The title compound, an hemisynthetic product, was obtained by the tosylation reaction of the naturally occurring meroterpene p-methoxythymol 1.
Keywords: crystal structure, organic synthesis, tosylation of alcohols, drug synthesis
Abstract
The title compound, C18H22O4S, an hemisynthetic product, was obtained by the tosylation reaction of the naturally occurring meroterpene p-methoxythymol. The molecule comprises a tetrasubstitued phenyl ring linked to a toluenesulfonate through one of its O atoms. In the crystal, C—H⋯O and C—H⋯π interactions link the molecules, forming a three-dimensional network.
Chemical context
Tosylation of alcohols is an important transformation in organic synthesis. This transformation is usually achieved with p-toluene sulfonyl chloride, which is very reactive (Greene & Wuts, 1999 ▸; Yoshida et al., 1999 ▸). Tosylate is an important functional group in organic synthesis as it makes a good leaving group (Wagner & Zokk, 1955 ▸; Sandler & Karo, 1983 ▸). Indeed, tosylates are used as intermediates in the synthesis of several drugs (Kim et al., 1995 ▸; Morgan et al., 1997 ▸). Furthermore, they have also been found to possess important biological activities (Kacem et al., 2002 ▸; Kaleemullah et al., 2012 ▸).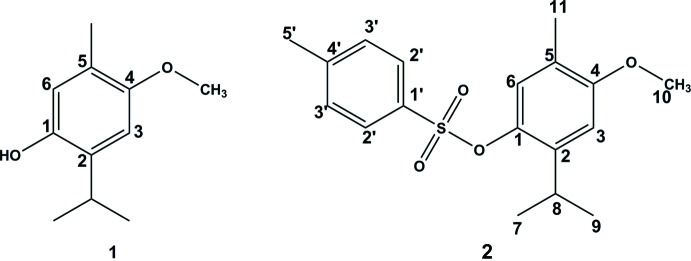
The hemisynthesis of 2-isopropyl-4-methoxy-5-methylphenyl 4-methylbenzenesulfonate 2 from naturally occurring p-methoxythymol 1 was undertaken with the aim of preparing meroterpenic tosylate. X-ray single-crystal structure analysis allowed its full structure to be confirmed unambiguously.
Structural commentary
Compound 2 is built up from a tetrasubstituted phenyl ring linked to a toluenesulfonate unit through one of its oxygen atoms (Fig. 1 ▸). The two phenyl rings form a dihedral angle of 60.03 (9)°. Atoms S1 and C5′ are coplanar with the C1′–C6′ phenyl ring, their distances from the plane being 0.057 (3) and 0.031 (3) Å, respectively. Considering the connected atoms of the four substituents on the C1–C6 phenyl ring, three of them O2, C8 and C11 are roughly in the plane of the phenyl ring, deviating by only 0.011 (3), 0.014 (3) and 0.012 (3) Å, respectively, from the mean plane, whereas atom O1 is displaced slightly out of the plane by 0.101 (3) Å. This slight distortion might be related to the occurrence of a weak C—H⋯π interaction between the C9 atom and the centroid Cg2 of the C1′–C6′ phenyl ring (Table 1 ▸).
Figure 1.
Molecular view of compound 2 with the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the phenyl rings C1–C6 and C1′–C6′, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C3′—H3′⋯O1i | 0.95 | 2.67 | 3.610 (2) | 169 |
| C5′—H5′3⋯O3ii | 0.98 | 2.66 | 3.589 (3) | 159 |
| C7—H7A⋯O4iii | 0.98 | 2.72 | 3.693 (3) | 175 |
| C10—H10B⋯O3iv | 0.98 | 2.61 | 3.317 (3) | 130 |
| C10—H10A⋯O4iii | 0.98 | 2.70 | 3.521 (3) | 142 |
| C5′—H5′2⋯Cg2v | 0.98 | 2.84 | 3.706 | 148 |
| C11—H11B⋯Cg1iv | 0.98 | 2.75 | 3.635 | 150 |
| C9—H9A⋯Cg2 | 0.98 | 2.70 | 3.5373 | 144 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Supramolecular features
In the crystal, pairs of molecules are linked though C—H⋯O interactions (Table 1 ▸), forming pseudo-dimer arranged around inversion centers (Fig. 2 ▸). Further C—H⋯O hydrogen bonds and C—H⋯π interactions (Table 1 ▸, Fig. 2 ▸) lead to the formation of a three-dimensional network.
