The structure of MAPK-interacting kinase 1 (Mnk1) in complex with a novel Mnk1-selective inhibitor was determined. It was found that the inhibitor stabilizes the auto-inhibited inactive state of Mnk1.
Keywords: kinase, kinase inhibitors, inactive conformations, transferases, Mnk1, MAPK-interacting kinase 1, DS12881479
Abstract
Mitogen-activated protein kinase (MAPK)-interacting kinases 1 (Mnk1) and 2 (Mnk2) modulate translation initiation through the phosphorylation of eukaryotic translation initiation factor 4E, which promotes tumorigenesis. However, Mnk1 and Mnk2 are dispensable in normal cells, suggesting that the inhibition of Mnk1 and Mnk2 could be effective in cancer therapy. To provide a structural basis for Mnk1 inhibition, a novel Mnk1 inhibitor was discovered and the crystal structure of Mnk1 in complex with this inhibitor was determined. The crystal structure revealed that the inhibitor binds to the autoinhibited state of Mnk1, stabilizing the Mnk-specific DFD motif in the DFD-out conformation, thus preventing Mnk1 from switching to the active conformation and thereby inhibiting the kinase activity. These results provide a valuable platform for the structure-guided design of Mnk1 inhibitors.
1. Introduction
Mitogen-activated protein kinase (MAPK)-interacting kinases 1 (Mnk1) and 2 (Mnk2) are protein serine/threonine kinases that are activated by extracellular signal-regulated kinase (Erk) or p38 MAPK (Waskiewicz et al., 1997 ▸; Fukunaga & Hunter, 1997 ▸). Either activated Mnk1 or Mnk2 phosphorylates Ser209 of eukaryotic translation initiation factor 4E (eIF4E; Waskiewicz et al., 1999 ▸), which regulates cap-dependent translation initiation (Scheper & Proud, 2002 ▸). Phosphorylation of eIF4E is crucial for the oncogenic activity of eIF4E, and constitutive active Mnk1 promotes tumorigenesis through the phosphorylation of eIF4E (Wendel et al., 2007 ▸). Additionally, Ser209Ala-mutated eIF4E knockin mice are resistant to tumorigenesis in a prostate cancer model (Furic et al., 2010 ▸), and Mnk1 and Mnk2 double-knockout mice show suppressed tumorigenesis in an Lck-Pten mouse model (Ueda et al., 2010 ▸). Notably, Mnk1 and Mnk2 double-knockout mice are viable and show normal development despite the absence of eIF4E phosphorylation (Ueda et al., 2004 ▸). These studies suggest that the phosphorylation of eIF4E by Mnk1 or Mnk2 is critical in cancer cells but not in normal cells; therefore, targeting Mnk1 and Mnk2 should be a promising strategy for cancer therapy.
Although both Mnk1 and Mnk2 phosphorylate eIF4E, the two Mnks seem to play different roles in cancer development. Mnk1 shows a low basal activity and is highly activated by cellular stimulation, while Mnk2 has a high basal activity (Parra et al., 2005 ▸), which suggests that the activity of Mnk1 is more affected by the MAPK signal pathway than that of Mnk2. It has also been reported that Mnk1 controls the translation of cancer-related mRNAs in glioblastoma (Grzmil et al., 2011 ▸).
The published crystal structures of Mnk1 and Mnk2 have revealed the characteristic features of the inactive state (Jauch et al., 2005 ▸, 2006 ▸). In both Mnk1 and Mnk2, the Mnk-specific DFD motif, which corresponds to the conserved DFG motif in other protein kinases, prefers the DFD-out conformation in the apo form and obstructs ATP binding. The Mnk-specific insertion in the activation segment packs against the kinase region and occupies the DFD-in pocket in the Mnk1 structure, which results in an autoinhibited state, whereas it protrudes away from the kinase region in the Mnk2 structure.
On the other hand, structural information on Mnk inhibitors is currently limited. Although the structure of mutated Mnk2 in complex with a broad kinase inhibitor, staurosporine, has been reported (Jauch et al., 2006 ▸), the structure of neither Mnk1 nor Mnk2 in complex with an Mnk-selective inhibitor is available. Given its importance in cancer biology, it is of great interest to determine the structure of Mnk with Mnk-selective inhibitors.
