Summary
Cassava is the second most important staple food crop in terms of per capita calories consumed in Africa and holds potential for climate change adaptation. Unfortunately, productivity in East and Central Africa is severely constrained by two viral diseases: cassava mosaic disease (CMD) and cassava brown streak disease (CBSD). CBSD was first reported in 1936 from northeast Tanzania. For approximately 70 years, CBSD was restricted to coastal East Africa and so had a relatively low impact on food security compared with CMD. However, at the turn of the 21st century, CBSD re‐emerged further inland, in areas around Lake Victoria, and it has since spread through many East and Central African countries, causing high yield losses and jeopardizing the food security of subsistence farmers. This recent re‐emergence has attracted intense scientific interest, with studies shedding light on CBSD viral epidemiology, sequence diversity, host interactions and potential sources of resistance within the cassava genome. This review reflects on 80 years of CBSD research history (1936–2016) with a timeline of key events. We provide insights into current CBSD knowledge, management efforts and future prospects for improved understanding needed to underpin effective control and mitigation of impacts on food security.
Keywords: CBSD, CBSV, CMD, food security, UCBSV, virology
Introduction
Cassava (Manihot esculenta Crantz, family Euphorbiaceae) produces carbohydrate‐rich storage roots, which are a staple food crop for approximately 800 million people worldwide [Food and Agriculture Organization (FAO), 2013]. In Africa, cassava is the second most important food staple in terms of per capita calories consumed (Nweke, 2004). Storage roots are used as a fresh carbohydrate source and can also be processed into flour, which may be consumed by the grower's family, sold in local markets or used to produce several industrial food products (Hillocks and Thresh, 2002). Subsistence farmers rely on cassava for a vital energy source, as it can be planted and harvested throughout the year, tolerates periods of unpredictable drought and grows on marginal soils (Hillocks and Thresh, 2002). Recent modelling has suggested that cassava may be highly resilient to future climate change and could provide Africa with adaptation opportunities, which are not offered by other staple food crops (Jarvis et al., 2012).
Cassava was introduced into Africa from Brazil by Portuguese traders in the 16th century and subsequently integrated into local agriculture in countries across the continent (Jones, 1959). Africa produces over one‐half of global cassava (57%) (Bennett, 2015); however, the continent's average fresh yield (9.9 tonnes/ha) lags behind potential yields (15–40 tonnes/ha) achieved under experimental conditions (Fermont et al., 2009). There are many reasons behind the reduced yields, including restricted access to labour, poor soil quality and premature harvesting (Fermont et al., 2009). Productivity in East and Central Africa is significantly constrained by two viral diseases, cassava mosaic disease (CMD) and cassava brown streak disease (CBSD), which together are estimated to cause annual losses worth US$1 billion [International Institute of Tropical Agriculture (IITA), 2014a] and adversely affect food security in the entire region (Patil et al., 2015).
In this article, we review CBSD research history, highlighting key events in a timeline (Fig. 1), and provide future prospects for further understanding and effective control. The review is split into two phases according to the geographical distribution of CBSD. The first phase covers the small number (n = 65) of reports published between 1936 and the early 1990s when CBSD was reported to be restricted to low‐altitude areas [<1000 m above sea level (masl)] along coastal East Africa and lakeshore districts of Malawi (Fig. 2) (Legg et al., 2011). The second phase examines CBSD re‐emergence after the mid‐1990s, when CBSD spread across East and Central Africa (Legg et al., 2011). We review the corresponding increased number (n = 277) of reports on CBSD geographical expansion, viral molecular characterization, host interactions, diagnostic techniques and control efforts.
Figure 1.
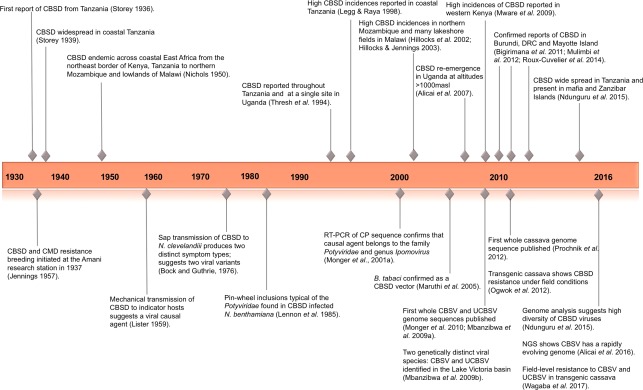
Key events in cassava brown streak disease (CBSD) geographical distribution and research history (1936–2016). CBSV, Cassava brown streak virus; CMD, cassava mosaic disease; NGS, next‐generation high‐throughput sequencing; RT‐PCR, reverse transcription polymerase chain reaction; UCBSV, Ugandan cassava brown streak virus.
Figure 2.
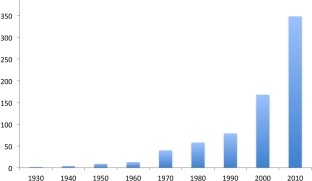
Cumulative number of scientific papers, reports or reviews which refer to cassava brown streak disease (CBSD), published in each decade between 1936 and 2016; dramatic increase in publications from the mid‐1990s following the re‐emergence of CBSD (accessed on Google scholar in December 2016).
We offer insights into what can be learnt from CBSD history, in particular the need for the application of knowledge to protect against and predict multiple biotic threats to staple food crops through improved understanding of CBSD epidemiology, diagnostics, surveillance and predictive modelling. This calls for effective international scientific collaborations across multiple areas of expertise and the rapid application of research and technologies to solve problems affecting farmers.
Initial Emergence and Symptom Description (1930s to the Early 1990s)
The first report of CBSD from northeast Tanzania (then called Tanganyika) describes distinctive foliar symptoms on lower mature cassava leaves and rot of storage roots (Storey, 1936). Nichols (1950) later reported that symptoms could be expressed on all parts of the plant and could include storage root necrosis (Fig. 3A), radial root constrictions (Fig. 3B), foliar chlorosis (Fig. 3C) and, occasionally, brown streaks or lesions on stems (Fig. 3D).
Figure 3.
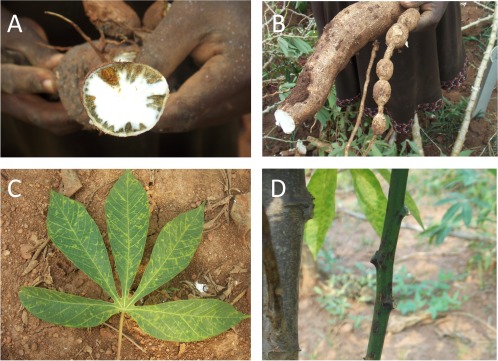
Cassava brown streak disease (CBSD) storage root necrosis (A), radial root constrictions (B), foliar chlorosis (C) and brown streaks or lesions on stems (D).
It was noted that two main types of foliar symptoms exist: (i) feathery chlorosis along secondary vein margins, which eventually coalesce to form blotches, and (ii) chlorotic mottling with no veinal association (Nichols, 1950). These distinctive symptoms lack the leaf distortion observed in CMD‐infected cassava plants. CBSD symptoms are variable in terms of severity, onset of symptom expression and parts of the plant affected, depending on the viral strain, cassava cultivar, environmental conditions and age of the plant when infected (Nichols, 1950). This variability makes diagnosis difficult for farmers (Nichols, 1950) and can result in farmers being unaware that their crop is affected until they harvest storage roots (Legg and Kanju, 2015). The difficulty in the diagnosis of CBSD has meant that infected stems have been transported to areas in which CBSD has previously been absent and used for planting material. Symptom variability has also hampered epidemiological studies, as the disease can go unnoticed in an area for long periods.
The surveying and symptom scoring of infected plants across different geographical areas have revealed that most plants with foliar symptoms usually also develop root necrosis (Hillocks et al., 2001). In the most sensitive cultivars, CBSD reduces root weight by up to 70% with necrosis developing at 6 months post‐planting (Hillocks et al., 2001). However, it has been reported that the local Tanzanian cultivar ‘Nachinyaya’ does not develop root necrosis and therefore is relatively tolerant to CBSD (Hillocks et al., 1996).
Early Geographical Distribution
Storey (1939) reported that CBSD was widespread in coastal Tanzania and, by 1950, the disease was endemic across coastal areas of East Africa from northeast Kenya, Tanzania to northern Mozambique at altitudes below 1000 masl (Nichols, 1950). The disease was reported in Uganda in 1945 and may have been introduced through infected cuttings sent from the Amani research station in Tanzania (Jameson, 1964; Nichols, 1950). Strict roguing of infected plants, replacement with non‐infected planting material and quarantine appear to have prevented the spread of CBSD in Uganda at this time (Nichols, 1950). Significantly, a lack of plant‐to‐plant vector transmission at higher altitudes was reported (Jennings, 1960; Nichols, 1950).
Causal Agent Characterization
Storey (1936) suspected a viral causal agent, as CBSD was successfully transmitted through the grafting of stem cuttings. Subsequently, Lister (1959) mechanically transmitted CBSD to indicator hosts, including Petunia hybrida, Datura stramonium, Nicotiana tabacum, N. rustica and N. glutinosa, which produce a range of symptoms depending on the sensitivity of the host and viral variant. In 1976, sap transmission of CBSD from infected cassava material to N. clevelandii produced two distinct symptom types, which suggested that two viral variants may be responsible for CBSD (Bock and Guthrie, 1976).
Virus particles were identified by electron microscopy analysis of CBSD‐infected N. debneyi (Bock, 1994). The infected samples contained 650‐nm filamentous particles with a similar morphology to viruses within the Carlavirus genus (Bock, 1994). However, pinwheel inclusions, typical of Potyviridae, were identified in CBSD‐infected N. benthamiana (Lennon et al., 1985). Pinwheel inclusions were subsequently found through more thorough electron microscopy of CBSD‐infected cassava samples, albeit at low concentrations (Were et al., 2004).
