Experimental structures of the archaeal RNase P protein PhoRpp38 in complex with K-turns of PhopRNA reveal how the protein recognizes the 3nt bulge and tandem G·A and A·G pairs. Moreover, the extended stem-loop containing P10-P12.2 in complex with PhoRpp21, PhoRpp29 and PhoRpp38 was crystallized.
Keywords: archaea, K-turn, L7Ae family, Pyrococcus horikoshii, ribonuclease P
Abstract
A characteristic feature of archaeal ribonuclease P (RNase P) RNAs is that they have extended helices P12.1 and P12.2 containing kink-turn (K-turn) motifs to which the archaeal RNase P protein Rpp38, a homologue of the human RNase P protein Rpp38, specifically binds. PhoRpp38 from the hyperthermophilic archaeon Pyrococcus horikoshii is involved in the elevation of the optimum temperature of the reconstituted RNase P by binding the K-turns in P12.1 and P12.2. Previously, the crystal structure of PhoRpp38 in complex with the K-turn in P12.2 was determined at 3.4 Å resolution. In this study, the crystal structure of PhoRpp38 in complex with the K-turn in P12.2 was improved to 2.1 Å resolution and the structure of PhoRpp38 in complex with the K-turn in P12.1 was also determined at a resolution of 3.1 Å. Both structures revealed that Lys35, Asn38 and Glu39 in PhoRpp38 interact with characteristic G·A and A·G pairs in the K-turn, while Thr37, Asp59, Lys84, Glu94, Ala96 and Ala98 in PhoRpp38 interact with the three-nucleotide bulge in the K-turn. Moreover, an extended stem-loop containing P10–P12.2 in complex with PhoRpp38, as well as PhoRpp21 and PhoRpp29, which are the archaeal homologues of the human proteins Rpp21 and Rpp29, respectively, was affinity-purified and crystallized. The crystals thus grown diffracted to a resolution of 6.35 Å. Structure determination of the crystals will demonstrate the previously proposed secondary structure of stem-loops including helices P12.1 and P12.2 and will also provide insight into the structural organization of the specificity domain in P. horikoshii RNase P RNA.
1. Introduction
A large number of RNA-binding proteins are present in both prokaryotic and eukaryotic cells and function in a variety of cellular processes in association with RNAs, such as regulation of gene expression, RNA processing and translation (Hall, 2017 ▸). Ribosomal protein L7Ae is a member of an RNA-binding protein family (L7Ae family) that specifically binds a kink-turn (K-turn), an RNA structural element that causes a tight kink in duplex RNA (Ban et al., 2000 ▸; Klein et al., 2001 ▸). The K-turn is a widespread structural motif and principally comprises a double-stranded structure with a three-nucleotide (3 nt) bulge followed on its 3′ side by tandem G·A and A·G pairs (Klein et al., 2001 ▸; Lilley, 2014 ▸). The helix containing the G·A pairs is termed a 3′ noncanonical (NC) helix, while that 5′ to the bulge is called a 5′ canonical (C) helix (Liu & Lilley, 2007 ▸). The K-turn introduces a sharp kink between the axes of the C and NC helices with an included angle of ∼60°. The L7Ae family proteins include the archaeal ribosomal protein L7Ae (Ban et al., 2000 ▸; Turner et al., 2005 ▸), human 15.5 kDa protein (Vidovic et al., 2000 ▸) and bacterial homologues such as YbxF (Baird et al., 2012 ▸), and thus they are involved in a variety of cellular functions in association with the K-turn motif in RNA species in all domains of life. Interestingly, the archaeal ribosomal protein L7Ae is a multifunctional protein, being present not only in ribosomes but also in box C/D as well as box H/ACA snoRNPs (Rozhdestvensky et al., 2003 ▸). Crystallographic and biochemical analysis revealed that the binding of ribosomal protein L7A to rRNA forms an appropriate conformation of rRNA (Ban et al., 2000 ▸), and that the binding of L7Ae family proteins to their cognate snoRNAs forms additional protein-binding sites on RNAs, thereby enabling the subsequent assembly of snoRNPs (Rashid et al., 2003 ▸; Moore et al., 2004 ▸). L7Ae has been shown to be a fifth protein component of archaeal RNase Ps (Fukuhara et al., 2006 ▸; Cho et al., 2010 ▸).
RNase P is an endoribonuclease that cleaves off a leader sequence to produce the mature 5′ ends of all tRNAs in an Mg2+ ion-dependent manner (Guerrier-Takada et al., 1983 ▸; Esakova & Krasilnikov, 2010 ▸). RNase P is a ribonucleoprotein (RNP) composed of a catalytic RNA and protein cofactors, although its chemical composition is distinct in the three domains of life. Previously, we found from reconstitution experiments that RNase P in the hyperthermophilic archaeon Pyrococcus horikoshii consists of RNA (PhopRNA) and five protein cofactors PhoPop5, PhoRpp21, PhoRpp29, PhoRpp30 and PhoRpp38, which are homologues of the human RNase P proteins hPop5, Rpp21, Rpp29, Rpp30 and Rpp38, respectively (Kouzuma et al., 2003 ▸; Fukuhara et al., 2006 ▸). Functional reconstitutions of the RNA component and archaeal homologues of human RNase P proteins have been reported for other archaeal RNase Ps (Jarrous & Gopalan, 2010 ▸).
