Summary
Syntrophobacter fumaroxidans is a sulfate‐reducing bacterium able to grow on propionate axenically or in syntrophic interaction with methanogens or other sulfate‐reducing bacteria. We performed a proteome analysis of S. fumaroxidans growing with propionate axenically with sulfate or fumarate, and in syntrophy with Methanospirillum hungatei, Methanobacterium formicicum or Desulfovibrio desulfuricans. Special attention was put on the role of hydrogen and formate in interspecies electron transfer (IET) and energy conservation. Formate dehydrogenase Fdh1 and hydrogenase Hox were the main confurcating enzymes used for energy conservation. In the periplasm, Fdh2 and hydrogenase Hyn play an important role in reverse electron transport associated with succinate oxidation. Periplasmic Fdh3 and Fdh5 were involved in IET. The sulfate reduction pathway was poorly regulated and many enzymes associated with sulfate reduction (Sat, HppA, AprAB, DsrAB and DsrC) were abundant even at conditions where sulfate was not present. Proteins similar to heterodisulfide reductases (Hdr) were abundant. Hdr/Flox was detected in all conditions while HdrABC/HdrL was exclusively detected when sulfate was available; these complexes most likely confurcate electrons. Our results suggest that S. fumaroxidans mainly used formate for electron release and that different confurcating mechanisms were used in its sulfidogenic metabolism.
Introduction
Syntrophobacter fumaroxidans is a sulfate‐reducing deltaproteobacterium able to grow on propionate in syntrophy with methanogens (Harmsen et al., 1998). It can also grow axenically by fermenting fumarate (Stams et al., 1993). To degrade propionate, it requires fumarate or sulfate as electron acceptors, or a H2‐ and formate‐consuming partner in the absence of an electron acceptor. S. fumaroxidans uses the methylmalonyl‐CoA (MMC) pathway to degrade propionate to acetate and CO2 (Plugge et al., 1993). Under standard conditions, propionate oxidation to H2, formate and acetate is an endergonic process. Reducing equivalents at the redox levels of reduced ferredoxin (Fdred) and NADH are released in the pyruvate and malate oxidation steps of the pathway respectively. Succinate oxidation via menaquinone is endergonic since the midpoint potential of succinate is more positive (+30 mV) than the menaquinone (−80 mV). Therefore, the reaction requires a transmembrane proton gradient to function (Plugge et al., 2012). For this, it has been proposed that S. fumaroxidans uses a periplasmic formate dehydrogenase, cytochrome b:quinone oxidoreductases, the menaquinone loop and a cytoplasmic fumarate reductase to drive energy dependent succinate oxidation (Müller et al., 2010). To keep the pathway functioning, the reduced electron mediators need to be re‐oxidized by reducing protons to H2 or CO2 to formate. Consequently, the role of the hydrogen/formate scavenger in the syntrophic association with S. fumaroxidans is to maintain H2 and formate at sufficiently low levels so that propionate degradation becomes energetically feasible (Stams and Dong, 1995). The minimal hydrogen partial pressure (pH2) that methanogens can maintain is between 1 and 10 Pa (Thauer et al., 2008). This level is not low enough to overcome the most energy‐consuming step in the MMC pathway, the oxidation of succinate to fumarate. To couple this step to proton or CO2 reduction would require a pH2 of 10−10 Pa and a formate concentration below 1 µM (Schink, 1997). Therefore, to drive this reaction, the input of metabolic energy is required. An investment of two‐thirds of an ATP via a mechanism known as reverse electron transport (RET) has been suggested by some authors (Van Kuijk et al., 1998a; Schink and Stams, 2013).
During RET energy is invested in the form of ATP to generate a proton gradient across the membrane which allows succinate oxidation to proceed (Stams and Plugge, 2009). Membrane‐associated proteins, such as ferredoxin:NAD+ oxidoreductases, cytochromes and periplasmic formate dehydrogenases and hydrogenases, have been reported to be involved in RET (Sieber et al., 2012; Grein et al., 2013). Moreover, novel energy conversion mechanisms have been discovered in anaerobic microorganisms, for instance flavin‐based electron bifurcation and its reversal, electron confurcation (Li et al., 2008; Buckel and Thauer, 2013; Schink, 2015). Genome analyses of S. fumaroxidans revealed membrane associated proteins, such as a fumarate reductase and a Rnf complex, as well as confurcating hydrogenases and formate dehydrogenases possibly involved in energy conservation mechanisms (Müller et al., 2010; Pereira et al., 2011; Plugge et al., 2012; Worm et al., 2014). Subsequently, transcriptomics studies with S. fumaroxidans in syntrophic and axenic cultures showed that a periplasmic formate dehydrogenase (Fdh2) and a hydrogenase (Hyn) play an important role to make the endergonic oxidation of succinate possible (Worm et al., 2011a). Moreover, it was found that confurcating hydrogenases and confurcating formate dehydrogenases (Hyd1, Hox and Fdh1) are important energy conversion enzymes required for propionate degradation (Worm et al., 2011a, 2011b).
In this study, a comparative proteomic analysis of S. fumaroxidans was made. Cells grown with propionate coupled to fumarate or sulfate reduction, or in syntrophic associations with Methanospirillum hungatei or Methanobacterium formicicum were compared. We aim to elucidate the main metabolic differences in lifestyles by identifying the key proteins used by S. fumaroxidans in interspecies electron transfer (IET), reverse electron transport (RET), electron confurcating processes and other energy conservation pathways.
In addition to the known syntrophic interactions of S. fumaroxidans with methanogens, our study was extended by including the proteomic profiling of S. fumaroxidans in coculture with a non‐methanogenic partner. Desulfovibrio desulfuricans has been studied before in cocultures with Syntrophobacter wolinii and S. fumaroxidans as a hydrogen‐ or formate‐scavenger in the oxidation of propionate (Boone and Bryant, 1980; Dong et al., 1994). However, the nature of the symbiotic interactions of such cocultures was not properly defined. S. wolinii and S. fumaroxidans are both able to couple propionate oxidation to sulfate reduction instead of proton reduction (Wallrabenstein et al., 1994; Van Kuijk and Stams, 1995). D. desulfuricans is a sulfate reducer that utilizes lactate, ethanol, hydrogen and formate in the presence of sulfate, but not acetate, propionate, butyrate or glucose (McInerney et al., 1979). Therefore, a syntrophic relationship with S. fumaroxidans, in which hydrogen and formate are produced, would be beneficial for D. desulfuricans. Nonetheless, it is intriguing why Syntrophobacter would engage in a syntrophic association while having sufficient sulfate to grow independently. By comparing the proteomic profile of S. fumaroxidans grown in coculture with D. desulfuricans with the proteomic profiles of the other known syntrophic lifestyles, and the sulfidogenic condition, we expect to be able to define the symbiotic relationship of S. fumaroxidans with D. desulfuricans.
Moreover, in a syntrophic coculture with Methanobrevibacter arboriphilus AZ, D. desulfuricans oxidized formate and transferred hydrogen to the methanogenic partner (Dolfing et al., 2008). The proteomic analysis of D. desulfuricans growing with hydrogen, formate and in coculture with S. fumaroxidans will reveal further insight into sulfate‐reducing syntrophic cocultures.
Results
Proteomic overview of S. fumaroxidans and most abundant proteins in all growth conditions
The genome of S. fumaroxidans contains 4098 protein coding genes (Plugge et al., 2012). Our proteomic analysis accurately identified a total of 813 proteins in the five studied conditions. Of these, 84 were designated as proteins with unknown function. About 514 proteins were detected in all the studied conditions. This core proteome represented slightly more than 60% of all the detected proteins (Supporting Information Fig. S1A). Principal component analysis (PCA) revealed that the protein abundance patterns were reproducible among triplicates of a given growth condition (Supporting Information Fig. S1B). Moreover, it shows that protein patterns of S. fumaroxidans differ depending on the electron acceptor or syntrophic partner used, clearly separating syntrophic methanogenic conditions from the axenic proteomic profiles. Statistical analysis indicated that 509 proteins significantly differed in at least one condition. This means that 304 proteins were constitutively produced in the five analysed conditions.
Total intensity‐based absolute quantification (iBAQ) revealed the most abundant proteins produced in the whole analysis (Supporting Information Table S1). Most of these proteins were involved in the methylmalonyl‐CoA pathway, sulfate reduction, electron transfer or energy conservation. Highly abundant proteins under all five conditions included chaperonins (GroEL and GroES), heat shock proteins and ribosomal proteins. Other abundant proteins had annotated functions involved in protection, signalling, transcription and ferrous ion transport. Rubrerythrins and proteins involved in the biosynthesis of cofactors like iron‐molybdenum and molybdopterin were also abundant.
Enzymes of the methylmalonyl CoA pathway
Previous genomic analyses of S. fumaroxidans predicted several genes coding for proteins involved in the MMC pathway (Müller et al., 2010; Plugge et al., 2012). Most of these proteins were abundant in our whole‐cell proteome analysis. For those predicted proteins that were not detected, paralogous proteins were found in high levels, which suggest that these proteins have a role in the MMC pathway. For instance, the predicted enzymes for propionate activation (Sfum_3926 to Sfum_3934) and for the conversion of acetyl‐CoA to acetate (Sfum_0388–0389, Sfum_0745–0746, Sfum_1278 and Sfum_3070) were not detected in the present study. Nevertheless, three CoA transferases were detected for the five conditions: CoA‐A (Sfum_0809–0810), CoA‐B (Sfum_0811–0812) and CoA‐S (Sfum_1132–1134) (Fig. 1). The amino acid sequences of these proteins indicate a relationship to coenzyme A transferase family I (InterPro IPR004165) and could therefore be involved in propionate activation and/or acetate formation.
