammation and eventually insulin resistance, possibly by altering the mechanisms of food intake regulation and fat mass development 18, 97, 98, 99. Since these first observations in rodent models, human data have confirmed the impact of high‐fat diet feeding on the onset of metabolic endotoxemia and have also shown an association among obesity, metabolic disorders, and circulating levels of LPS 100, 101, 102, 103, 104, 105. However, further studies in humans are warranted to determine whether this is always due to an active gut permeability or to other specific mechanisms 32, 103, 106, 107, 108, 109, 110. These observations clearly support the hypothesis that bacterial components may regulate metabolic functions.
Interestingly, TLRs that recognize bacterial products are also expressed on colonic epithelia and colocalize with enteroendocrine cells 111.
More importantly, TLRs are expressed on vagal afferent neurons and nodose ganglia, suggesting a possible direct sensing of bacterial products by visceral afferent nerves but also supporting additional mechanisms involved in the gut‐brain axis 112. For the sake of clarity, the vagus nerve is the X cranial nerve and innervates the larynx, pharynx, and visceral organ; it is composed of motor (descending) and sensory (ascending) fibers whose cellular bodies are located in the nodose ganglia. Sensory vagal inputs terminate in the nucleus of the solitary tract in the hindbrain. Intestinal vagal afferents, which are ninefold more abundant than efferents, terminate in the mucosal and muscular layers of the gut 113.
Thus, in pathological conditions with increased intestinal permeability (i.e., obesity, metabolic disorders, intestinal inflammation), bacterial metabolites could directly reach nerve endings. For example, de La Serre et al. 112 previously found that chronic LPS exposure (which mimics metabolic endotoxemia) induced higher food intake in rats (Figure 2). One of the key mechanisms was reduced vagal afferent leptin signaling after metabolic endotoxemia, which clearly shows that bacterial components can markedly affect the vagal afferent neurons and their functions 112. Conversely, in a “healthy gut,” bacterial metabolites mainly modulate the activity of epithelial transducers, such as enterochromaffin cells and enteroendocrine cells, which release mediators on the basal side that are able to activate nerve endings. Nevertheless, only a few animal studies have shown that the effect of probiotics is abolished after vagotomy 84, 114, supporting the hypothesis that microbes communicate to the brain via the vagus nerve. In this context, several studies have evaluated the effect of probiotics on stress‐ and anxiety‐related behavior 113, although we currently lack studies on the vagal‐mediated gut‐brain axis in the field of metabolic disorders (Figure 2).
In this context, two recent papers from the same team demonstrated that diet‐induced changes in microbiota altered the vagal gut‐brain communication by reducing vagal terminals in the gut and in the nucleus of the solitary tract and increasing inflammation markers in the nodose ganglia 115, 116.
Conclusion
Currently, several important mechanisms have been highlighted and clearly demonstrate that the gut microbiota communicates with host organs by using several pathways. For example, the role of metabolites produced by the degradation of nutrients or host products (e.g., SCFA, serotonin, BAs, bioactive lipids) and the production of specific hormones stimulated by the action of such metabolites on key receptors and, eventually, specific nervous routes have been described. Although not completely elucidated, the link between microbes and the onset of specific diseases such as obesity has led to the development of key methods devoted to using nutrients, probiotics, or next‐generation approaches to prevent or cure diseases.
Thus, this review offers additional mechanisms for important findings explaining how gut bacteria and metabolites or by‐products may contribute to the development of obesity and related metabolic disorders.
Acknowledgments
We thank Clara Depommier for the histological picture used in Figure 2.
Funding agencies: PDC was the recipient of grants from Fonds National de la Recherche Scientifique (FNRS). This work was supported by the Fonds de la Recherche Scientifique (FRS)/FNRS for the Fonds de Recherche Fondamentale Stratégique/Walloon Excellence in Life Sciences and Biotechnology Institute (FRFS/WELBIO) under grant WELBIO‐2017‐CGR. This work was supported in part by the Funds Baillet Latour (Grant for Medical Research 2015). PDC was the recipient of a European Research Council Starting Grant in 2013 (336452‐ENIGMO). CK was the recipient of grants from the Société Française de Nutrition (SFN), the Fondation Recherche Médicale (FRM) (ING20150532586), and the Société Francophone du Diabète (Allocations Exceptionnelles 2016).