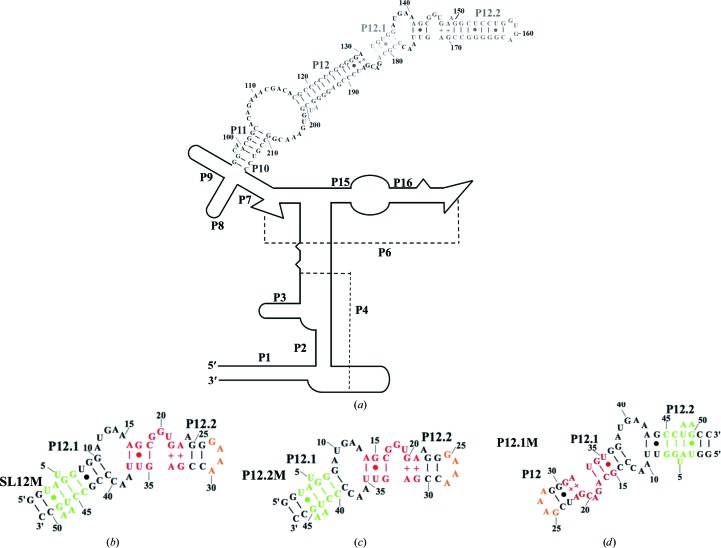
The fifth protein PhoRpp38 belonging to the ribosomal protein L7Ae family is predominantly involved in the elevation of the optimum temperature of reconstituted RNase P by binding to K-turn motifs in stem-loops containing helices P12.2 and P16 (Fukuhara et al., 2006 ▸; Oshima et al., 2012 ▸). Apart from P. horikoshii RNase P, the ribosomal protein L7Ae was identified as a fifth protein component in Methanococcus maripaludis RNase P, and it was reported that its addition increases the optimal reaction temperature and catalytic efficiency for pre-tRNA cleavage to the level of those observed for authentic RNase P (Cho et al., 2010 ▸). In order to gain insight into the structural basis for the recognition of K-turn motifs by PhoRpp38, we previously determined the crystal structure of PhoRpp38 in complex with the K-turn (SL12M) in P12.2 at a resolution of 3.1 Å (Oshima et al., 2016 ▸). This structural analysis, together with biochemical data, demonstrated the secondary structure of P12.2 and also suggested an additional K-turn motif at positions 129–133 and 180–187 in P12.1 (Fig. 1 ▸). In this study, we improved the crystal structure of PhoRpp38 in complex with the K-turn in P12.2 to a higher resolution of 2.1 Å and also determined the structure of PhoRpp38 in complex with the K-turn in P12.1 at 3.1 Å resolution.
Figure 1.
Secondary structures of PhopRNA and its fragments. (a) Secondary structure of PhopRNA. Single strands connected by broken lines indicate helices P4 and P6. (b, c, d) Secondary structures of SL12M (b), P12.2M (c) and P12.1M (d) used for crystallization. Watson–Crick base pairs are denoted by short lines, G–U pairs by dots and irregular AG and GA pairs by +. Residues are numbered in increments of five or ten. Helices are numbered according to the existing RNase P RNA nomenclature (Pace & Brown, 1995 ▸). The K-turn motifs, the GAAA tetraloop and its receptor are shown in red, orange and green, respectively.
2. Materials and methods
2.1. Materials
Oligonucleotides were purchased from Sigma–Aldrich (St Louis, USA) and Eurofins Genomics (Tokyo, Japan). Phusion High-Fidelity DNA polymerase was purchased from New England Biolabs (Ipswich, Massachusetts, USA). All other chemicals were of analytical grade for biochemical use.
2.2. Preparation of proteins and RNAs
His-tagged PhoRpp38 was prepared as described previously (Fukuhara et al., 2006 ▸; Oshima et al., 2016 ▸). Two sets of RNA constructs designated P12.2M (47 nt) and P12.1M (51 nt) were prepared and used for crystallization as follows (Fig. 1 ▸). The first construct, P12.2M (G135–G151/C168–C178), was designed based on the deduced secondary structure of P12.1 and P12.2 in PhopRNA (Gao et al., 2017 ▸), in which the 4 nt sequence (GUGA) at the tip of helix P12.2 and the 3 bp (U–A, G–C and U·G) in P12.1 were replaced by a GAAA tetraloop and its receptor sequence, respectively (Fig. 1 ▸ c; Pley et al., 1994 ▸; Zhang & Ferré-D’Amaré, 2014 ▸). Moreover, nucleotides 152–157 and 162–167 in P12.2 were deleted. Furthermore, GG and complementary CC sequences were added to enhance the transcription efficiency at the 5′-terminus and 3′-terminus, respectively (Fig. 1 ▸ c). The second set of constructs consisted of circularly permuted helices P12, P12.1 and P12.2 (Fig. 1 ▸ d). Thus, P12.1M (G127–G142/U173–C189) was designed based on the deduced secondary structure of P12, P12.1 and P12.2 in PhopRNA (Gao et al., 2017 ▸), in which nucleotides G127 and C189 in P12 were linked by the GAAA tetraloop and its receptor sequence was introduced instead of the K-turn in P12.2 (Fig. 1 ▸ d; Pley et al., 1994 ▸; Zhang & Ferré-D’Amaré, 2014 ▸). Furthermore, GG and complementary CC sequences were added to enhance the transcription efficiency at the 5′-terminus and 3′-terminus, respectively (Fig. 1 ▸ d). P12.2M and P12.1M were prepared by in vitro transcription with T7 RNA polymerase using the corresponding double-stranded DNA as a template and were purified as described previously (Oshima et al., 2016 ▸). The purified RNAs were annealed by heating to 90°C for 5 min and cooling to 4°C over 5 min, and were then used for crystallization.
2.3. Preparation of the PhopRNA fragment in complex with PhoRpp21, PhoRpp29 and PhoRpp38
The PhopRNA fragment (P10/12; G96–C216) containing helices P10, P11, P12, P12.1 and P12.2 was prepared by in vitro transcription with T7 RNA polymerase using the corresponding double-stranded DNA as a template and was purified as described above. The purified P10/12 was incubated with PhoRpp21 and PhoRpp29 in the presence of His-tagged PhoRpp38 in a buffer consisting of 50 mM Tris–HCl pH 8.0 containing 5 mM MgCl2 and 500 mM NaCl at 50°C for 10 min, and the complexes were applied onto an Ni–NTA column (Wako Pure Chemical Industries, Osaka, Japan). After incubation for 30 min at room temperature and extensive washing, the complexes bound to the column were eluted with a buffer containing 0.5 M imidazole and the effluent was subjected to SDS–PAGE using the WIDE RANGE Gel Preparation buffer and visualized by staining the gels with Coomassie Brilliant Blue solution.
