Abstract
Results from replica-exchange and regular room temperature molecular dynamics simulations of the Alzheimer’s amyloid monomer in an implicit solvent are reported. Our data indicate that at room temperature, the monomer assumes random-coil and soluble conformations. No beta content is observed which therefore seems to be a product of oligomerization and aggregation of monomers.
I. INTRODUCTION
Alzheimer’s disease (AD) is a neurological disorder that affects approximately people worldwide. Its molecular hallmark is the presence of senile plaques made up of insoluble deposits of amyloid beta peptides.1 These peptides, to , are released from amyloid precursor protein and have a chain length of 39–42 amino acid.2,3 Mass spectrometry and solid state NMR spectroscopy indicates the formation of either parallel or antiparallel strands (depending on the length of the fragment)4–7 that are placed perpendicular to the fibril axis. Less is known about the structure of monomer as it easily aggregates and has a noncrystalline nature. The NMR study of unmodified fragments of and monomers in water indicated the regions of localized stability, but they do not resolve the peptide at atomic resolution.8 Circular dichroism (CD) experiments show that monomeric is disordered, indicating 70% random, sheet, and 5% helix structure.9,10 Whether these frequencies correspond to distinct clusters of conformations is not known.
Computational approaches provide an alternate means to study the fibrillogenesis process. Ma and Nussinov11 suggested that at high temperature fragments and form a strand-loop-strand structure with parallel sheet, with an interior salt bridge between Asp23 and Lys28 residues as a major element of the fibril structure. On the other hand, computational studies using coarse-grained as well as atomistic atom models suggest the formation of antiparallel sheets by subpeptides.12 The relation between this antiparallel structure and the nucleation and fibril forming properties of in-register parallel structures formed by the full length and peptides is not clear, and no simulation has so far reproduced the experimentally measured secondary structure contents for peptides.
These discrepancies reflect the numerical difficulties of computational aggregation studies. Regular canonical simulations at low temperatures tend to be trapped in one of a huge number of local minimum energy states in a rough energy landscape. This difficulty is alleviated, in part, by the replica-exchange method, also known as parallel tempering.13,14 Here, a number of replicas are simulated at different temperatures and periodically swapped between the neighboring temperatures. Over the course of simulation, each replica walks along a ladder of temperatures. In that way, a replica will not only find local minima but can also escape from them. As a consequence, sampling at low temperature is much faster than when all computational resources are set solely in that temperature.13
Following earlier work,15 we report in this article replica-exchange molecular dynamics (MD) simulations of monomer. The prohibitive cost of simulations with explicit solvent molecules forces us to approximate the interaction between the peptide and the surrounding water by an implicit solvent model. We observe that at room temperature, solvated monomer does not assume a unique folded conformation but exists as a mixture of rapidly interconverting conformations. Three distinct clusters dominate, characterized by a random-coil-like structure and only limited helicity is present. The conformation with the lowest energy is part of one of these clusters and is characterized by turn regions spanning Ala21-Asn27 residues. Slightly higher in energy are conformations with solely a turn around Val12-Leu17 residues and suchconformers that have a helical region in the N-terminal, i.e., similar to the one observed in NMR studies done in trifluoroethanol (TFE)/water solution.16
II. MATERIAL AND METHODS
All calculations were carried out with the AMBER 9.0 (assisted model building with energy refinement)17 simulation package using the ff99 force field.18 The effect of solvation was approximated by a generalized Born solvent-accessible surface area19 implicit solvent model (bond radii of , solvent dielectric constant of 78.5, and surface tension of ). No attempt was made to modify the solvent parameters to capture their temperature dependence. The SHAKE algorithm was used to constrain all bond lengths to their equilibrium distances.20
The peptide consists of 39 amino-acidresidues: [Asp-Ala-Glu-Phe-Arg-His-Asp-Ser-Gly-Tyr-Glu-Val-His-His-Gln-Lys-Leu-Val-Phe-Phe-Ala-Glu-Asp-Val-Gly-Ser-Asn-Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met-Val-Gly-Gly-Val]. Canonical simulations were started both from NMR structures (PDB Id.: 1AML) and a completely extended conformation. 5000 steps of steepest decent minimization was followed by an initial equilibration run consisting of 50 000 steps of MD at time steps with a cutoff radius of .