The Potyviridae sequence identity was finally confirmed in 2001 through reverse transcription‐polymerase chain reaction (RT‐PCR) of CBSD‐infected N. benthamiana samples (Monger et al., 2001a). When the RT‐PCR product was sequenced, it aligned most closely to the coat protein sequence of Sweet potato mild mottle virus (SPMMV, genus Ipomovirus, family Potyviridae) (Monger et al., 2001a). The same RT‐PCR technique was used to detect Cassava brown streak virus (CBSV) in symptomless cassava leaves, highlighting the sensitivity of the RT‐PCR technique (Monger et al., 2001a).
Early Control Efforts
In the 1930s, a cassava breeding programme was launched in Tanzania, which included breeding for CBSD and CMD resistance at the Amani research station (Jennings, 1957; Nichols, 1946). Early breeding to develop virus‐resistant cultivars involved crossing cultivated cassava with wild relatives, including M. glaziovii, M. dichotoma, M. catingea, M. saxicola and M. melanobasis, which are believed to possess higher levels of CBSD resistance (Jennings, 1957; Kawuki et al., 2016). The breeding programme produced the M. esculenta–M. glaziovii hybrid, known as ‘Namikonga’ in Tanzania or ‘Kaleso’ in Kenya, which, for many years, offered relatively high levels of CBSD tolerance (Hillocks and Jennings, 2003; Kaweesi et al., 2014). However, ‘Namikonga’ was not widely distributed to farmers, which may be because of its susceptibility to CMD (Hillocks and Jennings, 2003; Kawuki et al., 2016).
Initial Vector Transmission Studies
Until relatively recently, very little was known about the vector transmission of CBSVs. It had been noted that CBSD outbreaks tended to coincide with increases in whitefly populations (Hillocks and Jennings, 2003; Storey, 1939). However, initial attempts to transmit CBSV with whitefly (Bemisia tabaci) or aphid (Myzus persicae) were unsuccessful (Bock, 1994).
Geographical Distribution in the Early 1990s
In the early 1990s, there were reports of high CBSD incidences in areas of Tanzania, Mozambique and Malawi (Hillocks and Jennings, 2003). Surveys revealed CBSD incidences reaching 36%–50% in cassava fields along coastal areas of Tanzania (Hillocks et al., 1999; Legg and Raya, 1998). Similarly, CBSD incidences in Malawi reached 75% in many fields surrounding Lake Malawi and nearly all plants inspected in northern coastal areas of Mozambique expressed CBSD symptoms (Hillocks and Jennings, 2003; Hillocks et al., 2002). In a control effort, virus‐free CBSD‐tolerant cultivars were distributed to farmers in Mozambique who depended heavily on CBSD‐sensitive cassava cultivars for food security (Hillocks and Jennings, 2003). CBSD was also re‐discovered in Uganda in 1994 at a site near Entebbe (Thresh et al., 1994). This led researchers to call for concerted efforts to understand CBSD through improved surveillance (Hillocks and Jennings, 2003).
Reflections on Initial Emergence (1930s to Early 1990s)
Despite CBSD being endemic across coastal East Africa during this period, relatively little work was done to understand and control CBSD. This is reflected by the small number of scientific papers, reports or reviews which feature CBSD published between 1936 and the early 1990s (n = 65) (Fig. 2). The slight increase in references to CBSD in the 1970s is a result of a small number of reports (n = 27) on the threat posed by CBSD. In hindsight, these reports should have served as a warning to take control actions, which may have prevented the later expansion of CBSD across the region.
There was a general lack of scientific interest in CBSD at this time as a result of many factors, including the restricted occurrence of CBSD to low‐altitude areas along coastal eastern Africa and the devastating impacts of CMD on food security. During this period, CMD was a greater priority because of its prevalence across all cassava‐growing areas of Africa, resulting in famines, higher economic losses and forcing many farmers to abandon the crop (Alabi et al., 2011; Thresh and Cooter, 2005; Thresh et al., 1994). To help control the disease, CMD‐resistant cultivars were distributed to areas severely affected (Legg and Thresh, 2000). Unfortunately, these cultivars showed varying levels of CBSD susceptibility (Legg et al., 2006). It is not known whether the deployment of these cultivars contributed to the increased distribution of CBSD in the field.
Re‐emergence and Expansion Across East and Central Africa (MID‐1990s to 2016)
In 2004, the apparent restriction of CBSD to coastal lowlands changed with the re‐emergence of CBSD at altitudes above 1000 masl (Alicai et al., 2007). Infections of cassava plants showing CBSD symptoms at higher altitudes in Uganda were confirmed by RT‐PCR. Coat protein sequences aligned to CBSV isolates from Mozambique and Tanzania with sequence identities from 77.0% to 82.9% (Alicai et al., 2007). It is not known whether CBSD had been re‐introduced to Uganda through infected cuttings or whether the disease had existed at a low level since it was first introduced in the 1940s (Alicai et al., 2007). Shortly after this first report, the overall incidence of CBSD in Uganda increased from 12% in 2008 to 27% in 2011 (T. Alicai, year of personal communication, 2017), and similar increases were reported in Tanzania and Kenya (Legg et al., 2011; Mware et al., 2009; Ntawuruhunga and Legg, 2007). There have since been CBSD reports from Burundi (Bigirimana et al., 2011), Rwanda (FAO, 2011), eastern Democratic Republic of Congo (Mulimbi et al., 2012), south Sudan (T. Alicai, year of personal communication, 2017) and Mayotte Island (Roux‐Cuvelier et al., 2014).
It is difficult to obtain a truly accurate estimation of the economic damage caused by CBSD; however, an overall loss of US$750 million a year has been estimated across Kenya, Tanzania, Uganda and Malawi (Hillocks and Maruthi, 2015). CBSD is now one of the leading causes of cassava losses in East Africa (Pennisi, 2010) and its ongoing spread threatens the major cassava‐growing areas of Central and West Africa (Legg et al., 2014).
The dramatic increase in the impact of CBSD on food security is reflected in the increase in the number of papers, reports and reviews which refer to CBSD published from the mid‐1990s to 2016 (n = 277) (Fig. 2). The expansion of the CBSD epidemic across the Great Lakes region of East and Central Africa has necessitated the rapid development and implementation of effective control strategies. Several important projects were initiated following CBSD re‐emergence, which aimed to develop research, extension and policy capacity in the countries affected. Key targets have been to breed or genetically engineer resistant cultivars, provide certified virus‐clean planting material and improve viral surveillance and diagnosis (Legg et al., 2014).
Recent Local and Regional CBSD Epidemiology
The reasons behind the sudden increase in CBSD incidence and geographical range remain poorly understood. Studies have shown that CBSD spread and development are enhanced by high disease pressure, the use of susceptible genotypes and high whitefly numbers (Katono et al., 2015). CBSD is dispersed locally and over long distances through the trade transportation of infected planting material, whereas whiteflies are only able to disperse and amplify CBSD locally (McQuaid et al., 2017).
The ability of B. tabaci to transmit CBSV from infected to healthy plants was confirmed under quarantine insectary and glasshouse conditions by Maruthi et al. (2005). It has since been shown that CBSD viruses are transmitted semi‐persistently, with whiteflies acquiring viruses in 5–10 min, retaining them for up to 48 h and transmitting them over relatively short distances of less than 17 m in a cropping season (Maruthi et al., 2017). CBSD outbreaks occur from 3 to 12 years after increases in whitefly numbers (Legg et al., 2011). Critically, one of the primary causes of the increases in both CMD and CBSD in the African Great Lakes region appears to be super‐abundant numbers of whiteflies (Fig. 4), which are able to thrive at altitudes above 1000 masl (Alicai et al., 2007; Jeremiah et al., 2015).
Figure 4.

Super‐abundant whiteflies on cassava in Uganda.
Survey data have revealed that the transportation of infected material to areas in which CBSD was previously absent has enabled the disease to spread from independent hot spots (Legg et al., 2011). This is because cassava stems used for vegetative planting material are exchanged by farmers across localities and transported over long distances. One report has concluded that plants can also be infected through the use of contaminated cutting tools, which could contribute to in‐field spread (Rwegasira and Chrissie, 2015); however, a similar study has shown that such practices do not result in the transmission of CBSVs (Maruthi et al., 2016).
CBSVs are found only in Africa and therefore it appears that these viruses evolved within East Africa on an unknown species and subsequently jumped host into cassava in a new encounter situation (Monger et al., 2010). Therefore, there may be other hosts for CBSVs, which could serve as viral inoculum sources in the field (Monger et al., 2010). CBSV has been detected in the wild perennial species M. glaziovii (Mbanzibwa et al., 2011b); the importance of this to CBSD epidemiology is not currently known.
Molecular Characterization of Unusual CBSVs Genome Features
CBSVs belong to the Ipomovirus genus of the Potyviridae family (Monger et al., 2001a). Ipomoviruses have positive‐sense, single‐stranded genomes, which are translated as large polyproteins and autocatalytically cleaved by virus‐encoded proteases into 10 mature proteins with an additional P3N‐PIPO (N terminal of protein three fused with the Pretty Interesting Potyviridae ORF) protein produced through ribosomal frameshifting (Valli et al., 2015). The genomic organization of CBSVs is shown in Fig. 5.
Figure 5.

Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV) genomes encode a large polyprotein which is autocatalytically cleaved at specific cleavage sequences by virus‐encoded proteases into 10 mature proteins with an additional P3N‐PIPO protein produced through a +2 ribosomal frameshift in the P3 region (Valli et al., 2015). CBSV proteins: P1, serine protease/silencing suppressor; P3, third protein; PIPO, pretty interesting Potyviridae open reading frame (ORF), 6K1 and 6K2, 6‐kDa proteins; CI, cylindrical inclusion protein; VPg, viral genome‐linked protein; NIaPro, main viral protease; NIb, viral RNA‐dependent RNA polymerase; Ham1, putative pyrophosphatase; CP, coat protein; UTR, untranslated region. Note unusual features: presence of single P1 protein, absence of helper‐component proteinase protein (HC‐Pro) and presence of novel Ham1 protein (Mbanzibwa et al., 2009a).
The molecular characterization of coat protein sequences has revealed that there are at least two genetically distinct species: Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV) (Monger et al., 2001b; Winter et al., 2010), which typically have 76%–78% nucleotide and 87%–90% amino acid identity (Mbanzibwa et al., 2009b).
Genomic analysis has revealed that CBSVs share unusual features (Mbanzibwa et al., 2009a; Monger et al., 2010). First, CBSVs lack the multifunctional helper‐component proteinase protein (HC‐Pro), which possesses silencing suppressor, vector transmission and long‐distance movement in planta activities in Potyviridae viruses (Valli et al., 2015). The HC‐Pro protein is found in all other known Potyviridae viruses, except for Squash vein yellowing virus (SqVYV) and Cucumber vein yellowing virus (CVYV) (Mbanzibwa et al., 2009a). In CBSVs, HC‐Pro appears to have been replaced by silencing suppressor activity of the P1 serine proteinase (Mbanzibwa et al., 2009a). CBSV and UCBSV P1 proteins are most closely related to P1 of SPMMV and P1b of SqVYV and CVYV, which are related to the tritomovirus P1 proteins (Mbanzibwa et al., 2009a). The CBSV and UCBSV P1 proteins both contain zinc finger and LXKA motifs (Mbanzibwa et al., 2009a). The zinc finger and LXKA motifs in CVYV P1b are essential for silencing suppressor activity through the binding and sequestering of small interfering RNA (siRNA) required for silencing (Valli et al., 2008). It is therefore likely that the same motifs are responsible for the silencing suppressor activities of CBSV and UCBSV P1 proteins (Mbanzibwa et al., 2009a).
CBSV and UCBSV also encode novel Ham1 proteins with conserved Maf/Ham1 motifs (Mbanzibwa et al., 2009a). Proteins with Maf/Ham1 domains are found across prokaryotic and eukaryotic organisms and have nucleoside triphosphate pyrophosphatase activities, which reduce mutation rates by preventing the incorporation of non‐canonical nucleotides into RNA and DNA (Galperin et al., 2006). The functions of CBSV and UCBSV Ham1 proteins are yet to be elucidated, but they are likely to provide essential functions in the lifecycles of CBSVs. For instance, Ham1 proteins may reduce mutation rates under oxidative stress conditions in mature cassava leaves where CBSV viruses are found at the highest concentrations within the plant (Ogwok et al., 2014). Euphorbia ring spot virus (EuRV, genus Potyvirus, family Potyviridae) also encodes a Ham1 protein with an uncharacterized function (Knierim et al., 2016). EuRV, CBSV and UCBSV are part of a small number of viruses which are able to infect plants in the Euphorbiaceae family, and so perhaps Ham1 proteins are a euphorbia host adaptation (Monger et al., 2010).
Differences Between CBSVs Infections and Genome Sequences
In 2010, CBSV was found in infected cassava samples from Mozambique and Tanzania and UCBSV in Kenya, Uganda, Malawi and northwestern Tanzania (Winter et al., 2010). However, recent phylogenetic analysis of whole genome sequences has revealed that the viral species are not limited to agro‐ecological zones, and that there may be three separate species within the UCBSV clade (Ndunguru et al., 2015).
CBSV and UCBSV produce distinctly different symptoms on cassava and indicator hosts. CBSV causes more severe root necrosis and feathery chlorosis along vein margins, which develops into chlorotic blotches, whereas UCBSV causes circular chlorotic blotches between veins in cassava (Mohammed et al., 2012; Nichols, 1950; Winter et al., 2010). CBSV tends to accumulate to higher titres than UCBSV in cassava (Kaweesi et al., 2014) and indicator plants (Mohammed et al., 2012; Ogwok et al., 2014).
Sequence differences between CBSV and UCBSV genomes should explain differences in symptom severity, viral load and host interactions observed between the two viral species. Key areas of CBSV and UCBSV genomes show relatively high levels of divergence, including the P1 and Ham1 regions, with only 59% and 47% amino acid identity, respectively (Winter et al., 2010). One suggestion for the low level of Ham1 sequence similarity is that Ham1 genes may have been acquired separately by CBSV and UCBSV from a eukaryotic host (Monger et al., 2010). Alternatively, CBSV and UCBSV Ham1 sequences may have been derived from a common ancestor, which may have diverged as a result of differential selection pressures on the genome sequences of the two species (Monger et al., 2010).
Evolution of CBSVs
Statistical analysis of CBSV and UCBSV genomes using the empirical Bayes approach has predicted amino acid sites in UCBSV and CBSV coat protein and UCBSV Ham1 sequences, which appear to have been under positive selection (Mbanzibwa et al., 2011a). It is possible that positive selection at these different amino acid positions may have enabled the adaptive evolution of the two viral species (Mbanzibwa et al., 2011a). Recent whole genome sequence analysis has revealed that there is a higher diversity of CBSV isolates relative to UCBSV (Alicai et al., 2016). This diversity may have enabled CBSV to rapidly adapt to overcome host resistance mechanisms, which breeders have been selecting for (Alicai et al., 2016).
Whole genome analysis has also identified putative homologous recombination sites within the genomes of CBSV and UCBSV isolates (Ndunguru et al., 2015). To date, there has been no evidence for recombination between CBSV and UCBSV isolates (Mbanzibwa et al., 2011a; Ndunguru et al., 2015). However, the analysis of more CBSV and UCBSV genome sequences should provide insights into the importance of recombination in CBSD viral evolution.
Potential Interactions Between CBSVs
There is potential for CBSV and UCBSV isolates to interact as RT‐PCR has revealed that mixed infections are common, making up 34%–50% of tested infections in Kenya (Kathurima et al., 2016), Tanzania (Mbanzibwa et al., 2011b) and Uganda (Ogwok et al., 2014). The potential interactions between the two viral species are not currently understood. Two of the CMD causal viruses, African cassava mosaic virus (ACMV) and East African cassava mosaic virus (EACMV), have been shown to interact synergistically, leading to increased viral titres (Vanitharani et al., 2004). It is therefore possible that similar synergistic interactions occur between CBSD viral isolates.
Breeding for CBSD Resistance
To date, there is no cassava cultivar with a high level of CBSD resistance available to farmers (Abaca et al., 2013). Breeding cassava is notoriously difficult because of the high heterozygosity and a challenging cross‐pollination process (Ceballos et al., 2012). Breeding is further complicated by cultivars showing variation in CBSD resistance across different environments, which necessitates the testing of cultivars in different agro‐ecological zones to ensure that their resistance is stable (Tumuhimbise et al., 2014).
Breeders and farmers across Tanzania, Kenya, Uganda and Malawi have been selecting cultivars which strongly express foliar symptoms, but develop low levels of storage root necrosis (Hillocks et al., 2016). Twenty‐five best‐bet clones from five countries across East and southern Africa were selected, virus‐cleaned, shared and regionally evaluated across diverse environments for sources of CBSD and CMD resistance under the 5CP project (IITA, 2014b). Breeding efforts also include a 7‐year evaluation process of Tanzanian and Ugandan germplasm, whereby extensive intraspecific hybridizations have generated tolerant clones which develop relatively low levels of root necrosis of 12%, compared with >80% in sensitive cultivars (Kawuki et al., 2016).
Although tolerant cultivars develop reduced symptoms, they remain susceptible to CBSD viruses and thereby their adoption does not remove viral inocula from the field. Therefore, considerable efforts have been made to screen and breed cassava cultivars for CBSD resistance, which are able to restrict CBSD viral replication and/or movement. Promisingly, protoplast studies have recently shown that the elite breeding line KBH2006/18 can inhibit CBSD viral replication, which offers exciting opportunities to characterize resistance and resistance‐breaking viral virulence factors (Anjanappa et al., 2016).
Responses of different cassava cultivars to cbsd
Cassava cultivars respond very differently to infection by CBSVs; they produce a range of symptoms and are associated with varying viral loads at different time points of infection (Kaweesi et al., 2014). Sensitive cultivars show severe shoot and root symptoms, whereas cultivars with higher tolerance tend to express foliar symptoms, but usually lack or exhibit mild root necrosis (Hillocks and Jennings, 2003). Cultivars such as NASE 3 show high levels of resistance to UCBSV infection, but remain susceptible to CBSV (Ogwok et al., 2016). It has been shown that cultivars, such as ‘Namikonga’, support lower viral titres than susceptible cultivars, such as Albert (Maruthi et al., 2014). However, symptom severity is not always correlated with viral load, as the cultivar NASE 1 supports a relatively high viral load, but produces no foliar or root necrosis symptoms, whereas the cultivar NASE 14 supports a low viral load, but expresses severe root necrosis (Kaweesi et al., 2014).