Figure 2.
Partial packing view showing the C—H⋯O and C—H⋯π interactions (dotted lines). Only H atoms involved in hydrogen bonding are shown.
Database survey
A search of the Cambridge Structural Database (CSD, version 5.38, last update May 2017; Groom et al., 2016 ▸) for a tosylate fragment bearing an organic substituent on one of its oxygen atoms revealed only three hits. Two of these compounds are closely related to compound 2. The first, 5-bromo-2,3-dimethylphenol-1-(4-methylphenylsulfonyloxy)benzene (KAWDAN; Niestroj et al., 1998 ▸), is built up from a tosylate attached to a phenyl ring substituted by two methyl groups and one bromine atom whereas the second, tetramethyl-p-phenylene p-ditoluenesulfonate (TMPDTS; Wieczorek et al., 1975 ▸), is built up from a tetramethyl-substituted phenyl ring attached to two tosylate units. A comparison of selected distances in compound 2 with those of two structures reveals that the geometries are very similar for all three compounds (Table 2 ▸). The most marked difference is the dihedral angle between the phenyl rings, 60.03 (9)° in 2 and 15.32 and 43.02° in KAWDAN and TMPDTS, respectively. The large dihedral angle in TMPDTS might be related to the occurrence of two bulky substituents on the central phenyl ring.
Table 2. Selected structural parameters of compound 2 compared with closely related structures.
| 2 | KAWDANa | TMPDTSb | |
|---|---|---|---|
| C1′—S1 | 1.749 (2) | 1.748 | 1.732 |
| S1—O1 | 1.597 (1) | 1.598 | 1.599 |
| O1—C1 | 1.428 (2) | 1.425 | 1.428 |
| C1′—S1—O1 | 104.76 (8) | 98.83 | 102.37 |
| S1—O1—C1 | 120.71 (11) | 116.07 | 119.84 |
| Dihedral angle | 60.03 (9) | 15.32 | 43.02 |
Synthesis and crystallization
In a 100mL flask, 430 mg (2.33mmol) of p-methoxythymol 1 were dissolved in 15 mL of pyridine and then 908 mg (4.66 mmol) of para-toluenesulfonyl chloride were added. The reaction mixture was heated to reflux for two h. The end of the reaction was controlled by TLC. The reaction mixture was washed with a hydrochloric acid solution (0.1 M) to neutral pH, extracted three times with ethyl ether (3 × 20 mL), dried over anhydrous Na2SO4 and concentrated under reduced pressure. The crude product was purified by silica gel column chromatography using hexane/ethyl acetate (94:6) as eluent to give 360 mg (1.07 mmol, 46% yield) of 2-isopropyl-4-methoxy-5-methylphenyl 4-methylbenzenesulfonate 2. X-ray quality colourless crystals were obtained by slow evaporation of a petroleum ether solution of the title compound.
NMR data for compound 2: 1H NMR (300 MHz, CDCl3): 6.66 (s, H-6), 6.84 (s, H-3), 3.45 (sept, H-8), 1.09 (d, H-9,H-10), 2.13 (s, H-7), 3.72 (s, H-10), 7.33 (d, H-3′), 7.75 (d, H-2′), 2.43 (s, H-5′) ppm. 13C NMR (75 MHz, CDCl3): 156.4 (C-1), 124.0 (C-2), 107.5 (C-3), 145.2 (C-4), 139.8 (C-5), 124.1 (C-6),15.7 (C-7), 26.8 (C-8), 23.1 (C-9, C-10), 55.4 (OCH3), 145.2 (C-1′), 129.7 (C-2′), 128.4 (C-3′), 139.7 (C-4′), 21.5 (C-5′) ppm.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. All H atoms were fixed geometrically and treated as riding with C—H = 1.0 (methine), 0.98 (methyl) or 0.95 Å (aromatic) with U iso(H) = 1.2U eq(CH and CH2) or U iso(H) = 1.5U eq(CH3).