The replica-exchange MD was implemented by using the Sander MD program. The 32 two replicas (fully extended configuration) were simulated over a range of temperatures from (200, 207, 215, 223, 232, 241, 250, 260, 270, 280, 291, 302, 313, 325, 338, 351, 364, 378, 392, 407, 423, 439, 456, 474, 492, 510, 530, 550, 571, 593, 616, 640). Exchange attempts are made after every . Replica temperatures are maintained at the appropriate temperatures by a combination of velocity reassignment and a Langevin thermostat with a collision frequency21 of . Initial are discarded and only the last are analyzed. For comparison, we have also performed five regular canonical MD simulations of length. The temperature is set by velocity reassignment from a Maxwell–Boltzmann distribution at and maintained at that temperature by using a Langevin thermostat with a collision frequency of .
Using multiscale modeling tools for structural biology (MMTSB),22 all structures collected in the replica-exchange MD production phase are grouped into clusters of similar geometry. Clustering is done according to the root mean square deviation (RMSD) over backbone atoms between configurations. Only clusters containing more than ten members are analyzed further. Final average structures are converted to the PDB format with program PYMOL.23 For calculations of molecular properties, all snapshot structures were used and analyzed with the program DSSP.24 Atoms within helical signatures (classes H alpha helix, G 3-helix ( helix), I 5 helix ( helix)) are counted and compared to the total number of atoms resulting in values for the relative helical content. For the calculation of the specific heat,25 we compute the heat capacities from finite differences, i.e., , with as the average total energy at some temperature . The radius of gyration and the fraction of helical content are calculated by using the AMBER ptraj module and DSSP.24
