Abstract
Human brain wrinkling has been implicated in neurodevelopmental disorders and yet its origins remain unknown. Polymer gel models suggest that wrinkling emerges spontaneously due to compression forces arising during differential swelling, but these ideas have not been tested in a living system. Here, we report the appearance of surface wrinkles during the in vitro development and self-organization of human brain organoids in a micro-fabricated compartment that supports in situ imaging over a timescale of weeks. We observe the emergence of convolutions at a critical cell density and maximal nuclear strain, which are indicative of a mechanical instability. We identify two opposing forces contributing to differential growth: cytoskeletal contraction at the organoid core and cell-cycle-dependent nuclear expansion at the organoid perimeter. The wrinkling wavelength exhibits linear scaling with tissue thickness, consistent with balanced bending and stretching energies. Lissencephalic (smooth brain) organoids display reduced convolutions, modified scaling and a reduced elastic modulus. Although the mechanism here does not include the neuronal migration seen in in vivo, it models the physics of the folding brain remarkably well. Our on-chip approach offers a means for studying the emergent properties of organoid development, with implications for the embryonic human brain.
Wrinkling and folding appears universally during growth of soft and natural systems1–5. During brain development, wrinkling occurs at two stages: First, the emergence of folds along the neural tube separates and defines different brain regions, such as the left and right hemispheres6,7. Second, folding of the cortex contributes to the expansion of brain surface area8–10. Reduced cortical folding (lissencephaly) is one of the manifestations of a severe neurodevelopmental disorder, which is accompanied with intellectual disability and reduced life expectancy11,12. From a physical perspective, surface wrinkles emerge as a mechanical instability when an elastic material is under compression13. At low forces, the material is compressed in the direction of the force and expands perpendicular to it, thus resulting in strain. Beyond a critical force, the material will exhibit folding and wrinkling to release the compression, without further increase in strain energy. Compression forces and wrinkling can arise internally during differential swelling of polymer gels, due to solvent absorption14–19. When the outer region of a gel is swelling faster than the interior, the mismatch leads to residual strain and results in periodic wrinkling. Similarly, wrinkling can arise during in vivo development both from non-homogenous tissue growth20, and localized cytoskeletal contraction5,21,22. Here, we study the emergence of folding during in vitro brain organoid development, characterize the physical mechanisms which lead to differential growth, and reveal the underlying critical and scaling behavior.
Recent breakthroughs in stem-cell technologies enable three-dimensional culturing of human stem cells, which develop into organ-like structures, and exhibit remarkable self-organization and collective behavior (‘organoids’)23–28. However, three-dimensional growth to a millimeter scale without vasculature leads to cell death in the organoid core, as diffusion becomes inefficient for nutrient supply. Furthermore, tissue thickness obstructs optical imaging and real-time microscopy is carried out in organoid slices. Here, we present an on-chip approach, which enables development of human brain organoids to millimeter diameter, together with efficient nutrient exchange by diffusion, and in situ whole-organ fluorescent live-imaging over several weeks.
Human embryonic stem cell aggregates were inserted into a micro-fabricated compartment29 of height h = 150 ± 10μm, and filled with collagen-laminin based hydrogel (Matrigel™) (Fig. 1a-d, Fig. S1, Methods). Within 1-3 days, the homogenous and isotropic cell aggregate self-organized into a spherical shell structure surrounding a small cavity (lumen) (Fig. 1e, Day 3, Fig. S2). During the first week, the organoid expanded by 20-fold to an area of 0.2 ± 0.05mm2 and thickness of t = 50 − 200μm (Fig. 1e Day 4-6, Figs. S3, S4). The cells in the organoid attained a bi-polar morphology, extending from the inner (apical) surface of the organoid, r = 0, to the outer (basal) surface, r = t (Fig. 1d). The cell nuclei were radially oriented (Fig. 1d), and performed an up-and-down radial motion coupled to the cell cycle, with cell division at the inner surface, r = 0 (Movie S1). This mimics the developing brain ventricular zone, which is composed of proliferating neuronal stem cells30. The developing organoid exhibited enrichment of cerebral cortex specific genes31 including FOXG1, PAX6, EMX2 and LHX2 as revealed by RNA sequencing (Fig. 1f, Fig. S5), and an increase in genes typical for radial glia cells and neurons, accompanied by a corresponding decrease in pluripotent cell markers (Figs. S5,6). By day 30, immunostaining revealed a layer of NEUN+ neurons surrounding the PAX6+ progenitor cells (Fig. S7). These findings indicate that the on-chip organoid approach successfully mimics the early developing cortex.
Fig. 1. Brain Organoid development and wrinkling.
(a) Illustration of the two-dimensional compartment, h = 150μm. The top membrane is coupled to a media reservoir, and the bottom coverslip enables in situ imaging. (b) Z-stack image of the organoid showing actin using Lifeact-GFP (green) and cell nuclei using H2B-mCherry (red). (c) Illustration of an organoid optical section. (d) Fluorescence image and illustration showing cell organization in the organoid. Dashed white line marks inner organoid surface surrounding a lumen. Cells exhibit a bi-polar morphology, stretched between the outer surface (r = t) and the inner surface (r = 0). Nuclei are distributed along the radial coordinate, and cell division occurs at the inner surface (r = 0). (e) Fluorescence images showing the development of the organoid during days 3-11 after Matrigel embedment, and the emergence of wrinkles. Arrows indicate initial wrinkling instability. (f) RNA sequencing expression data in the organoids for three developmental time points: embryonic stem cell culture, organoids 5 and 12 days after Matrigel embedment. Six samples are shown for each time point (3 wild-type, 3 LIS1 +/− mutants). Color bar indicates the row z-score. The list contains genes that are typical for telencephalon and cortical brain regions. Scale bars are 100μm (e), 50μm (b), 20μm (d). Error bars represent s.e.m.
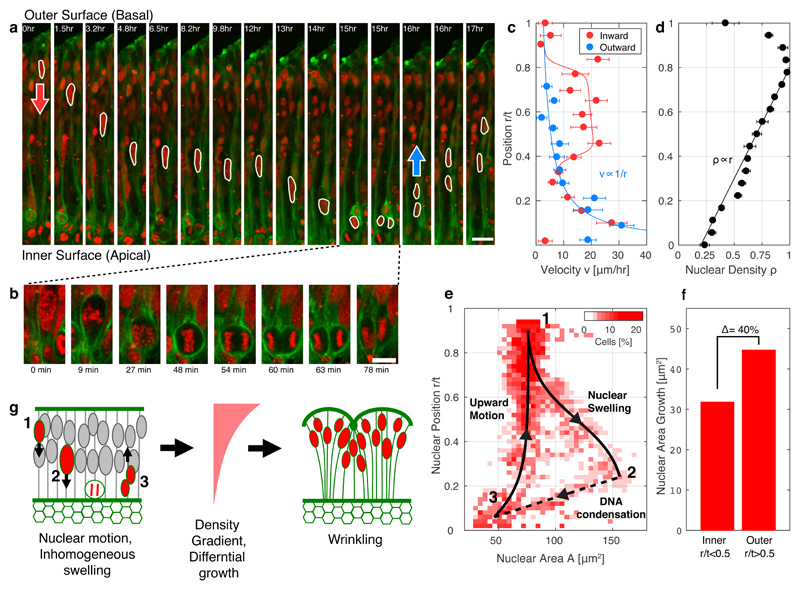
In the second week of development we observed the emergence of surface instabilities (Fig 1e; days 6-11). The two-dimensional wrinkling index, was computed as the contour length LG normalized by the length, LF, of an outer convex contour averaged over N = 14 organoids (Fig. 2a,c). The wrinkling dynamics exhibited an onset at days 6-8, followed by a maximal wrinkling rate at days 10-12, and saturation at a maximal average value < Wmax >= 2.1 ± 0.1 at day 15. Similar folds appeared in devices with a reduced hydrogel density, and with antiadhesive materials: PEG hydrogel and Lipidure (Fig. S8,9).