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C18H22O4S |
| M r | 334.41 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 173 |
| a, b, c (Å) | 8.2226 (6), 14.5382 (9), 14.7230 (8) |
| β (°) | 100.020 (6) |
| V (Å3) | 1733.17 (19) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.20 |
| Crystal size (mm) | 0.35 × 0.25 × 0.10 |
| Data collection | |
| Diffractometer | Rigaku Oxford Diffraction Xcalibur Eos Gemini ultra |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) |
| T min, T max | 0.791, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18172, 3532, 2777 |
| R int | 0.044 |
| (sin θ/λ)max (Å−1) | 0.625 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.042, 0.110, 1.06 |
| No. of reflections | 3532 |
| No. of parameters | 213 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.34, −0.34 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989018003110/xu5919sup1.cif
Supporting information file. DOI: 10.1107/S2056989018003110/xu5919Isup3.cml
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018003110/xu5919Isup3.hkl
CCDC reference: 1825267
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank Cadi Ayyad University for financial support.
supplementary crystallographic information
Crystal data
| C18H22O4S | F(000) = 712 |
| Mr = 334.41 | Dx = 1.282 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.2226 (6) Å | Cell parameters from 4433 reflections |
| b = 14.5382 (9) Å | θ = 3.8–28.5° |
| c = 14.7230 (8) Å | µ = 0.20 mm−1 |
| β = 100.020 (6)° | T = 173 K |
| V = 1733.17 (19) Å3 | Box, colourless |
| Z = 4 | 0.35 × 0.25 × 0.10 mm |
Data collection
| Rigaku Oxford Diffraction Xcalibur Eos Gemini ultra diffractometer | 3532 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2777 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.044 |
| Detector resolution: 16.1978 pixels mm-1 | θmax = 26.4°, θmin = 3.1° |
| ω scans | h = −10→10 |
| Absorption correction: multi-scan (CrysAlis PRO; Rigaku OD, 2015) | k = −18→16 |
| Tmin = 0.791, Tmax = 1.000 | l = −18→18 |
| 18172 measured reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H-atom parameters constrained |
| wR(F2) = 0.110 | w = 1/[σ2(Fo2) + (0.0459P)2 + 0.814P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max < 0.001 |
| 3532 reflections | Δρmax = 0.34 e Å−3 |
| 213 parameters | Δρmin = −0.34 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. All H atoms attached to C atoms were fixed geometrically and treated as riding with C—H = 0.95 Å (aromatic), 0.98 Å (methyl), 1.0Å (methine) with Uiso(H) = 1.2Ueq(CH and C=CH) or Uiso(H) = 1.5Ueq(CH3). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.0729 (2) | 0.19454 (12) | 0.12387 (12) | 0.0244 (4) | |