Figure 1.
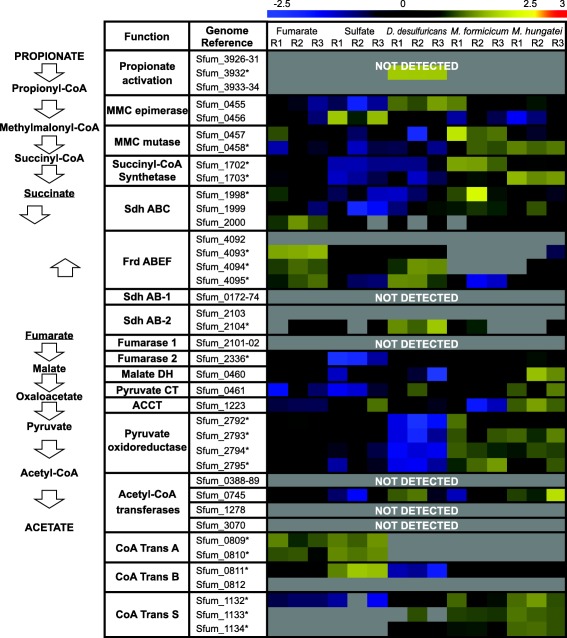
Relative expression levels of the proteins used in the methylmalonyl‐CoA pathway by Syntrophobacter fumaroxidans. Protein abundance levels are shown after Z‐score normalization. High relative expression is indicated in yellow and low relative expression is indicated in blue. Grey colour means not detected. In the left side the MMC steps are shown levelled to the associated proteins. The rows in the heat map show the detected proteins in five different growth conditions. The columns show from left to right, in triplicates, the electron acceptor used by S. fumaroxidans to couple propionate oxidation: fumarate, sulfate and interspecies compounds transferred to: Desulfovibrio desulfuricans, Methanobacterium formicicum and Methanospirillum hungatei. The asterisk indicates a statistically significant difference in at least one condition. MMC, methylmalonyl‐CoA; Sdh, succinate dehydrogenase; Frd, fumarate reductase; DH, dehydrogenase; CT, carboxyltransferase; ACCT, acetyl‐CoA carboxyltransferase; CoA Trans, coenzyme A transferase.
As predicted by previous genome studies (Müller et al., 2010; Plugge et al., 2012), the main protein complex responsible for the oxidation of succinate to fumarate was the membrane bound succinate dehydrogenase SdhABC (Sfum_1998–2000), which was abundant in all conditions. During axenic growth on propionate with fumarate, S. fumaroxidans converts propionate to succinate. Then, part of the fumarate in this growth condition is oxidized to acetate (Stams et al., 1993). This conversion is energy dependent, producing reducing equivalents during malate oxidation and pyruvate decarboxylation, and is only possible by coupling it to the energy yielding reduction of fumarate to succinate. The fumarate reductase FrdABEF (Sfum_4092–4095) complex was detected in higher levels during growth with fumarate. Except for a few subunits, the FrdABEF complex was not detected in cells grown with methanogens as expected since fumarate reduction only occurs when fumarate is provided. However, the complex was consistently detected in cells where sulfate was available, particularly in the coculture with D. desulfuricans. In the genome of S. fumaroxidans two additional gene clusters show similarity to succinate dehydrogenases SdhAB‐1 (Sfum_0172–0174) and SdhAB‐2 (Sfum_2103–2104). SdhAB‐1 was not detected in our study and only the alpha subunit of SdhAB‐2 showed a similar detection profile to FrdABEF. The predicted fumarase in the gene cluster Sfum_2101‐02 was not detected in any condition. Instead, a second fumarase from a non‐clustered gene (Sfum_2336) was abundant in all conditions. The amino acid sequence of this second fumarase corresponds to the previously isolated and characterized class I fumarase from S. fumaroxidans (Van Kuijk et al., 1996). Although this protein was abundant in all conditions, lower expression levels were measured in sulfate‐reducing cells. Finally, methylmalonyl‐CoA mutase (Sfum_0458) and succinyl‐CoA synthase (Sfum_1702–1703) were significantly more abundant in syntrophically grown cells, while the pyruvate oxidoreductase (Sfum_2792–2795) showed a lower relative expression during growth with Desulfovibrio.
Hydrogenases and formate dehydrogenases involved in electron transfer
The genome of S. fumaroxidans indicates the presence of six formate dehydrogenases and eight hydrogenases. Relative abundance levels of the hydrogenases and formate dehydrogenases produced by S. fumaroxidans during propionate degradation under different axenic or cocultured conditions are shown in Fig. 2. In this figure it can be seen that for most of the detected hydrogenases and formate dehydrogenases, the expression levels measured in syntrophic conditions with methanogens were higher than any of the axenic conditions. From the two predicted periplasmic hydrogenases, Hyn (Sfum_2952‐53) was detected in all conditions albeit more abundant during growth with fumarate and with D. desulfuricans, while Hyd2 (Sfum_0847‐48) was not detected in cells that were grown with D. desulfuricans and only in one triplicate of the sulfate condition.
Figure 2.
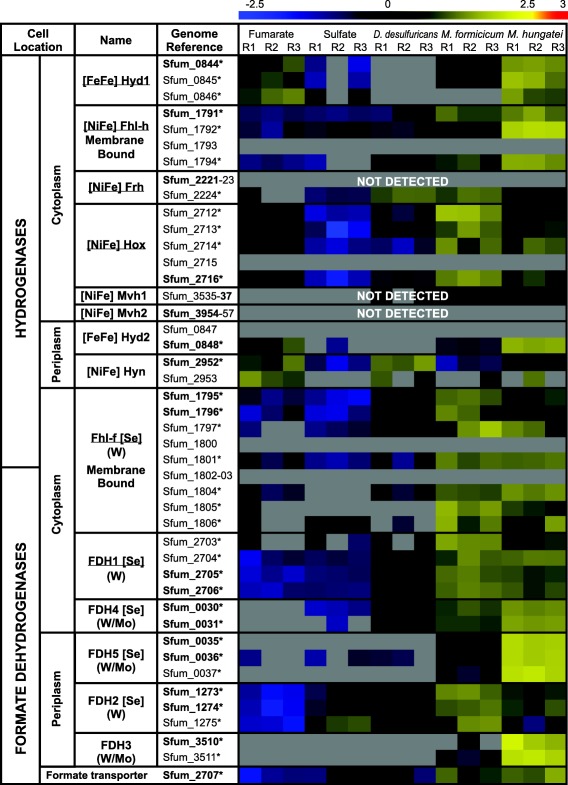
Relative abundance levels of hydrogenases and formate dehydrogenases in Syntrophobacter fumaroxidans during propionate oxidation. Protein abundance levels are shown after Z‐score normalization. The detected proteins are shown for five different growth conditions, in triplicates, according to the electron acceptor used by S. fumaroxidans to oxidize propionate; from left to right: fumarate, sulfate and interspecies compounds transferred to: Desulfovibrio desulfuricans, Methanobacterium formicicum and Methanospirillum hungatei. The colour intensity indicates the degree of protein up‐ or down regulation where high relative expression is indicated in red and low relative expression is indicated in blue; the grey colour represents not detected. Underlined complex names have been predicted to function as confurcating. Locus tags in bold font indicate the catalytic subunit of the complex. The asterisk indicates a statistical significant difference in at least one condition.
Proteins for Mvh2 (Sfum_3954‐57) were not found in our analysis, and only the subunits containing the FAD and NAD+‐binding oxidoreductase domains were detected for Mvh1 (Sfum_3535‐37) and Frh (Sfum_2221‐24) so these enzymes were classified as not detected. Of the three cytoplasmic hydrogenases detected, Hox (Sfum_2712‐16) and Fhl‐h (Sfum_1791‐94) were present in all conditions. Lastly, Hyd1 (Sfum_0844) was more abundant in syntrophically grown cells and cells grown with propionate and fumarate, but not when sulfate was present.
The three periplasmic formate dehydrogenases (Fdh2, Fdh3 and Fdh5) from S. fumaroxidans were abundant during growth in syntrophy with M. hungatei. However, for syntrophic growth with M. formicicum the detection levels of Fdh5 (Sfum_0035–37) and Fdh3 (Sfum_3509‐11) were significantly lower. Fdh3 was not detected in axenic conditions or in the coculture with D. desulfuricans, and Fdh5 was scarcely detected in such conditions.
Cytoplasmic Fdh1 (Sfum_2703‐06) and periplasmic Fdh2 (Sfum_1273‐75) were the most abundant formate dehydrogenases in all conditions. Moreover, significantly higher levels were measured during syntrophic growth. The membrane bound Fhl‐f (Sfum_1795–1806) was abundant in syntrophically grown cells but showed a lower relative expression during axenic growth. Fdh4 (Sfum_0030‐01) had very high relative abundance levels in syntrophic cultures. However, Fdh4 was not detected in the pure culture with fumarate, while only the lowest limits of detection were measured in sulfidogenic growth. The formate transporter (Sfum_2707) was detected in all conditions but more abundant in methanogenic cultures.