III. RESULTS AND DISCUSSION
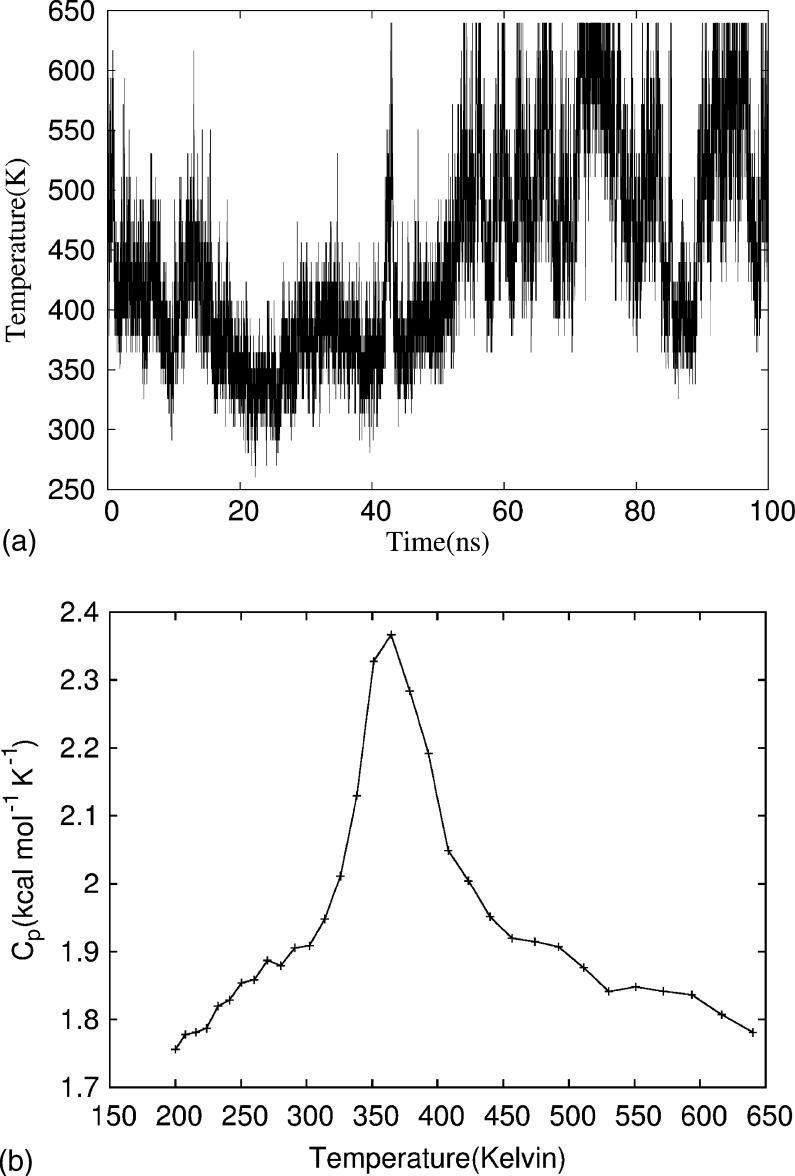
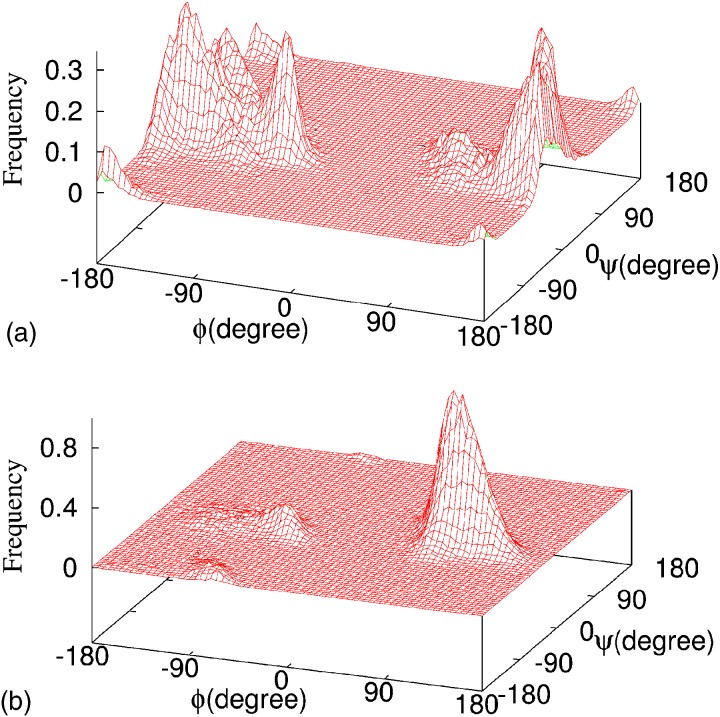
In order to probe the structure of monomer, we have performed multiple replica-exchange and regular canonical MD (at ) simulations starting with different initial configurations. Figure 1 shows the random walk for one replica in temperatures. Associated with it is a walk in energy, allowing the replica to escape local minima. As a consequence, reliable physical quantities can be calculated over the whole range of temperatures. An example is the specific heat plotted in Fig. 1, giving an approximate melting temperature of , i.e., above our temperature of interest . With the exception of a single canonical MD run, that got stuck in a local minimum with higher energy than found in all other runs, all simulations set at to similar U-shape structures, however, the energy is lower in the replica-exchange simulation. The improved sampling at room temperature is also seen in Fig. 2, where we compare the distribution of dihedral angles of Gly25. At room temperature, the replica-exchange simulation samples a much wider part of configuration space as the conventional canonical MD run.26
FIG. 1.
(a) Time series of temperature exchange for one of the replicas in the replica-exchange MD simulation. (b) Specific heat as a function of temperature. The line connecting the data points is guide for the eyes.
FIG. 2.
Distribution of dihedral angles of Gly25 for obtained (a) from a replica-exchange MD simulation and (b) from regular canonical MD simulation.