Fig. 2. Organoid wrinkling occurs at a critical nuclear density and maximal strain.
(a) A low magnification image of the wrinkled organoid. Dotted line marks the organoid contour of length LG. Solid line marks a convex contour of length LF. (b) Fluorescence images of H2B-mCherry showing nuclear aspect ratio and density in days 3,6. (c) Wrinkling index W = LG/LF as a function of time averaged over 14 organoids. (d) Nuclear density ρ and (e) aspect ratio R1/R2 as a function of time averaged over 250 nuclei sampled from five organoids, with 50 nuclei each. (f) Wrinkling index as a function of nuclear density ρ (Nuclei per area). Nuclear density was measured at the tissue outer region, r/t > 0.5, and normalized by the average nuclear area < a >= 89 ± 14μm2. A sharp wrinkling transition is observed at a critical density ρc = 0.85 ± 0.1 < a >. (g) Illustration showing organoid wrinkling at a critical density ρc. (h) Wrinkling wavelength λ as a function of thickness t exhibiting a linear scaling regime. Scale bars are 100μm (a), 10μm (b). Error bars represent s.e.m.
We next studied the cellular structure of the organoid, and observed that the number of nuclei per unit area ρ (nuclear density, Fig. 2b,d), as well as the nuclear aspect ratio R1/R2 (Fig. 2e) increased during days 1-6, before the onset of wrinkling. Nuclear data was averaged from 5 organoids, in each >50 nuclei were measured. Nuclear density was measured at the outer region, r/t > 0.5, and normalized by the average nuclear area < a >= 89 ± 14μm2 (Fig. S10). At the wrinkling onset, the nuclear aspect ratio reached a maximal value < R1/R2 >= 2.6 ± 0.1 (Day 8), followed by a decrease to a value of < R1/R2 >= 2.2 ± 0.1 (Day 16). The nuclear density exponentially saturated at a maximal value, at < ρmax >= 1.00 ± 0.05 < a >−1, suggesting that the nuclei physically encompass all the available area.
Plotting the wrinkling index as a function of nuclear density, we observed a sharp transition to wrinkling at a critical density ρc = 0.85 ± 0.1 < a >−1 (Fig. 2f). These data suggest that during organoid growth the nuclei undergo compression, as manifested by the increase in aspect ratio and density. As the compression reaches a critical value, the organoid surface wrinkles (Fig. 2g). We next exploited the range of organoid thickness, 50 < t < 200μm, to study the wrinkle wavelength λ as a function of thickness t (Fig. 2h). Strikingly, in the region t < 125μm we observed a linear scaling λ ∝ 0.8t. Beyond t > 125μm, the wavelength saturates at a maximal value, λ = 125 ± 8μm. A similar scaling behavior, was previously observed in polymer gel systems and theoretical models, in which a swelling outer layer of thickness t and elastic modulus Es, is constrained by a non-swelling bulk with elastic modulus EB13,17,18 (Fig. S11). In the following paragraphs, we provide evidence that the organoid undergoes differential swelling due to cell-cycle dependent nuclear swelling and motion, and due to cytoskeleton contraction in the organoid inner surface.
Nuclear position and area were analyzed in the growing organoid over 48 hours (Fig. 3, Movie S1). The nuclei performed a periodic in-out motion, which was coupled to the cell-cycle (Fig. 3a,b, Fig. S12). This phenomenon is known as interkinetic nuclear movement and is one of the hallmarks of the brain ventricular zone in vivo30,32–34. The nuclear motion included a stationary-phase close to the outer surface, rapid motion towards the inner surface, division into two daughter cells, followed by an outward motion. Nuclear velocities during motion reach ν = 10 − 30μm/hr consistent with in vitro 2D human cell culture measurements35 (Fig. 3c). The outward velocities decay as the nuclei move away from the inner (apical) surface, νout ∝ 1/r. The inward velocities follow the same trend with an additional high velocity phase in the range, 0.4 < r/t <0.8. At steady-state, we expect that nuclear density is inversely proportional to the nuclear velocities, such that nuclei accumulate at positions where their motion is slow. Indeed, the nuclear density profile exhibits a linear dependence, ρ ∝ r, which is inversely proportional to the slow outward velocity, ρ ∝ 1/νout (Fig. 3d). Overall, the active motility of the nuclei is a mechanism by which material is actively transported to the outer organoid regions, slows down, and accumulates. This implies that the outer regions of the organoid grow faster than the inner regions. Notably, the density profile becomes non-linear following the wrinkling onset (Fig. S13b).
Fig. 3. Nuclear motion and swelling during cell cycle lead to differential growth.
(a) Time-lapse fluorescence images of nuclear motion in the organoid during cell-cycle. H2B-mCherry (red) and Lifeact-GFP (green). (b) High-resolution images during cell mitosis at the inner surface. (c) nuclear velocities ν and (d) nuclear density ρ as a function of radial position r/t. Velocities were measured for both inward (red) and outward (blue) motions. (e) Two-dimensional phase-space diagram of nuclear area A and radial-position r/t. Red color intensity indicates the percentage of nuclei at each point. The nuclei distribution is limited to closed path, reflecting the periodic cell-cycle: (1 → 2) Nuclear swelling: two-fold increase in nuclear area, coupled with an inward nuclei motion, (2 → 3) DNA condensation: motion towards the mitosis region at the inner surface, and decrease in area during DNA condensation prior to cell division. (3 → 1) Upward motion of “newly-born” nuclei. Data was taken from analysis of 13 nuclei over 48 hours with a time step of 3 minutes. Black line and arrows were drawn to indicate progression with time. (f) Nuclear area growth, during a cell cycle period, in the outer half of the organoid (r/t > 0.5) compared to the inner half of the organoid (r/t < 0.5). (g) Illustration of a physical mechanism for wrinkling. Nuclear motion and position dependent nuclear swelling create density and growth gradients, which are maximal at the organoid outer regions. The differential growth leads to residual stress and wrinkling. Scale bars are 20μm (b) and 10μm (c). Error bars represent s.e.m.
