Abstract
Premise of the Study
Toona (Meliaceae, Sapindales) is a small genus of five species of trees native from southern and eastern Asia to New Guinea and Australia. Complete plastomes were sequenced for three Toona species to provide a basis for future plastome genetic studies in threatened species of Toona. In addition, plastome structural evolution and phylogenetic relationships across Sapindales were explored with a larger data set of 29 Sapindales plastomes (including members of six out of nine families).
Methods
The plastomes were determined using the Illumina sequencing platform; the phylogenetic analyses were conducted using maximum likelihood by RAxML.
Results
The lengths of three Toona plastomes range from 159,185 to 158,196 bp. A total of 113 unique genes were found in each plastome. Across Sapindales, plastome gene structure and content were largely conserved, with the exception of the contraction of the inverted repeat region to exclude ycf1 in some species of Rutaceae and Sapindaceae, and the movement of trnI‐GAU and trnA‐UGC to a position outside the inverted repeat region in some Rutaceae species.
Discussion
The three Toona plastomes possess the typical structure of angiosperm plastomes. Phylogenomic analysis of Sapindales recovered a mostly strongly supported phylogeny of Sapindales, including most of the backbone relationships, with some improvements compared to previous targeted‐gene analyses.
Keywords: phylogenomic analysis, plastome, Sapindales, structure, Toona
Toona (Endl.) M. Roem., commonly known as red cedar, is a small genus of trees in the mahogany family (Meliaceae subfam. Cedreloideae). It is distributed across southern and eastern Asia, New Guinea, and eastern Australia (Mabberley, 2008). Toona was previously treated as a section of Cedrela P. Browne (Meliaceae), but the latter is now circumscribed to include only species of the Neotropics (Muellner et al., 2009). Approximately five species of Toona are currently recognized following the treatment by J. M. Edmonds (1995): T. calantas Merr. & Rolfe, T. ciliata M. Roem., T. fargesii A. Chev., T. sinensis (A. Juss.) M. Roem., and T. sureni (Blume) Merr. (Fig. 1). Several of these species are economically important as timber trees (e.g., T. ciliata and T. sureni; Peng and Edmonds, 2008) or as ornamental, including T. sinensis, which is the most cold‐tolerant species in Meliaceae and the only member of the family that can be cultivated successfully in northern Europe (Rushforth, 1999). Wild populations of most Toona species are under threat due to habitat loss and logging, especially the extremely rare T. fargesii, which may be endemic to China (Peng and Edmonds, 2008).
Figure 1.
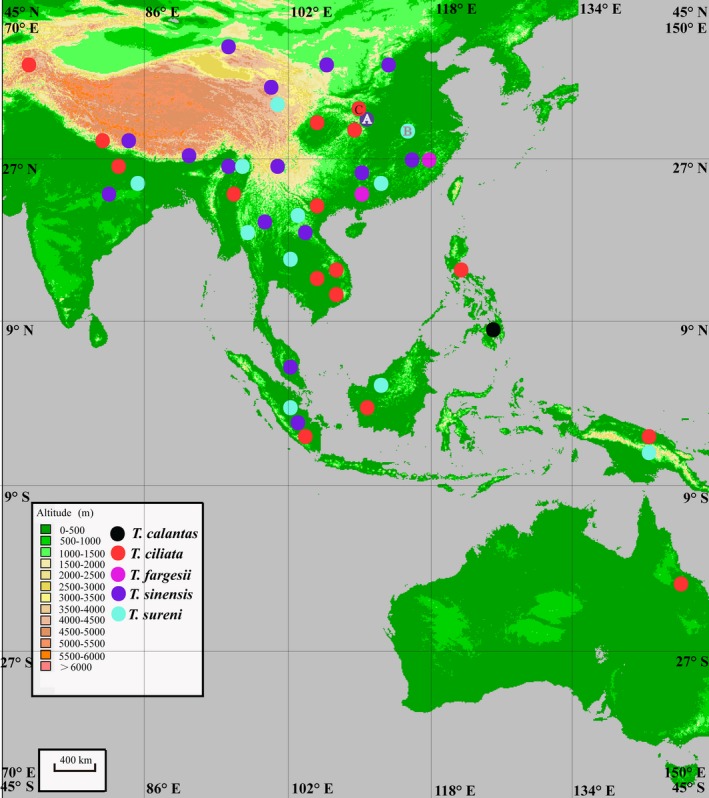
The distribution pattern of Toona. The colored dots represent the species range. A, B, and C indicate the sampling localities of three Toona species sequenced in the present study.
The large pantropical family Meliaceae is a member of the order Sapindales (Angiosperm Phylogeny Group, 2016) and consists of 50 genera and more than 650 species (Stevens, 2001 onwards). Meliaceae is strongly supported as monophyletic and consists of two subfamilies: Cedreloideae and Melioideae (Muellner et al., 2003). A recent phylogenetic study of Sapindales based on plastid rbcL, atpB, and trnL‐trnF sequences (Muellner‐Riehl et al., 2016) found that Simaroubaceae was sister to Meliaceae, with moderate support. Together, these two families formed a strongly supported clade with Rutaceae. Relationships among the remaining families of Sapindales were mostly moderately to strongly supported. Resolution and support found in Muellner‐Riehl et al. (2016) represent improvements over earlier studies based on fewer loci (e.g., Gadek et al., 1996; Muellner et al., 2007).
Phylogenetic data sets based on large numbers of plastid loci have the potential to resolve relationships that have resisted resolution using only a few loci, as has been demonstrated in many recent studies (e.g., Stull et al., 2015; Duvall et al., 2016). Plastomes are generally conserved in structure, gene content, and gene order (Green, 2011; Ruhlman and Jansen, 2014), although rearrangements and gene loss have been detected in a number of lineages and most differences in plastome gene number are related to fluctuations in the size of the inverted repeat (IR) region (e.g., Guisinger et al., 2011; Knox, 2014; Zhu et al., 2016). To date, complete plastomes of 26 species across six families are available for Sapindales, including one Meliaceae species (Azadirachta indica A. Juss., Melioideae). Although McPherson et al. (2013) sequenced the T. ciliata plastome for phylogeographical study of this species in Australia, the plastome structure of this species was not reported, and the assembled plastome sequences of this species are not openly available. Additional sequenced plastomes from Meliaceae as well as across Sapindales may help to improve our understanding of phylogenetic relationships within the order and would provide insight into plastome evolution in this clade. In this study, we sequenced and characterized the complete plastomes of three Toona species and downloaded all 26 available Sapindales plastomes from GenBank, with the following objectives: (1) to provide a basis for future plastome genetic studies in threatened species of Toona, (2) to determine whether plastomes can resolve phylogenetic relationships among families of Sapindales, and (3) to evaluate plastome structure evolution across Sapindales.