2.4. Crystallization
PhoRpp38 was mixed with P12.2M at a molar ratio of 0.9:1 in 20 mM Tris–HCl pH 8.0 containing 5 mM MgCl2 and 200 mM KCl and incubated at 50°C for 10 min. The mixture was crystallized after concentration to 3 mg ml−1 using Amicon Ultra centrifugal filters (Merck, Darmstadt, Germany). Crystals suitable for X-ray analysis were obtained by sitting-drop vapour diffusion at 20°C from a crystallization buffer consisting of 50 mM sodium cacodylate pH 6.5 containing 15% PEG 400 and 80 mM magnesium acetate, in which the sample was mixed with the same amount of the crystallization buffer. For crystallization of the PhoRpp38–P12.1M complex, purified His-tagged PhoRpp38 and P12.1M were mixed at a molar ratio of 2:1 in 20 mM Tris–HCl pH 8.0 containing 5 mM MgCl2 and 200 mM KCl, incubated at 50°C for 10 min and purified using an Ni–NTA column. The purified complex was concentrated to 2 mg ml−1 using Amicon Ultra centrifugal filters and crystallized. Crystals suitable for X-ray analysis were obtained by sitting-drop vapour diffusion at 20°C from a crystallization buffer consisting of 50 mM sodium cacodylate pH 6.0 containing 1.7 M ammonium sulfate and 15 mM magnesium acetate, in which the sample was mixed with the same amount of the crystallization buffer. P10/12 in complex with PhoRpp21, PhoRpp29 and PhoRpp38 was purified on an Ni–NTA column and concentrated to 2.5 mg ml−1 in 50 mM Tris–HCl pH 8.0 containing 5 mM MgCl2 and 800 mM NaCl using Amicon Ultra centrifugal filters. Crystals of the complex were grown using 0.2 M potassium chloride, 5 mM magnesium chloride hexahydrate, 50 mM sodium cacodylate trihydrate pH 6.5, 0.9–1.7 M 1,6-hexanediol. X-ray diffraction experiments were performed on beamline BL26B1 at SPring-8, Harima, Japan under proposal Nos. 2016A2549 and 2016B2549. Diffraction data were indexed, integrated, scaled and merged with XDS (Kabsch, 2010 ▸). The data statistics are given in Table 1 ▸.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| PhoRpp38–P12.2M | PhoRpp38–P12.1M | |
|---|---|---|
| Data collection | ||
| Space group | P21 | C2 |
| Unit-cell parameters (Å, °) | a = 52.1, b = 71.7, c = 91.3, β = 105.6 | a = 113.4, b = 176.6, c = 59.9, β = 105.1 |
| Beamline | BL26B1, SPring-8 | BL26B1, SPring-8 |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Resolution range (Å) | 39.36–2.10 | 48.38–3.10 |
| No. of reflections | ||
| Observed | 177058 (28014) | 78285 (12613) |
| Unique | 37804 (6007) | 20596 (3303) |
| Multiplicity | 4.8 (4.7) | 3.8 (3.8) |
| R meas (%)† | 9.6 (86.8) | 22.9 (86.7) |
| 〈I/σ(I)〉 | 13.70 (2.15) | 7.05 (1.96) |
| Completeness (%) | 99.5 (98.4) | 99.7 (99.0) |
| Refinement statistics | ||
| Resolution range (Å) | 39.36–2.10 | 48.38–3.10 |
| No. of reflections | ||
| Working set | 37786 | 20436 |
| Test set | 1876 | 1021 |
| Completeness (%) | 99.5 | 99.6 |
| R work/R free (%)‡ § | 19.69/22.21 | 22.49/24.72 |
| Root-mean-square deviations | ||
| Bond lengths (Å) | 0.002 | 0.001 |
| Bond angles (°) | 0.581 | 0.465 |
| No. of atoms | ||
| Protein | 1867 | 1928 |
| RNA | 2054 | 2264 |
| Ion | 24 [Mg2+] | 9 [SO4 2−] |
| Water | 306 | — |
| Ramachandran analysis | ||
| Favoured (%) | 98.75 | 97.97 |
| Allowed (%) | 1.25 | 2.03 |
| Outliers (%) | 0.0 | 0.0 |
R
meas = 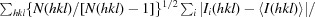
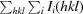 , where I
i(hkl) is the ith observed intensity of reflection hkl, 〈I(hkl)〉 is the average of symmetry-related observations of reflection hkl and N(hkl) is the multiplicity.
, where I
i(hkl) is the ith observed intensity of reflection hkl, 〈I(hkl)〉 is the average of symmetry-related observations of reflection hkl and N(hkl) is the multiplicity.
R
work = 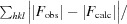
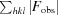 , where F
obs and F
calc are observed and calculated structure-factor amplitudes, respectively.
, where F
obs and F
calc are observed and calculated structure-factor amplitudes, respectively.
The R free value was calculated as for R work using only an unrefined randomly chosen subset of reflection data (5%).
2.5. Structure determination
The structure of PhoRpp38–P12.2M was solved by the molecular replacement method with Phaser (McCoy et al., 2007 ▸) using the structure of PhoRpp38 in complex with the PhopRNA fragment designated SL12M (PDB entry 5dcv; Oshima et al., 2016 ▸) as a search probe. The PhoRpp38 K-turn motif and GAAA tetraloop/receptor regions of the model were extracted before molecular replacement. To monitor the refinement, a random 5.0% subset of all unique reflections was set aside for R free evaluation, taking lattice symmetry into account, with PHENIX (Adams et al., 2010 ▸). Rigid-body refinement with phenix.refine (Afonine et al., 2012 ▸) was carried out. After several cycles of manual model fitting and building with Coot (Emsley & Cowtan, 2004 ▸), individual atomic coordinate refinement, individual ADP refinement and group ADP refinement were performed using phenix.refine (Afonine et al., 2012 ▸). The structure of PhoRpp38–P12.1M was solved by the molecular replacement method with Phaser (McCoy et al., 2007 ▸) using the structure of PhoRpp38 in complex with the 13 nt K-turn motif in the PhoRpp38–P12.2M complex as a search probe. Structural refinement was carried out in the same manner as described above. The coordinates of the PhoRpp38–P12.2M and PhoRpp38–P12.1M complexes were deposited in the Protein Data Bank as entries 5xtm and 5y7m, respectively.