We further studied nuclear growth during cell cycle, by measuring the nuclear area, A, as a function of the nuclear radial position, r/t (Fig. 3e). Nuclei undergo a two two-fold area increase as they move inward from the outer surface (r1/t ≈ 0.9, A1 ≈ 75μm2) towards the inner region (r2/t ≈ 0.25, A2 ≈ 150μm2). Cells undergo mitosis in the inner surface, preceded by rapid DNA condensation (r3/t < 0.1, A2 ≈ 50μm2). The two newly-born nuclei perform an outward motion with small change in area. The increase in nuclear area during the cell cycle period is 40% higher in the outer part of the organoid, r/t > 0.5, than inner part of the organoid, r/t < 0.5 (Fig. 3f). This is consistent with cell-cycle synthesis (S) phase, which occurs near the basal (outer) surface of the ventricular zone in vivo30. Thus, the cell-cycle dependent nuclear position and biosynthesis, leads to increased swelling in the outer region of the organoid.
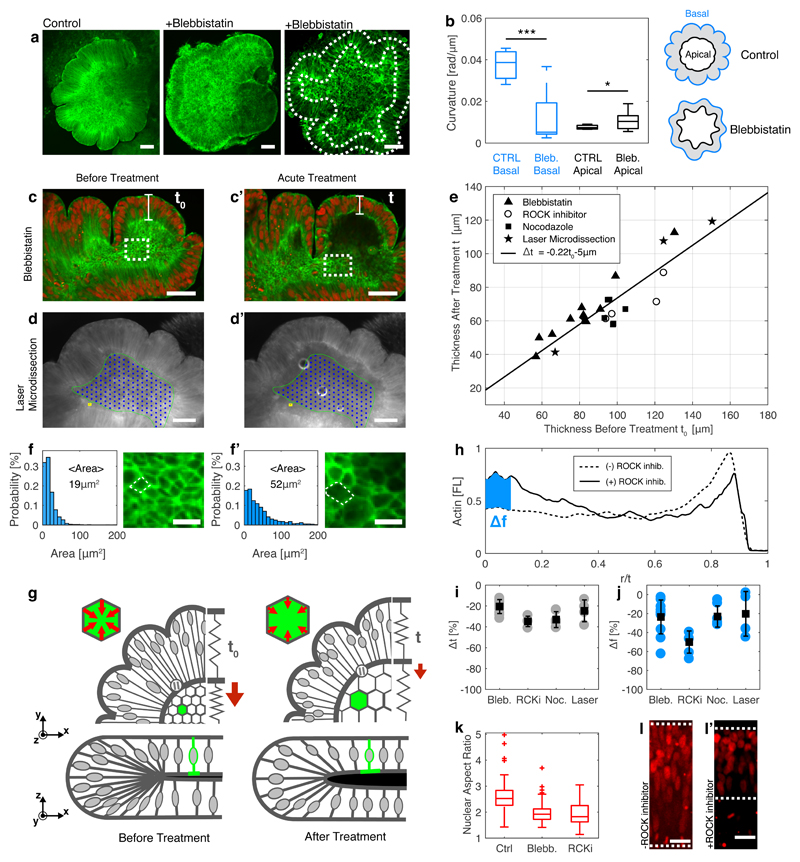
We next studied the role of the cell contractility in the organoid. We administrated a low concentration (5μM) of blebbistatin, which inhibits myosin contractility, during days 6-10 (Fig. 4a). The drug treated organoids had a smoother basal (outer) surface, in comparison to control organoids (Fig. 4b). In addition, the treated organoids exhibited a higher apical (inner) surface curvature than the control organoids. The curvature was measured by averaging the tangent angle θ(r) derivative along the surface perimeter contour < ∂rθ(r) > in 7-10 organoids. Notably, the effect of blebbistatin was irreversible. By comparing our results to the swelling gel model17, we conclude that the cytoskeleton inhibition drug effectively acts to soften the elasticity of the organoid core, EB. A soft core, EB < ES, is easily pulled and follows the outer surface folds, allowing long wavelength (small curvature) folds. In contrast, a stiff core, EB ≫ ES, retains its original shape during the swelling process and results in short wavelength folds and high curvature.
Fig. 4. Cytoskeletal forces maintain organoid core contraction and stiffness.
(a) Images of organoids treated with blebbistatin (+Blebbistatin) during days 6-10 and control. Dashed lines mark inner and outer surfaces. (b) Average curvature < |∂rθ(r)| > of outer (basal, blue) and inner (apical, black) organoid surfaces for control (CTRL) and treated (Bleb.) organoids (N=7-10. (c-d) Images of organoids before and after acute treatment with cytoskeleton disturbing drugs including (c,c’) blebbistatin and (d,d’) laser micro-dissection treatment of the organoid core (marked with green line and blue dots). (e) Organoid thickness before (t0) and after (t) cytoskeleton disturbing treatments including blebbistatin, ROCK inhibitor, nocodazole and laser microdissection. (f,f’) Magnified view of (c,c’) reveals that the inner surface area of cells is increased following drug treatment. (g) Illustration of the organoid during cytoskeleton inhibition, showing the reduction in thickness and increase in inner surface area. (h) Lifeact fluorescence profile along the radial coordinate r/t before (solid line) and after (dashed line) treatment with ROCK inhibitor. Blue area marks the difference in fluorescence intensity at the inner surface (0 < r/t < 0.1). (i) Reduction in inner surface fluorescence and (j) thickness for drug and microdissection treatments. Averages (squares) taken over 4-11 repeats (circles) for each condition. (k) Nuclear aspect ratio in control organoids (Ctrl) and after treatments. Data includes 40-70 nuclei from 3 organoids. (l,l’) H2B-mCherry fluorescence images before and after treatment. Dashed lines indicate inner and outer surfaces. Scale bars are 100μm (a,d), 50μm (c), 20μm (l) and 10μm (f). Error bars represent s.e.m.
Next, we studied the short term effects of cytoskeleton inhibition; using blebbistatin (Fig. 4c), nocodazole, which disturbs microtubules polymerization, and Rho-associated coiled-coil containing protein kinase (ROCK) inhibitor, which disrupts myosin contractility and actin polymerization22. Additionally, we used laser microdissection to irradiate cells in the organoid core, while keeping the perimeter cells intact (Fig. 4d, Fig. S14). All treatments resulted in expansion of the inner surface (Fig. 4c,d), and a reduction in perimeter thickness (Fig. 3e). Blebbistatin treatment resulted in an increase of the cellular inner surface area from 19 ± 0.5μm2 to 50 ± 2μm2 (Fig. 4f). Laser microdissection resulted in total area increase of 12 ± 2% (Fig. S14). The neuroepithelium thickness after the treatment, t, is described by a single linear function of the thickness before treatment, t0, for all drugs and including the microdissection (Fig. 4e,i). The reduction in thickness is Δt = t − t0 = −0.22t0 − 5μm (Fig. 4e). Lifeact-GFP fluorescence intensity along the inner surface was reduced (Fig. 4h,j), which is attributed to disassembly of actomyosin filaments. The outer surface fluorescence increased, which may be due to redistribution of actin within the cell, as the total amount of actin is preserved. Additionally, a decrease in the aspect ratio of the nucleus was observed from R1/R2 = 2.6 ± 0.3, before treatment, to R1/R2 = 1.9 ± 0.1 after blebbistatin treatment, and R1/R2 = 2.02 ± 0.16 after treatment with ROCK inhibitor (Fig. 4k,l).
