Abstract
Background
Evidence is overwhelming for sex differences in pain, with women representing the majority of the chronic pain patient population. There is a need to explore novel avenues to elucidate this sex bias in the development of chronic inflammatory pain conditions. Complex regional pain syndrome (CRPS) is a chronic neuropathic pain disorder, and the incidence of CRPS is greater in women than in men by ~4:1. Since neurogenic inflammation is a key feature of CRPS, dysregulation of inflammatory responses can be a factor in predisposing women to chronic pain.
Methods
Our studies investigating alterations in circulating microRNAs (miRNAs) in whole blood from female CRPS patients showed significant differential expression of miRNAs between responders and poor responders to ketamine treatment. Several of these miRNAs are predicted to target the long noncoding RNA, X-inactive-specific transcript (XIST). XIST mediates X-chromosome inactivation and is essential for equalizing the expression of X-linked genes between females and males. Based on the well-established role in inflammatory process, we focused on miR-34a, one of the miRNAs predicted to target XIST, and downregulated in CRPS patients responding poorly to ketamine.
Results
Our in vitro and in vivo models of acute inflammation and data from patients with CRPS showed that miR-34a can regulate XIST under inflammation directly, and through pro-inflammatory transcription factor Yin-Yang 1 (YY1). XIST was significantly upregulated in a subset of CRPS patients responding poorly to ketamine.
Conclusion
Since dysregulation of XIST can result in genes escaping inactivation or reactivation in female cells, further investigations on the role of XIST in the predominance of chronic inflammatory and pain disorders in women is warranted.
Keywords: long noncoding RNA, sex difference, miR-34a, inflammation, pain, XIST
Introduction
Chronic pain impacts millions of individuals and is extremely difficult to treat. Pain sensitivity differs between the sexes, and the majority of patients with chronic pain are women. Epidemiological and laboratory data provide overwhelming evidence for sex differences in pain, with women reporting more severe and more frequent bouts of pain that is more anatomically diffuse and longer-lasting compared with pain in men.1,2 This observation holds true even after sex-specific disorders are excluded from the analyses. However, the underlying basis for the sex bias observed in pain remains unclear. A number of factors ranging from experiential and sociocultural differences in pain between men and women, genetics, and hormone-driven sex differences in brain neurochemistry have been proposed to explain the predominance of chronic pain in women.1 Examples of highly prevalent chronic pain syndromes occurring predominantly in women include complex regional pain syndrome (CRPS), fibromyalgia, chronic fatigue syndrome, etc.
CRPS is characterized by intense pain, inflammation, altered autonomic function, abnormal motor function, and trophic changes, with an estimated 50,000 new cases in the USA each year.3 CRPS is a multifactorial disorder that cannot be explained by a single mechanism.4 The multitude and the complexity of the pathophysiological processes associated with CRPS are thought to be responsible for the heterogeneity of the clinical presentation and may underlie the difficulty in establishing evidence-based treatment regimens.5 Current therapies are limited in their effectiveness, and none are curative, leaving more than 80% of chronic CRPS patients severely disabled. The incidence of CRPS is greater in women than in men by ~4:1.6 We have previously identified differential expression of circulating miRNAs in whole blood from CRPS patients compared to control and between female CRPS patients with good and poor analgesic response to ketamine therapy.7,8 Studying the target genes of these miRNAs, and their role in regulating pain and inflammation can provide important insights on the functional significance underlying aberrant levels of circulating miRNAs. We extended our studies to understand if there is a common target for more than one miRNA, ie, to determine if a group of the altered miRNAs can target a common RNA molecule. In our bioinformatics analyses for the targets of the miRNAs differentially expressed in responders and poor responders prior to ketamine treatment using target prediction algorithms, we observed that many of these miRNAs are predicted to bind to X-inactive-specific transcript (XIST), a long noncoding RNA (lncRNA) involved in X chromosome inactivation. The inactivation of about a thousand genes on one of the two essentially identical X chromosomes in females is one of the well-known epigenetic gene silencing mechanism. However, not all the genes on the inactive X chromosome (Xi) are silenced. In humans, ~15% of X-linked genes escape from X chromosome inactivation (XCI).9 Thus, escape from XCI is not an absolute with either full or no expression from the Xi.10 In both humans and mice, there is variability in the extent of gene expression from Xi. There are also differences between X chromosomes and between tissues as to which genes are expressed. Therefore, escape from XCI will not only result in differences in expression between males and females, but also between tissues in the same individual (mosaic expression) and between females. This could have important implications for disease predispositions between men and women, or among women.10
Though several miRNAs are predicted to regulate XIST, we focused on miR-34a based on its role in regulating inflammatory response.11,12 miR-34a had an 11-fold downregulation in poor responders relative to responders prior to treatment.13 In our investigations of whether miR-34a can directly regulate XIST expression, we also studied if miR-34a can indirectly regulate the expression of XIST by targeting Yin-Yang 1 (YY1). YY1 is a transcription factor with diverse and complex biological functions14 and a validated miR-34a target gene.15 As its name “Yin-Yang” implies, YY1 either activates or represses gene transcription, depending on the stimuli received by the cells and its association with other cellular factors. YY1 regulates gene expression by directly binding to its target promoters and recruiting histone and DNA modifiers.14,16 YY1 is known to positively regulate XIST expression,17 but its role in regulating XIST under inflammatory states is unknown. We show that miR-34a can regulate XIST under inflammation directly and through proinflammatory transcription factor YY1.