Disclosure: PDC is the inventor on patent applications dealing with the use of Akkermansia muciniphila and its components in the treatment of obesity and related disorders. PDC is cofounder of A‐Mansia biotech SA. MR is a research fellow at FRS/FNRS, and PDC is a senior research associate at FRS‐FNRS. CK declared no conflict of interest.
References
- 1. GBD 2015 Obesity Collaborators . Health effects of overweight and obesity in 195 countries over 25 years. New Engl J Med 2017;377:13‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016;164:337‐340. [DOI] [PubMed] [Google Scholar]
- 3. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cani PD. Gut microbiota ‐ at the intersection of everything? Nat Rev Gastroenterol Hepatol 2017;14:321‐322. [DOI] [PubMed] [Google Scholar]
- 5. Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541‐546. [DOI] [PubMed] [Google Scholar]
- 6. Dore J, Blottiere H. The influence of diet on the gut microbiota and its consequences for health. Curr Opin Biotechnol 2015;32C:195‐199. [DOI] [PubMed] [Google Scholar]
- 7. Dao MC, Everard A, Aron‐Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 2016;65:426‐436. [DOI] [PubMed] [Google Scholar]
- 8. Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut 2016;65:2035‐2044. [DOI] [PubMed] [Google Scholar]
- 9. Bachmann R, Leonard D, Delzenne N, Kartheuser A, Cani PD. Novel insight into the role of microbiota in colorectal surgery. Gut 2017;66:738‐749. [DOI] [PubMed] [Google Scholar]
- 10. Gibson GR, Hutkins R, Sanders ME, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 2017;14:491‐502. [DOI] [PubMed] [Google Scholar]
- 11. Cani PD, Dewever C, Delzenne NM. Inulin‐type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon‐like peptide‐1 and ghrelin) in rats. Br J Nutr 2004;92:521‐526. [DOI] [PubMed] [Google Scholar]
- 12. Cani PD, Knauf C, Iglesias MA, Drucker DJ, Delzenne NM, Burcelin R. Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon‐like peptide 1 receptor. Diabetes 2006;55:1484‐1490. [DOI] [PubMed] [Google Scholar]
- 13. Cani PD, Possemiers S, Van de WT, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP‐2‐driven improvement of gut permeability. Gut 2009;58:1091‐1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027‐1031. [DOI] [PubMed] [Google Scholar]
- 15. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet‐induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008;3:213‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Everard A, Geurts L, Caesar R, et al. Intestinal epithelial MyD88 is a sensor switching host metabolism towards obesity according to nutritional status. Nat Commun 2014;5:5648. doi:10.1038/ncomms6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013;341:1241214. doi:10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia‐induced inflammation in high‐fat diet‐induced obesity and diabetes in mice. Diabetes 2008;57:1470‐1481. [DOI] [PubMed] [Google Scholar]
- 19. Carvalho BM, Guadagnini D, Tsukumo DM, et al. Modulation of gut microbiota by antibiotics improves insulin signaling in high‐fat fed mice. Diabetologia 2012;55:2823‐2834. [DOI] [PubMed] [Google Scholar]
- 20. Muccioli GG, Naslain D, Backhed F, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol 2010;6:392. doi:10.1038/msb.2010.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 2014;32:834‐841. [DOI] [PubMed] [Google Scholar]
- 22. Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513:59‐64. [DOI] [PubMed] [Google Scholar]
- 23. Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Costea PI, Zeller G, Sunagawa S, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol 2017;35:1069‐1076. [DOI] [PubMed] [Google Scholar]
- 25. Sinha R, Abu‐Ali G, Vogtmann E, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol 2017;35:1077‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cani PD, Knauf C. How gut microbes talk to organs: the role of endocrine and nervous routes. Mol Metab 2016;5:743‐752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cuomo R, D'Alessandro A, Andreozzi P, Vozzella L, Sarnelli G. Gastrointestinal regulation of food intake: do gut motility, enteric nerves and entero‐hormones play together? Minerva Endocrinol 2011;36:281‐293. [PubMed] [Google Scholar]