We next studied the effect of LIS1 heterozygous (+/−) mutation (Fig. 5a,b). The mutation is associated with lissencephaly, which is a severe smooth brain malformation11,36,37. The LIS1 protein is involved in several key functions including proliferation and neuronal migration, and is involved in regulation of molecular motors and the cell cytoskeleton38–45. Three isogenic mutant cell-line clones were generated, using CRISPR/Cas9 genome editing, and expressed reduced LIS1 protein levels (Fig. S15). The LIS1+/- organoids followed the same neuronal development pathway as the wild-type organoids, as observed by elevation of telencephalon related genes (Fig. S5,6, Pearson coefficient > 0.98). Cytoskeleton and extra cellular matrix (ECM) related genes exhibited significant differential expression between wild-type and LIS1+/- organoids (Fig. 5c, Fig. S5). ECM has been suggested to play an important role in the evolutionary expansion of the neocortex and its gyrification, especially within a unique subpopulation of basal or outer radial glia progenitors46,47.
Fig. 5. LIS1+/- mutation results in lissencephalic organoids, modified ECM and cytoskeleton, and reduced cell elasticity.
(a) Images of wild-type (WT) and (b) LIS1 mutant organoids (LIS1+/-). (c) Expression data of extra cellular matrix (ECM) and cytoskeleton related genes (N=3). (d) Distribution of organoid thickness and (e) wrinkling wavelength for WT and (f, g) LIS1+/- (N=5). (h) Wrinkle wavelength as a function of thickness. (i) Tangent correlation function, < Cos(θ(Δr)) >, between two points at a distance r along the organoid surface (N=9 WT, 10 LIS1+/-). At short distances the correlation decreases linearly, 1 − r/lp. WT organdies exhibit correlation peaks (arrowheads). Inset: correlation functions of healthy (blue) and lissencephalic (orange) human brains. (j) Average curvature < |∂rθ(r)| > of WT and three different and isogenic LIS1+/- clones (N=9 – 11). (k) Elastic modulus of WT and LIS1+/- embryonic stem cells (ES) and neuronal progenitors (NP) from several thousand force curves (N=7-10). (l) Nuclear velocities during apical (inward) and basal (outward) motion for WT and LIS1+/- (N=15). (m) Two-dimensional diagram of nuclear area A and radial-position r/t of LIS1+/- nuclei. Orange color intensity indicates the percentage of nuclei at each point. Black line and arrows indicate progression with time. (n) Nuclear area growth, over a cell-cycle period, in the outer (Out, r/t > 0.5) and inner (In, r/t < 0.5) parts of the organoid for WT (same as Fig. 2f) and LIS1+/−. Kolmogorov-Smirnov test was used to compare the WT and LIS1+/- distributions. Scale bars are 200μm (a,b), 50μm (c) and 5μm (n). Error bars represent s.e.m. Asterisks represent statistical significance (* p < 0.05, ** p < 0.01,***p < 0.001).
We compared the folding of wild-type and mutant organoids (days 6-8, Fig. 5d-g, N = 5 organoids). For the wild-type organoids, the mean wrinkle wavelength was similar to the mean thickness during wrinkling onset, < λ > ≈< t >≈ 100μm (Fig. 5d,e). We observed Gaussian distributions for the organoid thickness, σt = 24μm, and for the wavelength, σλ = 39μm. In the LIS1 +/− mutant we observed broadening of the Gaussian distribution for t, with no significant change in mean value (Fig. 5f). In contrast, the average wavelength < λ > ≈ 170μm was significantly longer (p < 0.001), and the distribution was wider σλ = 70μm (Fig. 5g). Notably, the LIS1 +/− organoids exhibited linear scaling of wavelength with thickness, similar to the wild-type organoids, but with an increased prefactor, λLIS1 ∝ 1.9t (Fig. 5h). Beyond t > 100μm, the wavelength saturated at a maximal value, λLIS1 = 200 ± 15μm.
The wrinkling morphology was further analyzed by measuring the surface tangent angle θ(r) of developed organoids (days 16-18, Fig. 5i,j). We computed the organoid curvature by averaging the tangent derivative along the organoid perimeter < ∂rθ(r) > (Fig. 5j, Fig. S16-18, N=9-11). We observed a significant reduction in the LIS1 +/− organoid curvature, for all three clones, in comparison to the wild-type. We further computed the tangent correlation function, < t̂(r)t̂(r + Δr) >=< Cos(Δθ(r)) > (Fig. 5i). The correlation exhibited sharp linear decrease at short distances, < Cos(Δθ(r)) >≈ 1 − r/lp, with persistence length lp = 148 ± 11μm (wild-type), and lp = 306 ± 11μm (mutant). At long distances we observed three correlation peaks for the wild-type organoids, r = 200μm, 380μm, 520μm. The peaks indicated a periodic structure λθ = 180 ± 15μm ≈< W >⋅< λ > consistent with the measured wavelength, < λ > ≈ 110, and the maximal wrinkling index of < W >= 2.1. In contrast, the mutant organoid did not exhibit correlation peaks. The main features of the wild-type correlation function persisted in an analysis of a healthy human brain, which exhibited multiple wavelength peaks (Fig. 5i, Inset). Analysis of a lissencephalic brain, resulted in a longer persistence length, and a lack of correlation peaks. The aberrant LIS1 +/− expression of ECM and cytoskeleton related genes, together with the modified wavelength scaling prefactor, suggested that the LIS1 +/− cells have different elastic properties than wild-type cells. To test this, individual cells were analyzed using atomic force microscopy (AFM) as embryonic stem cells (ES), and as neuronal progenitors (NP). The wild-type cells exhibited elastic modulus and similar to previous measurements48 (Fig. 5k, Figs. S19,20). Remarkably, the LIS1 +/− mutant cells were significantly softer than the wild-type cells by ~2.5 fold, (p < 0.005) and (p < 0.05). Thus, our findings indicate that the LIS1 +/− modified wrinkling stems from “softening” of the cytoskeleton, similar to the blebbistatin treatment (Fig. 4b), and is analogous to reduction of the bulk elastic modulus, EB, in the polymer gel model17.