METHODS
Fresh leaves of T. sinensis, T. sureni, and T. ciliata were obtained from Wuhan Botanical Garden (30.54°N, 110.42°E), Lushan Botanical Garden (29.55°N, 115.99°E), and the National Nature Reserve of Shi‐Ba‐Li valley (31.34°N, 109.92°E), respectively. Vouchers were deposited at the Herbarium of Wuhan Botanical Garden, Chinese Academy of Sciences (HIB) (Table 1). High‐quality plastid DNA was obtained following the plastid DNA extraction method of Shi et al. (2012). Approximately 30 g of fresh, young leaf tissue was used for each species, and for each plastome a DNA TruSeq Illumina (Illumina Inc., San Diego, California, USA) sequencing library, with 500‐bp insert sizes, was constructed at the Beijing Genomics Institute (BGI) in Wuhan, Hubei, China, using 2.5–5 ng of sonicated plastid DNA. An Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California, USA) and quantitative PCR were used to quantify DNA amounts in the libraries. Libraries were multiplexed by TruSeq adapter and 150‐bp paired‐end sequenced on an Illumina HiSeq 2000 platform at BGI (Wuhan, Hubei, China). The raw data are available from the National Center for Biotechnology Information Sequence Read Archive (accession no. SRR6146642, SRR6146640, and SRR6146641).
Table 1.
Taxa used in present study. Collection locality and voucher information are provided for newly sequenced plastomes
| Family | Species | Collection locality | Voucher information | GenBank accession no. |
|---|---|---|---|---|
| Anacardiaceae | Rhus chinensis Mill. | Yanggu, Korea | IM151120‐1 (Lee et al., 2016) | NC_033535 |
| Anacardiaceae | Spondias bahiensis P. Carvalho, Van den Berg & Machado | NA | NA | NC_030526 |
| Anacardiaceae | Spondias tuberosa L. | NA | NA | NC_030527 |
| Burseraceae | Boswellia sacra Flueck. | Natural Park | UC29 (Kohany et al., 2006) | NC_029420 |
| Meliaceae | Azadirachta indica A. Juss. | NA | NA | NC_023792 |
| Meliaceae | Toona ciliata M. Roem. | SBL | Nan.Lin‐521(HIB) | MF467523 |
| Meliaceae | Toona sinensis (A. Juss.) M. Roem. | WBG | Nan.Lin‐522 (HIB) | MF467522 |
| Meliaceae | Toona sureni (Blume) Merr. | LBG | Nan.Lin‐523 (HIB) | MF467521 |
| Rutaceae | Citrus aurantiifolia (Christm.) Swingle | Omani, Madha | Su et al., 2014 | KJ_865401 |
| Rutaceae | Citrus depressa Hayata | Okinawa, Japan | Ishikawa et al., 2016 | LC147381 |
| Rutaceae | Citrus platymamma Tanaka | Jeju Island, Korea | Lee et al., 2015 | NC_030194 |
| Rutaceae | Citrus sinensis (L.) Osbeck | USA | Bausher et al., 2006 | NC_008334 |
| Rutaceae | Clausena excavata Burm. f. | USDA | PI539715 (Shivakumar et al., 2016) | NC_032685 |
| Rutaceae | Glycosmis mauritiana (Lam.) Tanaka | USDA | PI600641 (Shivakumar et al., 2016) | KU949004 |
| Rutaceae | Glycosmis pentaphylla (Retz.) DC. | USDA | PI127866 (Shivakumar et al., 2016) | NC_032687 |
| Rutaceae | Merrillia caloxylon (Ridl.) Swingle | USDA | PI539733 (Shivakumar et al., 2016) | NC_032688 |
| Rutaceae | Micromelum minutum Wight & Arn. | USDA | PI539744 (Shivakumar et al., 2016) | NC_032689 |
| Rutaceae | Murraya koenigii (L.) Spreng. | USDA | PI539745 (Shivakumar et al., 2016) | NC_032684 |
| Rutaceae | Zanthoxylum bungeanum Maxim. | Fengxian, China | Liu and Wei, 2017 | KX497031 |
| Rutaceae | Zanthoxylum piperitum DC. | NA | Lee et al., 2015 | NC_027939 |
| Rutaceae | Zanthoxylum schinifolium Siebold & Zucc. | NA | IM2014_ZS (Lee et al., 2016) | NC_030702 |
| Sapindaceae | Acer buergerianum Miq. | NA | Sd0060 (Yang et al., 2014) | KF753631 |
| Sapindaceae | Acer davidii Franch. | Changan, China | EBL (Jia et al., 2016) | NC_030331 |
| Sapindaceae | Acer miaotaiense P. C. Tsoong | Shaanxi, China | MTQ20160406SAXHZ (Zhang et al., 2016) | NC_030343 |
| Sapindaceae | Acer morrisonense Hayata | Shaanxi, China | Amorr2015 (Li et al., 2017) | NC_029371 |
| Sapindaceae | Dipteronia dyeriana A. Henry | Shaanxi, China | Zhou et al., 2016 | NC_031899 |
| Sapindaceae | Dipteronia sinensis Oliv. | Shaanxi, China | Zhou et al., 2016 | NC_029338 |
| Sapindaceae | Sapindus mukorossi Gaertn. | NA | Yang et al., 2016 | NC_025554 |
| Simaroubaceae | Leitneria floridana Chapm. | NA | MO:MO 2008‐0670 (Yang et al., 2014) | NC_030482 |
HIB = Herbarium of Wuhan Botanical Garden, Chinese Academy of Sciences; LBG = Lushan Botanical Garden, Jiangxi, China; NA = not available; SBL = National Nature Reserve of Shi‐Ba‐Li valley, Shiyan, China; WBG = Wuhan Botanical Garden, Wuhan, China; USDA = United States Department of Agriculture.
The raw reads were subsequently filtered for high‐quality reads following the method described by Sun et al. (2016). Filtered reads were assembled into contigs with a minimum length of 1000 bp using CLC Genomics Workbench 9 (Girard et al., 2011) with default parameters, except that the k‐mer value was set to 60 for T. sinensis and T. sureni, and 64 for T. ciliata, to produce the highest N50 value. The assembly statistics are presented in Appendix 1. After trimming, the contigs were ordered according to the reference genome Azadirachta indica A. Juss. (NC_023792). Plastid genomes were annotated with DOGMA (Wyman et al., 2004), and gene start and stop codons were determined through comparison to start and stop codons in the homologous genes of A. indica. Annotation of tRNA genes was conducted using tRNAscan‐SE (Schattner et al., 2005). Junctions between large single‐copy regions (LSCs) and IRs and small single‐copy regions (SSCs) and IRs of the three plastomes were verified with PCR and Sanger sequencing. Physical maps of plastomes were generated using GenomeVx (Conant and Wolfe, 2008).
In total, 79 protein‐coding regions and the ycf15 region were identified from the plastomes of three Toona species and 26 other species of Sapindales, with two taxa of Malvales (Cytinus hypocistis (L.) L. and Hibiscus syriacus L.) as outgroups (Table 1). These sequences were then manually compiled into a single file of the 31‐taxon data set and aligned with MAFFT (Katoh et al., 2002) for phylogenetic analyses. GenBank information for all plastomes used for phylogenetic analyses are provided in Table 1. In order to further investigate the phylogenetic relationships within Sapindales, maximum likelihood (ML) analyses were conducted using RAxML version 7.4.2 (Stamatakis et al., 2008) under the general time‐reversible (GTR) substitution model. We conducted both unpartitioned and partitioned analyses. PartitionFinder version 1.1.1 (Lanfear et al., 2012) was employed to determine the best‐fit partition scheme for partitioned ML analysis. Bootstrap support was estimated with 1000 bootstrap replicates.