- 28. Mayer EA. Gut feelings: the emerging biology of gut‐brain communication. Nat Rev Neurosci 2011;12:453‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fournel A, Drougard A, Duparc T, et al. Apelin targets gut contraction to control glucose metabolism via the brain. Gut 2017;66:258‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weber C. Neurogastroenterology: improving glucose tolerance via the gut‐brain axis. Nat Rev Gastroenterol Hepatol 2016;13:4. doi:10.1038/nrgastro.2015.204 [DOI] [PubMed] [Google Scholar]
- 31. Hyland NP, Cryan JF. Microbe‐host interactions: influence of the gut microbiota on the enteric nervous system. Dev Biol 2016;417:182‐187. [DOI] [PubMed] [Google Scholar]
- 32. Monteiro‐Sepulveda M, Touch S, Mendes‐Sa C, et al. Jejunal T cell inflammation in human obesity correlates with decreased enterocyte insulin signaling. Cell Metab 2015;22:113‐124. [DOI] [PubMed] [Google Scholar]
- 33. Stenkamp‐Strahm CM, Nyavor YE, Kappmeyer AJ, Horton S, Gericke M, Balemba OB. Prolonged high fat diet ingestion, obesity, and type 2 diabetes symptoms correlate with phenotypic plasticity in myenteric neurons and nerve damage in the mouse duodenum. Cell Tissue Res 2015;361:411‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reichardt F, Baudry C, Gruber L, et al. Properties of myenteric neurones and mucosal functions in the distal colon of diet‐induced obese mice. J Physiol 2013;591:5125‐5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stenkamp‐Strahm C, Patterson S, Boren J, Gericke M, Balemba O. High‐fat diet and age‐dependent effects on enteric glial cell populations of mouse small intestine. Auton Neurosci 2013;177:199‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jo HJ, Kim N, Nam RH, et al. Fat deposition in the tunica muscularis and decrease of interstitial cells of Cajal and nNOS‐positive neuronal cells in the aged rat colon. Am J Physiol Gastrointest Liver Physiol 2014;306:G659‐G669. [DOI] [PubMed] [Google Scholar]
- 37. Neunlist M, Rolli‐Derkinderen M, Latorre R, et al. Enteric glial cells: recent developments and future directions. Gastroenterology 2014;147:1230‐1237. [DOI] [PubMed] [Google Scholar]
- 38. Kabouridis PS, Lasrado R, McCallum S, et al. The gut microbiota keeps enteric glial cells on the move; prospective roles of the gut epithelium and immune system. Gut Microbes 2015;6:398‐403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Everard A, Cani PD. Gut microbiota and GLP‐1. Rev Endocr Metab Disord 2014;15:189‐196. [DOI] [PubMed] [Google Scholar]
- 40. Greiner TU, Backhed F. Microbial regulation of GLP‐1 and L‐cell biology. Mol Metab 2016;5:753‐758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cani PD, Plovier H, Van Hul M, et al. Endocannabinoids ‐ at the crossroads between the gut microbiota and host metabolism. Nat Rev Endocrinol 2016;12:133‐143. [DOI] [PubMed] [Google Scholar]
- 42. Kellow NJ, Coughlan MT, Reid CM. Metabolic benefits of dietary prebiotics in human subjects: a systematic review of randomised controlled trials. Br J Nutr 2014;111:1147‐1161. [DOI] [PubMed] [Google Scholar]
- 43. Zakeri R, Batterham RL. Potential mechanisms underlying the effect of bariatric surgery on eating behaviour. Curr Opin Endocrinol Diabetes Obes 2017;25:3‐11. [DOI] [PubMed] [Google Scholar]
- 44. Svane MS, Jorgensen NB, Bojsen‐Moller KN, et al. Peptide YY and glucagon‐like peptide‐1 contribute to decreased food intake after Roux‐en‐Y gastric bypass surgery. Int J Obes (Lond) 2016;40:1699‐1706. [DOI] [PubMed] [Google Scholar]
- 45. Amato A, Cinci L, Rotondo A, Serio R, et al. Peripheral motor action of glucagon‐like peptide‐1 through enteric neuronal receptors. Neurogastroenterol Motil 2010;22:664‐e203. doi:10.1111/j.1365-2982.2010.01476.x [DOI] [PubMed] [Google Scholar]
- 46. Washington MC, Raboin SJ, Thompson W, Larsen CJ, Sayegh AI. Exenatide reduces food intake and activates the enteric nervous system of the gastrointestinal tract and the dorsal vagal complex of the hindbrain in the rat by a GLP‐1 receptor. Brain Res 2010;1344:124‐133. [DOI] [PubMed] [Google Scholar]
- 47. Grasset E, Puel A, Charpentier J, et al. A specific gut microbiota dysbiosis of type 2 diabetic mice induces GLP‐1 resistance through an enteric NO‐dependent and gut‐brain axis mechanism. Cell Metab 2017;26:278. doi:10.1016/j.cmet.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 48. Brooks L, Viardot A, Tsakmaki A, et al. Fermentable carbohydrate stimulates FFAR2‐dependent colonic PYY cell expansion to increase satiety. Mol Metab 2017;6:48‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jackerott M, Larsson LI. Immunocytochemical localization of the NPY/PYY Y1 receptor in enteric neurons, endothelial cells, and endocrine‐like cells of the rat intestinal tract. J Histochem Cytochem 1997;45:1643‐1650. [DOI] [PubMed] [Google Scholar]
