Abstract
The undertaking of large-scale DNA sequencing screens for somatic variants in human cancers requires accurate and rapid processing of traces for variants. Due to their often aneuploid nature and admixed normal tissue, heterozygous variants found in primary cancers are often subtle and difficult to detect. To address these issues, we have developed a mutation detection algorithm, AutoCSA, specifically optimized for the high throughput screening of cancer samples.
1. Introduction
Cancers arise due to the accumulation of mutations in critical target genes conferring growth/survival advantage in a clone of cells which eventually manifests as clinical disease. Whilst a proportion of these mutations can be inherited in the germline giving rise to cancer susceptibility syndromes, the majority are accumulated somatically. There has been considerable effort to identify the variants and hence the genes that cause cancer. Indeed, since the completion of the Human Genome Project it is now possible to systematically screen megabases of sequence for these somatic variants.
A number of software programs and protocols have been developed to identify sequence variants to a high sensitivity; PolyPhred (Nickerson et al., 1997) has been available for some time, while comparitive sequence analysis (CSA) (Mattocks et al., 2000), Mutation Surveyor (SoftGenetics), novoSNP (Weckx et al., 2005), InSNP (Manaster et al., 2005) and SNPdetector (Zhang et al., 2005) are more recent developments. In addition, PolyPhred has been enhanced to detect SNPs in PCR-amplified diploid samples (Stephens et al., 2006).
We have extended some of the concepts of CSA variant detection protocol developed by Mattocks et al. (2000) into a fully functional computer application, AutoCSA. CSA was initially developed to simplify and aid the detection of variants in DNA sequence traces. Briefly, CSA involves comparing raw trace profiles from each of the four channels (bases) between the sample under investigation and a reference sample by overlaying the traces using ABI Genescan software. Each channel is then manually inspected for the presence of a reduced peak height between the reference and the sample trace and also the presence of a novel peak indicating a possible variant. This key concept has been used in the development of AutoCSA which, unlike CSA, is capable of automatically analysing large numbers of sequence traces with minimal intervention.
In particular, AutoCSA has also been optimized to detect heterozygous substitutions present at less than 50% of wildtype signal that are frequently present in PCR-amplified templates from primary tumour samples. The software has been further developed to efficiently detect other classes of variants, notably small homozygous and heterozygous insertions and deletions.
2. Algorithm and Results
AutoCSA is split into three main components, pre-processing of the trace file, variant detection and a post-processing stage to remove false positives. Detailed information on the algorithm can be found on our website (http://www.sanger.ac.uk/genetics/CGP/Software/AutoCSA/detailed_algorithm.shtml). A training set of 161 somatic variants composed of 96 substitutions (84 heterozygous, 12 homozygous), 36 heterozygous insertions/deletions and 29 homozygous insertions/deletions was used to optimize the software.
2.1. Pre-processing
One of the important concepts of AutoCSA is that it uses raw data channels from the sequence trace file, which contains the absolute peak heights generated by the sequencing reaction. These data are likely to be more quantitative than the processed data generated by the software onboard the ABI sequencer, which equalizes peak heights across the trace (Mattocks et al., 2000). However, the raw data require some manipulation to render it suitable for analysis with AutoCSA, and a pre-processing step is required to produce a trace with an approximately uniform base spacing (mobility correction) and uniform base line intensity (baselining). The pre-processing stage also involves the identification of the position (scan index) and height (intensity) of the peaks in each of the four channels (bases) of the trace file. AutoCSA uses the known amplimer DNA sequence to identify the correct consecutive peaks in the sequence trace. A quality value is assigned to each base with a matched peak and defined as a signal (matched peak) to noise (unmatched peak) ratio of intensities.
2.2. Heterozygous substitutions
The primary discriminator for indicating the presence of a heterozygous substitution is a peak height drop ≥20% between the trace under investigation and a reference trace. In addition, the algorithm requires the presence of an additional mutant peak, which must satisfy a local peak height ratio test, by comparing height intensities of adjacent bases. Using these parameters, AutoCSA detected 81/84 (96.4%) heterozygous substitutions in the training set. Three substitutions were missed due to poor local quality issues in the traces.
Homozygous substitutions are identified by the absence of the wildtype base during the amplimer matching procedure. Each missing position is interrogated for the presence of a viable novel peak. Using these criteria, AutoCSA detected 12/12 (100%) homozygous somatic substitutions in the training set.
2.3. Homozygous insertions and deletions
Homozygous insertions are identified by interrogating the base-spacing between neighbouring nucleotides. A scan index gap is calculated between neighbouring bases that have been aligned to the amplimer sequence for the trace under investigation. If there is a homozygous insertion there will be a larger than expected scan gap. Homozygous deletions can be determined by failure to identify the expected peaks during the amplimer matching procedure. Using these criteria AutoCSA detected 28/29 (97%) of homozygous insertion/deletions in the training set.