In this study, we report the co-crystal structure of the kinase region of Mnk1 in complex with a novel Mnk1-selective inhibitor, DS12881479, which stabilizes the autoinhibited state of Mnk1. Our results provide structural insights into the inhibition of Mnk1 which could be useful for future drug design.
2. Materials and methods
2.1. Macromolecule production
For crystallographic experiments, the kinase region of human Mnk1 (Mnk1-KR; residues 37–341) with an N-terminal GST tag and HRV3c protease-cleavage site was expressed in Escherichia coli BL21(DE3) cells (Table 1 ▸). Protein expression was carried out in LB medium using Overnight Express Autoinduction System 1 (Novagen) at 25°C for 24 h. The cells were then harvested. After sonication and centrifugation, the supernatant was applied onto a GSTrap column (GE Healthcare). The eluate was pooled and mixed with PreScission protease (GE Healthcare) to cleave the GST tag, while being dialyzed against 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM DTT at 4°C overnight. The solution was again applied onto a GSTrap column to remove the GST tag and the protease, and the flowthrough fractions were collected. The pooled solution was further purified by ion-exchange chromatography on a Resource Q column (GE Healthcare) and subsequent gel filtration on a HiLoad 16/600 Superdex 75 pg column (GE Healthcare) equilibrated with buffer consisting of 20 mM Tris–HCl pH 7.5, 50 mM NaCl, 3 mM DTT. The fractions containing Mnk1-KR were collected and concentrated to 12 mg ml−1.
Table 1. Macromolecule-production information.
| Source organism | Homo sapiens |
| Expression vector | pGEX-6P-1 |
| Expression host | E. coli BL21(DE3) |
| Complete amino-acid sequence of the construct produced | MSPILGYWKIKGLVQPTRLLLEYLEEKYEEHLYERDEGDKWRNKKFELGLEFPNLPYYIDGDVKLTQSMAIIRYIADKHNMLGGCPKERAEISMLEGAVLDIRYGVSRIAYSKDFETLKVDFLSKLPEMLKMFEDRLCHKTYLNGDHVTHPDFMLYDALDVVLYMDPMCLDAFPKLVCFKKRIEAIPQIDKYLKSSKYIAWPLQGWQATFGGGDHPPKSDLEVLFQ/GPLGSTDSLPGKFEDMYKLTSELLGEGAYAKVQGAVSLQNGKEYAVKIIEKQAGHSRSRVFREVETLYQCQGNKNILELIEFFEDDTRFYLVFEKLQGGSILAHIQKQKHFNEREASRVVRDVAAALDFLHTKGIAHRDLKPENILCESPEKVSPVKICDFDLGSGMKLNNSCTPITTPELTTPCGSAEYMAPEVVEVFTDQATFYDKRCDLWSLGVVLYIMLSGYPPFVGHCGADCGWDRGEVCRVCQNKLFESIQEGKYEFPDKDWAHISSEAKDLISKLLVRDAKQRLSAAQVLQHPWVQGQAPEKG |
2.2. Crystallization
Mnk1-KR at a concentration of 12 mg ml−1 was mixed with 100 mM DS12881479 in a volume ratio of 19:1. Crystallization screens were then set up at room temperature and incubated at 277 K. The crystals were obtained by the hanging-drop vapour-diffusion method using a reservoir solution consisting of 12%(w/v) polyethylene glycol 3350, 200 mM ammonium sulfate, 100 mM sodium citrate buffer pH 6.2, 20 mM magnesium chloride (Table 2 ▸).