Redox proteins involved in dissimilatory sulfate reduction
A set of proteins required for dissimilatory sulfate reduction have previously been predicted in the genome of S. fumaroxidans (Pereira et al., 2011). Sulfate adenylyltransferase (Sat), proton‐translocating pyrophosphatase (HppA), APS reductase (AprAB), dissimilatory sulfite reductase (DsrAB) and DsrC complexes were among the most abundant proteins in all growth conditions. In contrast, neither of the two sulfate transporters (Sfum_0271 and Sfum_0653) predicted in the genome was detected in the analysis. Two periplasmic subunits of the QrcABCD complex (QrcB: Sfum_0610 and QrcC: Sfum_0609) were detected in all conditions and more abundant in syntrophic cultures (Fig. 3). The subunit QrcA (Sfum_0611) a membrane‐associated multihaem cytochrome c, was not detected. Sfum_4047 is the only other gene in S. fumaroxidans genome coding for a membrane‐anchored multihaem cytochrome c. The product of this gene was also detected in all conditions and more abundant in the cocultures with M. hungatei and D. desulfuricans.
Figure 3.
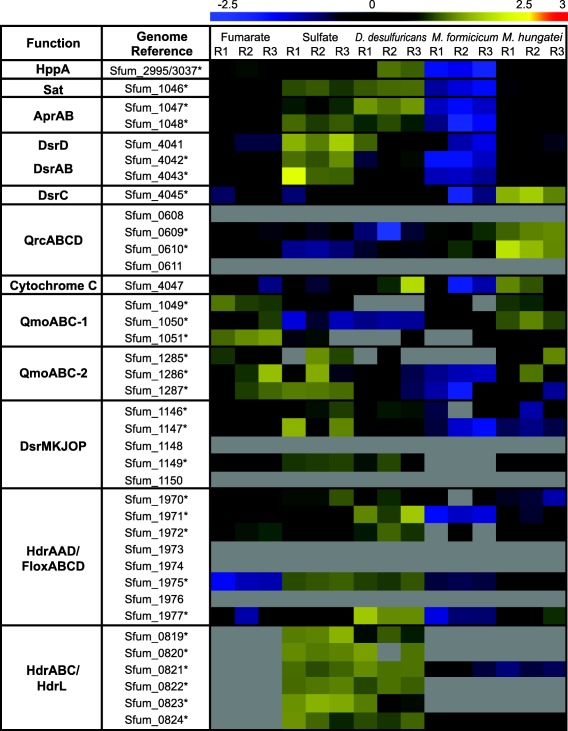
Relative abundance levels of proteins involved in sulfate reduction in Syntrophobacter fumaroxidans. Abundance levels after shown after Z‐score normalization. The columns show in triplicates, the electron acceptor used by S. fumaroxidans to couple propionate oxidation, from left to right: fumarate, sulfate and interspecies compounds transferred to: Desulfovibrio desulfuricans, Methanobacterium formicicum and Methanospirillum hungatei. High relative expression is indicated in red and low relative expression is indicated in blue. Grey colour means not detected. The asterisk indicates a statistical significant difference in at least one condition.
The genes coding for the trimeric complex QmoABC (Sfum_1049–1051) are well conserved in all known sulfate‐reducing bacteria (SRB) and are commonly located in a sat‐aprAB‐qmoABC cluster (Pereira et al., 2011). Surprisingly, the products of these genes were more abundant in cells grown with fumarate and in syntrophy than in cells grown with sulfate. However, a second QmoABC (Sfum_1285‐87) was detected in the proteome in all conditions. This complex was more abundant in cells grown axenically and in the coculture with M. hungatei. Similarly, the principal subunits of the DsrMKJOP (Sfum_1146–1150) complex were found in all conditions but more abundant in axenic conditions and in the coculture with D. desulfuricans.
Heterodisulfide reductases (Hdr) are enzymes present in methanogens and perform the reduction of CoM‐S‐S‐CoB heterodisulfide to CoM‐SH and CoB‐SH (Hedderich et al., 2005). Although the substrate of these enzymes CoM‐S‐S‐CoB heterodisulfide has only been found in methanogens, the high number of similar proteins (heterodisulfide reductases‐like) in SRB has been emphasized in several genome analyses (McInerney et al., 2007; Strittmatter et al., 2009; Pereira et al., 2011; Grein et al., 2013). Moreover, related enzymes have been purified from other non‐methanogenic archaea (Mander et al., 2004). An Hdr was detected in the proteome analysis of S. wolfei (Sieber et al., 2015), suggesting that the presence in the genome and production of such an enzyme complex is not dependent of a sulfate‐reducing lifestyle, but rather to microorganisms specialized in low energy metabolism. Two of the three predicted heterodisulfide reductases‐like enzymes in S. fumaroxidans were detected in this study, one associated with a flavin oxidoreductase complex Hdr/Flox (Sfum_1970–1977) and the other with a multicomplex that includes an HdrL, a MvhD and an FeS electron transfer protein: HdrABC/HdrL (Sfum_0819–0824). The Flox section of Hdr/Flox is produced in all conditions. HdrABC/HdrL was abundant when sulfate was present whereas only the subunits containing FAD/NAD‐binding domains were detected in syntrophic cultures. The fifth heterodisulfide reductase‐like found in the genome of S. fumaroxidans is associated with a pyruvate:Fd oxidoreductase, HdrAL/POR (Sfum_0012–0018); this complex was not detected.
Other proteins involved in energy conservation
The principle of electron bifurcation was originally proposed for a butyryl‐CoA dehydrogenase/electron transferring flavoprotein complex (Bcd‐Etf) in Clostridium kluyveri (Li et al., 2008). Since then three more flavin‐containing complexes capable of electron bifurcation from anaerobic bacteria and archaea have been described: [FeFe]‐hydrogenases (Hyd), transhydrogenases (NfnAB) and [NiFe]‐hydrogenase/heterodisulfide reductases (MvhADG–HdrABC) (Schut and Adams, 2009; Kaster et al., 2011; Huang et al., 2012; Buckel and Thauer, 2013).
Although S. fumaroxidans is not able to grow on butyrate or crotonate, complexes similar to Bcd/Etf have been predicted from the genome. The acyl‐CoA subunit (Sfum_1371) of one of these complexes was abundant in all conditions, while the Etf subunits (Sfum_1372 and Sfum_1373) were detected in lower levels, and the beta subunit was not detected at all in cells grown in cocultures. A second Etf complex from genes Sfum_0106 and Sfum_0107 was abundant in all conditions at similar levels than the acyl‐CoA subunit from gene Sfum_1371. (Supporting Information Fig. S2) Two additional paralogs coding for Acyl‐CoA/Etf complexes were found in the genome (Sfum_3686‐88 and Sfum_3929–3931), but not detected in our proteomic analysis. Finally, NfnAB (Sfum_2150–2151), another electron‐bifurcating iron‐sulfur flavoprotein commonly present in genomic analyses of sulfate reducers was exclusively detected during growth with fumarate.
Proteome generalities of Desulfovibrio desulfuricans
The complete genome of Desulfovibrio desulfuricans strain G11 has recently become available (Sheik et al., 2017). The genome counts with 2892 protein‐coding genes. Our proteome analysis successfully detected 779 proteins among the three growing conditions. The core proteome of D. desulfuricans consists of 317 proteins detected in all studied conditions (Supporting Information Fig. S3A). All these 317 proteins were detected in cells grown in coculture with S. fumaroxidans, while the cells growing with hydrogen or formate yielded more than 750 proteins each.
Differences in the proteome composition were explored using PCA (Supporting Information Fig. S3B). The first principal component (PC1; ∼ 70% of total variance) clearly separates growth in coculture from axenic growth in formate or hydrogen. However, PC1 did not establish a difference between growth on hydrogen or on formate. The second principal component (PC2) differentiates the three proteomic profiles, albeit PC2 accounts only for 10% of the variability of the data.
Although fewer D. desulfuricans proteins were detected in cells grown in coculture with S. fumaroxidans, proteins required for sulfate reduction such as AprA (G11_01440) were found among the ten most abundant proteins along with a periplasmic formate dehydrogenase (FDH3; G11_11530–11545) and a periplasmic [NiFe]‐hydrogenase (Hyd‐3; G11_06350–06355) (Supporting Information Fig S4). Periplasmic FDH3 and Hyd‐3 were in fact detected in all conditions, as well as cytoplasmic [NiFe]‐Hyd‐1 (G11_01905–01920) and FDH1 (G11_05250–05260) (Supporting Information Fig. S5). The cytoplasmic formate dehydrogenase FDH2 (G11_10090–10100) on the other hand, was detected only in cells grown with formate, while the periplasmic [FeFe]‐hydrogenase Hyd‐4 (G11_09530–09535) and cytoplasmic Ni‐Fe Hyd‐2 and Hyd‐6 (G11_02760‐80 and G11_10370‐75) were found in both axenic conditions but not in cells grown in coculture. Another [NiFe]‐hydrogenase (Hyd‐5; G11_10035‐45) with a cytochrome type‐b domain has been predicted from D. desulfuricans genome, but neither this protein nor the formate transporter (G11_05695) were detected in the proteomic results.
Discussion
The majority of the most abundant proteins detected in this study were involved in major processes such as propionate degradation, sulfate reduction, electron transfer, and energy conservation. Other abundant proteins, such as heat‐shock proteins, chaperonins, histones and transporters, emphasize the importance of protection, transport and stabilization of diverse macromolecules in the cell. These proteins have previously been reported as highly abundant in several proteomic analyses and identified as common stress‐induced molecules required for normal cell growth (Hemmingsen et al., 1988; Lu et al., 2007; Mancuso et al., 2012; Sieber et al., 2015).