2.4. Heterozygous insertions and deletions
To detect heterozygous insertions/deletions, AutoCSA first identifies an abrupt drop, or step in the quality of the sequence trace. The second criterion is a critical concentration of individual, closely spaced heterozygous substitutions from the start of the reduced quality step to the end of the trace. Using these criteria AutoCSA detected 36/36 (100%) heterozygous insertions/deletions in the training set.
2.5. Post-processing (variant flagging) and visualization
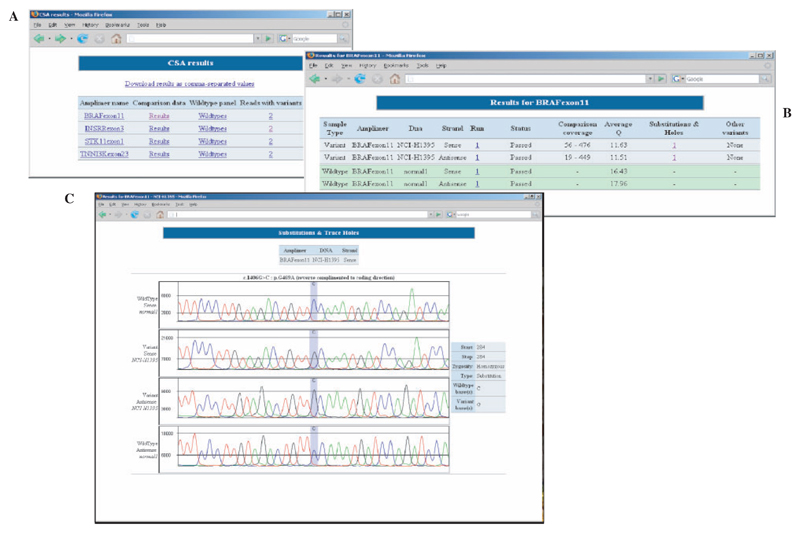
AutoCSA reduces the number of false calls displayed to users by using a series of novel filters that examine the global and local quality of a trace and the concentration of variants. Variants that pass the filters are ‘flagged’ for manual review or otherwise automatically rejected by the system. If bi-directional sequencing is used, a further set of rules can be applied by AutoCSA, which utilises information from both strands to reduce the false positive calls further. This second set of rules examines the corresponding base on the opposite strand to determine if an equivalent variant has been called and also to assess the noise level under the specific base to help rule out noise and sequencing artefacts. AutoCSA generates a series of web pages summarizing the resulting variants with images of each variant and associated protein annotation (Fig. 1).
Fig. 1. AutoCSA displays.
(A) Lists all sequences screened with a summary of variants found. (B) Lists traces screened with coverage information and number of variants on each trace. (C) Main substitution display, four traces are displayed with 20 bases either side of the potential substitution. The top two traces are the traces which were used to call the variant (reference first and trace under investigation second). The third and fourth traces are the reverse sequenced traces. The DNA and protein annotation of the variant are displayed above and to the right of the traces.
2.6. Testing of AutoCSA
To evaluate the performance of AutoCSA, Mutation Surveyor version 2.0 was used in a comparison analysis of 43 Mb of DNA. These data were obtained by resequencing the 518 protein kinase genes in the human genome in a series of 30 primary colorectal tumours and one colorectal cell line. A total of 105 somatic substitutions and 22 somatic heterozygous insertion/deletion mutations were identified in this set using a combination of AutoCSA and Mutation Surveyor. Ninety seven (92%) substitutions and 22 (100%) heterozygous insertion/deletions were identified by AutoCSA alone, 82 (78%) substitutions and 4 (18%) heterozygous insertion/deletions were identified by Mutation Surveyor alone. AutoCSA generated 0.21 false positives per sequence trace compared to 0.52 false positives per sequence trace generated by Mutation Surveyor.
3. Summary
In conclusion, we have developed a variant detection system which has been optimized to detect the often subtle heterozygous variants which are common in primary cancer samples. The software has been developed so it can automatically run over large numbers of trace files with minimal human intervention and can therefore be easily integrated into high throughput resequencing projects.
Acknowledgements
The authors would like to thank the Wellcome Trust for funding the Cancer Genome Project.
Footnotes
Conflict of Interest: none declared.
Availability: http://www.sanger.ac.uk/genetics/CGP/Software/AutoCSA.
References
- Manaster C, et al. InSNP: a tool for automated detection and visualization of SNPs and InDels. Hum Mutat. 2005;26:11–19. doi: 10.1002/humu.20188. [DOI] [PubMed] [Google Scholar]
- Mattocks C, et al. Comparative sequence analysis (CSA): a new sequence-based method for the identification and characterization of mutations in DNA. Hum Mutat. 2000;16:437–443. doi: 10.1002/1098-1004(200011)16:5<437::AID-HUMU9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Nickerson DA, et al. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, et al. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- Weckx S, et al. novoSNP, a novel computational tool for sequence variation discovery. Genome Res. 2005;15:436–442. doi: 10.1101/gr.2754005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]