| C1' | 0.2295 (2) | 0.11565 (13) | 0.32646 (12) | 0.0273 (4) | |
| C2' | 0.2423 (2) | 0.02563 (13) | 0.29895 (13) | 0.0319 (4) | |
| H2' | 0.150817 | −0.004394 | 0.262620 | 0.038* | |
| C2 | 0.2184 (2) | 0.20290 (12) | 0.08959 (12) | 0.0253 (4) | |
| C3' | 0.3907 (3) | −0.02031 (14) | 0.32514 (13) | 0.0351 (5) | |
| H3' | 0.400221 | −0.082269 | 0.306238 | 0.042* | |
| C3 | 0.2171 (2) | 0.16786 (13) | 0.00089 (12) | 0.0281 (4) | |
| H3 | 0.314049 | 0.172671 | −0.025740 | 0.034* | |
| C4 | 0.0786 (2) | 0.12650 (13) | −0.04881 (12) | 0.0283 (4) | |
| C4' | 0.5259 (2) | 0.02225 (14) | 0.37844 (12) | 0.0323 (4) | |
| C5 | −0.0681 (2) | 0.11891 (13) | −0.01288 (13) | 0.0293 (4) | |
| C5' | 0.6862 (3) | −0.02793 (16) | 0.40828 (15) | 0.0414 (5) | |
| H5'1 | 0.674385 | −0.091812 | 0.386905 | 0.062* | |
| H5'2 | 0.714962 | −0.026864 | 0.475724 | 0.062* | |
| H5'3 | 0.773618 | 0.002185 | 0.381667 | 0.062* | |
| C6 | −0.0682 (2) | 0.15452 (13) | 0.07405 (13) | 0.0284 (4) | |
| H6 | −0.166024 | 0.151593 | 0.100070 | 0.034* | |
| C6' | 0.5098 (2) | 0.11365 (15) | 0.40381 (13) | 0.0356 (5) | |
| H6' | 0.601735 | 0.144317 | 0.439007 | 0.043* | |
| C7 | 0.4269 (3) | 0.32911 (16) | 0.09104 (15) | 0.0451 (6) | |
| H7A | 0.459446 | 0.308452 | 0.033416 | 0.068* | |
| H7B | 0.520807 | 0.359505 | 0.129571 | 0.068* | |
| H7C | 0.334786 | 0.372516 | 0.077095 | 0.068* | |
| C7' | 0.3633 (2) | 0.16037 (14) | 0.37888 (13) | 0.0325 (4) | |
| H7' | 0.353664 | 0.222483 | 0.397271 | 0.039* | |
| C8 | 0.3736 (2) | 0.24634 (14) | 0.14252 (13) | 0.0310 (4) | |
| H8 | 0.349050 | 0.268405 | 0.203020 | 0.037* | |
| C9 | 0.5123 (3) | 0.17540 (18) | 0.16200 (16) | 0.0483 (6) | |
| H9A | 0.474804 | 0.122497 | 0.194085 | 0.072* | |
| H9B | 0.608770 | 0.203173 | 0.200759 | 0.072* | |
| H9C | 0.542309 | 0.155001 | 0.103658 | 0.072* | |
| C10 | 0.2226 (3) | 0.08648 (18) | −0.17038 (15) | 0.0526 (7) | |
| H10A | 0.261778 | 0.149089 | −0.178548 | 0.079* | |
| H10B | 0.203927 | 0.054579 | −0.229920 | 0.079* | |
| H10C | 0.305633 | 0.053107 | −0.126849 | 0.079* | |
| C11 | −0.2193 (3) | 0.07531 (15) | −0.06830 (14) | 0.0388 (5) | |
| H11A | −0.307373 | 0.073020 | −0.031237 | 0.058* | |
| H11B | −0.192880 | 0.012726 | −0.085726 | 0.058* | |
| H11C | −0.256290 | 0.111763 | −0.124059 | 0.058* | |
| O1 | 0.06333 (15) | 0.23611 (8) | 0.21065 (8) | 0.0269 (3) | |
| O2 | 0.07192 (18) | 0.09037 (10) | −0.13523 (9) | 0.0391 (4) | |