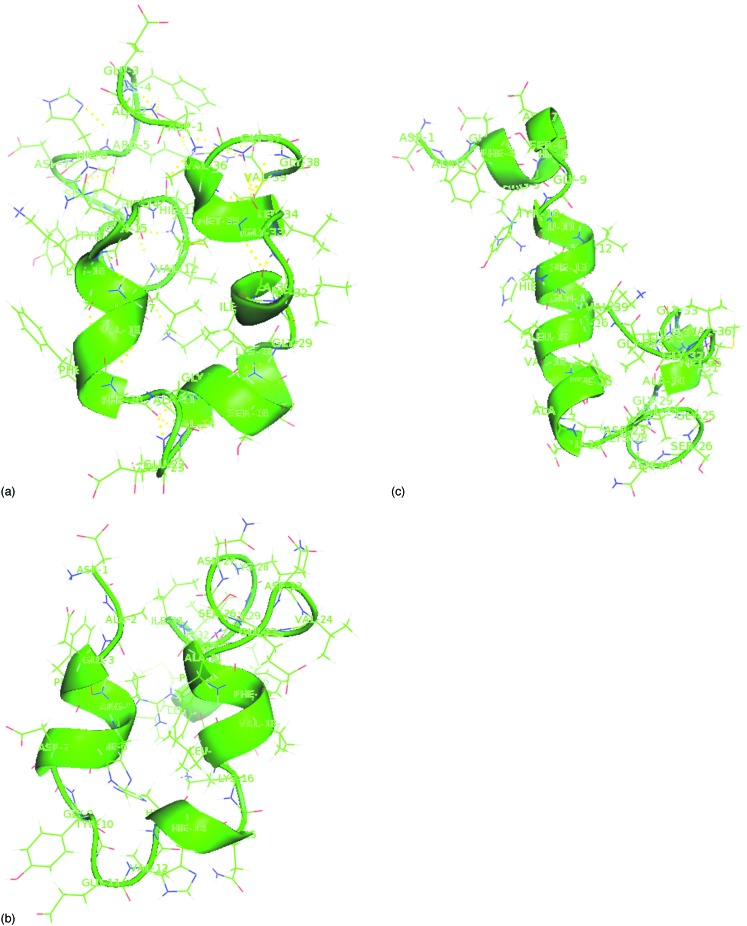
At room temperature, the monomer does not have a unique folded conformation in water but adopts one of the several low energy structures as seen, for instance, in Fig. 3. The root mean square displacement based clustering analysis22 shows that the ensemble at consists of 37 clusters. Of these, 26 can be further grouped by visual inspection into three super clusters. These three clusters differ little in their average potential energies [cluster A, ; cluster B, ; cluster C, ]. All three clusters share as a common theme the U-shape configurations that are strongly amphipathic. The longer arm is made mostly of hydrophilic and charged amino-acid residues, while the shorter arm is formed exclusively by hydrophobic branched residues. This “U-shape” is detrimental to the structure adopted by the sequence in the full protein. On the other hand, NMR results show conformations varying from -helical structure in a nonpolar solution8 to disordered N-terminal and C-terminal tails with consistent turn regions as determined by electrospray mass spectroscopy and implicit solvent MD.26,27 The side chain of most hydrophobic residues found in the shorter arm point outward, providing a hydrophobic platform for an association between monomer. This is consistent with the simulation by Tarus et al.28 of dimerization of using umbrella sampling and MD. They show also that hydrophobic interactions between monomers are the stabilizing forces and that the dimerization process involves substantial structure reorganization of both the C and N termini.
FIG. 3.
The three dominant clusters of configurations seen in the simulations.