In order to be convenient for subsequent population genetic study within Toona, simple sequence repeats (SSRs) were detected using MISA (Thiel et al., 2003) with thresholds of 10 repeat units for mononucleotide SSRs, five repeat units for di‐ and trinucleotide SSRs, and three repeat units for tetra‐, penta‐, and hexanucleotide SSRs. Additionally, repeat sequences were identified for each plastome using REPuter (Kurtz et al., 2001) with a minimum repeat size of 30 bp. Single‐nucleotide polymorphisms (SNPs) and insertion/deletion polymorphisms (indels) were also identified among three Toona plastomes with Geneious 7.0 (Kearse et al., 2012).
RESULTS
Within Toona, the plastome size of T. sureni was 159,371 bp, and those of T. sinensis and T. ciliata were 186 bp and 385 bp longer, respectively (Table 2). These three plastomes possess the typical quadripartite structure of angiosperm plastomes, comprising an LSC, an SSC, and two IR regions (Fig. 2). A total of 113 unique genes, including 30 tRNA genes, four rRNA genes, and 79 protein‐coding genes were found in each plastome. Nineteen genes were duplicated in the IR regions (Table 3). Additionally, 14 genes were found to possess one intron, and three genes (rps12, clpP, ycf3) were found to possess two introns (Appendix 2).
Table 2.
Plastome characteristics of Sapindales included in this study. Three Toona species were sequenced for the first time in this study, and other species were accessed from the National Center for Biotechnology Information database
| Family | Species | Total genome length (bp) | LSC length (bp) | SSC length (bp) | IR length (bp) | No. of genes within IR | Overall G/C content (%) |
|---|---|---|---|---|---|---|---|
| Anacardiaceae | Rhus chinensis | 149,011 | 96,882 | 18,647 | 16,741 | 18 | 37.8 |
| Anacardiaceae | Spondias bahiensis | 162,218 | 89,606 | 18,382 | 27,075 | 19 | 37.7 |
| Anacardiaceae | Spondias tuberosa | 162,039 | 89,453 | 18,368 | 27,139 | 19 | 37.7 |
| Burseraceae | Boswellia sacra | 160,543 | 88,054 | 18,962 | 26,764 | 20 | 37.6 |
| Meliaceae | Azadirachta indica | 160,737 | 88,137 | 18,624 | 26,983 | 19 | 37.5 |
| Meliaceae | Toona ciliata | 158,986 | 87,163 | 18,329 | 26,747 | 19 | 37.9 |
| Meliaceae | Toona sinensis | 159,185 | 87,358 | 17,933 | 26,947 | 19 | 37.9 |
| Meliaceae | Toona sureni | 159,371 | 87,505 | 18,472 | 26,697 | 19 | 37.9 |
| Rutaceae | Citrus aurantiifolia | 159,893 | 87,148 | 18,762 | 26,991 | 20 | 38.4 |
| Rutaceae | Citrus depressa | 160,120 | 87,794 | 18,376 | 26,955 | 20 | 38.5 |
| Rutaceae | Citrus platymamma | 160,121 | 87,732 | 18,393 | 26,998 | 20 | 38.5 |
| Rutaceae | Citrus sinensis | 160129 | 87,744 | 18,393 | 26,996 | 20 | 38.5 |
| Rutaceae | Clausena excavata | 161,172 | 88,055 | 18,295 | 27,411 | 17 | 38.3 |
| Rutaceae | Glycosmis mauritiana | 160,131 | 87,710 | 18,383 | 27,019 | 16 | 38.5 |
| Rutaceae | Glycosmis pentaphylla | 159,845 | 87,494 | 18,329 | 27,011 | 16 | 38.4 |
| Rutaceae | Merrillia caloxylon | 159,969 | 87,912 | 18,029 | 27,014 | 16 | 38.5 |
| Rutaceae | Micromelum minutum | 160,416 | 87,367 | 18,622 | 27,214 | 17 | 38.5 |
| Rutaceae | Murraya koenigii | 159,402 | 87,077 | 18,123 | 27,101 | 16 | 38.5 |
| Rutaceae | Zanthoxylum bungeanum | 158,401 | 85,898 | 17,611 | 27,446 | 19 | 38.5 |
| Rutaceae | Zanthoxylum piperitum | 158,154 | 85,340 | 17,526 | 27,644 | 19 | 38.5 |
| Rutaceae | Zanthoxylum schinifolium | 158,963 | 86,528 | 18,256 | 27,089 | 19 | 38.4 |
| Sapindaceae | Acer buergerianum | 156,911 | 85,314 | 18,093 | 26,752 | 18 | 37.9 |
| Sapindaceae | Acer davidii | 157,044 | 85,410 | 18,112 | 26,761 | 18 | 37.9 |
| Sapindaceae | Acer miaotaiense | 156,595 | 86,327 | 18,068 | 26,100 | 18 | 37.9 |
| Sapindaceae | Acer morrisonense | 157,197 | 85,655 | 18,086 | 26,728 | 18 | 37.8 |
| Sapindaceae | Dipteronia dyeriana | 157,071 | 85,529 | 18,082 | 26,730 | 19 | 38.0 |
| Sapindaceae | Dipteronia sinensis | 157,080 | 85,455 | 18,093 | 26,766 | 19 | 37.8 |
| Sapindaceae | Sapindus mukorossi | 160,481 | 85,649 | 18,874 | 27,979 | 21 | 37.7 |
| Simaroubaceae | Leitneria floridana | 158,763 | 85,689 | 18,186 | 27,444 | 20 | 37.6 |
IR = inverted repeat; LSC = large single copy; SSC = small single copy.
Figure 2.
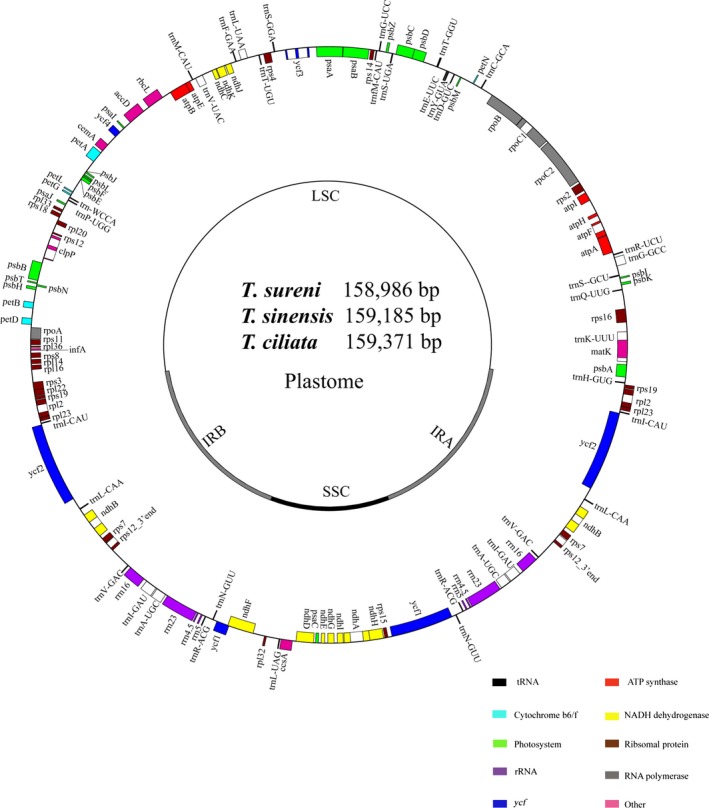
Physical maps of three Toona plastomes.