Table 2. Crystallization.
| Method | Hanging-drop vapour diffusion |
| Plate type | VDXm plate |
| Temperature (K) | 277 |
| Protein concentration (mg ml−1) | 11.4 |
| Buffer composition of protein solution | 20 mM Tris–HCl pH 7.5, 50 mM NaCl, 3 mM DTT |
| Composition of reservoir solution | 12%(w/v) polyethylene glycol 3350, 200 mM ammonium sulfate, 100 mM sodium citrate buffer pH 6.2, 20 mM magnesium chloride |
| Ratio of drop | 1:1 |
| Volume of reservoir (ml) | 0.4 |
2.3. Data collection and processing
Prior to data collection, the crystals were briefly soaked in precipitant solution containing an additional 30%(v/v) ethylene glycol as a cryoprotectant. X-ray diffraction data were collected from a single crystal on the NW12A beamline at the Photon Factory Advanced Ring (PF-AR), KEK, Tsukuba, Japan. The data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▸). The data-collection and processing statistics are shown in Table 3 ▸.
Table 3. Data collection and processing.
| Diffraction source | NW12A, PF-AR, KEK |
| Wavelength (Å) | 1.0000 |
| Temperature (K) | 95 |
| Detector | ADSC Quantum 210 CCD |
| Rotation range per image (°) | 1.0 |
| Total rotation range (°) | 90 |
| Exposure time per image (s) | 5 |
| Space group | P43212 |
| a, b, c (Å) | 92.64, 92.64, 174.44 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.26 |
| Resolution range (Å) | 50.000–3.000 (3.11–3.00) |
| Total No. of reflections | 379154 |
| No. of unique reflections | 15900 |
| Completeness (%) | 99.9 (100.0) |
| Multiplicity | 7.1 (7.2) |
| 〈I/σ(I)〉 | 18.6 (3.4) |
| R r.i.m. | 0.107 (0.501) |
| Overall B factor from Wilson plot (Å2) | 52.5 |
2.4. Structure solution and refinement
The initial phases were obtained by molecular replacement using the coordinates of Mnk1-KR (PDB entry 2hw6; Jauch et al., 2006 ▸) as a search model. Phaser (McCoy et al., 2007 ▸; Winn et al., 2011 ▸) found two molecules in the asymmetric unit, and the models were rebuilt with Coot (Emsley et al., 2010 ▸) and were refined with REFMAC (Murshudov et al., 2011 ▸). The DS12881479 molecule was built using AFITT (Wlodek et al., 2006 ▸) and fitted into positive electron density in the F o − F c map. Noncrystallographic symmetry (NCS) restraints were applied to both the N-terminal and the C-terminal lobes of Mnk1-KR. Structure refinement was iterated until the R and R free factors converged. A Ramachandran plot for the final model was calculated with RAMPAGE (Lovell et al., 2003 ▸). The refinement statistics are shown in Table 4 ▸. Figures were created with PyMOL (v.1.7; Schrödinger). The atomic coordinates and experimental structure factors have been deposited in the PDB as entry 5wvd.
Table 4. Structure refinement.
| Resolution range (Å) | 20.00–3.00 (3.08–3.00) |
| Completeness (%) | 99.7 (99.4) |
| σ Cutoff | 0.0 |
| No. of reflections, working set | 12684 |
| No. of reflections, test set | 1583 |
| Final R cryst | 0.239 (0.341) |
| Final R free | 0.271 (0.331) |
| Cruickshank DPI | 0.43 |
| No. of non-H atoms | |
| Protein | 3628 |
| Ion | 5 |
| Ligand | 42 |
| Total | 3675 |
| R.m.s. deviations | |
| Bonds (Å) | 0.01 |
| Angles (°) | 1.53 |
| Average B factors (Å2) | |
| Protein | 62.1 |
| Ion | 48.5 |
| Ligand | 48.7 |
| Ramachandran plot | |
| Favoured regions (%) | 97.5 |
| Additionally allowed (%) | 2.5 |
3. Results and discussion
3.1. Discovery of an Mnk1-selective inhibitor
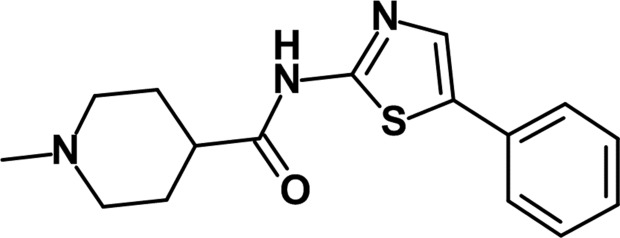
To identify novel Mnk1 inhibitors, we performed high-throughput screening (HTS) using full-length unphosphorylated (inactive) Mnk1, which was then subjected to the activation process, and found several hit compounds. Subsequent hit derivatization led to the potent Mnk1 inhibitor DS12881479 (Fig. 1 ▸), which exhibited an IC50 value for Mnk1 of 21 nM. Since our assay system selected compounds that interact with either inactive Mnk1 or active Mnk1, we tested DS12881479 in an assay using active Mnk1 to identify whether it inhibits active Mnk1. The IC50 value for DS12881479 against active Mnk1 was 416 nM. The modest inhibitory activity of DS12881479 in the assay suggested that it exhibits its potent inhibitory activity mainly by interacting with inactive Mnk1.