3. Results
3.1. Structural determination
We previously determined the structure of PhoRpp38 in complex with the K-turn (SL12M) in P12.2 (Figs. 1 ▸ a and 1 ▸ b; Oshima et al., 2016 ▸). Although the PhoRpp38–SL12M structure determined at a resolution of 3.4 Å demonstrated that PhoRpp38, like the L7Ae family proteins, interacts with characteristic G·A and A·G pairs and the 3 nt bulge (G-G-U) in SL12M, the detailed interaction between amino-acid side chains and nucleotides remained unclear owing to the low resolution of the data. In addition, electron densities for some nucleotides (U12–A15) in SL12M were not observed (Oshima et al., 2016 ▸). To understand the molecular recognition of K-turn motifs in PhopRNA by PhoRpp38 at the atomic level, we improved the crystal structure of PhoRpp38 in complex with the K-turn in P12.2 and also determined the structure of PhoRpp38 in complex with the K-turn in P12.1.
In order to improve the crystals of PhoRpp38–SL12M, a few base pairs were inserted or deleted in P12.1 and P12.2 in SL12M and the resulting RNAs were crystallized in complex with PhoRpp38. As a result, crystallization of the RNA fragment designated P12.2M (Fig. 1 ▸ c), in which two base pairs (U8–G43 and G9–C42) in SL12M were deleted, produced crystals of a complex that diffracted to a resolution of 2.1 Å. The crystals contained two copies of the complex, molecule A (PhoRpp38)–molecule B (P12.2M) and molecule C (PhoRpp38)–molecule D (P12.2M), in the asymmetric unit. The crystallographic data are summarized in Table 1 ▸. Despite extensive screening of conditions, all crystallization trials were unsuccessful for PhoRpp38 in complex with the K-turn in P12.1. Hence, the RNA fragment designated P12.1M was constructed (Fig. 1 ▸ d), in which the nucleotides in P12, P12.1 and P12.2 were circularly permuted as described in §2. For crystallization, a mixture of His-tagged PhoRpp38 and P12.1M was purified on an Ni–NTA column and the resulting complex was crystallized. The crystals diffracted to a resolution of 3.1 Å and contained two copies of the complex, molecule A (PhoRpp38)–molecule B (P12.1M) and molecule C (PhoRpp38)–molecule D (P12.1M), in the asymmetric unit. The crystallographic data are summarized in Table 1 ▸.
The PhoRpp38–P12.2M structure was primarily solved at a resolution of 2.1 Å by molecular replacement using the structure of PhoRpp38–SL12M (PDB entry 5dcv; Oshima et al., 2016 ▸) as a search model. The electron-density map calculated at 2.1 Å resolution was of extremely good quality and the refined model of the P12.2M coordinates was superimposed (Supplementary Fig. S1). The structure was refined to an R free of 22.21% (R work of 19.69%) with geometries close to ideal, with r.m.s.d.s of 0.002 Å and 0.581° from the ideal values for bond lengths and angles, respectively. The refined structures of molecules A, B, C and D contained 121 amino acids (Lys4–Lys124), 47 nucleotides (G1–C47), 123 amino acids (Met2–Lys124) and 47 nucleotides (G1–C47), respectively. When the structure was checked using phenix.refine (Afonine et al., 2012 ▸), 98.75% of the nonglycine and nonproline residues fell in the most favoured regions and 1.25% in the additionally allowed region of the Ramachandran plot (Table 1 ▸).
The PhoRpp38–P12.1M structure was similarly solved at a resolution of 3.1 Å by molecular replacement using the structure of the PhoRpp38–K-turn in the PhoRpp38–P12.2M complex (PDB entry 5xtm) as a search model. The structure was refined to an R free of 24.72% (R work of 22.49%) with geometries close to ideal, with r.m.s.d.s of 0.001 Å and 0.465° from the ideal values for bond lengths and angles, respectively. The refined structures of molecules A, B, C and D contained 121 amino acids (Lys4–Lys124), 52 nucleotides (G1–C52), 121 amino acids (Lys4–Lys124) and 52 nucleotides (G1–C52), respectively. The Ramachandran plot of the model shows that 97.97% of the residues lie within the most favoured regions and the remaining 2.03% of the residues lie within the additionally allowed region. The refinement statistics are summarized in Table 1 ▸.
3.2. Structures of PhoRpp38 in complex with P12.2M or P12.1M
The crystal structures of PhoRpp38 in complex with P12.2M or P12.1M are presented in Fig. 2 ▸. The structures observed in the asymmetric units are depicted in Supplementary Fig. S2. As expected, the GAAA tetraloop in P12.2M (molecule B) interacts with the tetraloop receptor in P12.2M (molecule D) and vice versa. Similarly, the GAAA tetraloop in P12.1M (molecule B) interacts with the tetraloop receptor in P12.1M (molecule B) in the neighbouring asymmetric unit (data not shown). When the two independent PhoRpp38–P12.2M complexes (molecules A–B and molecules C–D) in the asymmetric units were superimposed, they had a very fixed structure, as shown by the small r.m.s.d. values (0.20 Å for PhoRpp38 and 1.98 Å for P12.2M; Supplementary Fig. S3). For the PhoRpp38–P12.1M complex, superposition of the PhoRpp38–P12.1M complexes gave r.m.s.d.s of 0.25 Å for PhoRpp38 and 0.72 Å for P12.1M. The interaction of PhoRpp38 with the K-turns in P12.2M buried 582.0 Å2 of the PhoRpp38 solvent-accessible area for molecules A–B and 602.6 Å2 for molecules C–D. Similarly, the binding of P12.1M to PhoRpp38 buried 613.8 Å2 for molecules A–B and 599.7 Å2 for molecules C–D, which are consistent with the values for the interaction of L7Ae family proteins with a K-turn (Huang & Lilley, 2013 ▸).
Figure 2.