Table 3.
List of genes present in the plastomes of the three Toona species
| Function | Gene group | Gene name |
|---|---|---|
| Protein synthesis and DNA replication | Ribosomal RNAs | rrn4.5 (×2), rrn5 (×2), rrn16 (×2) c rrn23 (×2) |
| Transfer RNAs | trnH‐GUG, trnK‐UUU a, trnQ‐UUG, trnS‐GCU, trnG‐UCC a, trnR‐UCU, trnC‐GCA, trnD‐GUC, trnY‐GUA, trnE‐UUC, trnT‐GGU, trnS‐UGA, trnG‐UCC, trnfM‐CAU, trnS‐GGA, trnT‐UGU, trnL‐UAA a, trnF‐GAA, trnV‐UAC a, trnM‐CAU, trnW‐CCA, trnP‐UGG, trnI‐CAU a (×2), trnL‐CAA (×2), trnV‐GAC (×2), trnI‐GAU (×2), trnA‐UGC a (×2), trnR‐ACG (×2), trnN‐GUU (×2), trnL‐UAG | |
| Small subunit | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 a (×2), rps14, rps15, rps16, rps18, rps19 | |
| Ribosomal protein large subunit | rpl2 a (×2), rpl14, rpl16, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 | |
| RNA polymerase | rpoA, rpoB, rpoC1 a, rpoC2 | |
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Cytochrome b 6/f | petA, petB, petD, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF a, atpH, atpI | |
| NADH dehydrogenase | ndhA a, ndhB a (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Large subunit of RuBisCO | rbcL | |
| Miscellaneous proteins | Subunit of acetyl‐CoA‐carboxylase | accD |
| c‐type cytochrome synthesis gene | ccsA | |
| Envelope membrane protein | cemA | |
| Protease | clpP a | |
| Translational initiation factor | infA | |
| Maturase | matK | |
| Genes of unknown function | Hypothetical conserved coding frame | ycf1, ycf2 (×2), ycf3 a, ycf4 |
Genes with introns.
Across Sapindales, Spondias bahiensis P. Carvalho, Van den Berg & Machado (Anacardiaceae) and Rhus chinensis Mill. (Anacardiaceae) possessed the largest (162,218 bp) and smallest (149,011 bp) plastomes, respectively (Table 2). The latter also possessed the longest LSC and the shortest IR regions. Boswellia sacra Flueck. (Burseraceae) and Sapindus mukorossi Gaertn. (Sapindaceae) possessed the longest SSC and IR regions, respectively. Almost all 29 Sapindales plastomes contained 19 to 20 genes. Sapindus mukorossi of Sapindaceae possessed the longest IR region (21 genes). Among all 29 Sapindales plastomes, eight exhibited an IR expansion to rpl22 at the IR/LSC region boundaries and the IR region of S. mukorossi extended to rps3. In some Rutaceae (e.g., Clausena excavata Burm. f., Glycosmis mauritiana (Lam.) Tanaka, Glycosmis pentaphylla (Retz.) DC., Murraya koenigii (L.) Spreng., Merrillia caloxylon (Ridl.) Swingle, and Micromelum minutum Wight & Arn.) and Sapindaceae (e.g., Acer davidii Franch., A. morrisonense Hayata), the IR region was found to have contracted such that all of ycf1 is now within the SSC region. Moreover, in all of the above‐mentioned six Rutaceae plastomes, both trnI‐GAU and trnA‐UGC were present in the SSC region, while all rRNA genes were still located in the IR region. In Sapindales, infA was found as a pseudogene in several cases of Sapindaceae (e.g., B. sacra, A. davidii, A. morrisonense, and A. miaotaiense P. C. Tsoong). The G/C content of all plastomes was approximately 38% among 29 Sapindales plastomes (Table 2). The sequence divergence of 79 protein‐coding genes among all 29 genomes varied from 0.00361 (rps7) to 0.1582 (rps16). The genes rps16, ycf1, and matK had the highest sequence divergence (0.15582, 0.12381, and 0.09137, respectively; Fig. 3). Notably, rpl22 was found to have a high variation in length, from 171 bp (Micromelum minutum, Rutaceae) to 514 bp (Toona sureni, Meliaceae) (Appendix S1).
Figure 3.
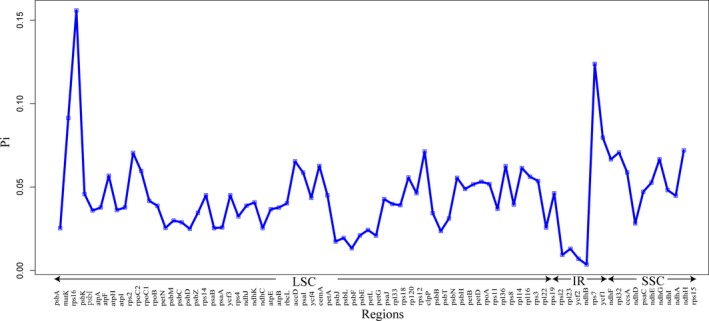
Plot of nucleotide variability (Pi) values among 29 Sapindales plastomes.
The alignment of the 31‐taxon data set was 63,597 bp in length. The best partition scheme determined by PartitionFinder contained 17 partitions (maximum likelihood score [ln L] = −229027.17027, Bayesian information criterion [BIC] = 460434.027954). The unpartitioned and partitioned ML analyses yielded identical tree topology, with slightly higher support values in the partitioned tree (Fig. 4; the unpartitioned tree is not shown). Most nodes had very high bootstrap support (Fig. 4), and Anacardiaceae, Sapindaceae, Rutaceae, and Meliaceae were recovered as monophyletic. The backbone of Sapindales was strongly supported except for one node that united Burseraceae, Rutaceae, and Sapindaceae (57%; Fig. 4). Meliaceae was sister to Simaroubaceae + Rutaceae.
Figure 4.
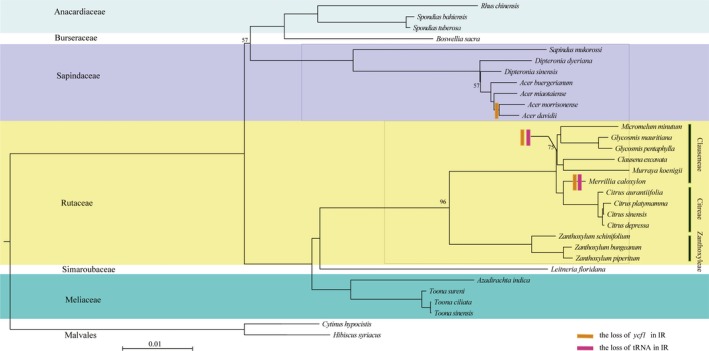
The best maximum likelihood tree of Sapindales based on the 17‐partition analysis of 79 plastid genes (and the ycf15 region). Numbers above branches are maximum likelihood bootstrap support values (unlabeled branches have bootstrap support of 100%).