Figure 1.
Chemical structure of DS12881479.
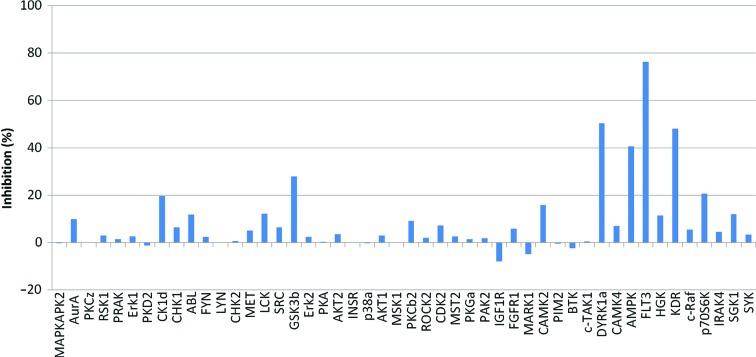
Next, the inhibitory activities of DS12881479 for a further 48 active kinases were examined. DS12881479 inhibited only two kinases, FLT3 and DYRK1a, by over 50% of their activity, even though the tests were performed at the rather high concentration of 5 µM, which is 250 and 12 times higher than the IC50 values for inactive and active Mnk1, respectively (Fig. 2 ▸). The data showed that DS12881479 has good selectivity for Mnk1 over other active kinases.
Figure 2.
Kinase-inhibition profile for DS12881479. The data were obtained at 5 µM DS12881479 using the ProfilerPro Kinase Selectivity Assay Kit (PerkinElmer Inc.) following the manufacturer’s protocol.
3.2. Overall structure of Mnk1-KR in complex with DS12881479
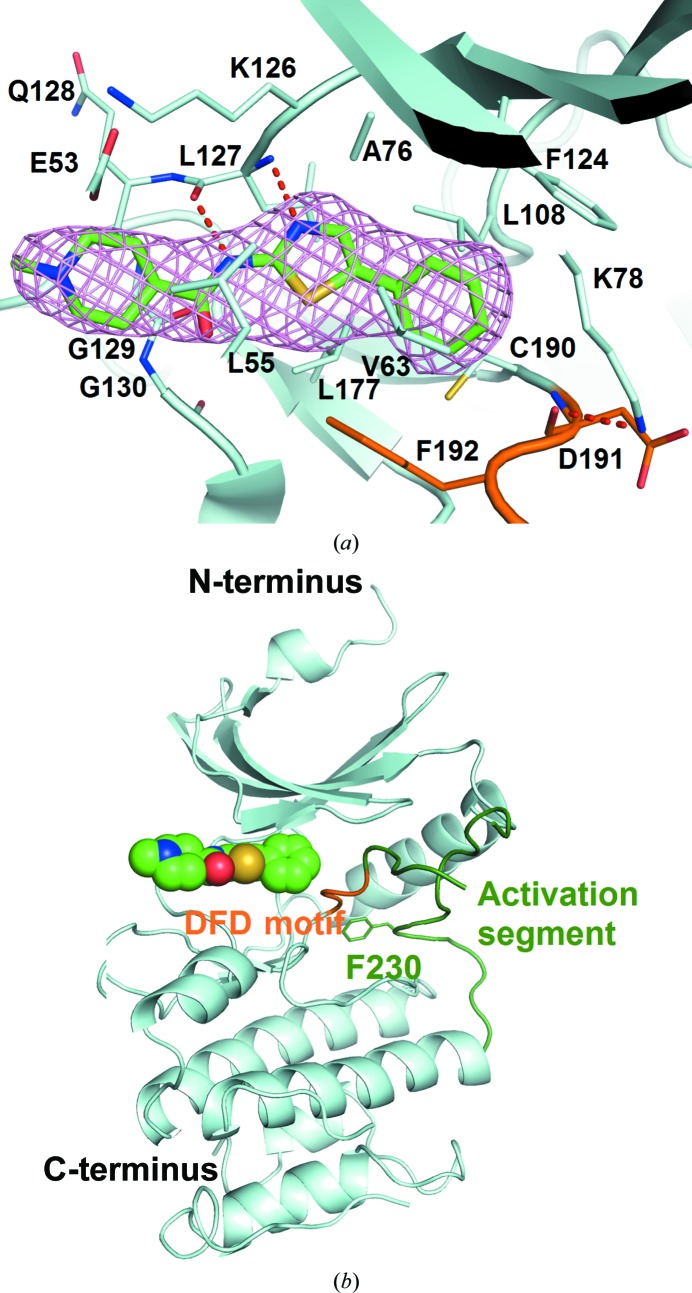
In an effort to understand the structural basis for the inhibition of Mnk1, we determined the crystal structure of Mnk1-KR in complex with DS12881479 at 3.0 Å resolution. Positive electron density for DS12881479 was observed in the F o − F c OMIT map (Fig. 3 ▸ a). The final model contains two Mnk1-KR molecules, two DS12881479 molecules and one sulfate ion in the asymmetric unit. The two Mnk1-KR molecules are nearly identical, with a root-mean-square deviation (r.m.s.d.) of 0.23 Å over 204 Cα atoms, and were therefore treated identically throughout this report.
Figure 3.
The structure of Mnk1-KR in complex with DS12881479. (a) DS12881479 covered with an F o − F c OMIT electron-density map shown as a pink mesh contoured at 2.5σ. The bound DS12881479 and the residues within 4 Å of DS12881479 are shown in stick representation. The C atoms of the DFD motif (residues 191–193) and