Energy‐dependent succinate oxidation in MMC
For propionate degradation with fumarate, S. fumaroxidans requires a fumarate reductase, whereas to oxidize propionate with sulfate, or in syntrophy, a succinate dehydrogenase is needed. The high levels of the fumarate reductase (FrdABEF) in cells grown with propionate and fumarate reflects the reduction of fumarate in this lifestyle. However, the abundance of this complex in cells growing with sulfate and in coculture with D. desulfuricans can only be explained by a reversible performance to succinate oxidation, since no succinate was accumulated in those conditions. Fumarate reductases and succinate dehydrogenases are functionally and structurally related enzymes (Mattevi et al., 1999). The membrane bound SdhABC of S. fumaroxidans has previously been purified, characterized and showed activity in both directions, fumarate reduction and succinate oxidation (Van Kuijk, 1998b). However, FrdABEF has not been purified and as such could not be tested for a reversible activity. Transcription experiments reported that FrdABEF was up‐regulated (> 2 log ratio) when fumarate was the electron acceptor in contrast with the gene transcription of cells gown in syntrophic cocultures with M. hungatei (Worm, 2010). Interestingly in such study FrdABEF was also up‐regulated in cells grown with sulfate as the electron acceptor and down‐regulated in cells cocultured with M. formicicum. Our proteomic study confirms the high expression levels of FrdABEF in propionate plus fumarate cultures. Moreover, FrdABEF was also present in conditions where propionate was oxidized with sulfate and in coculture with D. desulfuricans. These results might suggest a reversible function of the fumarate reductase FrdABEF toward succinate oxidation. Nevertheless, although in in‐vitro analysis the reversible activity of enzymes is possible, in vivo the enzymes are usually dedicated to one physiological function. Besides S. fumaroxidans has a succinate dehydrogenase (SdhABC) for succinate oxidation. A more likely possibility is that fumarate reduction occurred in the sulfidogenic condition. To pull the oxidation of succinate toward the formation of fumarate, hydrogen and formate, these products have to be efficiently removed. To maintain the levels of fumarate low, the fumarase has to convert fumarate efficiently to malate. This process is very important and as such fumarase is one of the most abundant proteins in S. fumaroxidans. However, cells grown with sulfate show the lowest expression levels of this enzyme. It might be that if fumarate is not removed efficiently in sulfate‐grown cells, the bacteria start to produce FrdABEF.
Hydrogen and formate in IET and RET
During syntrophic growth, S. fumaroxidans needs to transfer electrons via hydrogen and/or formate to a syntrophic partner. It has long been speculated that formate plays a more important role than hydrogen as an electron carrier in the syntrophic associations of this bacterium with methanogens (De Bok et al., 2002a, 2002b). Although slightly higher levels were measured in the formate transporter during syntrophic growth over the axenic conditions, S. fumaroxidans must rely on other mechanisms to transfer formate. Three formate dehydrogenases (Fdh2, Fdh3 and Fdh5) contain a twin‐arginine translocation (Tat) pathway conserved site, which points to the translocation of these proteins across the cytoplasmic membrane. Fdh3 and Fdh5 were detected only in syntrophically grown cells, while Fdh2 was detected in all conditions, but was more abundant during syntrophic growth. This suggests that periplasmic Fdh3 and Fdh5 are complexes specialized in transferring formate to the syntrophic partner, while Fdh2 is broadly used for energy conservation purposes as part of the reverse electron transport mechanism, possibly coupled to SdhABC or FrdABEF (Fig. 4).
Figure 4.
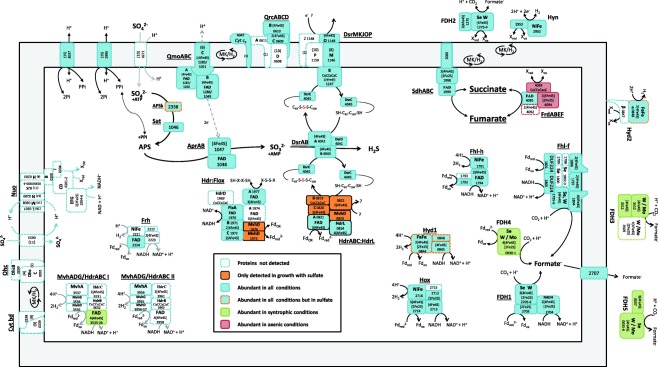
Schematic representation of energy converting complexes and proteins involved in sulfate reduction in Syntrophobacter fumaroxidans during propionate oxidation.
Among the cytoplasmic formate dehydrogenases, Fdh1 is homologous to the bifurcating [FeFe]‐hydrogenase of Thermotoga maritima (Schut and Adams, 2009). Furthermore, it contains a conserved site coding for a 51 kDa subunit of a NADH:ubiquinone oxidoreductase which makes this protein a very plausible candidate for a confurcating‐type of formate dehydrogenase. Fdh1 was detected in all conditions and higher levels were detected in syntrophic conditions. Similarly, the membrane associated Fhl‐f was also detected in all conditions and more abundant in syntrophically grown cells. The ubiquitous detection of Fdh1 and Fhl‐f indicates that their role is not restricted to IET, but that these complexes are essential for energy conservation and formate/hydrogen interconversion during propionate degradation. On the other hand, Fdh4 was not detected in cells grown with fumarate, scarcely detected in cells grown with sulfate and highly abundant in methanogenic conditions. This led us to speculate that Fdh4 has an exclusive role in IET. Furthermore, the genes coding for Fdh4 are located upstream in the genome of the periplasmic Fdh5 operon. Considering these observations, we propose that these neighbouring genes coding for cytoplasmic and periplasmic formate dehydrogenases are used mainly for interspecies formate transfer. Thus Fdh3, Fdh4 and Fdh5 seem to form a set of formate dehydrogenases used by S. fumaroxidans to transfer electrons to the syntrophic partner. It is conceivable that these formate dehydrogenases contain a molybdenum catalytic core (Mo‐FDH) in contrast to Fdh1 and Fdh2 whose structure has been characterized and were shown to have only tungsten‐containing active sites (W‐FDH) (De Bok et al., 2003). Further biochemical analysis of these formate dehydrogenases will give insight of the role of molybdenum in IET mechanisms in methanogenic environments (Plugge et al., 2009; Worm et al., 2011b).
Only five of the eight predicted hydrogenases of S. fumaroxidans were detected in the present analysis. Of the two periplasmic hydrogenases, Hyn was more abundant in cells grown with propionate and fumarate and in coculture with D. desulfuricans. Hyn has been proposed to be involved in reverse electron transport coupled with FrdABEF for fumarate reduction or SdhABC for succinate oxidation (Worm et al., 2011a). Considering the high levels of Hyn and FrdABEF in the coculture with D. desulfuricans, we suggest that indeed Hyn is involved in RET with FrdABEF, whether reducing fumarate in fumarate conditions or reversibly oxidizing succinate in the coculture with D. desulfuricans (Fig. 4).
Of the three cytoplasmic hydrogenases detected, Hox and Fhl‐h, which were detected in all conditions, were more abundant in cells grown with the methanogens. Hox is most probably a confurcating hydrogenase involved in energy conservation. The membrane‐bound Fhl‐h on the other hand, together with Fhl‐f might be involved in a cytoplasmic hydrogen‐formate interconversion during syntrophic growth to control electron release. Finally, the genes coding for Hyd1 and Hyd2 are adjacent in the genome, the products of these genes are produced only in the presence of fumarate and during syntrophic growth but not when sulfate was available. This might be due to the exclusive use of other confurcating energy‐conserving complexes in sulfidogenic conditions, for instance HdrABC/HdrL.
Although formate formation seems to prevail in the syntrophic lifestyle of S. fumaroxidans, our results indicate that hydrogen, via Hyd1, Hyd2, Hox and Hyn also plays an important role in energy conservation by RET. During growth with fumarate, when IET is not required, these hydrogenases were detected in higher abundance than any of the formate dehydrogenase in such growth condition.
Energy conservation mechanisms in the sulfate‐reducing metabolism
All the proteins necessary for sulfate reduction in S. fumaroxidans were abundant in this analysis, with the intriguing exception of the sulfate transporters that were not detected. In order to activate sulfate by sulfate adenylyltransferase, sulfate has to be transported into the cell. Therefore, another mechanism for transport of sulfate across the membrane must be used by S. fumaroxidans. Several transporters and unknown proteins were among the most abundant proteins in this study, it is possible that some of them could have played a role in the import of sulfate to the cytoplasm.
The abundance of HppA, Sat, Apr and DsrAB in our proteomic analysis in conditions where sulfate reduction was not observed indicates that the sulfate reduction pathway is not strictly regulated in S. fumaroxidans. However, all these enzymes were significantly more abundant in conditions where sulfate was available, indicating sulfidogenic activity in cells grown with sulfate and with D. desulfuricans. Similarly, for complexes such as Qmo‐2, DsrMKJOP and Hdr/Flox it is possible to observe an up‐regulation in axenic conditions and in some cases in coculture with D. desulfuricans, while for Qrc and Qmo‐1 higher levels are observed in syntrophically grown cells. These observations suggest that the use of these complexes in electron transfer is not constrained to a sulfidogenic lifestyle, and that they could for instance transfer electrons to periplasmic formate dehydrogenases for IET or to the FrdABEF for RET.