- 50. Mao YK, Wang YF, Ward G, Cipris S, Daniel EE, McDonald TJ. Peptide YY receptor in submucosal and myenteric plexus synaptosomes of canine small intestine. Am J Physiol 1996;271:G36‐G41. [DOI] [PubMed] [Google Scholar]
- 51. Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013;110:9066‐9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Geurts L, Everard A, Van Hul M, et al. Adipose tissue NAPE‐PLD controls fat mass development by altering the browning process and gut microbiota. Nat Commun 2015;6:6495. doi:10.1038/ncomms7495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cohen LJ, Esterhazy D, Kim SH, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017;549:48‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wong JM, de SR, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 2006;40:235‐243. [DOI] [PubMed] [Google Scholar]
- 55. Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987;28:1221‐1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Canfora EE, Jocken JW, Blaak EE. Short‐chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 2015;11:577‐591. [DOI] [PubMed] [Google Scholar]
- 57. Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 1980;21:793‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017;19:29‐41. [DOI] [PubMed] [Google Scholar]
- 59. Fukumoto S, Tatewaki M, Yamada T, et al. Short‐chain fatty acids stimulate colonic transit via intraluminal 5‐HT release in rats. Am J Physiol Regul Integr Comp Physiol 2003;284:R1269‐R1276. [DOI] [PubMed] [Google Scholar]
- 60. Soret R, Chevalier J, De Coppet P, et al. Short‐chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 2010;138:1772‐1782. [DOI] [PubMed] [Google Scholar]
- 61. Bhattarai Y, Schmidt BA, Linden DR, et al. Human‐derived gut microbiota modulates colonic secretion in mice by regulating 5‐HT3 receptor expression via acetate production. Am J Physiol Gastrointest Liver Physiol 2017;313:G80‐G87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nohr MK, Egerod KL, Christiansen SH, et al. Expression of the short chain fatty acid receptor GPR41/FFAR3 in autonomic and somatic sensory ganglia. Neuroscience 2015;290:126‐137. [DOI] [PubMed] [Google Scholar]
- 63. De Vadder F, Kovatcheva‐Datchary P, Goncalves D, et al. Microbiota‐generated metabolites promote metabolic benefits via gut‐brain neural circuits. Cell 2014;156:84‐96. [DOI] [PubMed] [Google Scholar]
- 64. Frost G, Sleeth ML, Sahuri‐Arisoylu M, et al. The short‐chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun 2014;5:3611. doi:10.1038/ncomms4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome‐brain‐beta‐cell axis to promote metabolic syndrome. Nature 2016;534:213‐217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bindels LB, Leclercq I. Colonic acetate in obesity: location matters! Clin Sci (Lond) 2016;130:2083‐2086. [DOI] [PubMed] [Google Scholar]
- 67. Mertens KL, Kalsbeek A, Soeters MR, Eggink HM. Bile acid signaling pathways from the enterohepatic circulation to the central nervous system. Front Neurosci 2017;11:617. doi:10.3389/fnins.2017.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Higashi T, Watanabe S, Tomaru K, et al. Unconjugated bile acids in rat brain: analytical method based on LC/ESI‐MS/MS with chemical derivatization and estimation of their origin by comparison to serum levels. Steroids 2017;125:107‐113. [DOI] [PubMed] [Google Scholar]
- 69. Kliewer SA, Mangelsdorf DJ. Bile acids as hormones: the FXR‐FGF15/19 pathway. Dig Dis 2015;33:327‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Marcelin G, Jo YH, Li X, et al. Central action of FGF19 reduces hypothalamic AGRP/NPY neuron activity and improves glucose metabolism. Mol Metab 2014;3:19‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Abbott CR, Monteiro M, Small CJ, et al. The inhibitory effects of peripheral administration of peptide YY(3‐36) and glucagon‐like peptide‐1 on food intake are attenuated by ablation of the vagal‐brainstem‐hypothalamic pathway. Brain Res 2005;1044:127‐131. [DOI] [PubMed] [Google Scholar]