the activation segment (residues 194–235) of Mnk1-KR are coloured orange and dark green, respectively. Those of DS12881479 are coloured green. (b) A ribbon representation of Mnk1-KR complexed with DS12881479. DS12881479 is depicted as a CPK model. The side chain of Phe230 is shown in stick representation. The colour scheme is the same as that used in (a).
Mnk1-KR consists of N- and C-terminal lobes, as is typical for protein kinases (Fig. 3 ▸ b). The N-terminal lobe includes a sheet of five antiparallel β-strands and a regulatory helix αC. The C-terminal lobe is mainly composed of α-helices, and involves a catalytic loop and the activation segment. The N-terminal nine residues, including the expression tag, and several parts of the C-terminal lobe (residues 198–220, 261–290 and 335–341 in chain A and residues 138–146, 182–183, 199–219, 259–307 and 334–341 in chain B) were omitted from the model owing to a lack of clear electron density. DS12881479 is located in the interlobal cleft between the N- and C-terminal lobes of Mnk1-KR.
The structure of Mnk1-KR with DS12881479 conserves the features of the autoinhibited state as seen in the apo Mnk1-KR structure (PDB entry 2hw6; Jauch et al., 2006 ▸). In brief, the Mnk-specific DFD motif (Asp191-Phe192-Asp193) adopts the DFD-out conformation, which is incompatible with ATP binding (Fig. 3 ▸ a). Asp191 forms a hydrogen bond to Lys78, hindering the ion pairing of Lys78 to Glu94 on helix αC which is crucial for kinase activation (Adams, 2001 ▸). The activation segment packs against the N-terminal lobe, and Phe230 in the Mnk-specific insertion (residues 228–232) lies in the DFD-in pocket (Fig. 3 ▸ b), preventing the DFD motif from switching to the DFD-in conformation.