Patients and methods
CRPS and control subjects
The study was approved by the Drexel University College of Medicine Institutional Review Board. All enrolled subjects gave written informed consent, and all patients met the clinical Budapest criteria for CRPS. For the control group, there were 14 females and 7 males. In the CRPS patient group there were 52 female patients including 19 CRPS female patients who received intravenous ketamine.7,8 Healthy control subjects were recruited from the general public. The demographics of control and CRPS patients are shown in Table S1. The exclusion criteria for all subjects included pregnancy, recent infection, lupus erythematosus, HIV/AIDS, rheumatoid arthritis, recent extracorporeal circulation (hemodialysis, bypass surgery, plasmapheresis), bone marrow transplant, immunosuppressive therapy, blood disorders (anemia, leukemia), thymectomy, or sarcoidosis. Eligibility for ketamine treatment, treatment protocol, and response assessment were discussed in detail before.7 Briefly, ketamine was infused for five and a half days in the intensive care unit of Hahnemann University Hospital (Philadelphia, PA, USA). The infusion rate began at 10 mg/h and was increased by 10 mg/h every 2 h until a maximum rate of 40 mg/h was reached. This rate was continued for 120 h and was then tapered off by 10 mg/h every 2 h. Patients were asked to rate their overall pain prior to and after the infusion using a 10-point Numerical Rating Scale and McGill Pain Questionnaire. Blood samples were collected prior to and after ketamine infusion. As reported previously, patients were grouped as responders if following treatment their average pain score decreased by at least 50% and poor responders if their average pain score either increased or decreased <50%.7
Complete Freund’s adjuvant (CFA)-induced inflammatory pain model
All studies were performed in accordance with the guidelines of the National Institutes of Health. The care and use of all mice were approved by the Institutional Animal Care & Use Committee of Drexel University College of Medicine. All behavioral tests were performed using 8-week-old C57BL/6 female mice purchased from Jackson labs (Bar Harbor, ME, USA). Mice were housed in 12 h light/dark cycles in a standard temperature and humidity-controlled room and had access to food and water ad libitum. Mice were habituated in the testing room 2–3 days before experiments. Baseline measurements were obtained before initiation of treatment. Twenty microliters of 50% CFA (Sigma-Aldrich, St. Louis, MO, USA) was administered by intraplantar injection into the right hind paw. Mechanical sensitivity was measured using a series of von Frey filaments (North Coast Medical). The smallest monofilament that evoked paw withdrawal responses on 3 of 5 trials was taken as the mechanical threshold.