This disparity between viral titres and symptom development has necessitated the use of viral load quantification during breeding to identify and select cultivars which support low CBSD viral titres. Until recently, the quantification of CBSD viruses in cassava was based on quantitative RT‐PCR, which measures the abundance of viral transcripts relative to the abundance of plant reference gene transcripts (Abarshi et al., 2012; Kaweesi et al., 2014; Moreno et al., 2011; Ogwok et al., 2014). However, the expression of plant reference genes can vary in different plant tissues, under varying developmental and environmental conditions (Brunner et al., 2004) and during viral infection (Liu et al., 2012). To overcome this, Shirima et al. (2017) have recently adapted the quantitative RT‐PCR technique to enable the absolute quantification of CBSV mRNA without normalization to plant reference genes. The higher levels of accuracy offered by this technique should be valuable in breeding efforts to generate cassava cultivars which support very low CBSD viral loads.
Identification of cbsd tolerance markers in cassava genomes
Despite the importance of cassava in developing countries, it has received relatively little scientific attention when compared with maize, rice and wheat (Varshney et al., 2012). Genomic studies of cassava are now enabling the identification of genetic markers associated with tolerance within the genomes of tolerant cultivars. In 2009, the first cassava genome assembly and annotation was publicly released (Prochnik et al., 2012). Since then, a large linkage map has been built using simple sequence repeats (SSRs) and single nucleotide polymorphisms (SNPs) to identify quantitative trait markers associated with CBSD tolerance across diverse African farmer‐preferred cultivars (Patil et al., 2015; Prochnik et al., 2012). This has revealed a number of putative CBSD tolerance alleles across different chromosomes in different cassava genotypes (Abaca et al., 2013; Nzuki et al., 2017). If validated, these alleles will be useful as markers in marker‐assisted breeding and could be combined into cultivars for effective and durable CBSD tolerance (Pariyo et al., 2013).
Transcriptional responses to cbsd viruses in different cassava cultivars
To date, very little is known about the function of these putative CBSD tolerance alleles. RNA sequencing analysis of transcripts, which are overexpressed during CBSD infection of the tolerant cultivar ‘Namikonga’, has implicated NAC transcription factors, as well as genes involved in jasmonic acid hormone signalling and the biosynthesis of phenylpropanoid, terpenoid and steroid secondary metabolites (Maruthi et al., 2014). In other plants, jasmonic acid and secondary metabolites are linked to abiotic and biotic stress responses (Izbiańska et al., 2014; Petrussa et al., 2013; Wasternack and Hause, 2013).
Transcriptional studies are also helping to gain an understanding of the mechanisms behind these different interactions between cassava cultivars and different CBSD viruses. Ogwok et al. (2016) have demonstrated recently that Dicer‐like proteins 2 and 4 (DCL2 and DCL4) and Argonaute 2 (AGO2) are differentially expressed during CBSV and UCBSV infections in different cassava cultivars. DCL and AGO proteins are integral to the plant antiviral defence mechanism of silencing viral RNA (Llave, 2010). Further studies are required to gain a fuller understanding of how the genes involved in host silencing of viral RNA are differentially expressed in different cultivars in response to different CBSVs.
Transcriptome analysis has also revealed that β‐1,3‐glucanase, which is involved in callose degradation at plasmodesmata, is up‐regulated during CBSD infection of the susceptible cultivar 60444, but not in the elite breeding line KBH2006/18, which shows relatively high levels of CBSD resistance (Anjanappa et al., 2017). The degradation of callose at plasmodesmata has been shown previously to promote viral movement (Zavaliev et al., 2011). Anjanappa et al. (2017) have suggested that enhanced callose degradation at plasmodesmata during CBSD infection of 60444 may promote viral movement, whereas the greater amount of callose present at plasmodesmata during KBH2006/18 infection is sufficient to limit systemic viral movement and thereby restrict infection.
CBSD resistance through genetic transformation
There have been promising attempts to introduce CBSD resistance into cassava through genetic engineering. The mechanism utilized involves the transgenic expression of inverted repeat CBSD viral sequences to trigger post‐transcriptional gene silencing (PTGS) of the corresponding sequences during infection, and hence confer viral resistance to the plant (Patil et al., 2011). The approach was successful in N. benthamiana; transgenic expression of UCBSV coat protein hairpin constructs resulted in high levels of resistance to six diverse CBSV and UCBSV isolates (Patil et al., 2011). The same construct was expressed in cassava and conferred resistance to CBSV and UCBSV under field conditions with high disease pressure (Ogwok et al., 2012; Yadav et al., 2011). Vegetative stem cuttings taken from transgenic plants retained CBSD resistance, enabling their use in vegetative propagation (Odipio et al., 2013).
To ensure that transgenic plants are resistant to both CBSV and UCBSV, the cultivar TME 204 was transformed with a construct (p5001) containing fused tandem repeat coat protein sequences from both CBSV and UCBSV to produce the transgenic line: TME 204 p5001 (Beyene et al., 2017). This transgenic line was resistant to CBSD when graft challenged (Beyene et al., 2017) and grown within confined field trials across different agro‐ecological locations in Uganda and Kenya, where plants were exposed to a range of both CBSV and UCBSV isolates over multiple vegetative propagation cycles (Wagaba et al., 2017).
It is vitally important that improved cultivars are resistant to both CBSD and CMD. Transgenic CBSD resistance was conferred to cultivars TME 7 and TME 204, which are naturally CMD resistant as a result of the presence of the single dominant CMD2 resistance locus (Beyene et al., 2016; Vanderschuren et al., 2012). Critically, however, these TME cultivars lost their CMD2 resistance through an unknown mechanism during somatic embryogenesis (Beyene et al., 2016). Work is ongoing to cross the CBSD‐resistant transgenic line TME 204 p5001 with a wild‐type CMD2 type cultivar to combine durable CBSD and CMD resistance into a single cultivar (Beyene et al., 2017).
Once biosafety issues have been addressed, the potential benefits of genetically modified cassava to smallholder famers will be substantial. It has been estimated that the net value of the release of CBSD‐resistant cultivars will be US$436 million for western Kenya and US$790 million for Uganda over a 35‐year period starting in 2025 (Taylor et al., 2016). The Virus Resistant Cassava for Africa (VIRCA Plus) project is working to deliver CMD‐ and CBSD‐resistant cassava cultivars to smallholder farmers in Uganda and Kenya, and so improve their livelihoods and food security (Taylor et al., 2016).
Distribution of Certified Virus‐clean Planting Material
The lack of cultivars highly resistant to CBSD makes the existence of a clean seed system critical for the effective management of CBSD. Clean cassava seed systems are non‐existent in most eastern Africa countries where CBSD is a problem. The Great Lakes Cassava Initiative was launched in 2008 with an overall goal to distribute certified virus‐clean CBSD‐tolerant cultivars to 1.15 million farmers across six East and Central African countries over a 4‐year period (Catholic Relief Services, 2010). As tolerant cultivars still retain viruses within their stems, planting material must be subjected to a cleaning process and highly sensitive diagnostic testing before it can be multiplied and supplied to farmers. This should reduce disease pressure in affected areas, as at least initially the majority of crops will be disease free (Mwangangi, 2014). The production of certified virus‐clean cassava germplasm is particularly important during the transportation of vegetative planting material because of the risks posed by CBSD to cassava‐growing areas which are currently unaffected (Legg et al., 2011). The cleaning process involves culturing meristem tissue in vitro, and subjecting it to thermo‐ and/or chemotherapy, which inactivates viruses and prevents viral replication or movement within tissues.
Mathematical modelling has shown that, in order for the clean seed system to be sustainable, multiplication sites should only be set up in areas with low disease pressure and low vector population density (McQuaid et al., 2015). Modelling has also shown that, to reduce CBSD dispersal and increase cassava yields, virus‐free planting material should be distributed to a number of different growers across a widespread area with restricted trade (McQuaid et al., 2017). Once certified virus‐clean material has been distributed, farmers must also be thoroughly trained in the identification of disease symptoms to enable sufficient roguing to further reduce CBSD spread (Legg et al., 2017; McQuaid et al., 2015). Cassava clean seed system projects have recently been piloted in Uganda and Tanzania. (Legg et al., 2017; McQuaid et al., 2015).
CBSVs Diagnostics
As many CBSD‐infected plants remain symptomless, highly sensitive diagnostic techniques are required in the production and transportation of material (Abarshi et al., 2010). There have been several important advancements in cassava disease diagnostic techniques, including the optimization of RT‐PCR to enable reliable simultaneous detection of CBSV and UCBSV (Mbanzibwa et al., 2011b), as well as cassava mosaic begomoviruses, in a single multiplex RT‐PCR (Abarshi et al., 2012). Next‐generation high‐throughput sequencing (NGS) has been used to screen large numbers of plants for the presence of CBSVs to ensure that they are virus free before dissemination as planting material. Adams et al. (2013) demonstrated that, with NGS, it was possible to detect 1% of infected plants out of a total of 300 plants with 95% probability. Although useful tools, to date, many of these techniques are relatively resource intensive, and so it is vitally important that affordable diagnostic tools are available in African countries to enable sensitive CBSD detection locally, even in cassava fields. One promising technique is reverse transcription loop‐mediated isothermal amplification (RT‐LAMP), which is able to detect and differentiate the presence of CBSV and UCBSV with lower consumables, resources and instrument costs than RT‐PCR (Tomlinson et al., 2013).
Conclusions
In the past 20 years, CBSD has become a major cause of food insecurity across East and Central Africa, and only since its recent geographical expansion has the disease received the scientific interest it deserves. Once the CBSD pandemic unravelled, it was largely too late to restrict the disease to limited outbreak areas. Lessons must be learnt from this to prevent similar disease outbreaks in the future. Critically, scientific interest should be applied to the prediction and prevention of future outbreaks before they are able to emerge and cause devastating yield losses across large areas.