A more thorough analysis reveals differences between the clusters. The dominant one, cluster A, is a random coil structure with turn around Ala21-Asn27 and appears with a frequency of 58%. It agrees well with the NMR studies of the fibril.5 The turn around of these residues are not unexpected as the Gly25, Ser26, and Asp227 residues have a high probability to be within a beta turn.29 Only marginal secondary content (3–10 helix, helix and turns as determined by DSSP program,24 but no structures) are observed for configurations of this cluster. The region around Leu17-Ala21 residues is well defined and its structure fluctuates considerably less than rest of the peptide. This is consistent with NMR studies,8,30,31 which suggest a central hydrophobic region, Leu17-Ala21, with well defined structure in the peptide, and this region is found to be critical for fibril formation.32 However, cluster A deviates from the ones found in NMR experiments of in TFE/water.16 The average main chain RMSD between cluster A structure and the 20 NMR models of the 1AML.PDB is , where structural superposition is done by using sequence alignment on 1–39 residues.
Configurations of cluster B are also random coil but show a turn around Val12-Leu17. This cluster appears at with a frequency of 29%. Only in these configurations, we observed the salt bridges between Asp23-Lys28/16 residues (Fig. 3) that have been reported in various experiments as controlling the aggregation rate of .33 In agreement with Baumketner and Shea,34 we find that this salt bridge is easily formed and stabilizes the U-type structure. Other polar contacts that stabilize the structure are the ones between carboxyl group of the side chain of Asp7 and the amino group in the side chain of Lys28, between the side chain of Glu1 and the side chain group of Arg5, and between the side chain amidegroup of Lys16 and the side chain of Asp7. The optimized structures have a short right handed helix region with dihedral angle extending around Gly15-Val18 residues. The type1 turns are present around Arg5-Ser8 residues with values of the residue and residue in degrees; and around Leu17-Phe20 residues with and values of residue and residue . turns regions are also observed, stabilized by an extensive although not a regular array of hydrogen bond between the group of the residue and the NH group of the residue. Carbonyl-carbonyl interactions are alsoobserved with various distances such as , , etc., and these distances lie in the range of .
Finally, 13% of configurations are rich in helicity and form cluster C. Configurations in this cluster are similar to the structure that was determined in a 40% TFE/water solution. The average main chain RMSD between cluster C configurations and the 20 NMR models of the 1AML entry in the protein data bank is . The structural comparison is done by using sequence alignment on 1–39 residues. Cluster C exhibits a helix around Gln11-Glu22 residues, another around Ile32-Met35 residues, and a coiled region around Val24-Ala30 residues. All three elements are in agreement with the NMR experiment16 in 40% TFE/watersolution.
Our results indicate that the experimentally measured secondary structure contents are thus the consequences of elemental contributions from three clusters. About 90% of configurations belong to either cluster A or B, and show characteristics found also in the fibrillar conformations. Cluster A exhibits the turn between residues around Asp23 and Lys28 found in NMR studies of the fibrillar structure.5 However, only the smaller cluster B has the salt bridges between Asp23-Lys28/16 residues that have been reported in various experiments33 as controlling the aggregation rate of . Following the comparison with the experimental results, we conjecture that configurations of this cluster are especially prone for oligomerization and aggregation. The remaining of configurations include the helix rich structures of cluster C. These are similar to the tertiary structure of 1AML.PDB.16 While that structure was obtained by NMR for the monomer in a TFE/water mixture, our results show that this structure appears also in the more physiologically relevant case of the monomer in water.
The percentage of ordered residues in aqueous in our simulations agrees reasonably with the NMR data35,36 that estimate the amount of disordered structure to be 60%–80%, and helices to be 10%. However, our simulations did not reveal the origin of the 25% content found in CD studies for disordered in water. We conjecture that the higher percentage of ordered structure (70% random, 25% , and 5% helical), and especially the content, in the CD experiment results from the fact that these experiments probe not only the monomer (as our work) but also a dynamic equilibrium between monomers and oligomers (i.e., dimer and trimer). If this assumption is correct, it would indicate that -structures are only formed in the processes of oligomerization, and are not a feature of the monomer. The brain microenvironmental influences are required to convert these configurations in sheets during the process of -amyloidosis in AD.