Cell culture
HEK293 cells (American Type Culture Consortium) were maintained in complete media (Dulbecco’s Modified Eagle’s Medium (DMEM)) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C in 5% CO2. J774A.1 cells (female mouse macrophages, ATCC) were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. SH-SY5Y cells, (ATCC) were maintained in a 1:1 mixture of F-12K Medium (Kaighn’s Modification of Ham’s F-12 Medium) (Corning Inc., New York, NY, USA) and DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
RNA isolation from human and mouse blood
Human whole blood was collected in PAXgene blood RNA tubes (BD Biosciences, Franklin Lakes, NJ, USA). RNA was isolated using a PAXgene blood miRNA kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Mouse blood was collected in 2 mL RNAlater tubes (Invitrogen, Carlsbad, CA, USA). RNA isolation from whole blood was performed using the mouse RiboPure-blood RNA kit (Ambion Inc., Foster city, CA, USA). RNA concentration was measured using Nanodrop 1000 (NanoDrop Technologies, Wilmington, DE, USA).
Quantitative real-time PCR (qPCR)
cDNA synthesis was performed from 100 to 500 ng total RNA using Maxima first strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA). All primer probes were purchased from Applied Biosystems (Foster city, CA, USA). Genes tested included XIST (Hs01079824_m1), Xist (Mm01232884_m1), YY1 (Hs00998747_m1), and Yy1 (Mm00456392_m1). GAPDH/Gapdh (4326317E/4352339E) was used for normalization.
qPCR for miRNA
cDNA synthesis was performed using 40 ng total RNA and qPCR for miR-34a was performed using TaqMan microRNA assay (Assay ID 002316, Applied Biosystems) as recommended by the vendor. U6 snRNA was used for normalization (Assay ID 001973, Applied Biosystems).
Luciferase reporter assay
Human XIST cDNA clone (Origene, Rockville, MD, USA) was subcloned using the Sac I and Xho I restriction sites into the pmirGLO vector (Promega, Madison, WI, USA). HEK293 cells were transfected with XIST pmirGLO plasmid and precursor miRNA clone miR-34a (HmiR0005, GeneCopoeia) or control miRNA using Lipofectamine 2000 (Invitrogen) at 1:1 ratio according to the manufacturer’s instructions for a final concentration of 100 ng DNA per well using 96 well plates. Firefly and Renilla luciferase activity were measured 24 h after transfection according to the manufacturer’s protocol (Promega).
Statistical analysis
All experiments were analyzed using Prism 5 (GraphPad Software Inc., La Jolla, CA, USA). Statistical differences were assessed using a two-tailed unpaired Student’s t-test, one-way analysis of variance, and Mann–Whitney U test. P-values of <0.05, <0.01, <0.001 indicated the significant difference between relevant groups. Data are represented as mean ± standard error of the mean.
Results
Decrease in miR-34a is associated with increase in XIST
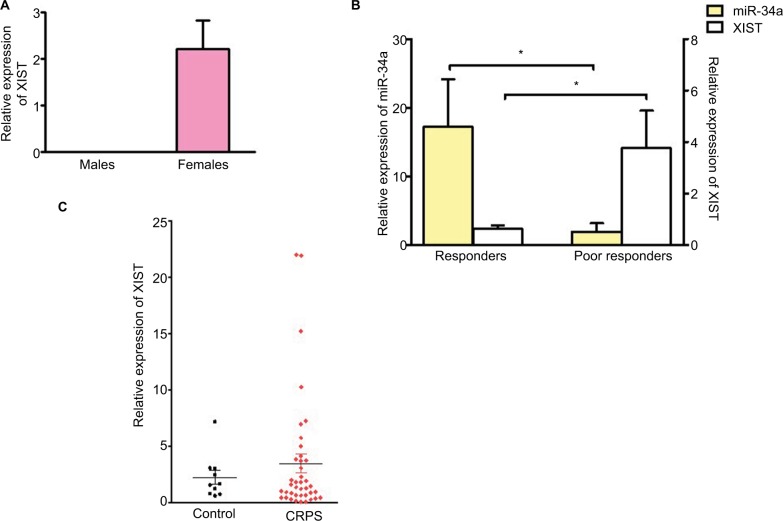
miR-34a was significantly downregulated in the blood of CRPS patients responding poorly to ketamine compared to responders. Based on the bioinformatics prediction that miR-34a can bind XIST, we performed a qPCR to determine if there is an inverse correlation in expression between XIST and miR-34a. We first validated the qPCR for XIST using RNA isolated from blood samples from control male and female individuals. Figure 1A shows the relative expression of XIST in blood from control male (n=6) and female (n=10) donors confirming XIST expression in females and not males. We then performed qPCR using CRPS patient samples grouped as responders and poor responders to ketamine. Figure 1B shows that there is an inverse correlation between expression of miR-34a and XIST in both groups, suggesting that a decrease in miR-34a could lead to an upregulation of XIST. We then investigated XIST expression using RNA samples from all female CRPS and control individuals (CRPS patients n=40, control females n=10) we had obtained from our previous study.8 Figure 1C shows that though there were some patients with increased levels of XIST, this was not statistically significant relative to female control group. CRPS patient population is heterogeneous and our data suggests that increased in XIST levels may be restricted to a subset of patients who may respond poorly to ketamine therapy.