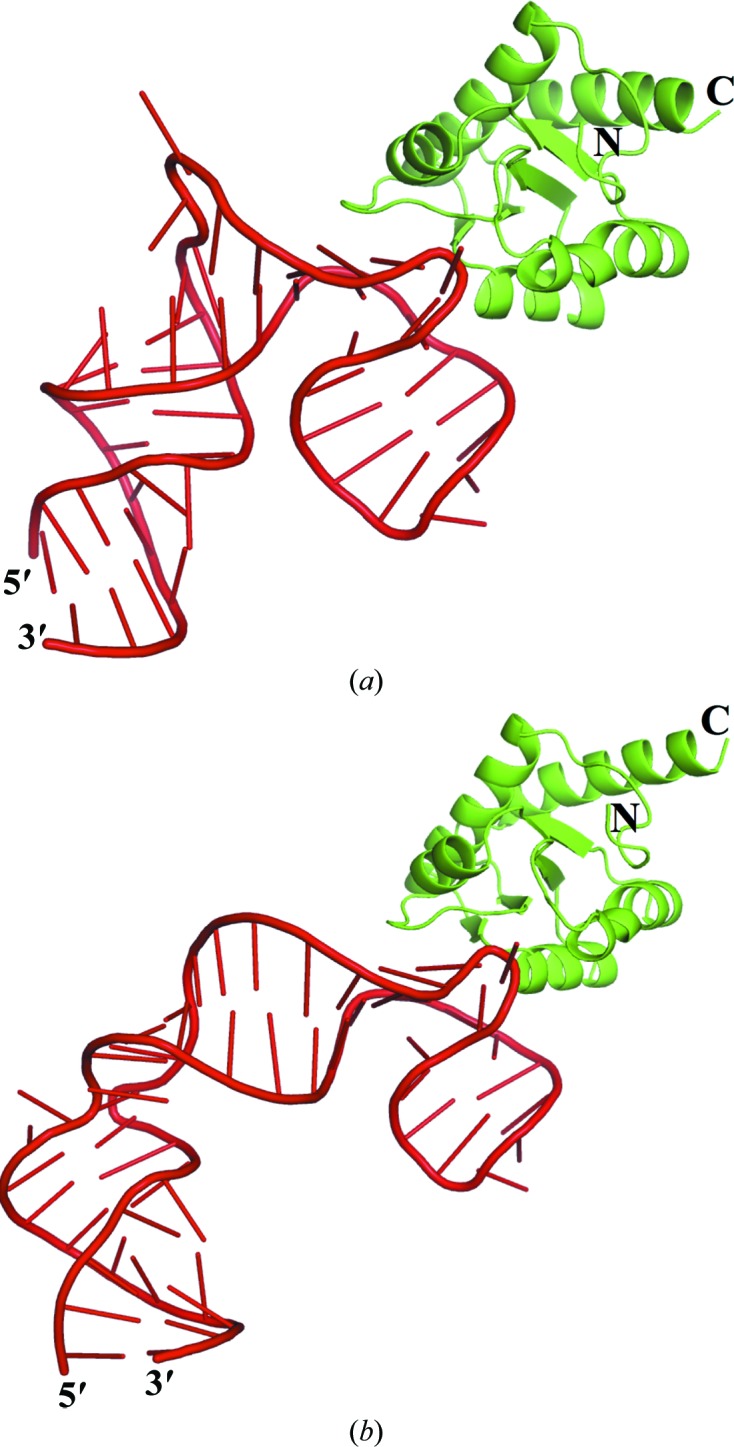
Structures of PhoRpp38 bound to P12.2M and P12.1M. The crystal structures are shown of PhoRpp38 in complex with P12.2M (a) and P12.1M (b). The protein is represented by a cartoon diagram coloured green, while P12.2M and P12.1M are drawn in cartoon form coloured red. The figures were drawn with PyMOL (http://pymol.sourceforge.net).
3.3. Protein–RNA interactions
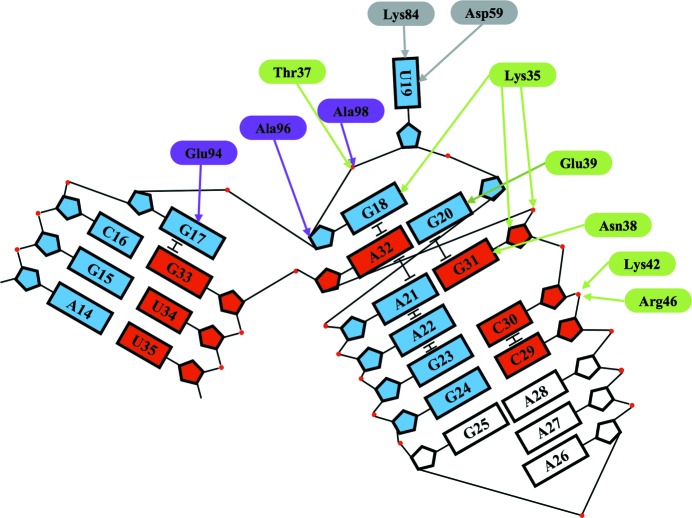
Since the crystal structure of the PhoRpp38–P12.2M complex was solved at a higher resolution than that of the PhoRpp38–P12.1M complex, the amino-acid side chains involved in the interaction with the K-turn were examined in the PhoRpp38–P12.2M structure. Although PhoRpp38 is an acidic protein (pI 5.2), the crystal structure revealed that the positively charged residues are concentrated mainly in the regions that are directly involved in interactions with the P12.2M RNA (Supplementary Fig. S4). PhoRpp38 interacts more extensively with the NC helix than with the C helix. The side chains of Asn38 and Glu39 interact with G31 and G20 in the NC helix, respectively (Figs. 3 ▸ and 4 ▸). In addition, the Lys35 side chain interacts with G18, which stabilizes the NC helix by a stacking interaction with A32, and also hydrogen-bonds to the phosphate backbone between G31 and A32, as well as to the ribose of G31. On the other hand, the Glu94 side chain hydrogen-bonds to G17, which stabilizes the C helix by stacking. In the 3 nt bulge structure (G17-G18-U19) in P12.2M the Glu94 side chain interacts with G17, which stabilizes the C helix by a stacking interaction, and the Ala96 and Ala98 side chains interact with the ribose of G18, which is stabilized by the Lys35 side chain, as described above (Figs. 3 ▸ and 4 ▸). Furthermore, U19 in the bulge structure is stabilized by interaction with the side chains of Asp59 and Lys84, as shown in Figs. 3 ▸ and 4 ▸.
Figure 3.