- 72. Orskov C, Poulsen SS, Moller M, Holst JJ. Glucagon‐like peptide I receptors in the subfornical organ and the area postrema are accessible to circulating glucagon‐like peptide I. Diabetes 1996;45:832‐835. [DOI] [PubMed] [Google Scholar]
- 73. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 2016;24:41‐50. [DOI] [PubMed] [Google Scholar]
- 74. Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 2015;17:565‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mittal R, Debs LH, Patel AP, et al. Neurotransmitters: the critical modulators regulating gut‐brain axis. J Cell Physiol 2017;232:2359‐2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Reigstad CS, Salmonson CE, Rainey JF, 3rd , et al. Gut microbes promote colonic serotonin production through an effect of short‐chain fatty acids on enterochromaffin cells. FASEB 2015;29:1395‐1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. O'Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain‐gut‐microbiome axis. Behav Brain Res 2015;277:32‐48. [DOI] [PubMed] [Google Scholar]
- 78. Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 2009;106:3698‐3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Clarke G, Grenham S, Scully P, et al. The microbiome‐gut‐brain axis during early life regulates the hippocampal serotonergic system in a sex‐dependent manner. Mol Psychiatry 2013;18:666‐673. [DOI] [PubMed] [Google Scholar]
- 80. Crumeyrolle‐Arias M, Jaglin M, Bruneau A, et al. Absence of the gut microbiota enhances anxiety‐like behavior and neuroendocrine response to acute stress in rats. Psychoneuroendocrinology 2014;42:207‐217. [DOI] [PubMed] [Google Scholar]
- 81. Diaz Heijtz R, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 2011;108:3047‐3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Asano Y, Hiramoto T, Nishino R, et al. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol 2012;303:G1288‐G1295. [DOI] [PubMed] [Google Scholar]
- 83. Hyland NP, Cryan JF. A gut feeling about GABA: Focus on GABA(B) receptors. Front Pharmacol 2010;1:124. doi:10.3389/fphar.2010.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bravo JA, Forsythe P, Chew MV, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A 2011;108:16050‐16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Barrett E, Ross RP, O'Toole PW, Fitzgerald GF, Stanton C. Gamma‐aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol 2012;113:411‐417. [DOI] [PubMed] [Google Scholar]
- 86. Mazzoli R, Pessione E. The neuro‐endocrinological role of microbial glutamate and GABA signaling. Front Microbiol 2016;7:1934. doi:10.3389/fmicb.2016.01934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ji XB, Hollocher TC. Reduction of nitrite to nitric oxide by enteric bacteria. Biochem Biophys Res Commun 1988;157:106‐108. [DOI] [PubMed] [Google Scholar]
- 88. Gusarov I, Starodubtseva M, Wang ZQ, et al. Bacterial nitric‐oxide synthases operate without a dedicated redox partner. J Biol Chem 2008;283:13140‐13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tennoune N, Chan P, Breton J, et al. Bacterial ClpB heat‐shock protein, an antigen‐mimetic of the anorexigenic peptide alpha‐MSH, at the origin of eating disorders. Transl Psychiatry 2014;4:e458. doi:10.1038/tp.2014.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Breton J, Tennoune N, Lucas N, et al. Gut commensal E. coli proteins activate host satiety pathways following nutrient‐induced bacterial growth. Cell Metab 2016;23:324‐334. [DOI] [PubMed] [Google Scholar]
- 91. Barbara G, Scaioli E, Barbaro MR, et al. Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease. Gut 2017;66:1252‐1261. [DOI] [PubMed] [Google Scholar]
- 92. Leclercq S, Matamoros S, Cani PD, et al. Intestinal permeability, gut‐bacterial dysbiosis, and behavioral markers of alcohol‐dependence severity. Proc Natl Acad Sci U S A 2014;111:E4485‐E4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab 2017;26:110‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pallister T, Jackson MA, Martin TC, et al. Hippurate as a metabolomic marker of gut microbiome diversity: modulation by diet and relationship to metabolic syndrome. Sci Rep 2017;7:13670. doi:10.1038/s41598-017-13722-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Piening BD, Zhou W, Contrepois K, et al. Integrative personal omics profiles during periods of weight fain and loss. Cell Syst 2018;6:157‐170.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Roager HM, Vogt JK, Kristensen M, et al. Whole grain‐rich diet reduces body weight and systemic low‐grade inflammation without inducing major changes of the gut microbiome: a randomised cross‐over trial [published online November 1, 2017]. Gut 2017. doi:10.1136/gutjnl-2017-314786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761‐1772. [DOI] [PubMed] [Google Scholar]