Figure 1.
XIST is upregulated in a subset of CRPS patients and inversely correlated with miR-34a.
Notes: (A) Relative expression of XIST in blood from control male (n=6) and female (n=10) donors. (B) Relative expression of miR-34a and XIST in CRPS patients receiving ketamine therapy. Responders n=7, poor responders n=6. (C) Relative expression of XIST in CRPS patients (n=40), control females (n=10) and CRPS patients receiving ketamine therapy, responders n=7, poor responders n=6. Expression levels of XIST normalized to GAPDH. miR-34a expression normalized to U6. One-way ANOVA and Student’s t-test were used for statistical analysis. *p<0.05, data represent mean ± SEM.
Abbreviations: ANOVA, analysis of variance; CRPS, complex regional pain syndrome; SEM, standard error of the mean; XIST, X-inactive-specific transcript.
miR-34a can bind to the lncRNA XIST
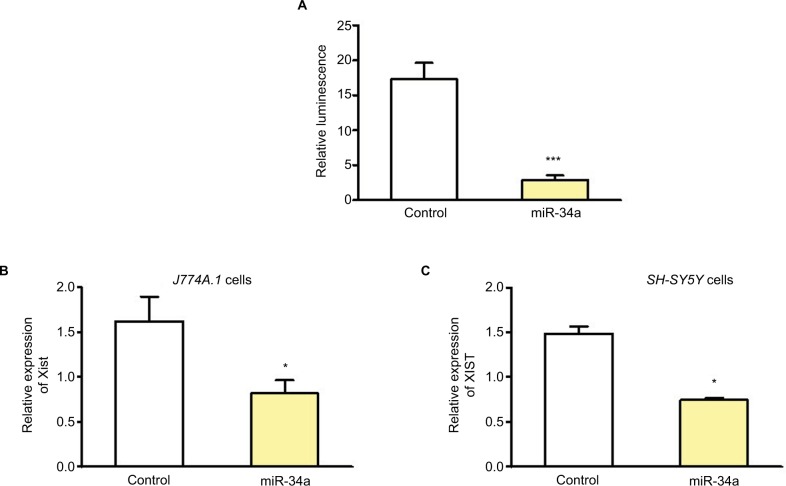
In addition to binding to 3′ untranslated region of mRNAs, miRNA can bind to lncRNAs, and lncRNAs can compete with mRNAs for miRNA binding. Potential binding of miRNAs to XIST was determined using the mirSVR method.18 Experimental validation was done using a reporter assay system designed to evaluate miRNA activity quantitatively by insertion of the sequence containing predicted miRNA binding sites downstream of the reporter gene. There are at least three miR-34a binding sites in this fragment derived from NR_001564.1 and their positions are 1055, 2045, and 2417. There are additional binding sites predicted along the whole length of XIST.19 We cloned 5 kb fragment from 5′XIST downstream of the firefly luciferase gene. Relative luminescence was determined after transfecting HEK293 cells with XIST pmirGLO plasmid and precursor miR-34a clone or control miRNA. There was a significant decrease in luciferase activity after miR-34a transfection but not the control miRNA, thereby confirming the binding of miR-34a to XIST (Figure 2A).
Figure 2.
miR-34a can bind and regulate XIST.
Notes: (A) Relative luminescence after transfecting HEK293 cells with XIST pmirGLO plasmid with 5 kb fragment from 5′XIST cloned downstream of the firefly luciferase gene, and precursor miR-34a or control miRNA (n=5). (B) and (C) Relative expression of Xist/XIST in mouse J774A.1 and human SH-SY5Y cells, respectively, after miR-34a transfection. Expression of Xist/XIST normalized to Gapdh/GAPDH. Student’s t-test used for statistical analysis *p<0.05, ***p<0.001. Data represent mean ± SEM.