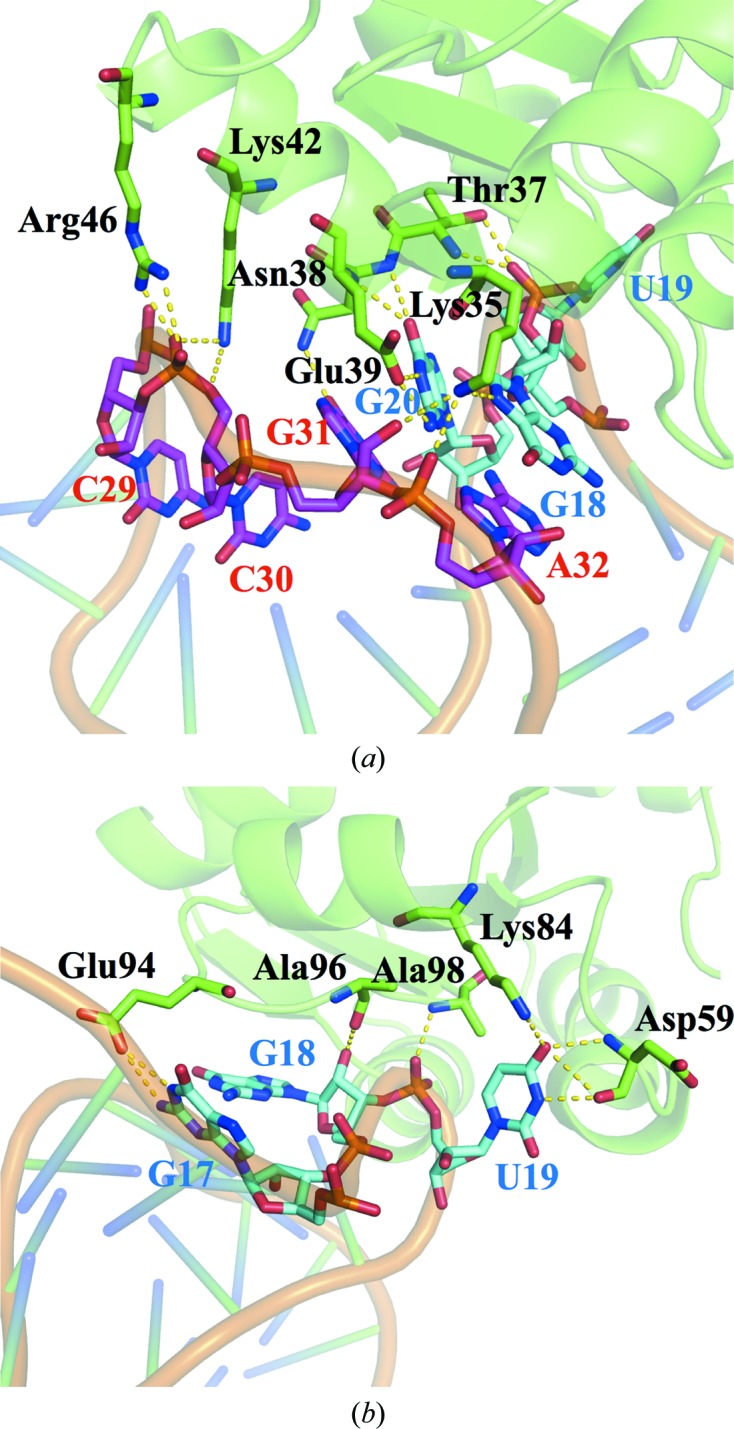
Interfaces for protein–RNA interactions. The hydrogen-bonding network for PhoRpp38 bound to the P12.2M K-turn. (a) Lys35 in β1 and Thr37, Asn38, Glu39 and Lys42 in α2 form a hydrogen-bond network with nucleotide residues in the NC helix. (b) Glu94, Ala96 and Ala98 in the loop between α4 and β4 bind G17 and G18 in the 3 nt bulge, and Asp59 and Lys84 hydrogen-bond U19 in the 3 nt bulge. PhoRpp38 and P12.2M are presented in cartoon representation, while the amino-acid residues involved in the interaction are shown in stick representation.
Figure 4.
Schematic representation of the interaction of PhoRpp38 with the K-turn in P12.2M. Nucleobases composing the K-turn are shown as boxes; nucleotides from A14 to G24 and from C29 to U35 are coloured blue and red, respectively. The amino acids located in β1–α2 and α4–β4 are coloured green and violet, respectively.
3.4. The structures of P12.2M and P12.1M in the complexes
P12.2M and P12.1M are folded into a standard K-turn structure, juxtaposing the two helices (C stem and NC stem; Liu & Lilley, 2007 ▸). Furthermore, a coaxial base stack consisting of G18, A32, A21, A22 and G23 constitutes a central hydrophobic core for organizing the K-turn structure (Figs. 4 ▸ and 5 ▸). The K-turn structures (C16–A21 and G31–G33) of P12.2M (molecule B) and P12.2M (molecule D) superimposed well, with an r.m.s.d. of 0.30 Å, and the bending angles for the K-turns in P12.2M (molecule B) and P12.2M (molecule D) are 49.1 and 52.6°, respectively (Supplementary Fig. S3), which are in accordance with the average calculated for other K-turn structures (Moore et al., 2004 ▸; Lilley, 2014 ▸). Similarly, the corresponding residues in P12.1M (molecule B) and P12.1M (molecule D) superimposed well, with an r.m.s.d. of 0.20 Å, and the bending angles for the K-turns in P12.1M (molecule B) and P12.1M (molecule D) are 53.9 and 51.9°, respectively (Supplementary Fig. S3). Furthermore, the superposition of the K-turn in the P12.2M structure (molecule B) onto those of P12.1M (molecule B) and SL12M (molecule B) gave r.m.s.d. values of 0.42 and 0.78 Å, respectively (Supplementary Fig. S5)
Figure 5.