| O3 | −0.08697 (16) | 0.10974 (10) | 0.27106 (10) | 0.0378 (3) | |
| O4 | 0.02806 (17) | 0.24132 (10) | 0.36634 (9) | 0.0386 (4) | |
| S1 | 0.04212 (6) | 0.17449 (3) | 0.29760 (3) | 0.02896 (14) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0260 (9) | 0.0203 (9) | 0.0269 (9) | 0.0030 (7) | 0.0041 (7) | 0.0025 (7) |
| C1' | 0.0316 (9) | 0.0231 (10) | 0.0270 (9) | 0.0023 (8) | 0.0049 (7) | 0.0001 (7) |
| C2' | 0.0354 (10) | 0.0247 (10) | 0.0349 (10) | −0.0030 (8) | 0.0038 (8) | −0.0036 (8) |
| C2 | 0.0248 (9) | 0.0208 (9) | 0.0298 (9) | −0.0004 (7) | 0.0029 (7) | 0.0022 (7) |
| C3' | 0.0445 (12) | 0.0226 (10) | 0.0392 (11) | 0.0032 (9) | 0.0098 (9) | −0.0032 (8) |
| C3 | 0.0297 (9) | 0.0242 (10) | 0.0313 (9) | −0.0007 (8) | 0.0084 (7) | 0.0022 (8) |
| C4 | 0.0363 (10) | 0.0210 (10) | 0.0262 (9) | 0.0000 (8) | 0.0016 (7) | 0.0025 (7) |
| C4' | 0.0379 (11) | 0.0306 (11) | 0.0287 (9) | 0.0052 (9) | 0.0067 (8) | 0.0033 (8) |
| C5 | 0.0301 (9) | 0.0206 (10) | 0.0349 (10) | −0.0003 (8) | −0.0013 (8) | 0.0063 (8) |
| C5' | 0.0415 (12) | 0.0441 (13) | 0.0398 (11) | 0.0126 (10) | 0.0103 (9) | 0.0081 (10) |
| C6 | 0.0230 (9) | 0.0255 (10) | 0.0366 (10) | 0.0030 (8) | 0.0045 (7) | 0.0067 (8) |
| C6' | 0.0355 (11) | 0.0355 (12) | 0.0331 (10) | 0.0002 (9) | −0.0019 (8) | −0.0039 (9) |
| C7 | 0.0464 (13) | 0.0481 (14) | 0.0418 (12) | −0.0235 (11) | 0.0107 (10) | −0.0079 (10) |
| C7' | 0.0395 (11) | 0.0236 (10) | 0.0327 (10) | 0.0021 (8) | 0.0017 (8) | −0.0047 (8) |
| C8 | 0.0251 (9) | 0.0385 (12) | 0.0297 (9) | −0.0050 (8) | 0.0057 (7) | −0.0050 (8) |
| C9 | 0.0276 (10) | 0.0672 (17) | 0.0477 (13) | 0.0069 (11) | 0.0003 (9) | −0.0045 (12) |
| C10 | 0.0641 (16) | 0.0617 (16) | 0.0368 (12) | −0.0238 (13) | 0.0219 (11) | −0.0168 (11) |
| C11 | 0.0359 (11) | 0.0347 (12) | 0.0416 (11) | −0.0050 (9) | −0.0046 (9) | 0.0020 (9) |
| O1 | 0.0272 (6) | 0.0225 (7) | 0.0322 (7) | 0.0011 (5) | 0.0083 (5) | 0.0012 (5) |
| O2 | 0.0473 (9) | 0.0394 (9) | 0.0305 (7) | −0.0112 (7) | 0.0062 (6) | −0.0057 (6) |
| O3 | 0.0309 (7) | 0.0380 (8) | 0.0450 (8) | −0.0052 (6) | 0.0079 (6) | 0.0068 (6) |
| O4 | 0.0411 (8) | 0.0403 (8) | 0.0376 (8) | 0.0094 (7) | 0.0153 (6) | −0.0038 (6) |
| S1 | 0.0277 (2) | 0.0285 (3) | 0.0323 (3) | 0.0020 (2) | 0.00963 (18) | 0.00179 (19) |
Geometric parameters (Å, º)
| C1—C2 | 1.382 (2) | C6—H6 | 0.9500 |
| C1—C6 | 1.388 (2) | C6'—C7' | 1.376 (3) |
| C1—O1 | 1.428 (2) | C6'—H6' | 0.9500 |
| C1'—C2' | 1.379 (3) | C7—C8 | 1.526 (3) |
| C1'—C7' | 1.390 (3) | C7—H7A | 0.9800 |
| C1'—S1 | 1.7486 (19) | C7—H7B | 0.9800 |
| C2'—C3' | 1.386 (3) | C7—H7C | 0.9800 |