A total of 193 SSRs were identified in the three plastomes of Toona. Among these, 70 were distributed in T. sureni, 57 in T. sinensis, and 66 in T. ciliata (Appendix 3). The majority of SSRs were A/T mononucleotides, a total of 14 AT dinucleotide repeats were found in the three plastomes, and one TA dinucleotide repeat was detected in T. sinensis, whereas the only AG dinucleotide repeat from T. sureni was located in the rpoB‐trnC‐GCA intergenic region. The other kinds of repeat units (e.g., six dinucleotide; four trinucleotide; three tetra‐, penta‐, and hexanucleotide) were not found in the three plastomes of Toona. Most SSRs were located in intergenic regions (72.5%), with few in introns (12.5%) and genes (15%). Overall, nine SSRs were shared by all three Toona species, including four in intergenic regions (trnE‐UCC/trnT‐GGU, trnT‐GGU/psbD, ccsA/ndhD, and ycf15/rps12), three in exons (rpoC2, rpoB, and psbF), and two in introns (trnL‐UAA and ndhB). In total, 23 repeats were detected in three Toona plastomes. A majority of the repeats (69.56%) were 30 to 40 bp in length, and 17.40% of the repeats were longer than 50 bp. Four repeats were shared by three Toona plastomes (Appendix S2). Additionally, we detected 466 SNPs (0.4%) and 90 indels among three plastomes, and we screened out four noncoding regions (psbZ‐trnG, psbA‐trnK, trnF‐ndhJ, trnK‐rps16) with potential to be loci for identification of Toona species (Appendix S3).
DISCUSSION
In most angiosperm plastomes, the IR/LSC boundary lies within the rps19 gene and the SSC/IR boundary lies within the ycf1 gene (Kumar et al., 2009). Among the 29 Sapindales plastomes, the LSC/IRB boundary of the majority lies within the rps19 gene, while nine of these 29 plastomes have experienced an IR region expansion. Obvious IR region expansion to the LSC region has been detected in many other taxa, e.g., in Pelargonium L'Hér. (Chumley et al., 2006), Tetracentron Oliv. (Sun et al., 2013), and Veronica nakaiana Ohwi (Choi et al., 2016). In contrast, within Sapindales, there have been at least eight cases where the SSC/IRA boundary has contracted to exclude all of ycf1 (Fig. 4). IR region contraction has been found to occur in several ways, ranging from complete IR loss (e.g., Geraniaceae [Blazier et al., 2011], Cephalotaxus oliveri Mast. [Yi et al., 2013], and Agathis dammara (Lamb.) Rich. & A. Rich. [Wu and Chaw, 2014]), to the loss of tRNA genes within the IR region (e.g., Epifagus virginiana (L.) W. P. C. Barton [Morden et al., 1991] and Bergera koenigii L. [Shivakumar et al., 2016]), to the rpl22 loss in rosids (Jansen et al., 2011), and to contraction at the IR/SSC boundaries reported in a number of early‐diverging angiosperms (e.g., Buxus L., Epimedium L., and Macadamia F. Muell.) (Hansen et al., 2007). Notably, in Rutaceae, all Clauseneae genera are characterized by the absence of trnI‐GAU and trnA‐UGC in the IR region. Tsuji et al. (2007) indicated that the tRNA loss may be caused by the RNA editing during the tRNA mutation. Pseudogenization of the infA gene has been detected in a number of angiosperm plastomes such as tobacco (Shinozaki et al., 1986), Arabidopsis Heynh. (Sato et al., 1999), and Oenothera elata Kunth (Hupfer et al., 2000), whereas among 29 Sapindales plastomes this was only detected in four plastomes (Boswellia sacra, Acer davidii, A. morrisonense, and A. miaotaiense) of Sapindaceae (Blazier et al., 2016). In some cases, the effect of plastid‐to‐nucleus gene transfer has been demonstrated to generate the pseudogenization of this gene (Millen et al., 2001).
As has been found in many other studies involving plastome‐scale phylogenetic analysis (Parks et al., 2009), we recovered improved phylogenetic support along the backbone of Sapindales compared to previous targeted gene analyses. We recovered Meliaceae as sister to the clade formed by Simaroubaceae (only one species included) + Rutaceae with maximal support, differing from the topology recovered by Muellner et al. (2007) and Muellner‐Riehl et al. (2016), where a moderately supported clade of Meliaceae + Simaroubaceae was sister to Rutaceae. Our result is consistent with the earlier work of Gadek et al. (1996) based on trnL‐F sequences, although they recovered only weak support. Unfortunately, the problem of the previously unsupported relationship of Sapindaceae with other Sapindales (Muellner‐Riehl et al., 2016) could also not be resolved by our plastome data analysis. It is important to emphasize caution for these results, however. Additional taxon sampling for complete plastomes, including additional lineages of already‐sampled families as well as the inclusion of the early‐diverging Sapindales families Biebersteiniaceae, Kirkiaceae, and Nitrariaceae may affect topology and support. Likewise, the plastome itself can be treated as a single locus for the purpose of phylogenetics, and genomic‐scale nuclear data may provide different estimates of phylogeny, especially for short branches.
Within Rutaceae, our results are highly congruent with those of the previous study (Shivakumar et al., 2016), which also found a clade of Citrus + Merrillia sister to a clade composed of (Micromelum + Glycosmis) + (Murraya + Clausena), although in the latter clade the bootstrap support was low. In our tree, all of the taxa sampled in Shivakumar et al. (2016) formed a clade, which is sister to Zanthoxylum. Our analysis suggests that tribe Clauseneae sensu Swingle and Reece (1967; Micromelum Blume, Glycosmis Corrêa, Clausena Burm. f., Murraya J. Koenig, and Merrillia Swingle) is not monophyletic because Merrillia is sister to Citrus L. of the tribe Citreae. The genera of Clauseneae are characterized by the absence of two tRNA genes (trnI‐GAU and trnA‐UGC), while this is not found in the genus Citrus (Fig. 4). Additionally, four genera (Micromelum + Glycosmis + Murraya + Clausena) in Rutaceae and two species (Acer davidii + Acer morrisonense) in Sapindales, characterized by the absence of ycf1 in the SSC region, each formed a clade in our phylogenetic tree (Fig. 4). This gene loss shared by multiple taxa shows a particularly strong case of homoplasy in the phylogeny. Within Sapindaceae, Sapindus L. is sister to a clade containing Dipteronia Oliv. and Acer L. Although the support value is weak (57%), the two species of Dipteronia do not form a clade, instead forming a grade with respect to Acer.
The plastome structure and gene content of Toona reported in the present study enrich the available plastome resources within Sapindales, the comparative analyses among 29 plastomes provide insight into the plastome evolution of Sapindales, and the phylogenomic analyses of Sapindales improve our understanding of phylogenetic relationships within this order. In addition, the SSRs detected in three Toona species could provide a basis for future plastome genetic studies in Toona, especially in the threatened species.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2017YFC0505200), the Major Program of the National Natural Science Foundation of China (31590823), and the National Natural Science Foundation of China (31370223).