- 98. Kim F, Pham M, Luttrell I, et al. Toll‐like receptor‐4 mediates vascular inflammation and insulin resistance in diet‐induced obesity. Circ Res 2007;100:1589‐1596. [DOI] [PubMed] [Google Scholar]
- 99. Tsukumo DM, Carvalho‐Filho MA, Carvalheira JB, et al. Loss‐of‐function mutation in Toll‐like receptor 4 prevents diet‐induced obesity and insulin resistance. Diabetes 2007;56:1986‐1998. [DOI] [PubMed] [Google Scholar]
- 100. Laugerette F, Vors C, Geloen A, et al. Emulsified lipids increase endotoxemia: possible role in early postprandial low‐grade inflammation. J Nutr Biochem 2011;22:53‐59. [DOI] [PubMed] [Google Scholar]
- 101. Amar J, Burcelin R, Ruidavets JB, et al. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr 2008;87:1219‐1223. [DOI] [PubMed] [Google Scholar]
- 102. Pussinen PJ, Havulinna AS, Lehto M, Sundvall J, Salomaa V. Endotoxemia is associated with an increased risk of incident diabetes. Diabetes Care 2011;34:392‐397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jayashree B, Bibin YS, Prabhu D, et al. Increased circulatory levels of lipopolysaccharide (LPS) and zonulin signify novel biomarkers of proinflammation in patients with type 2 diabetes. Mol Cell Biochem 2014;388:203‐210. [DOI] [PubMed] [Google Scholar]
- 104. Dewulf EM, Cani PD, Claus SP, et al. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin‐type fructans in obese women. Gut 2013;62:1112‐1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Malaguarnera M, Vacante M, Antic T, et al. Bifidobacterium longum with fructo‐oligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci 2012;57:545‐553. [DOI] [PubMed] [Google Scholar]
- 106. Magalhaes I, Pingris K, Poitou C, et al. Mucosal‐associated invariant T cell alterations in obese and type 2 diabetic patients. J Clin Invest 2015;125:1752‐1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chassaing B, Raja SM, Lewis JD, Srinivasan S, Gewirtz AT. Colonic microbiota encroachment correlates with dysglycemia in humans. Cell Mol Gastroenterol Hepatol 2017;4:205‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Casselbrant A, Elias E, Fandriks L, Wallenius V. Expression of tight‐junction proteins in human proximal small intestinal mucosa before and after Roux‐en‐Y gastric bypass surgery. Surg Obes Relat Dis 2015;11:45‐53. [DOI] [PubMed] [Google Scholar]
- 109. Zhang D, Zhang L, Zheng Y, Yue F, Russell RD, Zeng Y. Circulating zonulin levels in newly diagnosed Chinese type 2 diabetes patients. Diabetes Res Clin Pract 2014;106:312‐318. [DOI] [PubMed] [Google Scholar]
- 110. Wells JM, Brummer RJ, Derrien M, et al. Homeostasis of the gut barrier and potential biomarkers. Am J Pysiol Gastrointest Liver Pysiol 2017;312:G171‐G193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bogunovic M, Dave SH, Tilstra JS, et al. Enteroendocrine cells express functional Toll‐like receptors. A Am J Pysiol Gastrointest Liver Pysiol 2007;292:G1770‐G1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. de La Serre CB, de Lartigue G, Raybould HE. Chronic exposure to low dose bacterial lipopolysaccharide inhibits leptin signaling in vagal afferent neurons. Physiol Behav 2015;139:188‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Forsythe P, Kunze WA. Voices from within: gut microbes and the CNS. Cell Mol Life Sci 2013;70:55‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bercik P, Denou E, Collins J, et al. The intestinal microbiota affect central levels of brain‐derived neurotropic factor and behavior in mice. Gastroenterology 2011;141:599‐609, 609.e1‐3. [DOI] [PubMed] [Google Scholar]
- 115. Sen T, Cawthon CR, Ihde BT, et al. Diet‐driven microbiota dysbiosis is associated with vagal remodeling and obesity. Physiol Behav 2017;173:305‐317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Vaughn AC, Cooper EM, DiLorenzo PM, et al. Energy‐dense diet triggers changes in gut microbiota, reorganization of gutbrain vagal communication and increases body fat accumulation. Acta Neurobiol Exp (Wars) 2017;77:18‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]