Quinone reductase complexes (QrcABCD) are involved in the reduction of the quinone pool in D. vulgaris Hildenborough. Furthermore, it was shown that QrcABCD is reduced by periplasmic hydrogenases and formate dehydrogenase via the cytochrome c3 (subunit A of the complex) (Venceslau et al., 2010). Although in D. vulgaris the described role of QrcABCD is to reduce menaquinone with electrons gained from hydrogen or formate oxidation during sulfate reduction, we speculate that a reverse process is feasible. In D. desulfuricans G20, a mutant lacking the qrcB gene was unable to grow with H2 or formate as electron donor, while it grew similarly as the parent strain with lactate (Li et al., 2009). Moreover, this mutation also inhibited syntrophic growth with a methanogen in lactate. The higher levels of the QrcABCD of S. fumaroxidans in cells grown in syntrophy might be explained by its involvement in electron transfer to the periplasmic formate dehydrogenases Fdh3 and Fdh5 (Figs. 2 and 3).
Direct electron transfer from Qmo to Apr to facilitate the reduction of sulfate to sulfite has been reported in Desulfovibrio desulfuricans (Pires et al., 2003; Pereira, 2008; Duarte et al., 2016). In Syntrophobacter, the higher expression levels of the two Qmo complexes in cells grown with fumarate might be due to the use of this membrane bound complex in transferring electrons to FrdABEF for RET. FrdABEF lacks a transmembrane subunit, therefore it has been speculated that it receives electrons from menaquinone via cytochrome b and cytochrome b:quinone oxidoreductases (Müller et al., 2010), however these cytochromes were not detected in our study.
DsrMKJOP is another highly conserved membrane complex in SRB (Rabus et al., 2015). In many Gram‐positive SRB only the cytoplasmic‐facing DsrMK genes are present, suggesting that this is the minimal functional module (Pereira et al., 2011). Although in S. fumaroxidans the complete gene set of DsrMKJOP is present, only the essential subunits (DsrMK), and the periplasmic DsrO were detected in our proteomic study. In the heat map shown in Fig. 3 the expression profile of DsrMKO is similar to that of the Hdr/Flox complex. If Hdr/Flox is used in all conditions to confurcate electrons as will be discussed below, DsrMKO might be involved in electron transfer with this complex.
HdrABC/FloxABCD, a novel NADH dehydrogenase/heterodisulfide reductase widespread in anaerobic bacteria has been proposed to be involved in flavin‐based electron bifurcation in D. vulgaris Hildenborough (Ramos et al., 2015). The Flox proteins (Sfum_1970–1973) of the Hdr/Flox of S. fumaroxidans were constitutively present in all the conditions. Nevertheless, the Hdr‐like complex in the Hdr/Flox cluster have a composition different to the canonical HdrABC. For instance, HdrBC is replaced by the cysteine‐rich containing HdrD (Sfum_1969), which was not detected in our analysis. Furthermore, two hdrA genes are present (Sfum_1974 and Sfum_1977), but only the product of Sfum_1977 was detected. Hdr/Flox could be another confurcating system used by S. fumaroxidans to oxidize NADH during propionate degradation, possibly involved in recycling NAD+ during the reduction of fumarate. However, the conformational changes mentioned above might imply functional differences that need to be further investigated.
For the HdrABC/HdrL complex, the hdrABC genes (Sfum_0819–0821) are next to genes coding for a pyridine nucleotide‐disulphide oxidoreductase comprising an HdrL protein (Sfum_0824). HdrL is a large protein containing HdrA and one or two NADH binding domains (Strittmatter et al., 2009; Pereira et al., 2011). An MvhD protein is encoded in Sfum_0823, but the catalytic hydrogenase subunit MvhA is not present. The amino acid sequence of Sfum_0822 codes for iron‐sulfur domains (4Fe‐4S) commonly found in the beta subunits of hydrogenases or formate dehydrogenases (InterPro, December 2017). Moreover, a BlastP search of the amino acid sequence resulted in significant alignments with sequences of formate dehydrogenases in other SRB. We can only speculate if this HdrABC/HdrL complex is able to use hydrogen, formate or some other compound, but the high detection levels of the complete multimeric complex imply an important function in the sulfate‐reducing metabolism.
HdrABC/HdrL was detected only in conditions where sulfate was present, axenically or in the presence of D. desulfuricans. The soluble complex MvhADG/HdrABC has been shown to perform flavin‐based electron bifurcation in methanogens (Thauer et al., 2008; Kaster et al., 2011). We speculate that HdrABC/HdrL is preferred when sulfate is available, over the confurcating hydrogenase Hyd1 which in turn was highly abundant in cells grown with fumarate as electron acceptor and in syntrophy, but not detected when sulfate was in the medium (Fig. 2). The reason for the preference of HdrABC/HdrL under sulfidogenic conditions is unclear. However, it could be related to the substrates used by this complex. The MvhADG/HdrABC in methanogens uses H2 to reduce ferredoxin and heterodisulfide (Kaster et al., 2011). It is possible that the exclusive high levels of HdrABC/HdrL in our sulfidogenic conditions correspond to the need of reduction of the so called “bacterial heterodisulfide” DsrC (Venceslau et al., 2014).
It has been suggested (Venceslau et al., 2014), that the protein DsrC could serve as a redox hub, linking oxidation of several substrates to sulfate reduction. Our results with S. fumaroxidans show DsrC as one of the most abundant proteins present in all conditions and significantly more abundant in syntrophy with M. hungatei. The recent discoveries point to the role of DsrC as an electron carrier interacting with DsrAB, DsrMKJOP, Hdr/Flox and HdrABC/HdrL, but it could also connect other enzyme complexes like the fumarate reductase FrdABEF in our model bacterium S. fumaroxidans, which in turn would also explain the detection of FrdABEF in cells grown with sulfate.
Proteomic profiling of Desulfovibrio desulfuricans
The low amount of D. desulfuricans proteins detected from cells grown in coculture with S. fumaroxidans can be the result of low biomass in such condition. From microscopic observations we know that the ratio of S. fumaroxidans to D. desulfuricans was 2:1 (data not shown). Although normalization of the data performed with MaxQuant allowed us to compare the detected proteins with the other growth conditions where more proteins were identified, we rather focused in analysing the most abundant proteins detected in the coculture condition.
The abundance of the periplasmic Hyd‐3 and periplasmic FDH3 in cells grown with S. fumaroxidans indicates that interspecies electron transfer carried by formate and hydrogen was taking place in the coculture. The abundance of the proteins involved in sulfate reduction confirm that D. desulfuricans was actively reducing sulfate for which it certainly needed electron donors which could only come from S. fumaroxidans in such growth condition. This shows a remarkable metabolic tendency of S. fumaroxidans to engage in syntrophic interactions.
Conclusions
This study shows the importance of formate as electron carrier in IET and RET during syntrophic and axenic growth of Syntrophobacter fumaroxidans. S. fumaroxidans utilizes a specific set of enzymes (Fdh3, Fdh4 and Fdh5) to transfer electrons to the syntrophic partner. Previous isolation and characterization of Fdh1 and Fdh2 have revealed only tungsten‐containing active sites (W‐FDH). Biochemical analysis of the three above mentioned formate dehydrogenases could provide insight of the role of molybdenum‐dependent formate dehydrogenases in syntrophic growth.
Fdh2 and Hyn are the periplasmic enzymes used by S. fumaroxidans to recycle hydrogen and formate during RET. While Fdh2 is mainly coupled to Sdh during succinate oxidation, Hyn seems to be coupled to Frd for fumarate reduction in propionate plus fumarate but also for succinate oxidation in other growth conditions.
Although the sulfate‐reducing metabolism is poorly regulated, the abundance of membrane‐bound complexes like Qrc, Qmo and DsrMKJOP, consistently found in all conditions, as well as the absence of cytochromes in the present study (only two cytochromes detected from eight predicted in the genome), indicates that those membrane‐bound complexes might play a role in the transfer of electrons between cytoplasmic enzymes and the periplasmic formate dehydrogenases and hydrogen dehydrogenases.
HdrABC/HdrL is the most abundant putatively confurcating system in sulfidogenic conditions, possibly because of its probable connection to DsrC, an electron hub in sulfidogenic metabolism.
The proteomic profiles of both bacteria in the coculture of S. fumaroxidans with D. desulfuricans give insight in the metabolic flexibility of S. fumaroxidans. Results showed a proteomic profile of S. fumaroxidans in which sulfate reduction took place, while energy conservation and IET mechanisms were also used similarly as in the syntrophic associations with methanogens. The proteomic analysis of the partner D. desulfuricans confirmed IET via formate and hydrogen carried on by S. fumaroxidans in a sulfate rich environment.
Materials and methods
Organisms and growth conditions
Syntrophobacter fumaroxidans was grown in pure culture and in cocultures. Syntrophobacter fumaroxidans MPOBT (DSM 10017) was cultivated under anoxic conditions in basal medium as described previously (Stams et al., 1993). The medium for the pure cultures was supplemented with 20 mM propionate and 60 mM fumarate. Sulfidogenic cultures were grown on 20 mM propionate and 20 mM sulfate. Cocultures of S. fumaroxidans with Methanospirillum hungatei strain JF1T (DSM 864) or Methanobacterium formicicum MFT (DSM 1535) were grown with 30 mM of propionate without electron acceptor. A coculture of S. fumaroxidans with Desulfovibrio desulfuricans strain G11 (DSM 7057; Sheik et al., 2017) was grown with 20 mM propionate and 20 mM sulfate. Axenic cultures of D. desulfuricans were grown with 20 mM sulfate and 40 mM formate or hydrogen (1.7 atm H2/CO2 80:20 vol/vol). All organisms were batch cultured in triplicate at 37°C in 1 l flasks with 550 ml medium under anaerobic conditions provided by a pressurised (172 kPa; 1.7 atm) gas phase of N2/CO2 (80:20, vol/vol). Growth was monitored by measuring substrate consumption and product formation (propionate, sulfate, methane, acetate, succinate, malate and/or sulfide). Cells were harvested during mid‐exponential growth phase. The cultures for the experiment were inoculated with cells from cultures that were adapted to these conditions by transferring them at least five times in media with the respective substrates before the start of the experiment.