Appendix 1. Plastome assembly comparison among three Toona (Meliaceae) species.
| Species | Read length (bp) | No. of plastid reads | No. of plastid contigs | Total no. of bases in contigs | Average depth of coverage (×) | Percentage of reads mapping | Contigs N50 | Maximum/minimum contig size (bp) |
|---|---|---|---|---|---|---|---|---|
| T. sureni | 150 | 119,147 | 60 | 810,415 | 112.72 | 99.27 | 24,245 | 80,048/2049 |
| T. sinensis | 150 | 102,323 | 81 | 845,099 | 96.71 | 99.91 | 21,131 | 84,024/2015 |
| T. ciliata | 150 | 134,438 | 108 | 794,274 | 130.88 | 99.76 | 10,027 | 86,924/2043 |
Appendix 2. Exon and intron lengths (in base pairs) of genes in the three Toona (Meliaceae) plastomes.a
| Gene | Exon1 | Intron1 | Exon2 | Intron2 | Exon3 |
|---|---|---|---|---|---|
| trnK‐UUU | 28/28/28 | — | 36/36/36 | — | — |
| trnG‐UCC | 23/23/23 | 724/724/724 | 49/49/49 | — | — |
| trnL‐UAA | 36/36/36 | 530/530/530 | 49/49/49 | — | — |
| trnV‐UAC | 36/36/36 | 600/602/602 | 38/38/38 | — | — |
| trnI‐GAU | 41/41/41 | 956/956/954 | 33/33/33 | — | — |
| trnA‐UGC | 37/37/37 | 841/841/841 | 34/34/34 | — | — |
| atpF | 438/438/438 | 720/720/720 | 155/155/155 | — | — |
| ndhA | 536/536/536 | 1099/1098/1098 | 551/551/551 | — | — |
| ndhB | 756/756/756 | 682/682/682 | 775/775/775 | — | — |
| rpl2 | 433/433/433 | 671/671/671 | 390/390/390 | — | — |
| rps12 b | 113/113/113 | — | 25/25/25 | 537/537/537 | 231/231/231 |
| rpoC1 | 1619/1619/1619 | 753/753/753 | 434/434/434 | — | — |
| clpP | 198/198/198 | 690/690/690 | 290/290/290 | 860/860/860 | 67/67/67 |
| ycf3 | 152/152/152 | 791/789/789 | 227/227/227 | 628/628/628 | 728/728/728 |
Values presented correspond to T. sureni/T. sinensis/T. ciliata, respectively.
Intron 1 of rps12 is not shown because rps12 is trans‐spliced.
Appendix 3. Distribution of simple sequence repeats in the plastomes of three Toona (Meliaceae) species.
| Species/Base | Length (bp) | Position in plastid genome |
|---|---|---|
| Toona ciliata | ||
| A | 10 | 9258–9267, 34,518–34,527, 38,871–38,880, 54,296–54,305, 57,729–57,738, 62,577–62,586, 67,493–67,502, 68,780–68,789, 74,450–74,459, 116,277–116,286, 118,064–118,073, 118,119–118,128, 134,208–134,217 |
| 11 | 50,135–50,145, 73,952–73,962, 84,456–84,466, 116,647–116,657, 13,576–13,586, 147,115–147,125 | |
| 12 | 7044–7055, 31,827–31,838, 113,763–113,774 | |
| 13 | 143,677–143,689 | |
| 15 | 4738–4752 | |
| 16 | 78,707–78,722 | |
| T | 10 | 14,085–14,094, 27,614–27,623, 70,109–70,118, 73,655–73,664, 73,788–73,797, 83,409–83,418, 83,918–83,927, 119,623–119,632, 128,850–128,859, 130,504–130,513, 131,949–131,958 |
| 11 | 5882–5892, 7033–7043, 12,659–12,669, 31,412–31,422, 45,602–45,612, 54,222–54,232, 57,155–57,165, 57,236–57,246, 62,239–62,249, 63,742–63,752, 70,526–70,536, 99,025–99,035, 125,450–125,460 | |
| 12 | 9479–9490, 19,766–19,777, 52,802–52,813, 125,141–125,152, 132,376–132,387 | |
| 13 | 6828–6840, 74,806–74,818, 102,461–102,473 | |
| 15 | 48,905–48,919, 118,464–118,478 | |
| AT | 10 | 21,267–21,276, 33,424–33,433, 49,396–49,405, 49,786–49,795, 50,733–50,742, 54,811–54,820, 121,130–121,139 |
| Toona sureni | ||
| A | 10 | 34,799–34,808, 54,560–54,569, 57,999–58,008, 67,156–67,165, 67,798–67,807, 74,266–74,275, 116,568–116,577, 134,590–134,599 |
| 11 | 13,859–13,869, 50,428–50,438, 62,866–62,876, 74,770–74,780, 118,360–118,370, 147,491–147,501 | |
| 12 | 7220–7231, 84,776–84,787, 118,408–118,419 | |
| 13 | 79,023–79,035, 144,054–144,046 | |
| 14 | 9769–9782 | |
| 17 | 4886–4902, 39,136–39,152 | |
| T | 10 | 6989–6998, 9648–9657, 10,432–10,441, 12,954–12,963, 14,367–14,376, 27,928–27,937, 31,526–31,534, 32,502–32,511, 45,214–45,223, 54,487–54,496, 57,499–57,508, 60,086–60,095, 61,837–61,846, 62,536–62,545, 83,645–83,654, 112,278–112,287, 117,742–117,751, 119,996–120,005, 125,519–125,528, 129,225–129,234, 130,879–130,888, 132,330–132,339 |
| 11 | 14,446–14,456, 57,418–57,428, 64,050–64,060, 74,061–74,071, 99,376–99,386, 120,092–120,102, 125,826–125,836 | |
| 12 | 20,080–20,091, 45,861–45,872, 70,828–70,839, 73,966–73,977, 85,407–85,418, 132,757–132,768 | |
| 13 | 31,711–31,723, 53,075–53,087, 75,127–75,139, 102,811–102,823 | |
| 16 | 49,165–49,180, 118,836–118,851 | |
| AT | 10 | 21,581–21,590, 33,705–33,714, 49,669–49,678, 50,079–50,088, 55,074–55,083, 121,505–121,514 |
| AG | 12 | 29,112–29,123 |
| Toona sinensis | ||
| A | 10 | 34,737–34,746, 39,096–39,105, 57,923–57,932, 62,771–62,780, 66,759–66,768, 67,688–67,697, 68,975–68,984, 74,149–74,158, 74,646–74,655, 116,472–116,481, 134,407–134,416 |
| 11 | 13,706–13,716, 50,357–50,367, 84,652–84,662, 116,842–116,852, 147,314–147,324 | |
| 12 | 9384–9395, 32,046–32,057, 113,958–113,969 | |
| 13 | 143,876–143,878 | |
| 15 | 78,903–78,917 | |
| 18 | 4864–4881 | |
| T | 10 | 14,215–14,224, 27,768–27,777, 31,065–31,074, 45,827–45,836, 49,496–49,505, 70,304–70,313, 73,985–73,994, 84,114–84,123, 112,128–112,137, 119,821–119,830, 127,827–127,836, 129,049–129,058, 130,703–130,712, 132,148–132,157 |
| 11 | 6010–6020, 6956–6966, 31,631–31,641, 62,433–62,443, 63,936–63,946, 70,721–70,731, 73,851–73,861, 99,220–99,230, 125,648–125,658 | |
| 12 | 9607–9618, 12,788–12,799, 19,896–19,907, 53,002–53,013, 125,339–125,350, 132,575–132,586 | |
| 13 | 75,002–75,014, 49,128–49,141 | |
| 14 | 49,128–49,141 | |
| AT | 10 | 33,644–33,653 |
| TA | 10 | 49,617–49,626, 50,954–50,963 |
Lin, N. , Moore M. J., Deng T., Sun H., Yang L.‐S., Sun Y.‐X., and Wang H.‐C.. Complete plastome sequencing from Toona (Meliaceae) and phylogenomic analyses within Sapindales. Applications in Plant Sciences 6(4): e1040.