Since the molecule does not contain any regular secondary structure element (except a very short small region), most of the potential hydrogen bonding sites are free, and it is the hydration of these moieties and other hydrophilic groups that makes them soluble. Solubility is also supported by the free hydrogen bonding sites at the ends of both the arms.
IV. CONCLUSIONS
Our study demonstrates that increased computational power and sophisticated sampling techniques allow now a detailed analysis of the folding and interactions of peptides. Replica-exchange MD simulations illustrate that monomer at room temperature does not have a unique structure in water but adopt one of the several possible low-energy conformations. Three distinct families of mostly random coil-like structures dominate, with one cluster having a turn around residues of 21–27, another with few helical elements present, and a third having a turn around residues of 12–17. The exposed free hydrogen bonding sites indicate that the monomer is soluble. However, no stable -structure has been observed. This indicates that the sheets observed in fibrillar form only later in the process of oligomerization and aggregation while at the onset of the process are monomers that exist as soluble random coil configurations. We hope that further analysis of the interactions stabilizing the “U-like” structure in the monomer, with a longer arm rich in hydrophilic and charged residues and the shorter arm dominated by hydrophobic residues, will help to understand the process of dimerization and oligomerization of .
ACKNOWLEDGMENTS
This work was supported by DAAD grants (Desk:422 and PKZ:A/07/93172). We would like to thank Sandipan Mohanty for his useful discussion throughout this study. All calculations were done on computers of the John von Neumann Institute for Computing, Research Center Jülich, Jülich, Germany.
REFERENCES
- 1.Alzheimer A., Centralblatt fur Nervenheilkunde und Psychiatric 30, 177 (1907).; [Google Scholar]; Stelzmann Rainulf A., Schnitzlein H. Norman, and Murtagh F. Reed, Clinical Anatomy 8, 429 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D. J., Nature (London) 10.1038/19866 399, A23 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Dahlgren K. N., Manelli A. M., Stine W. B., Baker L. K., Krafft G. A., and LaDu M. J., J. Biol. Chem. 10.1074/jbc.M201750200 277, 32046 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Tycko R., Prog. Nucl. Magn. Reson. Spectrosc. 10.1016/S0079-6565(03)00003-7 42, 53 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petkova A., Ishii T. Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., and Tycko R., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.262663499 99, 16742 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torok M., Milton S., Kayed R., Wu P., McIntiri T., Glabe C. G., and Langen R., J. Biol. Chem. 10.1074/jbc.M205659200 277, 40810 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Serpell L. C., Biochim. Biophys. Acta 10.1016/S0925-4439(00)00029-6 1502, 16 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Hou L., Shao H., Zhang Y., Li H., Menon N. K., Neuhaus E. B., Brewer J. M., Byeona I. J. L., Ray D. G., Vitek M. P., Iwashita T., Makula R. A., Przybyla A. B., and Zagorski M. G., J. Am. Chem. Soc. 10.1021/ja036813f 126, 1992 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Fezori Y. and Teplow D. B., J. Biol. Chem. 10.1074/jbc.M204168200 277, 36948 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Chen Y. R., Huang H. B., and Chyan C. L., J. Biochem. (Tokyo) 139, 733 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Ma B. Y. and Nussinov R., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.212206899 99, 14126 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favrin G., Irbäck A., and Mohanty S., Biophys. J. 10.1529/biophysj.104.046839 87, 3657 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansmann U. H. E., Chem. Phys. Lett. 10.1016/S0009-2614(97)01198-6 281, 140 (1997). [DOI] [Google Scholar]
- 14.Sugita Y. and Okamoto Y., Chem. Phys. Lett. 10.1016/S0009-2614(99)01123-9 314, 141 (1999). [DOI] [Google Scholar]
- 15.Banerjee Soma, Conformational analysis and molecular modeling of AB assembly; implications for Alzheimer's disease, MSc Thesis, Panjab University Chandigarh (2003).