Furthermore, LIS1 +/− organoids exhibited significantly reduced velocities during inward motion of the nuclei, νLis = −9.6 ± 1, compared to wild-type, νWT = −6.8 ± 0.5, p < 0.05 (Fig. 5l, Movie S2). Additionally, the LIS1 +/− nuclear swelling profile was different from that of the wild-type, p < 10−3 (Fig. 5m,n). The reduction in differential growth may contribute to the reduction in LIS1+/- wrinkling. Finally, the response of LIS1 +/− organoids to the drug treatments was similar to that observed in the wild-type organoids (Fig S21).
Taken together, our data suggest that the organoid wrinkling is driven by a mechanical instability, which is universal for differentially swelling materials. The increased growth in the organoid outer regions, together with the actively contracting organoid inner surface, both contribute to the emergent wrinkling pattern. The presented three-dimensional organization of the organoid, gene expression data, and nuclei motion, indicate that the on-chip organoid approach successfully mimics the early developing cortex.
We next compare the organoid wrinkling to cortical gyrification. The periodic folding morphology and temporal dynamics are remarkably similar to MRI images of fetal brains18,20,49. The nuclei motion provides a differential growth mechanism, analogous to the way radial neuronal migration may play a role in brain folding50. However, cortical gyrification occurs around week 30 of embryonic development, when the cell population is largely composed of neurons. This is in contrast to the organoids, which are composed of neuronal progenitors, and model an earlier developmental time point. Thus, while organoid wrinkling results from differential growth of progenitor cells, and includes cell-cycle dependent changes in nuclear size, cortical folding are driven by crowding of non-dividing neurons which are migrating towards the surface of the brain. In addition, apical contraction seems to play a significant role in organoid wrinkling, which has not been observed in cortical folding, and may be opposed by the hydrostatic pressure of the cerebral fluid in-vivo. Finally, a LIS1 +/− mutation led to reduction in organoid wrinkling, involving changes in nuclear motion, ECM and cytoskeleton. However, smooth brains of LIS1 +/− patients are widely accepted to reflect a defect in neuronal migration, a process not modelled here. Thus, while the organoid wrinkling is remarkably analogous to cortical gyrification, there are significant biological and physical differences. Overall, the on-chip organoids provide a novel model system to study the physics and biology of early human brain development, and reveal the mechanism of wrinkling in a living system.
Methods
Device fabrication
The device was fabricated on a commercial 6cm polystyrene tissue culture dish (Falcon). 11 holes of diameter 1.5mm were drilled at the dish bottom (Fig. S1, Step 1). A UV-curable adhesive was spread on the bottom surface using a sharp blade (NOA81, Norland Products, Fig. S1, step 2). A semi-permeable polycarbonate membrane (Whatman® Nuclepore Track-Etched Membranes, pore diameter 0.1μm) was placed on the adhesive layer (Fig. S1, step 3). The device was cured under UV-light for 2 minutes (365nm 20W LED, Shenzen Yonton Opto). The membrane covered 9 drilled holes, thus leaving 2 holes uncovered, which will serve as inlets. A circular Polydimethylsiloxane (PDMS) stamp of 150μm thickness and 20 mm diameter was prepared using a metal mold of the same dimensions. The PDMS stamp was placed on a 24x24 mm2 coverslip (Fig. S1, Step 4). The UV-curable adhesive was flown between the coverslip and PDMS by capillary, thus forming a spacer of thickness 150μm (Fig. S1, Step 5). The spacer was half-cured by UV-exposure, and the PDMS was pealed off (Fig. S1, Step 6).
Human ESC aggregates
Human embryonic stem cells were cultured on Matrigel coated plates in Naïve Human Stem-Cell Media (NHSM,Table S1) as described51 (Fig. S1, Step 7). Cells were dissociated using Trypsin-EDTA (0.05%, ThermoFisher), and diluted in serum containing media to inhibit trypsin activity. Cells were centrifuged and re-suspended in NHSM with 40μM Rho Kinase inhibitor Y − 27632. The cells were counted and diluted to a final concentration of 2 ⋅ 104 cells/ml. About 600 cells (30μl) were dispensed into ultra-low cell-attachment 96-plate (Fig. S1, Step 8, S-BIO, Hudson, NH, USA). Within several hours cells aggregated at the bottom of the wells (Fig. S1, Step 9). After 24 hours, 150μl of Neural Induction Media (Table S1) was added.
Device assembly
After 48-72 hours cell aggregates were collected by a pipette and placed on the fabricated culture dish (Fig. S1, Step 10). Nine aggregates were placed on top of the membrane covered holes. The fabricated coverslip was placed on top of the aggregates, thus sealing the device (Fig. S1, Step 11). The device was inverted, and the UV-adhesive spacer was fully cured using a mask to protect the cell aggregates. The device was then filled with media and kept in a cell incubator at 37°C and 5% CO2 (Fig. S1, Step 11). Media was exchanged every other day. Several days after Matrigel injection, the homogenous and isotropic cell aggregate self-organized into a spherical shell structure surrounding a small cavity (lumen) (Fig. 1e, Day 3, Fig. S2). Each organoid contained a single or several lumens.
Hydrogel embedment
Five days following the device assembly, collagen-laminin based hydrogel (100% Matrigel) was injected into the device, and thus embedded the aggregates in a gel environment (Fig. S1, Step 13). First, the device was dried. Then, a drop of cool liquid Matrigel was placed on one of the inlets, and a tissue paper was placed on the second inlet. The Matrigel entered the fabricated compartment by capillary force. The device was inserted into the incubator for 25 min for gelification. Finally, Neural Differentiation media (Table S1) supplemented with EGF (20ng/ml) and FGF2 (20ng/ml) was added to the device25 (Fig. S1, Step 14). The device was kept in the cell incubator and ready for imaging. Media was exchanged every other day.
PEG Coating
A ~10μm thick polyethylene glycoel (PEG) hydrogel can be assembled on the glass cover slip to exclude surface adhesion, or control surface stiffness52. Glass slides were plasma treated (1min, 100W, 1bar Argon, 2bar Oxygen), and immediately coated with TMSMA solution (10ml ethanol, 300μL 1:10 glacial acetic acid:ddw, and 0.5ml 3-(trimethoxysilyl) propyl methacrylate Sigma, 440159), incubated for 1hr, double rinsed with ethanol, dried with Nitrogen and stored in the dark. PEG mixture: Photoinitiator stock solution was prepared as 100mg 2,2-Dimethoxy-2-phenylacetophenone (Sigma, 196118) to 1ml 70% ethanol. PEG solution was prepared as 200mg PEG-6000 diacrylate (Sigma, 701963) to 1ml PBS. The photoinitiator was mixed into the PEG solution to a final 0.1% concentration and stored at 4C. Gel curing: A 2.5 μL drop of PEG mixture was placed on the surface treated slide, and carefully closed from the top with a 18mm diameter circular coverslip. The PEG mixture uniformly spread between the two surfaces, thus reaching an average thickness of ~10μm. The solution was cross-linked by exposure to UV light. The 18mm coverslip is removed gently using tweezers. The PEG coated coverslip is washed and incubated over night with sterile PBS. Before device assembly the slide is dried with nitrogen, and the PEG gel is clearly visible as an opaque disc on the clear coverslip.