Contributor Information
Yan‐xia Sun, Email: sunyanxia@wbgcas.cn.
Heng‐chang Wang, Email: hcwang@wbgcas.cn.
LITERATURE CITED
- Angiosperm Phylogeny Group . 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1–20. [Google Scholar]
- Bausher, M. G. , Singh N. D., Lee S. B., Jansen R. K., and Daniell H.. 2006. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: Organization and phylogenetic relationships to other angiosperms. BMC Plant Biology 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier, J. C. , Guisinger M. M., and Jansen R. K.. 2011. Recent loss of plastid‐encoded ndh genes within Erodium (Geraniaceae). Plant Molecular Biology 76: 263–272. [DOI] [PubMed] [Google Scholar]
- Blazier, J. C. , Jansen R. K., Mower J. P., Govindu M., Zhang J., Weng M. L., and Ruhlman T. A.. 2016. Variable presence of the inverted repeat and plastome stability in Erodium . Annals of Botany 117: 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K. S. , Chung M. G., and Park S.. 2016. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): Comparative analysis and highly divergent regions. Frontiers in Plant Science 7: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley, T. W. , Palmer J. D., Mower J. P., Fourcade H. M., Calie P. J., Boore J. L., and Jansen R. K.. 2006. The complete chloroplast genome sequence of Pelargonium × hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Molecular Biology and Evolution 23: 2175–2190. [DOI] [PubMed] [Google Scholar]
- Conant, G. C. , and Wolfe K. H.. 2008. GenomeVx: Simple web‐based creation of editable circular chromosome maps. Bioinformatics 24: 861–862. [DOI] [PubMed] [Google Scholar]
- Duvall, M. R. , Fisher A. E., Columbus J. T., Ingram A. L., Wysocki W. P., Burke S. V., Clark L. G., and Kelchner S. A.. 2016. Phylogenomics and plastome evolution of the chloridoid grasses (Chloridoideae: Poaceae). International Journal of Plant Sciences 177: 235–246. [Google Scholar]
- Edmonds, J. M. 1995. Toona In Mabberley D. J., Pannell C. M., and Sing A. M. [eds.], Flora Malesiana, ser. 1, vol. 12, 358–371. Erven P. Noordhoff, Groningen, The Netherlands. [Google Scholar]
- Gadek, P. A. , Fernando E. S., Quinn C. J., Hoot S. B., Terrazas T., Sheahan M. C., and Chase M. W.. 1996. Sapindales: Molecular delimitation and infraordinal groups. American Journal of Botany 83: 802–811. [Google Scholar]
- Girard, S. L. , Gauthier J., Noreau A., Xiong L., Zhou S., Jouan L., Dionne‐Laporte A., et al. 2011. Increased exonic de novo mutation rate in individuals with schizophrenia. Nature Genetics 43: 860–863. [DOI] [PubMed] [Google Scholar]
- Green, B. R. 2011. Chloroplast genomes of photosynthetic eukaryotes. Plant Journal 66: 34–44. [DOI] [PubMed] [Google Scholar]
- Guisinger, M. M. , Kuehl J. V., Boore J. L., and Jansen R. K.. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Molecular Biology and Evolution 28: 583–600. [DOI] [PubMed] [Google Scholar]
- Hansen, D. R. , Dastidar S. G., Cai Z., Penaflor C., Kuehl J. V., Boore J. L., and Jansen R. K.. 2007. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early‐diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Molecular Phylogenetics and Evolution 45: 547–563. [DOI] [PubMed] [Google Scholar]
- Hupfer, H. , Swiatek M., Hornung S., Herrmann R. G., Maier R. M., Chiu W. L., and Sears B.. 2000. Complete nucleotide sequence of the Oenothera elata plasti chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Molecular and General Genetics 263: 581–585. [DOI] [PubMed] [Google Scholar]
- Ishikawa, R. , Badenoch N., Miyagi K., Medoruma K., Osada T., and Onishi M.. 2016. Multi‐lineages of Shiikuwasha (Citrus depressa Hayata) evaluated by using whole chloroplast genome sequences and its bio‐diversity in Okinawa, Japan. Breeding Science 66: 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. K. , Saski C., Lee S., Hansen A. K., and Daniell H.. 2011. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Molecular Biology and Evolution 28: 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y. , Yang J., Li Z. H., He Y. L., Niu C., and Gong L. L.. 2016. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conservation Genetic Resources 8: 141–143. [Google Scholar]
- Katoh, K. , Misawa K., Kuma K., and Miyata T.. 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir R., Wilson A., Stones‐Havas S., Cheung M., and Sturrock S.. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox, E. B. 2014. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proceedings of the National Academy of Sciences USA 111: 11097–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany, O. , Gentles A. J., Hankus L., and Jurka J.. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Hahn F. M., McMahan C. M., Cornish K., and Whalen M. C.. 2009. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biology 9: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz, S. , Choudhuri J. V., Ohlebusch E., Schleiermacher C., Stoye J., and Giegerich R.. 2001. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Research 29: 4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, R. , Calcott B., Ho S. Y., and Guindon S.. 2012. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Lee, M. , Park J., Lee H., Sohn S. H., and Lee J.. 2015. Complete chloroplast genomic sequence of Citrus platymamma determined by combined analysis of Sanger and NGS data. Horticulture Environment and Biotechnology 56: 704–711. [Google Scholar]
- Lee, Y. S. , Kim I., Kim J. K., Park J. Y., Joh H. J., Park H. S., Lee H. O., and Lee S. C.. 2016. The complete chloroplast genome sequence of Rhus chinensis Mill. (Anacardiaceae). Mitochondrial DNA Part B 1: 696–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Xie Y., Zhou T., Jia Y., He Y., and Yang J.. 2017. The complete chloroplast genome sequence of Acer morrisonense (Aceraceae). Mitochondrial DNA Part A 28: 309–310. [DOI] [PubMed] [Google Scholar]
- Liu, Y. L. , and Wei A. Z.. 2017. The complete chloroplast genome sequence of an economically important plant, Zanthoxylum bungeanum (Rutaceae). Conservation Genetic Resources 9: 25–27. [Google Scholar]
- Mabberley, D. 2008. Mabberley's plant‐book, 863. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- McPherson, H. , van der Merwe M., Delaney S. K., Edwards M. A., Henry R. J., McIntosh E., Rymer P. D., et al. 2013. Capturing chloroplast variation for molecular ecology studies: A simple next generation sequencing approach applied to a rainforest tree. BMC Ecology 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen, R. S. , Olmstead R. G., Adams K. L., Palmer J. D., Lao N. T., Heggie L., Kavanagh T. A., et al. 2001. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morden, C. W. , Wolfe K. H., dePamphilis C. W., and Palmer J. D.. 1991. Plastid translation and transcription genes in a nonphotosynthetic plant: Intact, missing and pseudo genes. EMBO Journal 10: 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner, A. N. , Samuel R., Johnson S. A., Cheek M., Pennington T. D., and Chase A. M. W.. 2003. Molecular phylogenetics of Meliaceae based on nuclear and plastid DNA sequences. American Journal of Botany 90: 471–480. [DOI] [PubMed] [Google Scholar]