3.3. The binding mode of DS12881479
The details of the interaction between DS12881479 and Mnk1-KR were investigated. DS12881479 is composed of three main parts: a phenyl group, a 2-aminothiazole moiety and an N-methylpiperidinyl group. Each part interacts with Mnk1-KR (Fig. 3 ▸ a). Firstly, the phenyl group occupies the hydrophobic pocket, which consists of Val63, Leu108, Phe124, Leu177, Cys190 and Phe192. In the pocket, the phenyl ring of DS12881479 makes edge-to-face interactions with Phe124 and Phe192, while Val63 and Leu108 form CH–π interactions with this phenyl ring. Secondly, the 2-aminothiazole moiety makes two hydrogen bonds to the backbone N and O atoms of Leu127 in the hinge loop. The intramolecular interaction between the S atom of the thiazole ring and the O atom of an amide group makes the 2-aminothiazole moiety conformationally rigid and helps the moiety to maintain a hinge-binder conformation. Additionally, Ala76 and Leu177 support this moiety by making CH–π interactions from both sides of the thiazole ring. Thirdly, the N-methylpiperidinyl group faces towards the solvent area, making van der Waals contacts with Glu53, Leu55, Lys126, Gln128, Gly129 and Gly130.
3.4. Comparison with apo Mnk1-KR
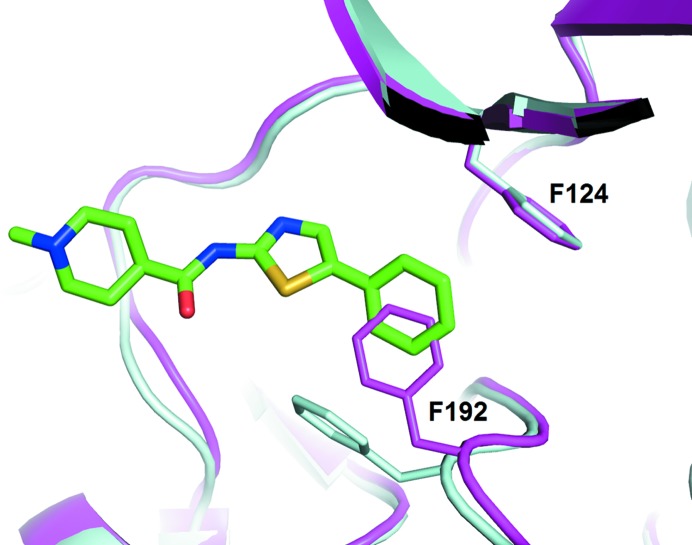
A comparison of DS12881479-bound Mnk1-KR with apo Mnk1-KR revealed the mechanism of the inhibition of Mnk1 activity by DS12881479. Although the overall fold of DS12881479-bound Mnk1-KR resembles that of apo Mnk1-KR, with an r.m.s.d. value of 0.45 Å over 235 Cα atoms, there is a significant induced-fit conformational change at the side chain of Phe192 in the DFD motif (Fig. 4 ▸). In apo Mnk1-KR in the autoinhibited state there appeared to be no space for DS12881479 to fit around the ATP-binding site where most kinase inhibitors bind. In DS12881479-bound Mnk1-KR, the induced fit of Phe192 has created a binding space for the phenyl group of DS12881479. As described earlier, the crystal structure of Mnk1-KR with DS12881479 is in the inactive state, in which the DFD motif adopts the DFD-out conformation. The phenyl group of DS12881479 stabilizes this DFD-out conformation by forming an edge-to-face interaction with the side chain of Phe192, preventing the DFD motif of Mnk1-KR from switching to the active DFD-in conformation.
Figure 4.
The conformational change of Mnk1-KR upon binding DS12881479. DS12881479-bound Mnk1-KR is superposed on apo Mnk1-KR (PDB entry 2hw6). Mnk1-KR in complex with DS12881479 is coloured cyan and apo Mnk1-KR is coloured magenta. The side chains of residues 124 and 192 of Mnk1-KR are shown in stick representation. DS12881479 is also shown in stick representation with the same colour scheme as used in Fig. 3 ▸.