- 16.Stricht H., Bayer P., Willbold D., Dames S., Hilbich C., Beyreuther K., Frank R. W., and Rosch P., Eur. J. Biochem. 10.1111/j.1432-1033.1995.293_1.x 233, 293 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Case D. A., Darden T. A., Cheatham T. E. III, Simmerling C. L., Wang J., Duke R. E., Luo R., Merz K. M., Pearlman D. A., Crowley M., Walker R. C., Zhang W., Wang B., Hayik S., Roitberg A., Seabra G., Wong K. F., Paesani F., Wu X., Brozell S., Tsui V., Gohlke H., Yang L., Tan C., Mongan J., Hornak V., Cui G., Beroza P., Mathews D. H., Schafmeister C., Ross W. S., and Kollman P. A., AMBER 9, University of California, San Francisco (2006).
- 18.Wang J., Cieplak P. and Kollman P. A., J. Comput. Chem. 21, 1049 (2000). [DOI] [Google Scholar]
- 19.Hawkins G. D., Cramer C. J., and Truhlar D. G., J. Phys. Chem. 10.1021/jp961710n 100, 19824 (1996). [DOI] [Google Scholar]
- 20.Gunsteren W. F. and Berendsen H. J. C., Mol. Phys. 10.1080/00268977700102571 34, 1311 (1977). [DOI] [Google Scholar]
- 21.Izaguirre J. A., Catarello D. P., Wozniak J. M., and Skeel R. D., J. Chem. Phys. 10.1063/1.1332996 114, 2090 (2001). [DOI] [Google Scholar]
- 22.Feig M., Karanicolas J., and Brooks C. L. III, J. Mol. Graphics Modell. 10.1016/j.jmgm.2003.12.005 22, 377 (2004). [DOI] [PubMed] [Google Scholar]
- 23.DeLano W. L., The PYMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA, 2002.
- 24.Kabsch W. and Sander C., Biopolymers 10.1002/bip.360221211 22, 2577 (1983). [DOI] [PubMed] [Google Scholar]
- 25.Pitera J. W. and Swope W., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.1330954100 100, 7587 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao H., Jao S., Ma K., and Zagorski M. G., J. Mol. Biol. 10.1006/jmbi.1998.2348 285, 755 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Crescenzi O., Tomaselli S., Guerrini R., Salvadori S., Ursi A. M. D., Temussi P. A., and Picone D., Eur. J. Biochem. 10.1046/j.1432-1033.2002.03271.x 269, 5642 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Tarus B., Straub J. E., and Thirumalai D., J. Mol. Biol. 10.1016/j.jmb.2004.11.022 345, 1141 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Chou P. Y. and Fasman G. D., Biochemistry 10.1021/bi00699a002 13, 222 (1974). [DOI] [PubMed] [Google Scholar]
- 30.Riek R., Guntert P., Dobeli H., Wipf B., and Wüthrich K., Eur. J. Biochem. 10.1046/j.0014-2956.2001.02537.x 268, 5930 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Zhang S., Iwata K., Lachenmann M. J., Peng J. W., Li S., Stimson E. R., Lu Y. A., Felix A. M., Maggio J. E., and Lee J. P., J. Struct. Biol. 10.1006/jsbi.2000.4288 130, 130 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Fraser P. E., Nguyen J. T., Surewicz W. K., and Kirschner D. A., Biophys. J. 60, 1190 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sciarretta K. L., Gordan D. L., Petkova A. T., Tycko R., and Meredith S. C., Biochemistry 10.1021/bi0474867 44, 6003 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Baumketner A. and Shea J. E., J. Phys. Chem. B 10.1021/jp051325a 109, 21322 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Walsh D. M., Hartley D. M., Kusumoto Y., Fezoui Y., Condron M. M., Lomakin A., Benedek G. B., Selkoe D. J., and Teplow D. B., J. Biol. Chem. 10.1074/jbc.274.36.25945 274, 25945 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Kirkitadze M., Condron M. M., and Teplow D. B., J. Mol. Biol. 10.1006/jmbi.2001.4970 312, 1103 (2001). [DOI] [PubMed] [Google Scholar]