Lipidure® Coating
Glass slides and membrane assembled petri-dish were coated with antiadhesive Lipidure®-CM5206 (NOF CORPORATION) according to manufacture protocol53. A 0.5wt% solution was prepared by dissolving 0.05gr Lipidure powder to 10ml ethanol. A ~200ul Lipidure solution was placed on the coverslip/membrane surface for 1-2 min. The solution was aspirated. The process was repeated twice. The surfaces were dried with nitrogen followed by 50°C oven for 1hr.
Imaging and analysis
Imaging was carried out in a spinning disk confocal microscope based on an OLYMPUS IX83 inverted microscope, VisiScope CSU-W1-T1 confocal system (Visitron systems, Germany) and an sCMOS 4.2 MPixel camera. Imaging was performed using VisiView software. Image and Data analysis were carried out in FIJI and Matlab 2016b.
Cell line and CRISPR genome editing
In this work, we used early passages of NIH approved embryonic stem cell line NIHhESC-10-0079 (WIBR3). Human ES cells were grown in optimal conditions as described51. Three isogenic mutant cell-line clones were generated, by partial deletion of the LIS1 gene from one allele of the wild-type cell-line. To insert the LIS1 mutation we used the Clustered regularly interspaced short palindromic repeats (CRISPR) genome editing method. We used double Cas9 nickase to reduce the probability of off-targets effects. The two guide-RNA sequences: GGACGGGGAATCCATTCTTTTGG and AATATGCATTGAGTGGTCACAGG were designed according to http://crispr.mit.edu/,and targeted the 4th coding exon region of LIS1 gene. The corresponding oligonucleotides were cloned into pX335 vector, which contains an expression vector for mutant Cas9 (nickase). Cells were transfected using electroporation. In addition to the plasmid encoding the gRNA and Cas9, tracer amounts of a GFP expression vector was added. Three days after transfection, the cells were subject to fluorescence-activated cell sorting (FACS) and plated at a density of 2000 cells per plate, allowing for the growth of single-cell derived colonies. Colonies were picked, and screened by RNA and protein levels. Since LIS1 knock-outs are embryonic lethal we expected only LIS1 +/- heterozygous deletions54. Three LIS1 +/- were used in the study: 10F, 9G, 45 respectively referred to as clones 1,2,3 in main text. Reduced LIS1 protein levels were verified by western blot (Fig. S16). Cell lines were regularly checked for mycoplasma contamination. Spectral karyotyping test was performed to cell lines WIBR3 (100% normal metaphases, N=10), and LIS1 +/− 10F (12 normal metaphases, 1 metaphase with chromosome 17 duplication, total N=13).
Cell electroporation and fluorescence reporters
Human embryonic stem cells were stably labeled with a fluorescent H2B-mCherry reporter, labeling chromosomes, and a Lifeact-GFP reporter labeling actin. Cell transfection was carried out in a NEPA21 electroporation system according to the company protocol (Nepa Gene, Japan). To ensure stable expression over the whole cell lineage we used the PiggyBac transposase method, which allows DNA integration into the cell genome at random sites55,56. We co-electroporated pCAG:H2B-mCherry and pCAG:LiveAct-GFP plasmids together with a pCAG-PBase plasmid expressing the PiggyBac transposase. The reporter plasmids carried inverted terminal repeat sequences (ITRs), recognized by PiggyBac transposase, before and after the promoter:gene sequence. Following electroporation cells were plated, passaged once after several days and selected using FACS.
AFM sample preparation
Atomic force microscopy (AFM) experiments were carried out on cells cultured on 5cm tissue-culture dishes. Cells were cultured in two conditions: (i) as embryonic stem cell colonies (ES)51 and (ii) differentiated to neuronal progenitors (NP) as described57. For the ES sample, ES cells were plated onto a Matrigel coated plate, cultured for about a week in NHSM media (Table S1) and taken for AFM analysis. For the NP sample, ES cells were plated onto a 10cm plate coated with Matrigel, cultured until they reached confluency, and then transferred to a 5cm plate. After 24hrs, the NHSM media was replaced with Neuronal Differentiation media (Table S1) supplemented with TGFB inhibitor (SB431542, 5μM), Noggin (0.5 μg/ml) and Vitamin A (present in B27 supplement, Table S1). Cells were cultured for 14 days, with daily media exchange, and were taken for AFM analysis.
AFM experiments
AFM studies were performed using a JPK Nanowizard 3 and Nanosensors qp-BioAC probes (probe 1, force constant of approximately 50 pN/nm) in QI™ mode. This mode allows acquisition of topographic images simultaneously with mechanical data (e.g., stiffness and adhesion). It also allows collection of force/distance curves at each pixel which can subsequently be converted to elastic modulus after calibration of optical lever sensitivity and cantilever spring constant, by applying a contact mechanical model. Initially, regions of approximately 20 x 20 μm2 were scanned at moderate resolution to locate the positions of the cells and their boundaries. Then, zoomed areas centering on selected cells were acquired, approximately 5 x 5 μm2 at 32 x 32 pixels together with the pixel-resolved force curves. Ultimate force and pull-back distance were tuned to ensure penetration into the cell of nominally 300-800 nm and complete disengagement from the cell for each cycle. The tip speed was approached to surface at a speed of 50 μm/s. The traces showed no evidence of significant adhesion and had good repeatability in a single location, and were analyzed using a Herzian model presuming Poisson ratio of 0.5, and a conical probe with opening half-angle of 20 degrees. For analysis, force curves from the cell were selected using the maps, avoiding cell-cell border regions. Several thousand force curves were combined from 7-10 different cells for the analysis of each cell-line (Fig. 5k, Figs. S19-20).
Cytoskeleton drug experiments
Cytoskeleton drug experiments were carried out directly in the organoid device. For the long-term experiments (Fig. 4a,b) 5μM blebbistatin was added during Days 6-10 after Matrigel embedding. In the short-term experiments (Fig. 4c-e) drugs were added to the media, and the organoids were incubated for 3hrs in a cell incubator. Organoids were imaged before and after the addition of the drugs. Drugs were used at the following concentrations: Rock Inhibitor (Y-27632) 20μM, blebbistatin 50μM and nocodazole 10μM.
Laser microdissection experiments
Laser Microdissection experiments were carried out directly on the organoid device, using the Palm MicroBeam Zeiss system. The system works with a pulsed 337nm UV Nitrogen Laser. The irradiation area was chosen using Lifeact fluorescence imaging with a 10X objective (Fig. S14, green line). Laser irradiation was performed using a 40X air objective, in Laser Pressure Catapulting (LPC) mode. In this mode, the laser pulses are localized in discrete locations within the irradiation area (Fig. S14, blue dots). The following parameters were used: 60% power, 100% focus, and speed 1. The organoids were fluorescently imaged before and after the laser microdissection, which lasted about a minute.