Harvesting cells and Percoll gradient centrifugation
Cells were aerobically harvested by centrifugation at 16,000g for 16 min at 4°C. The pellet was washed twice with TE buffer (10 mM Tris‐HCl, pH 7.5; 1 mM EDTA). Only cells from the syntrophic coculture of S. fumaroxidans and M. hungatei were separated by Percoll gradient centrifugation (Percoll®, Sigma‐Aldrich, Missouri) as described elsewhere (De Bok et al., 2002b). The separated layers, containing Syntrophobacter cells in the upper layer and Methanospirillum cells in the lower layer, were collected and subjected to Percoll gradient separation a second time. Cells were then washed twice with 10 mM sodium phosphate buffer (pH 7.5).
Protein extraction and SDS‐PAGE
Cells were resuspended in lysis buffer (100 mM Tris‐HCl, pH 7.5; 4% w/v SDS; 50 mM dithiothreitol and SIGMAFAST™ Protease Inhibitor Cocktail Tablet—Sigma‐Aldrich, Missouri), and passed three times through a French press (French® Type Pressure Cell Disrupter, Stansted Fluid Power, Harlow, UK) at 2 MPa (40 K cell). Cell debris and undisrupted cells were removed by centrifugation at 18,000g for 10 min at 4°C. The supernatant was collected in Eppendorf™ LoBind Protein Microcentrifuge Tubes and stored at −80°C. Still in the lysis buffer, proteins were denatured by heating at 95°C for 5 min. Samples were loaded on a 10% polyacrylamide separation gel (Precise™ Tris‐HEPES Gels, Thermo Scientific, Rockford) using the Mini‐PROTEAN Tetra Cell (Bio‐Rad Laboratories B.V, Veenendaal, The Netherlands). The electrophoresis procedure was according to the precast gels manufacturer's instructions. Gels were stained using Coomassie Brilliant Blue (CBB) R‐250. Protein concentration was normalized among triplicates and samples in a qualitative way by analysing the gel pictures taken with G:BOX Chemi XT4 (Syngene, Cambridge, UK) and using the software GeneSys version 1.5.5.0 (GeneTools version 4.03.01).
In‐gel trypsin digestion
In‐gel digestion of proteins and purification of peptides was done following a modified version of a previously described protocol (Rupakula et al., 2013). Disulfide bridges in proteins were reduced by covering the gels with reducing solution (10 mM dithiothreitol, pH 7.6, in 50 mM NH4HCO3), and the gels were incubated at 60°C for 1 h. Alkylation was performed in darkness and shaking (100 rpm) for 1 h by adding 25 ml of iodoacetamide solution (10 mM iodoacetamide in 100 mM Tris‐HCl, pH 8.0). Gels were thoroughly rinsed with demineralized water in between steps. Each gel lane was cut into three slices, and the slices were cut into approximately 1 mm3 cubes and transferred to a separate 0.5 ml protein LoBind tube (Eppendorf, Hamburg, Germany). Enzymatic digestion was done with trypsin sequencing grade (Roche, Mannheim, Germany). About 100 µl of trypsin solution (5 ng µl−1 trypsin in 50 mM NH4HCO3) were added to each tube and incubated 2 h at 45°C with gentle shaking. To stop trypsin digestion, trifluoroacetic acid (10%) was added to the supernatant to lower the pH below 5. The digested protein mixture was purified and concentrated using an in‐house made SPE pipette tip (Lu et al., 2011).To recover hydrophobic peptides, 50 µl acetonitrile (vol/vol in 0.1% formic acid) was passed through the column. Finally, the volume was reduced to 20 µl using a SpeedVac concentrator and then adjusted to 50 µl with 0.1% formic acid. Samples were analysed using nLC–MS/MS with a Proxeon EASY nLC and a LTQ‐Orbitrap XL mass spectrometer as previously described (Lu et al., 2011).
LC–MS data analysis
The obtained MS/MS spectra were processed with MaxQuant v. 1.5.2.8. Database with the protein sequences of S. fumaroxidans was downloaded from UniProt (http://www.uniprot.org). The protein database of Desulfovibrio desulfuricans strain G11 was downloaded from GenBank accession number CP023415. An additional dataset with protein sequences of common contaminants (trypsin, human keratins and bovine serum albumin) was included. False discovery rates (FDR) of < 1% were set at peptide and protein levels. Modifications for acetylation (Protein N‐term), deamidation (N, Q) and oxidation (M) were allowed to be used for protein identification and quantification. All other quantification settings were kept default. Filtering and further bioinformatics and statistical analysis were performed with Perseus v.1.5.3.0. Proteins included in our analysis contain at least two identified peptides of which at least one is unique and at least one unmodified. Reversed hits and contaminants were filtered out. Protein groups were filtered to require three valid values in at least one experimental group. Label‐free quantification (LFQ) intensities (values normalized with respect to the total amount of protein and all of its identified peptides) were used to analyse the abundance of proteins in the fractions and further statistical comparisons among conditions. LFQ intensities were transformed to logarithmic values base 10. Missing values were imputed with random numbers from a normal distribution, the mean and standard deviation of which were chosen to best simulate low abundance values close to noise level (Width: 0.3 and downshift 1.8 times). A multiple‐sample test (ANOVA) with permutation‐based FDR statistics (250 permutations, FDR = 0.01 and S0 = 1) was applied to filter significant proteins. PCA were performed with default settings and without category enrichment in components. Z‐score normalization in which the mean of each row (where each row is a protein in triplicate and in different conditions) is subtracted from each value and the result divided by the standard deviation of the row was applied before clustering. Hierarchical clustering of rows, using Euclidean distances, produced a heat map representation of the clustered data matrix. Row clusters were automatically defined (100) and exported to a new matrix. Imputed values were then replaced back to missing values and previously defined clusters were displayed in a new heat map. For D. desulfuricans the Z‐score and hierarchical clustering was done for columns instead of rows in order to compare the most abundant proteins detected in each condition.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. A. Venn diagram of the 813 proteins detected in Syntrophobacter fumaroxidans growth on propionate with five different (biological or chemical) electron acceptors. B. Principal Component Analysis performed for S. fumaroxidans protein profiles obtained from each triplicate grown under five different conditions. Symbols: Orange diamonds, sulfate reducing; Red crosses, growth with fumarate; Grey squares, in coculture with Desulfovibrio desulfuricans in a sulfate rich environment; Green triangles, in syntrophy with Methanospirillum hungatei; Blue circles, in syntrophy with Methanobacterium formicicum.
Fig. S2. Normalized expression matrix of energy conservation mechanisms predicted for Syntrophobacter fumaroxidans. Proteins are shown for five different growth conditions, in triplicates; from left to right: fumarate, sulfate and interspecies compounds transferred to: Desulfovibrio desulfuricans, Methanobacterium formicicum and Methanospirillum hungatei. The colour scale illustrates the relative detection level of each protein across the 5 samples; blue (log ratio −2.5) and yellow (log ratio 2.5) indicate lower and higher levels compared with the average level value (in black) respectively. Not detected proteins in a specific condition appear in grey. The asterisk indicates a statistical significant difference in at least one condition.
Fig. S3. A. Venn diagram of the 779 proteins detected in Desulfovibrio desulfuricans growing in sulfate rich medium in coculture with Syntrophobacter fumaroxidans or axenically on H2/CO2 or formate. B. PCA performed for D. desulfuricans protein profiles. Symbols: red diamonds, hydrogenotrophic conditions; black squares, growth with formate and filled grey squares correspond to the cocultured partnership of D. desulfuricans with S. fumaroxidans.
Fig. S4. Heat map of hierarchical clustered proteins produced by Desulfovibrio desulfuricans. The proteins are shown in a clustered matrix after column Z‐score normalization and automatic hierarchical columns clustering. Three growth conditions, in triplicates, are shown according to the electron donor used; from left to right: formate, hydrogen and compounds transferred from Syntrophobacter fumaroxidans. The colour scale represents the relative detection level of each protein across the samples; blue log ratio −3, yellow log ratio 3, red log ratio 4 and green log ratio 5 indicate lower and higher levels compared with the average level value 0 (in black) respectively. The colour intensity indicates the degree of protein up‐ or down regulation; the grey colour represents not detected.
Fig. S5. Normalized expression matrix of hydrogenases and formate dehydrogenases of Desulfovibrio desulfuricans. The rows in the heat map show proteins levels after row Z‐score standardization in three different growth conditions. The columns show from left to right, in triplicates, the electron donor used by D. desulfuricans: formate, hydrogen and interspecies compounds transferred from Syntrophobacter fumaroxidans. The colour scale indicates the degree of protein down‐ or up regulation ranging from blue (−2.2 log ratio), to yellow (2.2 log ratio). The colour intensities indicate lower and higher levels compared with the average level 0 value (in black); the grey colour represents not detected. Subunits, twin‐arginine translocation (TAT) pathway signal and selenocysteine insertion (Sec) sequences are indicated after the locus tag.