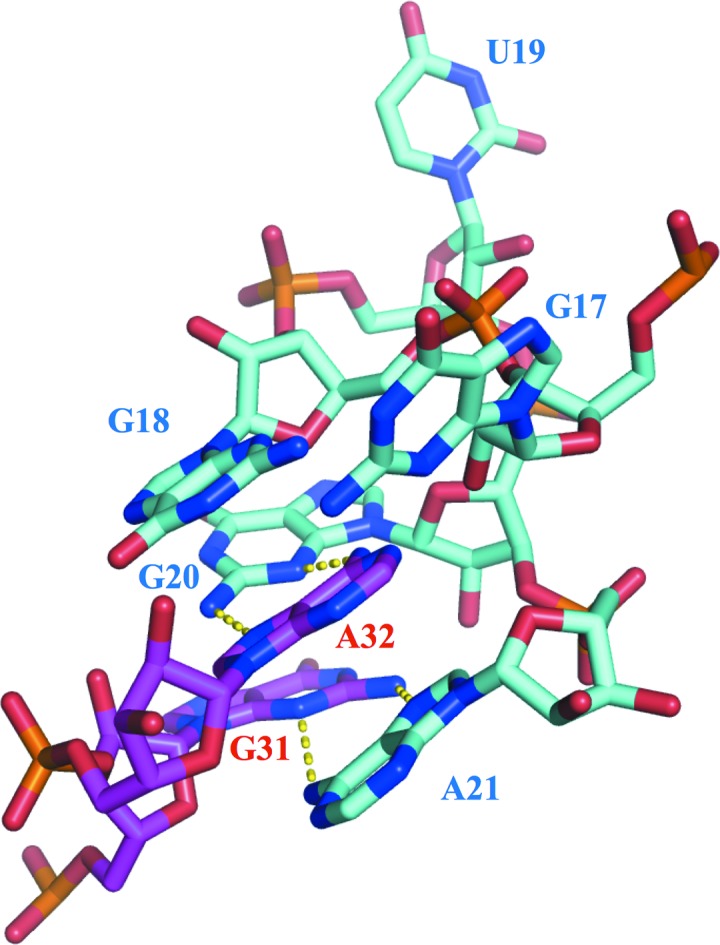
Structure of the K-turn. Nucleobases composing the K-turn in P12.2M are shown in stick representation. Hydrogen bonds between G20 and A32 and between A21 and G31 are shown by dashed lines.
When the crystal structure of the PhoRpp38–SL12M complex was determined (Oshima et al., 2016 ▸), no structural information about the single-stranded loops (A11–A15 and A38–C40) connecting P12.1 and P12.2 was available (Fig. 1 ▸ b). The presently identified structures of full-length P12.2M show a distinct conformation of the loop structure in P12.2M (molecule B) and P12.2M (molecule D) (Supplementary Fig. S3a). The nucleotide residues G8-A9-U10, A12-A14, U35-A36, and A37-C38-C39 in P12.2M (molecule B) are stabilized by a stacking interaction, and G11, A12 and A36 interact with each other forming a base triple (Supplementary Fig. S6a and S6b). In molecule D the loop structure is maintained by stacking interactions of the nucleobases of G8-A9-U10, G11-A12, and the bases of A37-C38-C39 similarly stack on each other (Supplementary Fig. S6c). The superposition of nucleotides G7–C16 and G33–C40 in P12.2M (molecule B) onto the corresponding structures in P12.2M (molecule D) and P12.1M (molecule B) gave larger r.m.s.d. values of 1.76 and 5.98 Å, respectively (Fig. 6 ▸). This finding suggests a flexible structure of the single loops connecting P12.1 and P12.2 in PhopRNA.
Figure 6.
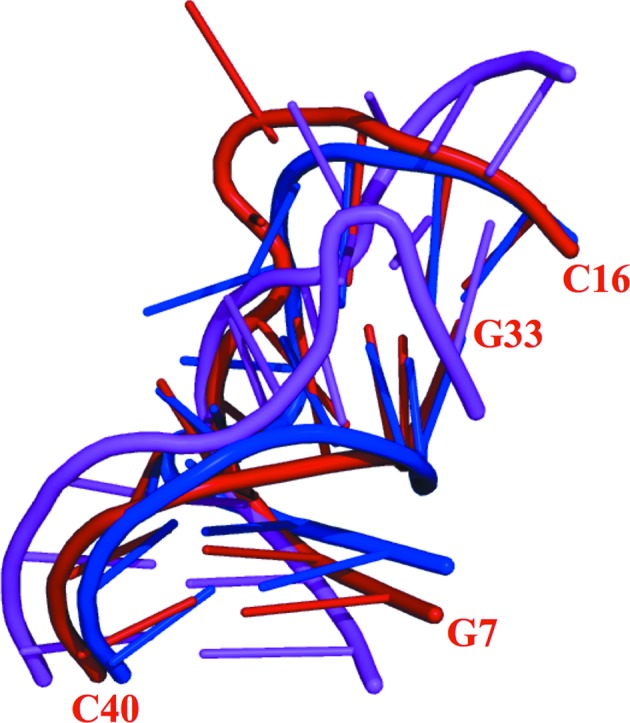
Structure of the single-stranded loops connecting P12.1 and P12.2. Superposition of the backbone atoms of nucleotides G7–C16 and G33–C40 in P12.2M (molecule B, red), P12.2M (molecule D, blue) and P12.1M (molecule B, magenta).
3.5. Crystallization of the extended helices in complex with PhoRpp21, PhoRpp29 and PhoRpp38
Recently, Lai and coworkers reported from biochemical studies that archaeal RNase P RNAs have a novel double K-turn module in helix P12 where the first (KT2) and second (KT3) K-turns are interconnected by the NC helix of KT2, which also serves as the C helix of KT3 (Lai et al., 2017 ▸). The present structural analysis predicts a distinct secondary structure of P12.1 and P12.2 in PhopRNA. However, as described in §2, GUGA at the tip of the helix P12.2 and three base pairs were replaced by a GAAA tetraloop and its receptor sequence, respectively, and the GAAA tetraloop in close proximity to the K-turn in P12.2 interacted with the tetraloop receptor in P12.1 as expected. Hence, we cannot exclude the possibility that the modifications of P12.1 and P12.2 may result in an artificial folding into the K-turns in P12.1M and P12.2M. Our previous studies showed that in a stem-loop (P10/12) containing P10–P12.2 in complex with PhoRpp21 and PhoRpp29 was pulled down by His-tagged PhoRpp38 (Oshima et al., unpublished results). Therefore, in order to confirm the K-turns in P12.1 and P12.2, and also to localize the binding region of the PhoRpp21–PhoRpp29 complex on the PhopRNA S-domain, we prepared a P10/12 RNA fragment in complex with PhoRpp21, PhoRpp29 and PhoRpp38 and crystallized it. The crystals were grown in 0.2 M potassium chloride, 5 mM magnesium chloride hexahydrate, 50 mM sodium cacodylate trihydrate pH 6.5, 0.9–1.7 M 1,6-hexanediol (Supplementary Fig. S7). SDS–PAGE analysis of the crystals clearly detected PhoRpp21, PhoRpp29, PhoRpp38 and P10/P12 (data not shown). The conditions produced crystals that diffracted to a resolution of 6.35 Å (Supplementary Fig. S7). Crystallographic analysis is now under way in our laboratory.