Immunostaining
Immunostaining was carried out directly in the organoid device. Organoids were fixed with PFA 2.5% for two hours, washed 3 times in PBS and permeabilized using PBS-triton 0.1% for 15 minutes, followed by 30 minutes blocking (30% Horse serum, 10% Fetal Calf Serum, 0.1% Triton X-100 in PBS). The organoids were incubated for two hours of primary anti-body diluted in blocking solution (Rabbit anti-PAX6 1:200 Covance PRB-278P, Mouse anti-NeuN 1:200 MILLIPORE mab377), washed in PBS, incubated for twenty minutes with secondary anti-body 1:200 in blocking, followed by incubation with 4',6-diamidino-2-phenylindole (DAPI) for 10 min and PBS washing. Organoid devices were imaged following the immunostaining.
Contour analysis of human brains
Contour analysis of a healthy human brain (Fig. 5i, inset) was done on a single post-mortem section image (Brain Biodiversity Bank, Michigan State University, https://msu.edu/~brains/, coronal section #2000). Analysis of lissencephalic brain was done on patient MRI scan image (Courtesy of Dr. Nadia Bahi-Buisson, French Institute of Health and Medical Research).
Bulk RNA-Seq Library Construction and Sequencing
Total RNA from was extracted using the RNeasy Mini kit (Qiagen) under manufacturer protocols. RNA was extracted at three time points: (i) human embryonic stem cells, (ii) organoids at days 5 and (iii) 12 after Matrigel embedment. Three repeats were taken for each time point. Organoid RNA was extracted directly from the device, with total of 20-40 organoids for each repeat. RNA concentration and integrity were measure using Nanodrop (Thermo Scientific) and an Agilent Tapestation, respectively. Libraries were prepared from 50 ng of total RNA using Bulk MARS-seq, a modified version of directional gene expression through 3’ end developed for single-cell RNA-seq (MARS seq), to produce expression libraries58. The quality of the libraries was assessed by tapestation and qPCR, and high-quality libraries were sequenced by the Illumina NextSeq 500 sequencer to obtain single read of 75bp.
Transcriptome Assembly and analysis
Raw data files were processed by removing the adapters from the raw sequencing reads using Cutadapt59 after quality-assessment of FASTQ files using the FastQC tool60. Reads were then aligned to the reference genome GRCh37/hg38 using STAR61. Differential expression was tested by fitting raw counts on a negative binomial distribution by using DESeq262. Genes were sorted according to their log2-transformed fold-change values after shrinkage in DESeq2. The clusters of pathways that shared many leading-edge genes were detected using GeneAnalytics63, and manually curated these clusters to elucidate the important phenotype-associated pathway groups visualized on the heatmaps. Hierarchical clustering was performed using R software. First, all the differentially expressed genes for a genotype and/or developmental time point were identified using the methods listed above and clustering was subsequently performed using the hclust and dendsort package in R. Gene set enrichment analysis (GSEA) was carried out using the R package edgeR64, and ROAST65 function from the R package Limma (Fig. S5). Differentially expressed genes were manually divided to categories of radial glia and neuronal categories according to previous works66–73 (Fig. S6).
Supplementary Material
Acknowledgements
We are grateful for the help of Sergey Viukov, Talia Levy, Tamar Sapir, and for fruitful discussions with Sam Safran, Alexandra Tayar and Roy Bar-Ziv from the Weizmann Institute of Science. Device fabrication was carried out with Alex Jahanfard, Laser microdissection with Yael Fried, RNA sequencing and analysis with advise of Hadas Keren-Shaul, Refael Kohen, and Tsviya Olender. Plasmid gifts from Michael Davidson, Florida State University and Joseph LoTurco, University of Connecticut. MRI scans were provided by Nadia Bahi-Buisson, French Institute of Health and Medical Research. OR is the incumbent of the Bernstein-Mason Chair of Neurochemistry. EK is a Koshland fellow. The research has been supported by the Legacy Heritage Biomedical Program of the Israel Science Foundation (grant no. 2041/16), ERA-NET Neuron with support of the IMOH (grant no. 3-0000-12276), European Cooperation on Science and Technology (COST Action CA16118), Weizmann-FAPESP supported by a research grant from Sergio and Sonia Lozinsky, Nella and Leon Benoziyo Center for Neurological Diseases, Jeanne and Joseph Nissim Foundation for Life Sciences Research, Wohl Biology Endowment Fund, Lulu P. & David J. Levidow Fund for Alzheimers Diseases and Neuroscience Research, the Helen and Martin Kimmel Stem Cell Research Institute, the Kekst Family Institute for Medical Genetics, the David and Fela Shapell Family Center for Genetic Disorders Research. J.H.H. is a New York Stem Cell Foundation (NYSCF)–Robertson Investigator and is supported by research grants from the European Research Council (ERC-CoG2016 CellNaivety), Flight Attendant Medical Research Council (FAMRI), Israel Science Foundation Morasha program, Nella and Leon Benoziyo Center for Neurological Diseases.
Footnotes
Data availability. The data that support the plots within this paper and other findings of this study are available from the corresponding author upon request. RNA sequencing data was submitted to GEO (GSE106821).
Author Contributions
E.K., A.K., and O.R. planned and conducted experiments. S.C. designed and conducted AFM experiments. J.H. planned and assisted in hES related experiments. The manuscript was prepared with inputs by all the authors.