Abbreviations: SEM, standard error of the mean; XIST, X-inactive-specific transcript.
miR-34a can downregulate endogenous XIST
To confirm miR-34a can modulate endogenous XIST levels, this miRNA was overexpressed in female cell lines and qPCR for XIST was performed 24–48 h after transfection. We used female mouse J774A.1 cells and human SH-SY5Y cells for miR-34a transfection and used mouse- or human-specific primer probe, respectively, for qPCR. Figure 2B shows that overexpression of miR-34a can decrease Xist in J774A.1 cells, and Figure 2C shows downregulation of XIST in SHSY-5Y cells. These experiments provided in vitro confirmation of the inverse correlation of the expression of miR-34a and XIST.
Acute inflammation induces a decrease in miR-34a and an increase in Xist expression
To determine if inflammation can alter the expression of miR-34a, J774A.1 cells were stimulated with lipopolysaccharide (LPS) (1 μg/mL for 4 h). LPS treatment induced a decrease in miR-34a and an increase in Xist in J774A.1 cells, confirming the reciprocal relationship between miR-34a and Xist under acute inflammation (Figure 3).
Figure 3.
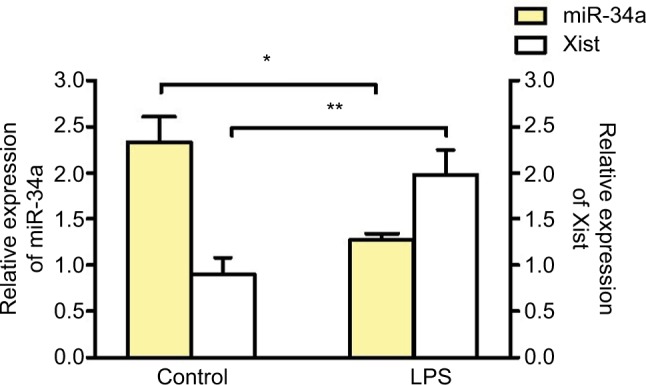
miR-34a regulates Xist under inflammation.
Notes: Relative expression of Xist and miR-34a in J774A.1 (mouse macrophage cell line) after LPS treatment (1 μg/mL for 4 h) (n≥3). Expression of Xist normalized to Gapdh. miR-34a expression normalized to U6 (n≥3). One-way ANOVA and Student’s t-test were used for statistical analysis *p<0.05, **p<0.01. Data represent mean ± SEM.
Abbreviations: ANOVA, analysis of variance; LPS, lipopolysaccharide; SEM, standard error of the mean; Xist, X-inactive-specific transcript.
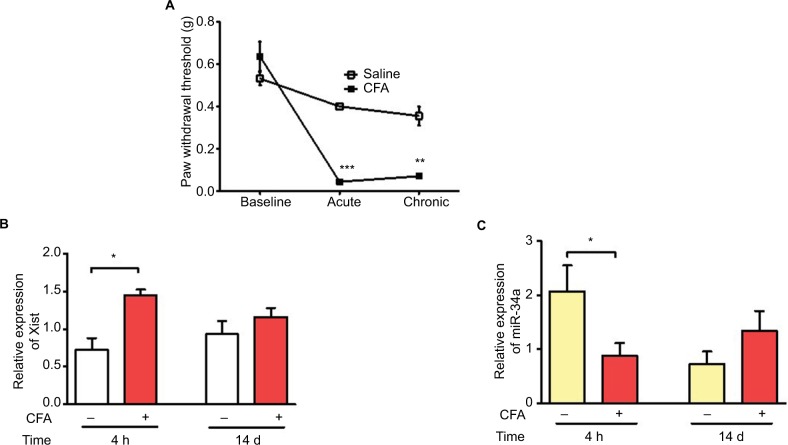
Xist is upregulated and miR-34a is downregulated in mouse model of acute inflammation
To further investigate how inflammation can modulate Xist expression in vivo, we used the CFA model of acute inflammatory pain. CFA model was established in 8-week old female C57BL/6 mice by intraplantar injection of 20 μL of 50% CFA into the right hind paw. Behavioral assays assessing tactile allodynia were performed before and after CFA administration, to confirm increased sensitivity (Figure 4A). Blood samples were collected for RNA isolation and qPCR analysis at two time points representing acute and chronic inflammation. Xist RNA was upregulated in whole blood from CFA treated mice compared to saline injected control mice at 4 h but not after 14 days post CFA (Figure 4B). miR-34a was also downregulated in the acute inflammatory phase of the CFA model compared to blood samples from saline-treated mice. There was no significant difference miR-34a expression 14 days after CFA administration (Figure 4C).