In terms of understanding CBSD, recent studies have begun to show that CBSVs are diverse and that CBSV has a high evolutionary capacity (Alicai et al., 2016). Many control efforts are being aided by advancing molecular techniques, including marker‐assisted breeding, development of genetically modified resistant lines, provision of certified virus‐clean planting material and the use of sensitive diagnostics. Despite this progress, there are still many areas of CBSD biology and epidemiology which remain poorly understood and offer opportunities to further understand and control the disease.
Future Prospects
Understanding key drivers in CBSD epidemiology
Relatively little is known about the complex interactions between viral variants, vectors, cassava cultivars and environmental conditions, and how they may influence the spread of CBSD. Therefore CBSD incidence, prevalence and whitefly populations in farmers' fields need to be regularly monitored in major cassava‐producing areas to track periodic changes in the general status of the disease in affected countries and those at risk. Where control interventions are deployed, they should be evaluated for their impact in the control of CBSD. The availability of this information is required for the development of predictive models that will provide an evidence base for disease control decisions and resource allocation. The effectiveness of CBSD control strategies is also heavily dependent on the level of farmer engagement and awareness. In Uganda, extension work includes efforts to raise farmer awareness of CBSD and to deliver information on its management (Kumakech et al., 2013).
Gaining insights into viral populations
We currently know very little about viral populations within wild hosts, which may serve as important sources of viral inoculum and enable the evolution of CBSD and other emerging viral diseases. Next‐generation deep sequencing can be used to detect viral populations about which very little sequence information is known (Prabha et al., 2013). It would be fascinating to apply this to cassava and to characterize viral populations within CBSD‐infected cassava and wild hosts surrounding cassava crops. This could shed light on viral evolution and the contribution of wild hosts to epidemiology. It may also help to identify potential unknown viral diseases, against which pre‐emptive control could be taken in anticipation of emerging diseases (Newbery et al., 2016).
Measures to restrict CBSD spread into unaffected areas
To date, CBSD viruses are only found in East and Central Africa. However, CBSD distribution could increase should infected material be transported to other cassava‐growing areas of Africa, Latin America and Asia, which would result in huge economic losses and food insecurity (Legg et al., 2014). Therefore, the movement of cassava material from CBSD‐affected countries should be subject to strict quarantine measures to ensure that planting material is virus free before transportation. Such measures will facilitate the movement of superior cultivars for production or breeding purposes.
Utilizing diverse cultivars for genomic resources
It is important to continue to maintain and investigate diverse cassava germplasm from across Africa and Latin America and their wild relatives for potential sources of disease resistance and other beneficial agronomic traits (Turyagyenda et al., 2012). This will enable farmers to adapt to changing environmental, socio‐cultural and market conditions (Pautasso et al., 2013).
Surveillance of viral diseases
To target control efforts, it is vitally important to accurately survey viral disease distribution. The IITA has recently launched the Cassava Disease Surveillance Platform in Nigeria, which offers opportunities for cassava breeders and extension workers to upload images of plants suspected to be infected with CBSD and other diseases. The images are analysed by a team of experts to enable rapid diagnosis and coordination of emergency control responses (IITA, 2016). Similarly, the West African Virus Epidemiology Project launched in 2015 aims to use field surveys to gain a clear understanding of the viruses which affect cassava in West Africa to predict viral emergence and inform policy decisions. Structured surveys under the Cassava Virus Diagnostics Project in eastern and southern Africa are tracking area‐wide changes in cassava viral diseases over time. This will provide the basis for disease control intervention decision‐making and impact assessment.
Predicted effects of climate change on cassava production
Cassava demonstrates relatively high levels of resilience to temperature and rainfall fluctuations predicted in climate change models (El‐Sharkawy, 2004). A model based on temperature and rainfall projections across Africa has predicted that, compared with other staple food crops, overall cassava is the least likely to be adversely affected by climate change (Jarvis et al., 2012). This makes cassava an attractive food security crop for climate change adaptation in Africa. However, climate change is also predicted to affect the distribution and abundance of cassava pests and diseases, including B. tabaci (Jarvis et al., 2012). Recent ecological niche modelling has predicted that, with climate change, the potential distribution of CBSD‐ and CMD‐carrying B. tabaci will spread over West, Central and southwestern coastal Africa where cassava production is high and CBSD is currently absent (Herrera Campo et al., 2011). Therefore, the monitoring and control of B. tabaci populations are major priorities. The deep sequencing technique could be extended to B. tabaci, enabling the mapping of the most active and abundant viral species carried by B. tabaci populations across different agricultural regions (Ng et al., 2011).
Understanding CBSVs infection mechanisms and virulence determinants
Despite the increasing number of sequenced CBSV genomes, very little is known about the virulence determinants within CBSV and UCBSV genomes responsible for key functions during infection, and their effect on disease symptomatology. To date, only the silencing suppression activity of the UCBSV P1 protein has been characterized (Mbanzibwa et al., 2009a). The construction of infectious clones will enable the targeted mutagenesis of key viral sequences to identify the functions of viral proteins and the host proteins with which they interact, which should serve as potential targets to restrict viral infection. Current work to develop and manipulate CBSV infectious clones is ongoing at various institutions.
Collaborative sharing of information and resources
There are many opportunities to exploit the recent progress made in understanding CBSD through progress in cassava, viral and vector research. There is a need for this research to be integrated into a central, easily accessible platform (Ayling et al., 2012). This will require experts across diverse backgrounds and countries to openly communicate, engage, share data and collaborate through networks, such the Global Cassava Partnership for the 21st Century. Such partnerships should help to generate solutions to control CBSD and enable cassava to fulfil its potential of feeding billions of people by 2050 (Legg et al., 2014).
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgements
This work was made possible by the South West Training Partnership, funded by the Biotechnology and Biological Sciences Research Council, UK. All photographs were taken by Katie Tomlinson.
References
- Abaca, A. , Kawuki, R. , Tukamuhabwa, P. , Baguma, Y. , Pariyo, A. , Alicai, T. , Omongo, A.C.C. , Abidrabo, P. , Katono, K. and Bua, A. (2013) Genetic relationships of cassava genotypes that are susceptible or tolerant to cassava brown streak disease in Uganda. J. Agric. Sci. 5, 107. [Google Scholar]
- Abarshi, M.M. , Mohammed, I.U. , Wasswa, P. , Hillocks, R.J. , Holt, J. , Legg, J.P. , Seal, S.E. and Maruthi, M.N. (2010) Optimization of diagnostic RT‐PCR protocols and sampling procedures for the reliable and cost‐effective detection of Cassava brown streak virus . J. Virol. Methods, 163, 353–359. [DOI] [PubMed] [Google Scholar]
- Abarshi, M.M. , Mohammed, I.U. , Jeremiah, S.C. , Legg, J.P. , Kumar, P.L. , Hillocks, R.J. and Maruthi, M.N. (2012) Multiplex RT‐PCR assays for the simultaneous detection of both RNA and DNA viruses infecting cassava and the common occurrence of mixed infections by two cassava viruses in East Africa. J. Virol. Methods, 179, 176–184. [DOI] [PubMed] [Google Scholar]
- Adams, I.P. , Abidrabo, P. , Miano, D.W. , Alicai, T. , Kinyua, Z.M. , Clarke, J. , Macarthur, R. , Weekes, R. , Laurenson, L. , Hany, U. , Peters, D. , Potts, M. , Glover, R. , Boonham, N. and Smith, J. (2013) High throughput real‐time RT‐PCR assays for specific detection of Cassava brown streak disease causal viruses, and their application to testing of planting material. Plant Pathol. 62, 233–242. [Google Scholar]
- Alabi, O. , Kumar, P. and Naidu, R. (2011) Cassava mosaic disease: a curse to food security in sub‐Saharan Africa. APSnet Features. doi: 10.1094/APSnetFeature-2011-0701. [DOI] [Google Scholar]
- Alicai, T. , Omongo, C.A. , Maruthi, M.N. , Hillocks, R.J. , Baguma, Y. , Kawuki, R. , Bua, A. , Otim‐Nape, G.W. and Colvin, J. (2007) Re‐emergence of Cassava brown streak disease in Uganda. Plant Dis. 91, 24–29. [DOI] [PubMed] [Google Scholar]
- Alicai, T. , Ndunguru, J. , Sseruwagi, P. , Tairo, F. , Okao‐Okuja, G. , Nanvubya, R. , Kiiza, L. , Kubatko, L. , Kehoe, M.A. and Boykin, L.M. (2016) Characterization by next generation sequencing reveals the molecular mechanisms driving the faster evolutionary rate of Cassava brown streak virus compared with Ugandan cassava brown streak virus . Sci. Rep. 6, 36164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjanappa, R.B. , Mehta, D. , Maruthi, M.N. , Kanju, E. , Gruissem, W. and Vanderschuren, H. (2016) Characterization of virus‐resistant cassava. Mol. Plant–Microbe Interact. 29, 527–534. [DOI] [PubMed] [Google Scholar]
- Anjanappa, R.B. , Mehta, D. , Okoniewski, M.J. , Szabelska‐Berȩsewicz, A. , Gruissem, W. and Vanderschuren, H. (2017), Molecular insights into Cassava brown streak virus susceptibility and resistance by profiling of the early host response. Mol Plant Pathol. doi: 10.1111/mpp.12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayling, S. , Ferguson, M. , Rounsley, S. and Kulakow, P. (2012) Information resources for cassava research and breeding. Trop. Plant Biol. 5, 140–151. [Google Scholar]
- Bennett, B. (2015) Guest editorial: smallholder cassava production and the cassava processing sector in Africa. Food Chain, 5, 1–3. [Google Scholar]
- Beyene, G. , Chauhan, R.D. , Wagaba, H. , Moll, T. , Alicai, T. , Miano, D. , Carrington, J.C. and Taylor, N.J. (2016) Loss of CMD2‐mediated resistance to Cassava mosaic disease in plants regenerated through somatic embryogenesis. Mol. Plant Pathol. 17, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyene, G. , Chauhan, R.D. , Ilyas, M. , Wagaba, H. , Fauquet, C.M. , Miano, D. , Alicai, T. and Taylor, N.J. (2017) A virus‐derived stacked RNAi construct confers robust resistance to Cassava brown streak disease. Front. Plant Sci. 7, 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigirimana, S. , Barumbanze, P. , Ndayihanzamaso, P. , Shirima, R. and Legg, J.P. (2011) First report of Cassava brown streak disease and associated Ugandan cassava brown streak virus in Burundi. New Dis. Rep. 24, 26. [Google Scholar]
- Bock, K. (1994) Studies on Cassava brown streak virus disease in Kenya. Trop. Sci. 34, 134–145. [Google Scholar]
- Bock, K.R. and Guthrie, E.J. (1976) Recent advances in research on cassava viruses in East Africa. East Afr. Agric. Forest. J. 11–16. https://idl-bnc-idrc.dspacedirect.org/bitstream/handle/10625/18934/IDL-18934.pdf?sequence=1 [Google Scholar]
- Brunner, A.M. , Yakovlev, I.A. and Strauss, S.H. (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant. 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catholic Relief Services . (2010) Final Report on the Great Lakes Cassava Initiative https://www.crs.org/sites/default/files/tools-research/final-report-great-lakes-cassava-initiative.pdf
- Ceballos, H. , Kulakow, P. and Hershey, C. (2012) Cassava breeding: current status, bottlenecks and the potential of biotechnology tools. Trop. Plant Biol. 5, 73–87. [Google Scholar]
- Clean Seed System Report . (2016) Commercializing clean cassava planting material delivery system in Uganda. The second annual review and planning meeting held at the National Crops Resources Research Insitute, 28th–30th June 2016.