Fig. S6. Heat map of hierarchical clustered proteins produced by Syntrophobacter fumaroxidans for propionate degradation. The proteins are shown in a clustered matrix after automatic hierarchical cluster of rows from row Z‐score normalization values. Proteins appear from left to right, in triplicates, according to the growth conditions defined by the electron acceptor used by S. fumaroxidans to oxidize propionate: fumarate, sulfate and interspecies compounds transferred to: Desulfovibrio desulfuricans, Methanobacterium formicicum and Methanospirillum hungatei. The colour scale illustrates the relative detection level of each protein across the samples; blue (log ratio −2.5), yellow (log ratio 2.5) and red (log ratio 3) indicate lower and higher levels compared with the average level value 0 (in black). The colour intensity indicates the degree of protein up‐ or down regulation; the grey colour represents not detected.
Table S1. iBAQ values of proteins detected in axenic and cocultured conditions in Syntrophobacter fumaroxidans and Desulfovibrio desulfuricans.
Supporting Information Dataset 1
Supporting Information Dataset 2
Acknowledgements
This research was supported by the Dutch Technology Foundation (STW) (project 11603), which is part of the Netherlands Organization for Scientific Research (NWO), and which is partly funded by the Ministry of Economic Affairs. Research of AJMS is supported by the European Research Council (ERC grant 323009) and the Gravitation grant (024.002.002) of the Netherlands Ministry of Education, Culture and Science.
References
- Boone, D.R. , and Bryant, M.P. (1980) Propionate‐degrading bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from methanogenic ecosystems. Appl Environ Microbiol 40: 626–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel, W. , and Thauer, R.K. (2013) Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta 1827: 94–113. [DOI] [PubMed] [Google Scholar]
- De Bok, F.A.M. , Roze, E.H. , and Stams, A.J.M. (2002a) Hydrogenases and formate dehydrogenases of Syntrophobacter fumaroxidans . Antonie Van Leeuwenhoek 81: 283–291. [DOI] [PubMed] [Google Scholar]
- De Bok, F.A.M. , Luijten, M.L.G.C. , and Stams, A.J.M. (2002b) Biochemical evidence for formate transfer in syntrophic propionate‐oxidizing cocultures of Syntrophobacter fumaroxidans and Methanospirillum hungatei . Appl Environ Microbiol 68: 4247–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bok, F.A.M. , Hagedoorn, P.‐L. , Silva, P.J. , Hagen, W.R. , Schiltz, E. , Fritsche, K. , and Stams, A.J.M. (2003) Two W‐containing formate dehydrogenases (CO2‐reductases) involved in syntrophic propionate oxidation by Syntrophobacter fumaroxidans . Eur J Biochem 270: 2476–2485. [DOI] [PubMed] [Google Scholar]
- Dolfing, J. , Jiang, B. , Henstra, A.M. , Stams, A.J.M. , and Plugge, C.M. (2008) Syntrophic growth on formate: a new microbial niche in anoxic environments. Appl Environ Microbiol 74: 6126–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. , Plugge, C.M. , and Stams, A.J.M. (1994) Anaerobic degradation of propionate by a mesophilic acetogenic bacterium in coculture and triculture with different methanogens. Appl Environ Microbiol 60: 2834–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, A.G. , Santos, A.A. , and Pereira, I.A. (2016) Electron transfer between the QmoABC membrane complex and adenosine 5'‐phosphosulfate reductase. Biochim Biophys Acta 1857: 380–386. [DOI] [PubMed] [Google Scholar]
- Grein, F. , Ramos, A.R. , Venceslau, S.S. , and Pereira, I.A. (2013) Unifying concepts in anaerobic respiration: insights from dissimilatory sulfur metabolism. Biochim Biophys Acta 1827: 145–160. [DOI] [PubMed] [Google Scholar]
- Harmsen, H.J.M. , Van Kuijk, B.L.M. , Plugge, C.M. , Akkermans, A.D.L. , de Vos, W.M. , and Stams, A.J.M. (1998) Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate‐degrading sulfate‐reducing bacterium. Int J Syst Bacteriol 48: 1383–1387. [DOI] [PubMed] [Google Scholar]
- Hedderich, R. , Hamann, N. , and Bennati, M. (2005) Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron‐sulfur cluster. Biol Chem 386: 961–970. [DOI] [PubMed] [Google Scholar]
- Hemmingsen, S.M. , Woolford, C. , van der Vies, S.M. , Tilly, K. , Dennis, D.T. , Georgopoulos, C.P. , et al (1988) Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333: 330–334. [DOI] [PubMed] [Google Scholar]
- Huang, H. , Wang, S. , Moll, J. , and Thauer, R.K. (2012) Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2 . J Bacteriol 194: 3689–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaster, A.K. , Moll, J. , Parey, K. , and Thauer, R.K. (2011) Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci 108: 2981–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Hinderberger, J. , Seedorf, H. , Zhang, J. , Buckel, W. , and Thauer, R.K. (2008) Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl‐CoA dehydrogenase/Etf complex from Clostridium kluyveri . J Bacteriol 190: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Luo, Q. , Wofford, N.Q. , Keller, K.L. , McInerney, M.J. , Wall, J.D. , and Krumholz, L.R. (2009) A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J Bacteriol 191: 2675–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P. , Vogel, C. , Wang, R. , Yao, X. , and Marcotte, E.M. (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25: 117–124. [DOI] [PubMed] [Google Scholar]
- Lu, J. , Boeren, S. , de Vries, S.C. , van Valenberg, H.J.F. , Vervoort, J. , and Hettinga, K. (2011) Filter‐aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J Proteom 75: 34–43. [DOI] [PubMed] [Google Scholar]
- Mancuso, F. , Bunkenborg, J. , Wierer, M. , and Molina, H. (2012) Data extraction from proteomics raw data: an evaluation of nine tandem MS tools using a large Orbitrap data set. J Proteom 75: 5293–5303. [DOI] [PubMed] [Google Scholar]
- Mander, G.J. , Pierik, A.J. , Huber, H. , and Hedderich, R. (2004) Two distinct heterodisulfide reductase‐like enzymes in the sulfate‐reducing archaeon Archaeoglobus profundus . Eur J Biochem 271: 1106–1116. [DOI] [PubMed] [Google Scholar]
- Mattevi, A. , Tedeschi, G. , Bacchella, L. , Coda, A. , Negri, A. , and Ronchi, S. (1999) Structure of L‐aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Structure 7: 745–756. [DOI] [PubMed] [Google Scholar]
- McInerney, M.J. , Bryant, M.P. , and Pfennig, N. (1979) Anaerobic bacterium that degrades fatty‐acids in syntrophic association with methanogens. Arch Microbiol 122: 129–135. [DOI] [PubMed] [Google Scholar]
- McInerney, M.J. , Rohlin, L. , Mouttaki, H. , Kim, U. , Krupp, R.S. , Rios‐Hernandez, L. , et al (2007) The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci 104: 7600–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, N. , Worm, P. , Schink, B. , Stams, A.J.M. , and Plugge, C.M. (2010) Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanisms. Environ Microbiol Rep 2: 489–499. [DOI] [PubMed] [Google Scholar]
- Pereira, I.A.C. (2008) Respiratory membrane complexes of Desulfovibrio In Microbial Sulfur Metabolism. Dahl C., and Friedrich C.G. (eds). Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 24–35. [Google Scholar]
- Pereira, I.A. , Ramos, A.R. , Grein, F. , Marques, M.C. , da Silva, S.M. , and Venceslau, S.S. (2011) A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front Microbiol 2: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, R.H. , Lourenco, A.I. , Morais, F. , Teixeira, M. , Xavier, A.V. , Saraiva, L.M. , and Pereira, I.A. (2003) A novel membrane‐bound respiratory complex from Desulfovibrio desulfuricans ATCC 27774. Biochim Biophys Acta 1605: 67–82. [DOI] [PubMed] [Google Scholar]
- Plugge, C.M. , Dijkema, C. , and Stams, A.J.M. (1993) Acetyl‐CoA cleavage pathway in a syntrophic propionate oxidizing bacterium growing on fumarate in the absence of methanogens. FEMS Microbiol Lett 110: 71–76. [Google Scholar]
- Plugge, C.M. , Jiang, B. , de Bok, F.A.M. , Tsai, C. , and Stams, A.J.M. (2009) Effect of tungsten and molybdenum on growth of a syntrophic coculture of Syntrophobacter fumaroxidans and Methanospirillum hungatei . Arch Microbiol 191: 55–61. [DOI] [PubMed] [Google Scholar]
- Plugge, C.M. , Henstra, A.M. , Worm, P. , Swarts, D.C. , Paulitsch‐Fuchs, A.H. , Scholten, J.C.M. , et al (2012) Complete genome sequence of Syntrophobacter fumaroxidans strain (MPOBT). Stand Genom Sci 7: 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus, R. , Venceslau, S.S. , Wohlbrand, L. , Voordouw, G. , Wall, J.D. , and Pereira, I.A. (2015) A post‐genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate‐reducing prokaryotes. Adv Microb Physiol 66: 55–321. [DOI] [PubMed] [Google Scholar]
- Ramos, A.R. , Grein, F. , Oliveira, G.P. , Venceslau, S.S. , Keller, K.L. , Wall, J.D. , and Pereira, I.A. (2015) The FlxABCD‐HdrABC proteins correspond to a novel NADH dehydrogenase/heterodisulfide reductase widespread in anaerobic bacteria and involved in ethanol metabolism in Desulfovibrio vulgaris Hildenborough. Environ Microbiol 17: 2288–2305. [DOI] [PubMed] [Google Scholar]
- Rupakula, A. , Kruse, T. , Boeren, S. , Holliger, C. , Smidt, H. , and Maillard, J. (2013) The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics. Philos Trans R Soc Lond B 368: 20120325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink, B. (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61: 262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink, B. (2015) Electron confurcation in anaerobic lactate oxidation. Environ Microbiol 17: 543. [DOI] [PubMed] [Google Scholar]