Figure 4.
Upregulation of Xist and downregulation of miR-34a in blood samples from CFA model of inflammatory pain.
Notes: (A) CFA-induced mechanical hypersensitivity assessed using von Frey filaments in female C57BL/6 mice (n=3–4). Paw withdrawal threshold dropped significantly by 4 h (acute inflammatory phase) and remained low at 14 days (chronic inflammatory phase). (B) Relative expression of Xist in female C57BL/6 mice injected with CFA or saline into the hind paw (n=3–4). Blood samples collected at 4 h and 14 days were used to determine the expression of Xist during acute and chronic inflammatory phases respectively. (C) Relative expression of miR-34a in blood under acute and chronic inflammation in female C57BL/6 mice injected with CFA or saline (n=3–4). One-way ANOVA and Student’s t-test were used for statistical analysis except for mice mechanical hypersensitivity in which Mann–Whitney U test was used *p<0.05, **p<0.01, ***p<0.001, data represent mean ± SEM.
Abbreviations: ANOVA, analysis of variance; CFA, complete Freund’s adjuvant; SEM, standard error of the mean; Xist, X-inactive-specific transcript.
miR-34a can contribute to the upregulation of XIST indirectly by regulating the expression of YY1
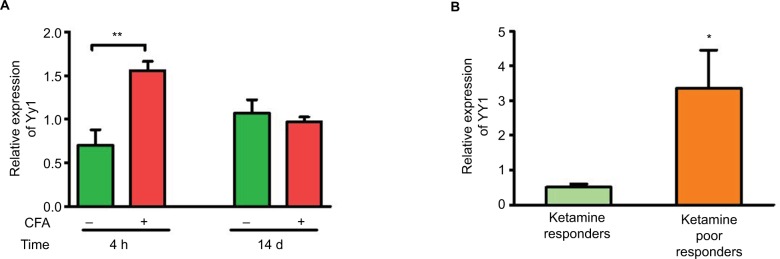
YY1, a transcriptional activator of XIST,17 is a validated target of miR-34a. Hence, we investigated YY1 expression in blood samples from CFA model. Figure 5A shows that similar to Xist levels, YY1 was upregulated in the acute phase of inflammation in the CFA model (Figure 5A). We then investigated expression of YY1 in CRPS patients, and there was significant upregulation of YY1 in patients grouped as poor responders to ketamine compared to responders (Figure 5B), indicating YY1 expression paralleled XIST. Thus, downregulation of miR-34a can play a dual role in XIST increase by regulating XIST directly, and indirectly by modulating YY1.
Figure 5.
Upregulation of Yy1 parallels Xist.
Notes: (A) Relative expression of Yy1 in female C57BL/6 mice injected with CFA or saline (n=3–4). (B) Relative expression of YY1 in CRPS patients receiving ketamine therapy, responders n=7, poor responders n=6. Expression of Yy1/YY1 normalized to Gapdh/GAPDH. One-way ANOVA and Student’s t-test were used for statistical analysis *p<0.05, **p<0.01. Data represent mean ± SEM.
Abbreviations: ANOVA, analysis of variance; CFA, complete Freund’s adjuvant; CRPS, complex regional pain syndrome; SEM, standard error of the mean; Xist, X-inactive-specific transcript; YY1, Yin-Yang 1.