| C2'—H2' | 0.9500 | C7'—H7' | 0.9500 |
| C2—C3 | 1.400 (2) | C8—C9 | 1.527 (3) |
| C2—C8 | 1.513 (2) | C8—H8 | 1.0000 |
| C3'—C4' | 1.389 (3) | C9—H9A | 0.9800 |
| C3'—H3' | 0.9500 | C9—H9B | 0.9800 |
| C3—C4 | 1.380 (3) | C9—H9C | 0.9800 |
| C3—H3 | 0.9500 | C10—O2 | 1.424 (3) |
| C4—O2 | 1.369 (2) | C10—H10A | 0.9800 |
| C4—C5 | 1.403 (3) | C10—H10B | 0.9800 |
| C4'—C6' | 1.393 (3) | C10—H10C | 0.9800 |
| C4'—C5' | 1.504 (3) | C11—H11A | 0.9800 |
| C5—C6 | 1.381 (3) | C11—H11B | 0.9800 |
| C5—C11 | 1.503 (3) | C11—H11C | 0.9800 |
| C5'—H5'1 | 0.9800 | O1—S1 | 1.5967 (12) |
| C5'—H5'2 | 0.9800 | O3—S1 | 1.4216 (14) |
| C5'—H5'3 | 0.9800 | O4—S1 | 1.4221 (14) |
| C2—C1—C6 | 122.74 (16) | C8—C7—H7B | 109.5 |
| C2—C1—O1 | 118.38 (15) | H7A—C7—H7B | 109.5 |
| C6—C1—O1 | 118.64 (15) | C8—C7—H7C | 109.5 |
| C2'—C1'—C7' | 120.86 (18) | H7A—C7—H7C | 109.5 |
| C2'—C1'—S1 | 120.13 (15) | H7B—C7—H7C | 109.5 |
| C7'—C1'—S1 | 119.00 (14) | C6'—C7'—C1' | 119.26 (18) |
| C1'—C2'—C3' | 118.97 (18) | C6'—C7'—H7' | 120.4 |
| C1'—C2'—H2' | 120.5 | C1'—C7'—H7' | 120.4 |
| C3'—C2'—H2' | 120.5 | C2—C8—C7 | 111.27 (16) |
| C1—C2—C3 | 116.24 (16) | C2—C8—C9 | 110.51 (17) |
| C1—C2—C8 | 123.71 (16) | C7—C8—C9 | 110.87 (17) |
| C3—C2—C8 | 120.05 (16) | C2—C8—H8 | 108.0 |
| C2'—C3'—C4' | 121.44 (18) | C7—C8—H8 | 108.0 |
| C2'—C3'—H3' | 119.3 | C9—C8—H8 | 108.0 |
| C4'—C3'—H3' | 119.3 | C8—C9—H9A | 109.5 |
| C4—C3—C2 | 121.63 (17) | C8—C9—H9B | 109.5 |
| C4—C3—H3 | 119.2 | H9A—C9—H9B | 109.5 |
| C2—C3—H3 | 119.2 | C8—C9—H9C | 109.5 |
| O2—C4—C3 | 123.76 (17) | H9A—C9—H9C | 109.5 |
| O2—C4—C5 | 114.99 (16) | H9B—C9—H9C | 109.5 |
| C3—C4—C5 | 121.26 (17) | O2—C10—H10A | 109.5 |
| C3'—C4'—C6' | 118.20 (18) | O2—C10—H10B | 109.5 |
| C3'—C4'—C5' | 121.55 (18) | H10A—C10—H10B | 109.5 |
| C6'—C4'—C5' | 120.25 (18) | O2—C10—H10C | 109.5 |
| C6—C5—C4 | 117.33 (16) | H10A—C10—H10C | 109.5 |
| C6—C5—C11 | 121.84 (18) | H10B—C10—H10C | 109.5 |
| C4—C5—C11 | 120.83 (18) | C5—C11—H11A | 109.5 |
| C4'—C5'—H5'1 | 109.5 | C5—C11—H11B | 109.5 |
| C4'—C5'—H5'2 | 109.5 | H11A—C11—H11B | 109.5 |
| H5'1—C5'—H5'2 | 109.5 | C5—C11—H11C | 109.5 |
| C4'—C5'—H5'3 | 109.5 | H11A—C11—H11C | 109.5 |
| H5'1—C5'—H5'3 | 109.5 | H11B—C11—H11C | 109.5 |
| H5'2—C5'—H5'3 | 109.5 | C1—O1—S1 | 120.71 (11) |
| C5—C6—C1 | 120.79 (17) | C4—O2—C10 | 117.16 (15) |
| C5—C6—H6 | 119.6 | O3—S1—O4 | 119.91 (9) |
| C1—C6—H6 | 119.6 | O3—S1—O1 | 109.37 (8) |
| C7'—C6'—C4' | 121.25 (18) | O4—S1—O1 | 102.77 (8) |
| C7'—C6'—H6' | 119.4 | O3—S1—C1' | 109.08 (9) |
| C4'—C6'—H6' | 119.4 | O4—S1—C1' | 109.79 (9) |