In summary, the present study reports the structure of Mnk1-KR in complex with a novel and selective Mnk1 inhibitor. The crystal structure revealed that the inhibitor binds to the autoinhibited state of Mnk1 and stabilizes the inactive state by interacting with the DFD-out conformation. With the growing interest in Mnk1 and Mnk2 as targets for cancer therapy, our results could be helpful in developing inhibitors of Mnk.
Supplementary Material
PDB reference: Mnk1 in complex with DS12881479, 5wvd
Details of kinase assay.. DOI: 10.1107/S2053230X18002108/nw5069sup1.pdf
Acknowledgments
We would like to thank the staff at the Photon Factory for their excellent support during the use of the synchrotron beamlines. We also thank Dr Shuichiro Ito for his help in preparing the manuscript.
References
- Adams, J. A. (2001). Chem. Rev. 101, 2271–2290. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fukunaga, R. & Hunter, T. (1997). EMBO J. 16, 1921–1933. [DOI] [PMC free article] [PubMed]
- Furic, L., Rong, L., Larsson, O., Koumakpayi, I. H., Yoshida, K., Brueschke, A., Petroulakis, E., Robichaud, N., Pollak, M., Gaboury, L. A., Pandolfi, P. P., Saad, F. & Sonenberg, N. (2010). Proc. Natl Acad. Sci. USA, 107, 14134–14139. [DOI] [PMC free article] [PubMed]
- Grzmil, M., Morin, P. Jr, Lino, M. M., Merlo, A., Frank, S., Wang, Y., Moncayo, G. & Hemmings, B. A. (2011). Cancer Res. 71, 2392–2402. [DOI] [PubMed]
- Jauch, R., Cho, M.-K., Jäkel, S., Netter, C., Schreiter, K., Aicher, B., Zweckstetter, M., Jäckle, H. & Wahl, M. C. (2006). EMBO J. 25, 4020–4032. [DOI] [PMC free article] [PubMed]
- Jauch, R., Jäkel, S., Netter, C., Schreiter, K., Aicher, B., Jäckle, H. & Wahl, M. C. (2005). Structure, 13, 1559–1568. [DOI] [PubMed]
- Lovell, S. C., Davis, I. W., Arendall, W. B., de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S. & Richardson, J. S. (2003). Proteins, 50, 437–450. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Parra, J. L., Buxadé, M. & Proud, C. G. (2005). J. Biol. Chem. 280, 37623–37633. [DOI] [PubMed]
- Scheper, G. C. & Proud, C. G. (2002). Eur. J. Biochem. 269, 5350–5359. [DOI] [PMC free article] [PubMed]
- Ueda, T., Sasaki, M., Elia, A. J., Chio, I. I. C., Hamada, K., Fukunaga, R. & Mak, T. W. (2010). Proc. Natl Acad. Sci. USA, 107, 13984–13990. [DOI] [PMC free article] [PubMed]
- Ueda, T., Watanabe-Fukunaga, R., Fukuyama, H., Nagata, S. & Fukunaga, R. (2004). Mol. Cell. Biol. 24, 6539–6549. [DOI] [PMC free article] [PubMed]
- Waskiewicz, A. J., Flynn, A., Proud, C. G. & Cooper, J. A. (1997). EMBO J. 16, 1909–1920. [DOI] [PMC free article] [PubMed]
- Waskiewicz, A. J., Johnson, J. C., Penn, B., Mahalingam, M., Kimball, S. R. & Cooper, J. A. (1999). Mol. Cell. Biol. 19, 1871–1880. [DOI] [PMC free article] [PubMed]
- Wendel, H. G., Silva, R. L., Malina, A., Mills, J. R., Zhu, H., Ueda, T., Watanabe-Fukunaga, R., Fukunaga, R., Teruya-Feldstein, J., Pelletier, J. & Lowe, S. W. (2007). Genes Dev. 21, 3232–3237. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wlodek, S., Skillman, A. G. & Nicholls, A. (2006). Acta Cryst. D62, 741–749. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Mnk1 in complex with DS12881479, 5wvd
Details of kinase assay.. DOI: 10.1107/S2053230X18002108/nw5069sup1.pdf