Discussion
Differences in miRNA signature in CRPS patients provided molecular insights distinguishing responders from poor responders to ketamine. Investigating genes targeted by these differentially regulated miRNAs can be beneficial in obtaining insight into previously unexplored mediators in CRPS. We wanted to investigate if an individual transcript can be targeted by multiple circulating miRNAs altered in CRPS. Based on bioinformatics prediction, we hypothesized that circulating miRNAs downregulated in pretreatment samples of CRPS patients responding poorly to ketamine therapy can target XIST resulting in upregulation of XIST in poor responders. XIST is a 17-kb spliced, polyadenylated transcript that is retained in the nucleus coating the X chromosome in cis, rendering it inactive. Since CRPS is a female-predominant disorder, we sought to investigate if XIST, a lncRNA expressed only in female cells, has a role beyond X-chromosome inactivation, and whether differential expression of miRNAs in female CRPS patients can contribute to differences in target gene expression between responders and poor responders to ketamine.
It has been shown that miRNAs and lncRNAs have a regulatory role over each other.20 We pursued target validation studies focusing on miR-34a. As previously reported, CRPS patients responding poorly to ketamine showed a downregulation of miR-34a,7 and is involved in regulating inflammatory and immune functions.11,21 Our previous studies have shown a role for miR-34a in the negative regulation of CRHR1 and CRH-mediated pro-opiomelanocortin expression.13 To further elucidate the different ways miR-34a could be influencing molecular underpinnings observed in CRPS, we investigated miR-34a-mediated regulation of XIST. We confirmed that miR-34a can bind and regulate the expression of XIST in both human and mouse female cell lines. We observed an upregulation of XIST in blood samples from poor responders indicating inverse correlation in expression of miRNA and its target gene. Based on these observations, we conclude that miR-34a is a potential regulator of XIST in a subset of CRPS patients. Additional studies in larger patient cohort are needed to further elucidate the biomarker utility of XIST expression as a predictor for ketamine response.
Our studies in J774A.1 female mouse macrophage cell line showed an upregulation of Xist and a corresponding decrease in miR-34a upon LPS stimulation. miR-34a is known to be involved in many inflammatory and immune functions. miR-34a level was reduced in LPS-treated RAW 264.7 macrophages, and transfection of miR-34a mimics diminished proinflammatory responses, evidenced by lower levels of M1 cytokines TNF and IL-6. Mechanistically, miR-34a targeted Notch1, which is needed for LPS-mediated production of proinflammatory cytokines in macrophages.11 A previous study reporting the binding of XIST with miR-34a suggested that XIST can act as a sponge for miR-34a allowing the upregulation of other miR-34a targets such as the transcription factor E2F3.22 Our studies in CFA model also showed a decrease in miR-34a and an upregulation of Xist under acute inflammation. Collectively, these data indicate that miR-34a dysregulation can inversely affect XIST expression under acute inflammation in vitro and in mouse model, and under chronic inflammatory pain in a subset of CRPS patients resistant to ketamine.
Several factors regulate Xist expression during initiation of XCI.23 It has been shown that YY1 upregulates XIST expression17 and miR-34a negatively regulate YY1.24 YY1 plays an important role in analgesia and inflammation. YY1 has a role in the initiation of transcription of the mu opioid receptor gene25 and COX-226 in lymphocytes. Transcriptional regulation of miRNA genes expressed during monocytic differentiation is mediated by YY1.27 In addition, YY1 plays a crucial role in the B-cell development and immune function.28 YY1 binds to the promoter regions of several cytokine genes in T-cells.29 Studies also showed that YY1 mutant mice displayed reduced inflammatory pain sensitivity on the formalin test, increased sensitivity to systemic and spinal (but not supraspinal) morphine analgesia, and greatly increased endogenous (swim stress-induced) opioid analgesia.30 YY1 mRNA expression and protein was upregulated in rheumatoid arthritis patients and in mice with collagen-induced arthritis. Inhibition of YY1 reduced inflammation in the collagen-induced arthritis model.31 This recent study also showed that YY1 can positively regulate IL-6 transcription by binding to the promoter region of the IL-6 gene. It has been shown that NF-κB regulates the expression of YY1,32 which in turn can regulate the transcription of XIST.17 In CRPS, there was significant upregulation of YY1 in patients grouped as poor responders to ketamine compared to responders. Thus, downregulation of miR-34a can play a dual role in XIST increase by regulating XIST directly, and indirectly by modulating YY1. XIST is predominantly a nuclear lncRNA and mature miRNAs are usually present in the cytoplasm. It is known that reciprocal regulations exist between nuclear lncRNA and miRNA.33,34 This reciprocal interaction underscores the rich and complex regulation of gene expression mediated by ncRNAs.20 Though miR-34a can regulate XIST directly, YY1 can be an important factor in the signaling cascade. Although the well-studied function of XIST is related to its fundamental role in XCI, some reports suggest that it can have other functions unrelated to the silencing of the second X chromosome. XIST expression is higher in several tumors, including ovarian cancer,35 non-small-cell lung cancer,36 glioblastoma,37 breast cancer,38 and hepatocellular carcinoma,39 indicating that XIST might act as a potential diagnostic biomarker for these cancers. Gene expression profiling of rheumatoid arthritis synovial cells showed a 40-fold overexpression of XIST compared to control.40 This was reversed by treatment with prednisolone, suggesting a role for XIST in rheumatoid arthritis, a disease predominantly affecting women.