- Schink, B. , and Stams, A.J.M. (2013) Syntrophism among prokaryotes In The Prokaryotes. Rosenberg E., DeLong E., Lory S., Stackebrandt E., and Thompson F. (eds). Berlin, Heidelberg: Springer, pp. 471–493. [Google Scholar]
- Schut, G.J. , and Adams, M.W. (2009) The iron‐hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 191: 4451–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik, C.S. , Sieber, J.R. , Badalamenti, J.P. , Carden, K. , and Olson, A. (2017) Complete genome sequence of Desulfovibrio desulfuricans strain G11, a model sulfate‐reducing, hydrogenotrophic, and syntrophic partner organism. Genome Announce 5: e01207‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber, J.R. , McInerney, M.J. , and Gunsalus, R.P. (2012) Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66: 429–452. [DOI] [PubMed] [Google Scholar]
- Sieber, J.R. , Crable, B.R. , Sheik, C.S. , Hurst, G.B. , Rohlin, L. , Gunsalus, R.P. , and McInerney, M.J. (2015) Proteomic analysis reveals metabolic and regulatory systems involved in the syntrophic and axenic lifestyle of Syntrophomonas wolfei . Front Microbiol 6: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stams, A.J.M. , and Dong, X. (1995) Role of formate and hydrogen in the degradation of propionate and butyrate by defined suspended cocultures of acetogenic and methanogenic bacteria. Antonie Van Leeuwenhoek 68: 281–284. [DOI] [PubMed] [Google Scholar]
- Stams, A.J.M. , and Plugge, C.M. (2009) Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7: 568–577. [DOI] [PubMed] [Google Scholar]
- Stams, A.J.M. , Van Dijk, J.B. , Dijkema, C. , and Plugge, C.M. (1993) Growth of syntrophic propionate‐oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol 59: 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter, A.W. , Liesegang, H. , Rabus, R. , Decker, I. , Amann, J. , Andres, S. , et al (2009) Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ Microbiol 11: 1038–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer, R.K. , Kaster, A.K. , Seedorf, H. , Buckel, W. , and Hedderich, R. (2008) Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6: 579–591. [DOI] [PubMed] [Google Scholar]
- Van Kuijk, B.L.M. , and Stams, A.J.M. (1995) Sulfate reduction by a syntrophic propionate‐oxidizing bacterium. Antonie Van Leeuwenhoek 68: 293–296. [DOI] [PubMed] [Google Scholar]
- Van Kuijk, B.L.M. , van Loo, N.D. , Arendsen, A.F. , Hagen, W.R. , and Stams, A.J.M. (1996) Purification and characterization of fumarase from the syntrophic propionate‐oxidizing bacterium strain MPOB. Arch Microbiol 165: 126–131. [DOI] [PubMed] [Google Scholar]
- Van Kuijk, B.L.M. , Schlosser, E. , and Stams, A.J.M. (1998a) Investigation of the fumarate metabolism of the syntrophic propionate‐oxidizing bacterium strain MPOB. Arch Microbiol 169: 346–352. [DOI] [PubMed] [Google Scholar]
- Van Kuijk, B.L.M. (1998b) Isolation and properties of the oxygen‐sensitive fumarate reductase of the syntrophic propionate‐oxidizing bacterium strain MPOB. PhD thesis Wageningen University, The Netherlands.
- Venceslau, S.S. , Lino, R.R. , and Pereira, I.A. (2010) The Qrc membrane complex, related to the alternative complex III, is a menaquinone reductase involved in sulfate respiration. J Biol Chem 285: 22774–22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venceslau, S.S. , Stockdreher, Y. , Dahl, C. , and Pereira, I.A. (2014) The “bacterial heterodisulfide” DsrC is a key protein in dissimilatory sulfur metabolism. Biochim Biophys Acta 1837: 1148–1164. [DOI] [PubMed] [Google Scholar]
- Wallrabenstein, C. , Hauschild, E. , and Schink, B. (1994) Pure culture and cytological properties of ‘Syntrophobacter wolini ’. FEMS Microbiol Lett 123: 249–254. [Google Scholar]
- Worm, P. (2010) Formate Dehydrogenases and Hydrogenases in Syntrophic Propionate‐Oxidizing Communities: Gene Analysis and Transcriptional Profiling Wageningen, The Netherlands: Wageningen University, pp. 67–81.
- Worm, P. , Stams, A.J.M. , Cheng, X. , and Plugge, C.M. (2011a) Growth‐ and substrate‐dependent transcription of formate dehydrogenase and hydrogenase coding genes in Syntrophobacter fumaroxidans and Methanospirillum hungatei . Microbiology 157: 280–289. [DOI] [PubMed] [Google Scholar]
- Worm, P. , Fermoso, F.G. , Stams, A.J.M. , Lens, P.N.L. , and Plugge, C.M. (2011b) Transcription of fdh and hyd in Syntrophobacter spp. and Methanospirillum spp. as a diagnostic tool for monitoring anaerobic sludge deprived of molybdenum, tungsten and selenium. Environ Microbiol 13: 1228–1235. [DOI] [PubMed] [Google Scholar]
- Worm, P. , Koehorst, J.J. , Visser, M. , Sedano‐Nunez, V.T. , Schaap, P.J. , Plugge, C.M. , et al (2014) A genomic view on syntrophic versus non‐syntrophic lifestyle in anaerobic fatty acid degrading communities. Biochim Biophys Acta 1837: 2004–2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's web‐site:
Fig. S1. A. Venn diagram of the 813 proteins detected in Syntrophobacter fumaroxidans growth on propionate with five different (biological or chemical) electron acceptors. B. Principal Component Analysis performed for S. fumaroxidans protein profiles obtained from each triplicate grown under five different conditions. Symbols: Orange diamonds, sulfate reducing; Red crosses, growth with fumarate; Grey squares, in coculture with Desulfovibrio desulfuricans in a sulfate rich environment; Green triangles, in syntrophy with Methanospirillum hungatei; Blue circles, in syntrophy with Methanobacterium formicicum.
Fig. S2. Normalized expression matrix of energy conservation mechanisms predicted for Syntrophobacter fumaroxidans. Proteins are shown for five different growth conditions, in triplicates; from left to right: fumarate, sulfate and interspecies compounds transferred to: Desulfovibrio desulfuricans, Methanobacterium formicicum and Methanospirillum hungatei. The colour scale illustrates the relative detection level of each protein across the 5 samples; blue (log ratio −2.5) and yellow (log ratio 2.5) indicate lower and higher levels compared with the average level value (in black) respectively. Not detected proteins in a specific condition appear in grey. The asterisk indicates a statistical significant difference in at least one condition.
Fig. S3. A. Venn diagram of the 779 proteins detected in Desulfovibrio desulfuricans growing in sulfate rich medium in coculture with Syntrophobacter fumaroxidans or axenically on H2/CO2 or formate. B. PCA performed for D. desulfuricans protein profiles. Symbols: red diamonds, hydrogenotrophic conditions; black squares, growth with formate and filled grey squares correspond to the cocultured partnership of D. desulfuricans with S. fumaroxidans.
Fig. S4. Heat map of hierarchical clustered proteins produced by Desulfovibrio desulfuricans. The proteins are shown in a clustered matrix after column Z‐score normalization and automatic hierarchical columns clustering. Three growth conditions, in triplicates, are shown according to the electron donor used; from left to right: formate, hydrogen and compounds transferred from Syntrophobacter fumaroxidans. The colour scale represents the relative detection level of each protein across the samples; blue log ratio −3, yellow log ratio 3, red log ratio 4 and green log ratio 5 indicate lower and higher levels compared with the average level value 0 (in black) respectively. The colour intensity indicates the degree of protein up‐ or down regulation; the grey colour represents not detected.
Fig. S5. Normalized expression matrix of hydrogenases and formate dehydrogenases of Desulfovibrio desulfuricans. The rows in the heat map show proteins levels after row Z‐score standardization in three different growth conditions. The columns show from left to right, in triplicates, the electron donor used by D. desulfuricans: formate, hydrogen and interspecies compounds transferred from Syntrophobacter fumaroxidans. The colour scale indicates the degree of protein down‐ or up regulation ranging from blue (−2.2 log ratio), to yellow (2.2 log ratio). The colour intensities indicate lower and higher levels compared with the average level 0 value (in black); the grey colour represents not detected. Subunits, twin‐arginine translocation (TAT) pathway signal and selenocysteine insertion (Sec) sequences are indicated after the locus tag.
Fig. S6. Heat map of hierarchical clustered proteins produced by Syntrophobacter fumaroxidans for propionate degradation. The proteins are shown in a clustered matrix after automatic hierarchical cluster of rows from row Z‐score normalization values. Proteins appear from left to right, in triplicates, according to the growth conditions defined by the electron acceptor used by S. fumaroxidans to oxidize propionate: fumarate, sulfate and interspecies compounds transferred to: Desulfovibrio desulfuricans, Methanobacterium formicicum and Methanospirillum hungatei. The colour scale illustrates the relative detection level of each protein across the samples; blue (log ratio −2.5), yellow (log ratio 2.5) and red (log ratio 3) indicate lower and higher levels compared with the average level value 0 (in black). The colour intensity indicates the degree of protein up‐ or down regulation; the grey colour represents not detected.
Table S1. iBAQ values of proteins detected in axenic and cocultured conditions in Syntrophobacter fumaroxidans and Desulfovibrio desulfuricans.
Supporting Information Dataset 1
Supporting Information Dataset 2