| C8—C7—H7A | 109.5 | O1—S1—C1' | 104.76 (8) |
Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of the phenyl rings C1–C6 and C1'–C6', respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C3′—H3′···O1i | 0.95 | 2.67 | 3.610 (2) | 169 |
| C5′—H5′3···O3ii | 0.98 | 2.66 | 3.589 (3) | 159 |
| C7—H7A···O4iii | 0.98 | 2.72 | 3.693 (3) | 175 |
| C10—H10B···O3iv | 0.98 | 2.61 | 3.317 (3) | 130 |
| C10—H10A···O4iii | 0.98 | 2.70 | 3.521 (3) | 142 |
| C5′—H5′2···Cg2v | 0.98 | 2.84 | 3.706 | 148 |
| C11—H11B···Cg1iv | 0.98 | 2.75 | 3.635 | 150 |
| C9—H9A···Cg2 | 0.98 | 2.70 | 3.5373 | 144 |
Symmetry codes: (i) −x+1/2, y−1/2, −z+1/2; (ii) x+1, y, z; (iii) x+1/2, −y+1/2, z−1/2; (iv) −x, −y, −z; (v) −x+1, −y, −z+1.
References
- Altomare, A., Burla, M. C., Camalli, M., Cascarano, G. L., Giacovazzo, C., Guagliardi, A., Moliterni, A. G. G., Polidori, G. & Spagna, R. (1999). J. Appl. Cryst. 32, 115–119.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Greene, T. W. & Wuts, P. G. M. (1999). Protecting Groups in Organic Synthesis, 3rd ed. New York: Wiley.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Kacem, Y., Kraiem, J., Kerkeni, E., Bouraoui, A. & Ben Hassine, B. (2002). Eur. J. Pharm. Sci. 16, 221–228. [DOI] [PubMed]
- Kaleemullah, T., Ahmed, M. & Sharma, H. K. (2012). J. Chem. Pharm. Res. 4, 483–490.
- Kim, H. S., Oh, S. H., Kim, D., Kim, I. C., Cho, K. H. & Park, Y. B. (1995). Bioorg. Med. Chem. 3, 367–374. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Morgan, B., Dodds, D. R., Zaks, A., Andrews, D. R. & Klesse, R. (1997). J. Org. Chem. 62, 7736–7743.
- Niestroj, A. J., Bruhn, C. & Maier, M. E. (1998). J. Prakt. Chem. 340, 175–177.
- Rigaku OD (2015). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Sandler, S. R. & Karo, W. (1983). Organic Functional Group Preparations, Vol. 1. New York: Academic Press.
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Wagner, R. B. & Zokk, H. D. (1955). Synthetic Organic Chemistry, Vol. 3, edited by E. C. Horning. New York: John Wiley and Sons.
- Wieczorek, M. W., Bokiy, N. G. & Struchkov, Yu. T. (1975). Acta Cryst. B31, 2603–2606.
- Yoshida, Y., Shimomishi, K., Sakakura, Y., Okada, S., Aso, N. & Tanabe, Y. (1999). Synthesis, 1633–1636.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989018003110/xu5919sup1.cif
Supporting information file. DOI: 10.1107/S2056989018003110/xu5919Isup3.cml
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018003110/xu5919Isup3.hkl
CCDC reference: 1825267
Additional supporting information: crystallographic information; 3D view; checkCIF report