- El‐Sharkawy, M.A. (2004) Cassava biology and physiology. Plant Mol. Biol. 56, 481–501. [DOI] [PubMed] [Google Scholar]
- Fermont, A.M. , van Asten, P.J.A. , Tittonell, P. , van Wijk, M.T. and Giller, K.E. (2009) Closing the cassava yield gap: an analysis from smallholder farms in East Africa. Field Crop. Res. 112, 24–36. [Google Scholar]
- Food and Agriculture Organization (FAO) . (2011) Cassava Virus on Verge of Epidemic in East Africa: Experts Urge Funding, Swift Action to Protect Staple Food Crop. Rome: Food and Agricultural Organization. [Google Scholar]
- Food and Agriculture Organization (FAO) . (2013) Save and Grow Cassava: A Guide to Sustainable Production Intensification. Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Galperin, M.Y. , Moroz, O.V. , Wilson, K.S. and Murzin, A.G. (2006) House cleaning, a part of good housekeeping. Mol. Microbiol. 59, 5–19. [DOI] [PubMed] [Google Scholar]
- Herrera Campo, B.V. , Hyman, G. and Bellotti, A. (2011) Threats to cassava production: known and potential geographic distribution of four key biotic constraints. Food Secur. 3, 329–345. [Google Scholar]
- Hillocks, R.J. and Jennings, D. (2003) Cassava brown streak disease: a review of present knowledge and research needs. Int. J. Pest Manag. 49, 225–234. [Google Scholar]
- Hillocks, R.J. and Maruthi, M.N. (2015) Post‐harvest impact of Cassava brown streak disease in four countries in eastern Africa. Food Chain, 5, 116–122. [Google Scholar]
- Hillocks R.J. and Thresh J.M. (eds) (2002) Cassava: Biology, Production and Utilization. Wallingford, CT: CABI. [Google Scholar]
- Hillocks, R.J. , Raya, M. and Thresh, J.M. (1996) The association between root necrosis and above‐ground symptoms of brown streak virus infection of cassava in southern Tanzania. Int. J. Pest Manag. 42, 285–289. [Google Scholar]
- Hillocks, R.J. , Raya, M. and Thresh, J.M. (1999) Factors affecting the distribution, spread and symptom expression of Cassava brown streak disease in Tanzania. Int. J. Pest Manag. 42, 285–289. [Google Scholar]
- Hillocks, R.J. , Raya, M.D. , Mtunda, K. and Kiozia, H. (2001) Effects of brown streak virus disease on yield and quality of cassava in Tanzania. J. Phytopathol. 149, 389–394. [Google Scholar]
- Hillocks, R.J. , Thresh, J.M. , Tomas, J. , Botao, M. , Macia, R. and Zavier, R. (2002) Cassava brown streak disease in northern Mozambique. Int. J. Pest Manag. 48, 178–181. [Google Scholar]
- Hillocks, R. , Maruthi, M. , Kulembeka, H. , Jeremiah, S. , Alacho, F. , Masinde, E. , Ogendo, J. , Arama, P. , Mulwa, R. , Mkamilo, G. , Kimata, B. , Mwakanyamale, D. , Mhone, A. and Benesi, I. (2016) Disparity between leaf and root symptoms and crop losses associated with Cassava brown streak disease in four countries in eastern Africa. J. Phytopathol. 164, 86–93. [Google Scholar]
- International Institute of Tropical Agriculture (IITA) . (2014a) IITA Bulletin, Issue 2215. Ibadan: International Institute of Tropical Agriculture.
- International Institute of Tropical Agriculture (IITA) . (2014b) ITA‐led 5CP project reports great strides in regional exchange of improved cassava varieties. IITA News, Ibadan. [Google Scholar]
- International Institute of Tropical Agriculture (IITA) . (2016) IITA Bulletin, Issue 2324. IITA and NAQS Pilot Digital Cassava Disease Surveillance Platform.
- Izbiańska, K. , Arasimowicz‐Jelonek, M. and Deckert, J. (2014) Phenylpropanoid pathway metabolites promote tolerance response of lupine roots to lead stress. Ecotoxicol. Environ. Saf. 110, 61–67. [DOI] [PubMed] [Google Scholar]
- Jameson, J.D. (1964) Cassava mosaic disease in Uganda. East Afr. Agric. Forest. J. 29, 208–213. [Google Scholar]
- Jarvis, A. , Ramirez‐Villegas, J. , Herrera Campo, B.V. and Navarro‐Racines, C. (2012) Is cassava the answer to African climate change adaptation? Trop. Plant Biol. 5, 9–29. [Google Scholar]
- Jennings, D.L. (1957) Further studies in breeding cassava for virus resistance. East Afr. Agric. J. 22, 213–219. [Google Scholar]
- Jennings, D.L. (1960) Observations on virus diseases of cassava in resistant and susceptible varieties. II. Brown streak disease. Empire J. Exp. Agric. 28, 261–270. [Google Scholar]
- Jeremiah, S.C. , Ndyetabula, I.L. , Mkamilo, G.S. , Haji, S. , Muhanna, M.M. , Chuwa, C. , Kasele, S. , Bouwmeester, H. , Ijumba, J.N. and Legg, J.P. (2015) The dynamics and environmental influence on interactions between Cassava brown streak disease and the whitefly, Bemisia tabaci . Phytopathology, 105, 646–655. [DOI] [PubMed] [Google Scholar]
- Jones, W. (1959) Manioc in Africa. Stanford, CA: Stanford University Press. [Google Scholar]
- Kathurima, T. , Nyende, A. , Kiarie, S. and Ateka, E. (2016) Genetic diversity and distribution of Cassava brown streak virus and Ugandan cassava brown streak virus in major cassava‐growing regions in Kenya. Annu. Res. Rev. Biol. 10, 1–9. [Google Scholar]
- Katono, K. , Alicai, T. , Baguma, Y. , Edema, R. , Bua, A. and Omongo, C. (2015) Influence of host plant resistance and disease pressure on spread of Cassava brown streak disease in Uganda. Am. J. Exp. Agric. 7, 284–293. [Google Scholar]
- Kaweesi, T. , Kawuki, R. , Kyaligonza, V. , Baguma, Y. , Tusiime, G. and Ferguson, M.E. (2014) Field evaluation of selected cassava genotypes for Cassava brown streak disease based on symptom expression and virus load. Virol. J. 11, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawuki, R.S. , Kaweesi, T. , Esuma, W. , Pariyo, A. , Kayondo, I.S. , Ozimati, A. , Kyaligonza, V. , Abaca, A. , Orone, J. , Tumuhimbise, R. , Nuwamanya, E. , Abidrabo, P. , Amuge, T. , Ogwok, E. , Okao, G. , Wagaba, H. , Adiga, G. , Alicai, T. , Omongo, C. , Bua, A. , Ferguson, M. , Kanju, E. and, Baguma, Y. (2016) Eleven years of breeding efforts to combat Cassava brown streak disease. Breed. Sci. 66, 560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knierim, D. , Menzel, W. and Winter, S. (2016) Analysis of the complete genome sequence of Euphorbia ringspot virus, an atypical member of the genus Potyvirus . Arch. Virol. 162, 1–3. [DOI] [PubMed] [Google Scholar]
- Kumakech, A. , Acipa, A. , Tumwine, V. and Maiteki, G.A. (2013) Knowledge on cassava disease management: the case of cassava brown streak disease awareness in Northern Uganda. Afr. J. Plant Sci. 7, 597–601. [Google Scholar]
- Legg, J.P. and Kanju, E. (2015) Virus Diseases of Tropical and Subtropical Crops. Wallingford, CABI Plant Protection Series. [Google Scholar]
- Legg, J.P. and Raya, M.D. (1998) Survey of cassava virus diseases in Tanzania. Int. J. Pest Manag. 44, 17–23. [Google Scholar]
- Legg, J.P. and Thresh, J.M. (2000) Cassava mosaic virus disease in East Africa: a dynamic disease in a changing environment. Virus Res. 71, 135–149. [DOI] [PubMed] [Google Scholar]
- Legg, J.P. , Owor, B. , Sseruwagi, P. and Ndunguru, J. (2006) Cassava mosaic virus disease in East and Central Africa: epidemiology and management of a regional pandemic. Adv Virus Res. 67, 355–418. [DOI] [PubMed] [Google Scholar]
- Legg, J.P. , Jeremiah, S.C. , Obiero, H.M. , Maruthi, M.N. , Ndyetabula, I. , Okao‐Okuja, G. , Bouwmeester, H. , Bigirimana, S. , Tata‐Hangy, W. , Gashaka, G. , Mkamilo, G. , Alicai, T. and Lava Kumar, P. (2011) Comparing the regional epidemiology of the Cassava mosaic and Cassava brown streak virus pandemics in Africa. Virus Res. 159, 161–170. [DOI] [PubMed] [Google Scholar]
- Legg, J.P. , Somado, E.A. , Barker, I. , Beach, L. , Ceballos, H. , Cuellar, W. , Elkhoury, W. , Gerling, D. , Helsen, J. , Hershey, C. , Jarvis, A. , Kulakow, P. , Kumar, L. , Lorenzen, J. , Lynam, J. , McMahon, M. , Maruthi, G. , Miano, D. , Mtunda, K. , Natwuruhunga, P. , Okogbenin, E. , Pezo, P. , Terry, E. , Thiele, G. , Thresh, M. , Wadsworth, J. , Walsh, S. , Winter, S. , Tohme, J. and Fauquet, C. (2014) A global alliance declaring war on cassava viruses in Africa. Food Secur. 6, 231–248. [Google Scholar]
- Legg, J.P. , Ndalahwa, M. , Yabeja, J. , Ndyetabula, I. , Bouwmeester, H. , Shirima, R. and Mtunda, K. (2017) Community phytosanitation to manage cassava brown streak disease. Virus Res. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon, A. , Aiton, M. and Harrison, B. (1985) Cassava viruses from Africa: third country quarantine. Report for 1985, pp. 168–169. Invergowrie: Scottish Crop Research Institute.