4. Conclusion
The crystal structures of PhoRpp38 in complex with the K-turns predicted in helices P12.1 and P12.2 of PhopRNA were determined at resolutions of 3.1 and 2.1 Å, respectively. The present structures reveal how the archaeal RNase P protein PhoRpp38 recognizes the 3 nt bulge and tandem G·A and A·G pairs featured in K-turn motifs in PhopRNA. Moreover, the extended stem-loop containing P10–P12.2 in complex with PhoRpp21, PhoRpp29 and PhoRpp38 was crystallized. Crystallographic analysis of the improved crystals will demonstrate the secondary structure of the K-turns and will also provide information about the tertiary structure of the S-domain in archaeal RNase P RNAs.
5. Related literature
The following reference is cited in the Supporting Information for this article: Baker et al. (2001 ▸).
Supplementary Material
PDB reference: PhoRpp38–P12.2M, 5xtm
PDB reference: PhoRpp38–P12.1M, 5y7m
Supplementary Figures.. DOI: 10.1107/S2053230X17018039/tb5124sup1.pdf
Acknowledgments
We are grateful to Dr K. Kato (Hokkaido University) for his active interest and helpful discussions. The synchrotron-radiation experiments were performed on beamline BL26B1 at SPring-8, Harima, Japan with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) under proposal Nos. 2016A2549 and 2016B2549.
Funding Statement
This work was funded by Japan Society for the Promotion of Science grant JP15H04487 to Makoto Kimura.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Baird, N. J., Zhang, J., Hamma, T. & Ferré-D’Amaré, A. R. (2012). RNA, 18, 759–770. [DOI] [PMC free article] [PubMed]
- Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001). Proc. Natl Acad. Sci. USA, 98, 10037–10041. [DOI] [PMC free article] [PubMed]
- Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000). Science, 289, 905–920. [DOI] [PubMed]
- Cho, I.-M., Lai, L. B., Susanti, D., Mukhopadhyay, B. & Gopalan, V. (2010). Proc. Natl Acad. Sci. USA, 107, 14573–14578. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Esakova, O. & Krasilnikov, A. S. (2010). RNA, 16, 1725–1747. [DOI] [PMC free article] [PubMed]
- Fukuhara, H., Kifusa, M., Watanabe, M., Terada, A., Honda, T., Numata, T., Kakuta, Y. & Kimura, M. (2006). Biochem. Biophys. Res. Commun. 343, 956–964. [DOI] [PubMed]
- Gao, X., Oshima, K., Ueda, T., Nakashima, T. & Kimura, M. (2017). Biochem. Biophys. Res. Commun. 493, 1063–1068. [DOI] [PubMed]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. (1983). Cell, 35, 849–857. [DOI] [PubMed]
- Hall, K. B. (2017). F1000Res. 6, 345. [DOI] [PMC free article] [PubMed]
- Huang, L. & Lilley, D. M. J. (2013). RNA, 19, 1703–1710. [DOI] [PMC free article] [PubMed]
- Jarrous, N. & Gopalan, V. (2010). Nucleic Acids Res. 38, 7885–7894. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Klein, D. J., Schmeing, T. M., Moore, P. B. & Steitz, T. A. (2001). EMBO J. 20, 4214–4221. [DOI] [PMC free article] [PubMed]
- Kouzuma, Y., Mizoguchi, M., Takagi, H., Fukuhara, H., Tsukamoto, M., Numata, T. & Kimura, M. (2003). Biochem. Biophys. Res. Commun. 306, 666–673. [DOI] [PubMed]
- Lai, L. B., Tanimoto, A., Lai, S. M., Chen, W.-Y., Marathe, I. A., Westhof, E., Wysocki, V. H. & Gopalan, V. (2017). Nucleic Acids Res. 45, 7432–7440. [DOI] [PMC free article] [PubMed]
- Lilley, D. M. J. (2014). Biochim. Biophys. Acta, 1839, 995–1004. [DOI] [PMC free article] [PubMed]
- Liu, J. & Lilley, D. M. J. (2007). RNA, 13, 200–210. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Moore, T., Zhang, Y., Fenley, M. O. & Li, H. (2004). Structure, 12, 807–818. [DOI] [PubMed]
- Oshima, K., Kakiuchi, Y., Tanaka, Y., Ueda, T., Nakashima, T., Kimura, M. & Yao, M. (2016). Biochem. Biophys. Res. Commun. 474, 541–546. [DOI] [PubMed]
- Oshima, K., Nakashima, T., Kakuta, Y., Tsumoto, K. & Kimura, M. (2012). Biosci. Biotechnol. Biochem. 76, 1252–1255. [DOI] [PubMed]
- Pace, N. R. & Brown, J. W. (1995). J. Bacteriol. 177, 1919–1928. [DOI] [PMC free article] [PubMed]
- Pley, H. W., Flaherty, K. M. & McKay, D. B. (1994). Nature (London), 372, 111–113. [DOI] [PubMed]
- Rashid, R., Aittaleb, M., Chen, Q., Spiegel, K., Demeler, B. & Li, H. (2003). J. Mol. Biol. 333, 295–306. [DOI] [PubMed]
- Rozhdestvensky, T. S., Tang, T. H., Tchirkova, I. V., Brosius, J., Bachellerie, J.-P. & Hüttenhofer, A. (2003). Nucleic Acids Res. 31, 869–877. [DOI] [PMC free article] [PubMed]
- Turner, B., Melcher, S. E., Wilson, T. J., Norman, D. G. & Lilley, D. M. J. (2005). RNA, 11, 1192–1200. [DOI] [PMC free article] [PubMed]
- Vidovic, I., Nottrott, S., Hartmuth, K., Lührmann, R. & Ficner, R. (2000). Mol. Cell, 6, 1331–1342. [DOI] [PubMed]
- Zhang, J. & Ferré-D’Amaré, A. R. (2014). Curr. Opin. Struct. Biol. 26, 9–15. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PhoRpp38–P12.2M, 5xtm
PDB reference: PhoRpp38–P12.1M, 5y7m
Supplementary Figures.. DOI: 10.1107/S2053230X17018039/tb5124sup1.pdf