- Muellner, A. N. , Vassiliades D. D., and Renner S. S.. 2007. Placing Biebersteiniaceae, a herbaceous clade of Sapindales, in a temporal and geographic context. Plant Systematics and Evolution 266: 233–252. [Google Scholar]
- Muellner, A. N. , Pennington T. D., and Chase A. M. W.. 2009. Molecular phylogenetics of Neotropical Cedreleae (mahogany family, Meliaceae) based on nuclear and plastid DNA sequences reveal multiple origins of “Cedrela odorata”. Molecular Phylogenetics and Evolution 52: 461–469. [DOI] [PubMed] [Google Scholar]
- Muellner‐Riehl, A. N. , Weeks A., Clayton J. W., Buerki S., Nauheimer L., Chiang Y. C., Cody S., and Pell S. K.. 2016. Molecular phylogenetics and molecular clock dating of Sapindales based on plastid rbcL, atpB and trnL‐trnF DNA sequences. Taxon 65: 1019–1036. [Google Scholar]
- Parks, M. , Cronn R., and Liston A.. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, H. , and Edmonds J. M.. 2008. Toona In Wu Z. Y., Raven P. H., and Hong D. Y. [eds.], Flora of China, vol. 11, 113–115. Science Press, Beijing, China, and Missouri Botanical Garden Press, St. Louis, Missouri, USA. [Google Scholar]
- Ruhlman, T. A. , and Jansen R. K.. 2014. The plastid genomes of flowering plants In Maliga P. [ed.], Methods in molecular biology, vol. 1132: Chloroplast biotechnology: Methods and Protocols, 3–38. Humana Press, Totowa, New Jersey, USA. [DOI] [PubMed] [Google Scholar]
- Rushforth, K. 1999. Trees of Britain and Europe. HarperCollins, London, United Kingdom. [Google Scholar]
- Sato, S. , Nakamura Y., Kaneko T., Asamizu E., and Tabata S.. 1999. Complete structure of the chloroplast genome of Arabidopsis thaliana . DNA Research 6: 283–290. [DOI] [PubMed] [Google Scholar]
- Schattner, P. , Brooks A. N., and Lowe T. M.. 2005. The tRNAscan‐SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Research 33: 686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, C. , Hu N., Huang H., Gao J., Zhao Y. J., and Gao L. Z.. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One 7: e31468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K. , Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO Journal 5: 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar, V. S. , Appelhans M. S., Johnson G., Carlsen M., and Zimmer E. A.. 2016. Analysis of whole chloroplast genomes from the genera of the Clauseneae, the curry tribe (Rutaceae, Citrus family). Molecular Phylogenetics and Evolution 117: 135–140. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. , Hoover P., and Rougemont J.. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–71. [DOI] [PubMed] [Google Scholar]
- Stevens, P. F. 2001. onwards. Angiosperm phylogeny website, version 14, July 2017 [more or less continuously updated]. Website http://www.mobot.org/MOBOT/research/APweb/ [accessed 30 March 2018].
- Stull, G. W. , Duno de Stefano R., Soltis D. E., and Soltis P. S.. 2015. Resolving basal lamiid phylogeny and the circumscription of Icacinaceae with a plastome‐scale data set. American Journal of Botany 102: 1794–1813. [DOI] [PubMed] [Google Scholar]
- Su, H. J. , Hogenhout S. A., Al‐Sadi A. M., and Kuo C. H.. 2014. Complete chloroplast genome sequence of Omani lime (Citrus aurantiifolia) and comparative analysis within the rosids. PLoS One 9: e113049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. X. , Moore M. J., Meng A. P., Soltis P. S., Soltis D. E., Li J. Q., and Wang H. C.. 2013. Complete plastid genome sequencing of Trochodendraceae reveals a significant expansion of the inverted repeat and suggests a Paleogene divergence between the two extant species. PLoS One 8: e60429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. X. , Moore M. J., Zhang S., Soltis P. S., Soltis D. E., and Zhao T.. 2016. Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early‐diverging eudicots, including an angiosperm‐wide analysis of IR gene content evolution. Molecular Phylogenetic Evolution 96: 93–101. [DOI] [PubMed] [Google Scholar]
- Swingle, W. T. , and Reece P. C.. 1967. The botany of Citrus and its wild relatives of the orange subfamily In Reuther W., Webber H. J., and Batchelor L. D. [eds.], The Citrus industry: History, world distribution, botany, and varieties (revised 2nd edition), 190–430. University of California Press, Berkeley, California, USA. [Google Scholar]
- Thiel, T. , Michalek W., Varshney R. K., and Graner A.. 2003. Exploiting EST databases for the development and characterization of gene‐derived SSR‐markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 106: 411–422. [DOI] [PubMed] [Google Scholar]
- Tsuji, S. , Ueda K., Nishiyama T., Hasebe M., Yoshikawa S., Konagaya A., Nishiuchi T., and Yamaguchi K.. 2007. The chloroplast genome from a lycophyte (microphyllophyte), Selaginella uncinata, has a unique inversion, transpositions and many gene losses. Journal of Plant Research 120: 281–290. [DOI] [PubMed] [Google Scholar]
- Wu, C. S. , and Chaw S. M.. 2014. Highly rearranged and size‐variable chloroplast genomes in conifers II clade (cupressophytes): Evolution towards shorter intergenic spacers. Plant Biotechnology Journal 12: 344–353. [DOI] [PubMed] [Google Scholar]
- Wyman, S. K. , Jansen R. K., and Boore J. L.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. [DOI] [PubMed] [Google Scholar]
- Yang, B. , Li M., Ma J., Fu Z., Xu X., Chen Q., and Tian J.. 2016. The chloroplast genome sequence of Sapindus mukorossi . Mitochondrial DNA Part A 27: 1825–1826. [DOI] [PubMed] [Google Scholar]
- Yang, J. B. , Li D. Z., and Li H. T.. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Molecular Ecology Resources 14: 1024–1031. [DOI] [PubMed] [Google Scholar]
- Yi, X. , Gao L., Wang B., Su Y. J., and Wang T.. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biology and Evolution 5: 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Li B., Chen H., and Wang Y.. 2016. Characterization of the complete chloroplast genome of Acer miaotaiense (Sapindales: Aceraceae), a rare and vulnerable tree species endemic to China. Conservation Genetics Resources 8: 383–385. [Google Scholar]
- Zhou, T. , Chen C., Wei Y., Chang Y. X., Bai G. Q., Li Z. H., Kanwal N., and Zhao G. F.. 2016. Comparative transcriptome and chloroplast genome analyses of two related Dipteronia species. Frontiers in Plant Science 7: 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, A. , Guo W., Gupta S., Fan W., and Mower J. P.. 2016. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytologist 209: 1747–1756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