References
- 1.Hannezo E, Prost J, Joanny J-F. Mechanical Instabilities of Biological Tubes. Phys Rev Lett. 2012;109:18101. doi: 10.1103/PhysRevLett.109.018101. [DOI] [PubMed] [Google Scholar]
- 2.Shraiman BI. Mechanical feedback as a possible regulator of tissue growth. Proc Natl Acad Sci U S A. 2005;102:3318–23. doi: 10.1073/pnas.0404782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein Y, Efrati E, Sharon E. Shaping of Elastic Sheets by Prescription of Non-Euclidean Metrics. Science (80- ) 2007;315 doi: 10.1126/science.1135994. [DOI] [PubMed] [Google Scholar]
- 4.Savin T, et al. On the growth and form of the gut. Nature. 2011;476:57–62. doi: 10.1038/nature10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HY, Varner VD, Nelson CM. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development. 2013;140:3146–3155. doi: 10.1242/dev.093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- 7.Taber LA. Morphomechanics: transforming tubes into organs. Curr Opin Genet Dev. 2014;27:7–13. doi: 10.1016/j.gde.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. 2014;15:217–32. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–94. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 10.Mota B, Herculano-Houzel S. Cortical folding scales universally with surface area and thickness, not number of neurons. Science (80- ) 2015;349:74–77. doi: 10.1126/science.aaa9101. [DOI] [PubMed] [Google Scholar]
- 11.Reiner O, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–21. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 12.Reiner O, Sapir T. LIS1 functions in normal development and disease. Curr Opin Neurobiol. 2013;23:951–956. doi: 10.1016/j.conb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Groenewold J. Wrinkling of plates coupled with soft elastic media. Phys A Stat Mech its Appl. 2001;298:32–45. [Google Scholar]
- 14.Tanaka T, et al. Mechanical instability of gels at the phase transition. Nature. 1987;325:796–798. [Google Scholar]
- 15.Bowden N, et al. Spontaneous formation of ordered structures in thin films of metals supported on an elastomeric polymer. Nature. 1998;393:146–149. [Google Scholar]
- 16.Cerda E, Mahadevan L. Geometry and physics of wrinkling. Phys Rev Lett. 2003;90:74302. doi: 10.1103/PhysRevLett.90.074302. [DOI] [PubMed] [Google Scholar]
- 17.Dervaux J, Couder Y, Guedeau-Boudeville MA, Ben Amar M. Shape transition in artificial tumors: From smooth buckles to singular creases. Phys Rev Lett. 2011;107:1–4. doi: 10.1103/PhysRevLett.107.018103. [DOI] [PubMed] [Google Scholar]
- 18.Tallinen T, Chung JY, Biggins JS, Mahadevan L. Gyrification from constrained cortical expansion. Proc Natl Acad Sci U S A. 2014;111:12667–72. doi: 10.1073/pnas.1406015111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tallinen T, et al. On the growth and form of cortical convolutions. Nat Phys. 2016;12:588–593. [Google Scholar]
- 20.Budday S, Raybaud C, Kuhl E. A mechanical model predicts morphological abnormalities in the developing human brain. Sci Rep. 2014;4:5644. doi: 10.1038/srep05644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutzman JH, Graeden EG, Lowery LA, Holley HS, Sive H. Formation of the zebrafish midbrain-hindbrain boundary constriction requires laminin-dependent basal constriction. Mech Dev. 2008;125:974–83. doi: 10.1016/j.mod.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecuit T, Lenne P-F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–44. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 23.Eiraku M, et al. Self-Organized Formation of Polarized Cortical Tissues from ESCs and Its Active Manipulation by Extrinsic Signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paşca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clevers H. Modeling Development and Disease with Organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 27.Bershteyn M, et al. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell. 2017;0 doi: 10.1016/j.stem.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morel M, Galas J-C, Dahan M, Studer V. Concentration landscape generators for shear free dynamic chemical stimulation. Lab Chip. 2012;12:1340–6. doi: 10.1039/c2lc20994b. [DOI] [PubMed] [Google Scholar]
- 30.Miyata T, Okamoto M, Shinoda T, Kawaguchi A. Interkinetic nuclear migration generates and opposes ventricular-zone crowding: insight into tissue mechanics. Front Cell Neurosci. 2014;8:473. doi: 10.3389/fncel.2014.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hébert JM, Fishell G. The genetics of early telencephalon patterning: some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dantas TJ, Carabalona A, Hu DJK, Vallee RB. Emerging roles for motor proteins in progenitor cell behavior and neuronal migration during brain development. Cytoskeleton. 2016;73:566–576. doi: 10.1002/cm.21293. [DOI] [PubMed] [Google Scholar]
- 33.Tsai J-W, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170 doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziv O, et al. Quantitative Live Imaging of Human Embryonic Stem Cell Derived Neural Rosettes Reveals Structure-Function Dynamics Coupled to Cortical Development. PLOS Comput Biol. 2015;11:e1004453. doi: 10.1371/journal.pcbi.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–77. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiner O. LIS1 and DCX: Implications for Brain Development and Human Disease in Relation to Microtubules. Scientifica (Cairo) 2013;2013 doi: 10.1155/2013/393975. 393975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapir T. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faulkner NE, et al. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 40.Feng Y, et al. LIS1 Regulates CNS Lamination by Interacting with mNudE, a Central Component of the Centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- 41.Niethammer M, et al. NUDEL Is a Novel Cdk5 Substrate that Associates with LIS1 and Cytoplasmic Dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 42.Huang L, Seker E, Landers JP, Begley MR, Utz M. Molecular Interactions in Surface-Assembled Monolayers of Short Double-Stranded DNA. Langmuir. 2010;26:11574–11580. doi: 10.1021/la100860d. [DOI] [PubMed] [Google Scholar]
- 43.Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE Induce a Persistent Dynein Force-Producing State. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fietz SA, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc Natl Acad Sci U S A. 2012;109:11836–41. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–70. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 48.Guz N, Dokukin M, Kalaparthi V, Sokolov I. If Cell Mechanics Can Be Described by Elastic Modulus: Study of Different Models and Probes Used in Indentation Experiments. Biophys J. 2014;107:564–575. doi: 10.1016/j.bpj.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gholipour A, et al. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Sci Rep. 2017;7:476. doi: 10.1038/s41598-017-00525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reillo I, de Juan Romero C, García-Cabezas MÁ, Borrell V. A Role for Intermediate Radial Glia in the Tangential Expansion of the Mammalian Cerebral Cortex. Cereb Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 51.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 52.Cuchiara MP, Allen ACB, Chen TM, Miller JS, West JL. Multilayer microfluidic PEGDA hydrogels. Biomaterials. 2010;31:5491–5497. doi: 10.1016/j.biomaterials.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 53.Kim J-Y, et al. 3D spherical microtissues and microfluidic technology for multi-tissue experiments and analysis. J Biotechnol. 2015;205:24–35. doi: 10.1016/j.jbiotec.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 54.Hirotsune S, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–9. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 55.Wu SC-Y, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A. 2006;103:15008–13. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siddiqi F, et al. Fate mapping by piggyBac transposase reveals that neocortical GLAST+ progenitors generate more astrocytes than Nestin+ progenitors in rat neocortex. Cereb Cortex. 2014;24:508–20. doi: 10.1093/cercor/bhs332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y, Kirwan P, Smith J, Robinson HPC, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–86. doi: 10.1038/nn.3041. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–9. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10. [Google Scholar]
- 60.A S. FastQC: a quality control tool for high throughput sequence data. 2010 at < http://www.bioinformatics.babraham.ac.uk/projects/fastqc>.
- 61.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ben-Ari Fuchs S, et al. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. Omi A J Integr Biol. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu D, et al. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 67.Lui JH, et al. Radial glia require PDGFD–PDGFRβ signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Y, et al. Medial ganglionic eminence–like cells derived from human embryonic stem cells correct learning and memory deficits. Nat Biotechnol. 2013;31:440–447. doi: 10.1038/nbt.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson MB, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Close JL, et al. Single-Cell Profiling of an In Vitro Model of Human Interneuron Development Reveals Temporal Dynamics of Cell Type Production and Maturation. Neuron. 2017;93:1035–1048.e5. doi: 10.1016/j.neuron.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Camp JG, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112:15672–7. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Onorati M, et al. Molecular and functional definition of the developing human striatum. Nat Neurosci. 2014;17:1804–1815. doi: 10.1038/nn.3860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.