- Lister, R.M. (1959) Mechanical transmission of Cassava brown streak virus . Nature, 183, 1588–1589. [DOI] [PubMed] [Google Scholar]
- Liu, D. , Shi, L. , Han, C. , Yu, J. , Li, D. , Zhang, Y. and Vinatzer, B.A. (2012) Validation of reference genes for gene expression studies in virus‐infected Nicotiana benthamiana using quantitative real‐time PCR. PLoS One, 7, e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C. (2010) Virus‐derived small interfering RNAs at the core of plant–virus interactions. Trends Plant Sci. 15, 701–707. [DOI] [PubMed] [Google Scholar]
- Maruthi, M.N. , Hillocks, R.J. , Mtunda, K. , Raya, M.D. , Muhanna, M. , Kiozia, H. , Rekha, A.R. , Colvin, J. and Thresh, J.M. (2005) Transmission of Cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 153, 307–312. [Google Scholar]
- Maruthi, M.N. , Bouvaine, S. , Tufan, H.A. , Mohammed, I.U. , Hillocks, R.J. and Fang, D.D. (2014) Transcriptional response of virus‐infected cassava and identification of putative sources of resistance for Cassava brown streak disease. PLoS One, 9, e96642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthi, M.N. , Jeremiah, S.C. , Mohammed, I.U. and Legg, J.P. (2017) The role of the whitefly, Bemisia tabaci (Gennadius), and farmer practices in the spread of cassava brown streak ipomoviruses. J Phytopathol. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y. , Mukasa, S.B. and Valkonen, J.P. (2009a) Cassava brown streak virus (Potyviridae) encodes a putative Maf/HAM1 pyrophosphatase implicated in reduction of mutations and a P1 proteinase that suppresses RNA silencing but contains no HC‐Pro. J. Virol. 83, 6934–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y.P. , Tugume, A.K. , Mukasa, S.B. , Tairo, F. , Kyamanywa, S. , Kullaya, A. and Valkonen, J.P. (2009b) Genetically distinct strains of Cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch. Virol. 154, 353–359. [DOI] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y.P. , Tugume, A.K. , Patil, B.L. , Yadav, J.S. , Bagewadi, B. , Abarshi, M.M. , Alicai, T. , Changadeya, W. , Mkumbira, J. , Muli, M.B. , Mukasa, S.B. , Tairo, F. , Baguma, Y. , Kyamanywa, S. , Kullaya, A. , Maruthi, M.N. , Fauquet, C.M. and Valkonen, J.P.T. (2011a) Evolution of Cassava brown streak disease‐associated viruses. J. Gen. Virol. 92, 974–987. [DOI] [PubMed] [Google Scholar]
- Mbanzibwa, D.R. , Tian, Y.P. , Tugume, A.K. , Mukasa, S.B. , Tairo, F. , Kyamanywa, S. , Kullaya, A. and Valkonen, J.P.T. (2011b) Simultaneous virus‐specific detection of the two Cassava brown streak‐associated viruses by RT‐PCR reveals wide distribution in East Africa, mixed infections, and infections in Manihot glaziovii . J. Virol. Methods, 171, 394–400. [DOI] [PubMed] [Google Scholar]
- McQuaid, C.F. , Sseruwagi, P. , Pariyo, A. and van den Bosch, F. (2015) Cassava brown streak disease and the sustainability of a clean seed system. Plant Pathol. 65, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaid, C.F. , van den Bosch, F. , Szyniszewska, A. , Alicai, T. , Pariyo, A. , Chikoti, P.C. and Gilligan, C.A. (2017) Spatial dynamics and control of a crop pathogen with mixed‐mode transmission. PLoS Comput. Biol. 13, e1005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed, I.U. , Abarshi, M.M. , Muli, B. , Hillocks, R.J. and Maruthi, M.N. (2012) The symptom and genetic diversity of Cassava brown streak viruses infecting cassava in East Africa. Adv. Virol. 2012, 795697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monger, W.A. , Seal, S. , Isaac, A.M. and Foster, G.D. (2001a) Molecular characterization of the Cassava brown streak virus coat protein. Plant Pathol. 50, 527–534. [Google Scholar]
- Monger, W.A. , Seal, S. , Cotton, S. and Foster, G.D. (2001b) Identification of different isolates of Cassava brown streak virus and development of a diagnostic test. Plant Pathol. 50, 768–775. [Google Scholar]
- Monger, W.A. , Alicai, T. , Ndunguru, J. , Kinyua, Z.M. , Potts, M. , Reeder, R.H. , Miano, D.W. , Adams, I.P. , Boonham, N. , Glover R.H. and Smith, J. (2010) The complete genome sequence of the Tanzanian strain of Cassava brown streak virus and comparison with the Ugandan strain sequence. Arch. Virol. 155, 429–433. [DOI] [PubMed] [Google Scholar]
- Moreno, I. , Gruissem, W. and Vanderschuren, H. (2011) Reference genes for reliable potyvirus quantitation in cassava and analysis of Cassava brown streak virus load in host varieties. J. Virol. Methods, 177, 49–54. [DOI] [PubMed] [Google Scholar]
- Mulimbi, W. , Phemba, X. , Assumani, B. , Kasereka, P. , Muyisa, S. , Ugentho, H. , Reeder, R. , Legg, J.P. , Laurenson, L. , Weekes, R. and Thom, F.E.F. (2012) First report of Ugandan cassava brown streak virus on cassava in Democratic Republic of Congo. New Dis. Rep. 26, 11. [Google Scholar]
- Mware, B.O. , Ateka, E.M. , Songa, J.M. , Narla, R.D. , Olubayo, F. and Amata, R. (2009) Transmission and distribution of cassava brown streak virus disease in cassava growing areas of Kenya. J. Appl. Biosci. 16, 864–870. [Google Scholar]
- Mwangangi, M., Ateka , E., Nyende, A. and Kagundu, A. (2014) Elimination of Cassava Brown Streak Virus from Infected Cassava. Journal of Biology, Agriculture and Healthcare. 4, 13. [Google Scholar]
- Ndunguru, J. , Sseruwagi, P. , Tairo, F. , Stomeo, F. , Maina, S. , Djinkeng, A. , Kehoe, M. , Boykin, L.M. and Melcher, U. (2015) Analyses of twelve new whole genome sequences of Cassava brown streak viruses and Ugandan cassava brown streak viruses from East Africa: diversity, supercomputing and evidence for further speciation. PLoS One, 10, e0139321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbery, F. , Qi, A. and Fitt, B.D. (2016) Modelling impacts of climate change on arable crop diseases: progress, challenges and applications. Curr. Opin. Plant Biol. 32, 101–109. [DOI] [PubMed] [Google Scholar]
- Ng, T.F.F. , Duffy, S. , Polston, J.E. , Bixby, E. , Vallad, G.E. , Breitbart, M. and Qiu, J. (2011) Exploring the diversity of plant DNA viruses and their satellites using vector‐enabled metagenomics on whiteflies. PLoS One, 6, e19050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, R.F.W. (1946) Breeding cassava for virus resistance. East Afr. Agric. J. 12, 184–194. [Google Scholar]
- Nichols, R.F.W. (1950) The brown streak disease of cassava. East Afr. Agric. J. 15, 154–160. [Google Scholar]
- Ntawuruhunga, P. and Legg, J. (2007). New spread of Cassava brown streak virus disease and its implications for the movement of cassava germplasm in the East and Central African region. Crop Crisis Control C3P Project Report http://c3project.iita.org/Doc/A25-CBSDbriefMay6.pdf
- Nzuki, I., Katari, M.S., Bredeson, J.V., Masumba, E., Kapinga, F., Salum, K. and Ferguson, M.E. (2017). QTL Mapping for Pest and Disease Resistance in Cassava and Coincidence of Some QTL with Introgression Regions Derived from Manihot glaziovii. Front Plant Sci. 8, 1168 10.3389/fpls.2017.01168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nweke, F. (2004) New challenges in the cassava transformation in Nigeria and Ghana. Int. Food Pol. Res. Inst. 118. [Google Scholar]
- Odipio, J. , Ogwok, E. , Taylor, N.J. , Halsey, M. , Bua, A. , Fauquet, C.M. and Alicai, T. (2013) RNAi‐derived field resistance to Cassava brown streak disease persists across the vegetative cropping cycle. GM Crops Food, 5, 16–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogwok, E. , Odipio, J. , Halsey, M. , Gaitán‐Solís, E. , Bua, A. , Taylor, N.J. , Fauquet, C.M. and Alicai, T. (2012) Transgenic RNA interference (RNAi)‐derived field resistance to Cassava brown streak disease. Mol. Plant Pathol. 13, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogwok, E. , Alicai, T. , Rey, M.E.C. , Beyene, G. and Taylor, N.J. (2014) Distribution and accumulation of Cassava brown streak viruses within infected cassava (Manihot esculenta Crantz) plants. Plant Pathol. 64, 1235–1246. [Google Scholar]
- Ogwok, E. , Ilyas, M. , Alicai, T. , Rey, M.E.C. and Taylor, N.J. (2016) Comparative analysis of virus‐derived small RNAs within cassava (Manihot esculenta Crantz) infected with Cassava brown streak viruses . Virus Res. 215, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariyo, A. , Tukamuhabwa, P. , Baguma, Y. , Kawuki, R.S. , Alicai, T. , Gibson, P. , Kanju, E. , Wanjala, B.W. , Harvey, J. , Nzuki, I. and Rabbi, I.Y. (2013) Simple sequence repeat (SSR) diversity of cassava in South, East and Central Africa in relation to resistance to Cassava brown streak disease. Afr. J. Biotechnol. 12, 4453–4464. [Google Scholar]
- Patil, B.L. , Ogwok, E. , Wagaba, H. , Mohammed, I.U. , Yadav, J.S. , Bagewadi, B. , Taylor, N.J. , Kreuze, J.F. , Maruthi, M.N. , Alicai, T. and Fauquet. C.M. (2011) RNAi‐mediated resistance to diverse isolates belonging to two virus species involved in Cassava brown streak disease. Mol. Plant Pathol. 12, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, B.L. , Legg, J.P. , Kanju, E. and Fauquet, C.M. (2015) Cassava brown streak disease: a threat to food security in Africa. J. Gen. Virol. 96, 956–968. [DOI] [PubMed] [Google Scholar]
- Pautasso, M. , Aistara, G. , Barnaud, A. , Caillon, S. , Clouvel, P. , Coomes, O.T. , Delêtre, M. , Demeulenaere, E. , De Santis, P. , Döring, T. and Eloy, L. (2013) Seed exchange networks for agrobiodiversity conservation. A review. Agron. Sustain. Dev. 33, 151–175. [Google Scholar]
- Pennisi, E. (2010) Armed and dangerous. Science, 327, 804–805. [DOI] [PubMed] [Google Scholar]
- Petrussa, E. , Braidot, E. , Zancani, M. , Peresson, C. , Bertolini, A. , Patui, S. and Vianello, A. (2013) Plant flavonoids: biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 14, 14 950–14 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabha, K. , Baranwal, V.K. and Jain, R.K. (2013) Applications of next generation high throughput sequencing technologies in characterization, discovery and molecular interaction of plant viruses. Indian J. Virol. 24, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik, S. , Marri, P.R. , Desany, B. , Rabinowicz, P.D. , Kodira, C. , Mohiuddin, M. , Rodriguez, F. , Fauquet, C. , Tohme, J. , Harkins, T. , Rokhsar, D.S. and Rounsley, S. (2012) The Cassava genome: current progress, future directions. Trop. Plant Biol. 5, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux‐Cuvelier, M. , Teyssedre, D. , Chesneau, T. , Jeffray, C. , Massé, D. , Jade, K. , Abdoul Karime, A.L. , Hostachy, B. , Reynaud, B. , Legg, J.P. and Lett, J.M. (2014) First report of Cassava brown streak disease and associated Ugandan cassava brown streak virus in Mayotte Island. New Dis. Rep. 30, 28. [Google Scholar]
- Rwegasira, G. and Chrissie, M. (2015) Efficiency of non‐vector methods of Cassava brown streak virus transmission to susceptible cassava plants. Afr. J. Food Agric. Nutr. Dev. 15, 10 335–10 351. [Google Scholar]
- Shirima, R.R. , Maeda, D.G. , Kanju, E. , Ceasar, G. , Tibazarwa, F.I. and Legg, J.P. (2017) Absolute quantification of Cassava brown streak virus mRNA by real‐time qPCR. J. Virol. Methods, 245, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, H.H. (1936) Virus diseases of East African plants. East Afr. Agric. J. 1, 333–337. [Google Scholar]
- Storey, H. (1939) Report of the Plant Pathologist . Annual Report East African Agricultural Research. East African Agricultural (Amani), Tanzania. [Google Scholar]
- Taylor, N.J. , Sekabira, H.A. , Sibiko, K.W. , Bua, A. and Lynam, J.K . (2016) Disease‐resistant GM cassava in Uganda and Kenya during a pandemic. B4FA. Banson: Cambridge.
- Thresh, J.M. and Cooter, R.J. (2005) Strategies for controlling Cassava mosaic virus disease in Africa. Plant Pathol. 54, 587–614. [Google Scholar]
- Thresh, J.M. , Fargette, D. and Otim‐Nape, G.W. (1994) The viruses and virus diseases of cassava in Africa. Afr. Crop Sci. J. 2, 459–478. [Google Scholar]
- Tomlinson, J.A. , Ostoja‐Starzewska, S. , Adams, I.P. , Miano, D.W. , Abidrabo, P. , Kinyua, Z. , Alicai, T. , Dickinson, M.J. , Peters, D. , Boonham, N. and Smith, J. (2013) Loop‐mediated isothermal amplification for rapid detection of the causal agents of Cassava brown streak disease. J. Virol. Methods, 191, 148–154. [DOI] [PubMed] [Google Scholar]
- Tumuhimbise, R. , Shanahan, P. , Melis, R. and Kawuki, R. (2014) Combining ability analysis of storage root yield and related traits in cassava at the seedling evaluation stage of breeding. J. Crop Improv. 28, 530–546. [Google Scholar]
- Turyagyenda, L. , Kizito, E.B. , Ferguson, M.E. , Baguma, Y. , Harvey, J.W. , Gibson, P. , Wanjala, B.W. and Osiru, D.S.O. (2012) Genetic diversity among farmer‐preferred cassava landraces in Uganda. Afr. Crop Sci. J. 20, 15–30. [Google Scholar]
- Valli, A. , Dujovny, G. and García, J.A. (2008) Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor p1b from Cucumber vein yellowing ipomovirus . J. Virol. 82, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli, A. , López‐Moya, J.J. and García, J.A. (2015) Potyviridae In: ELS, pp. 1–10. Chichester: John Wiley. [Google Scholar]
- Vanderschuren, H. , Moreno, I. , Anjanappa, R.B. , Zainuddin, I.M. , Gruissem, W. and Zhang, T. (2012) Exploiting the combination of natural and genetically engineered resistance to Cassava mosaic and Cassava brown streak viruses impacting cassava production in Africa. PLoS One, 7, e45277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitharani, R. , Chellappan, P. , Pita, J.S. and Fauquet, C.M. (2004) Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78, 9487–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, R.K. , Ribaut, J.‐M. , Buckler, E.S. , Tuberosa, R. , Rafalski, J.A. and Langridge, P. (2012) Can genomics boost productivity of orphan crops? Nat. Biotechnol. 30, 1172–1176. [DOI] [PubMed] [Google Scholar]
- Wagaba, H. , Beyene, G. , Aleu, J. , Odipio, J. , Okao‐Okuja, G. , Chauhan, R.D. , Munga, T. , Obiero, H. , Halsey, M.E. , Ilyas, M. , Raymond, P. , Bua, A. , Taylor, N.J. , Miano, D. and Alicai, T. (2017) Field level RNAi‐mediated resistance to Cassava brown streak disease across multiple cropping cycles and diverse East African agro‐ecological locations. Front. Plant Sci. 7, 2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann. Bot. 6, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Were, H.K. , Winter, S. and Maiss, E. (2004) Viruses infecting cassava in Kenya. Plant Dis. 88, 17–22. [DOI] [PubMed] [Google Scholar]
- Winter, S. , Koerbler, M. , Stein, B. , Pietruszka, A. , Paape, M. and Butgereitt, A. (2010) Analysis of Cassava brown streak viruses reveals the presence of distinct virus species causing Cassava brown streak disease in East Africa. J. Gen. Virol. 91, 1365–1372. [DOI] [PubMed] [Google Scholar]
- Yadav, J. , Ogwok, E. , Wagaba, H. , Patil, B.L. , Bagewadi, B. , Alicai, T , Gaitan‐Solis, E. , Taylor, N.J. and Fauquet, C. M. (2011) RNAi‐mediated resistance to Cassava brown streak Uganda virus in transgenic cassava. Mol. Plant Pathol. 12, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev, R. , Ueki, S. , Epel, B.L. and Citovsky, V. (2011) Biology of callose (β‐1,3‐glucan) turnover at plasmodesmata. Protoplasma, 248, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]