Our observation raises the question as to why the upregulation of Xist in CFA model was limited to 4 h but not at 14 days post CFA administration. In a separate study, we investigated the molecular mechanisms underlying Xist upregulation in mouse macrophage cell line. Our unpublished data show that under acute inflammation XIST can have a protective anti-inflammatory effect. These findings indicate that the role of XIST differ under acute and chronic inflammation. Based on the protective anti-inflammatory role for XIST under acute inflammation, we postulate that a greater incidence of acute inflammation in men could be due to the absence of XIST in male cells. However, in chronic painful disorders, XIST may have a proinflammatory role. A recent report showed upregulation of Xist in spinal cord from rat chronic constriction injury model of neuropathic pain, and silencing Xist alleviated both mechanical and thermal hyperalgesia.41
X chromosome is known to contain the largest number of immune-related genes in the whole human genome and a higher density of miRNAs compared to autosomes and the Y-chromosome.42–44 This highlights the importance of tight control and complex regulation of gene expression from the X chromosome. Any aberration in X-linked gene inactivation9 from either abnormal escape or increased inactivation, mediated by lncRNAs or epigenetic modifications, can result in aberrant immune and inflammatory response. We postulate that epigenetic changes including histone modifications and DNA methylation in female cells and miRNA alterations may predispose immune cells to heightened chronic inflammatory response. Dysregulation of XIST can result in genes escaping inactivation or reactivation in female cells.45 We postulate that under chronic inflammatory pain disorders, aberrant expression of XIST could affect the sites it coats on the inactive X chromosome. This in turn can influence the silencing or inactivation of genes encoded by the X chromosome. Mechanistic studies on how the expression of proinflammatory genes and transcription factors can be influenced by changes in XIST levels and localization under acute and chronic inflammation are needed to validate this. Understanding the tissue- or cell-specific aberrations and temporal regulation is crucial for further understanding the role of XIST in acute inflammation and chronic pain. We propose a dual and opposing role for XIST in acute and chronic inflammatory and pain disorders predominant in women that warrants further investigations. Though it is early to assume that miR-34a-induced XIST regulation would directly help in treating or preventing pain, our studies link lncRNA XIST to ketamine treatment resistance in a female-predominant chronic inflammatory pain disorder CRPS. XIST is essential for dosage compensation in female cells, and thus manipulating XIST directly or downregulating it completely will not be a feasible therapeutic intervention strategy. However, higher levels of XIST as we observed in a subset of CRPS patients, in addition to being a potential biomarker, can be modulated by altering miRNAs such as miR-34a. miRNAs are fine tuners of gene regulation and can induce nuanced knockdown of its target mRNAs. Downregulation of Xist by siRNAs was recently shown to decrease pain hypersensitivity41 in mouse model of neuropathic pain. miR-34a is a tumor suppressor and currently in Phase 1 clinical trials for cancer.46 Thus, miRNA-mediated downregulation of XIST could be a potential avenue for pain relief.
Supplementary material
Table S1.
The demographics of control and CRPS patients
| Controls | Sex | Age (years) | Weight (lb) | Height (cm) | BMI |
|---|---|---|---|---|---|
| C1 | F | 42 | 120 | 63 | 21.25 |
| C2 | F | 37 | 134 | 67 | 20.99 |
| C3 | F | 40 | 240 | 67 | 37.59 |
| C4 | F | 24 | 113 | 62 | 20.67 |
| C5 | F | 34 | 124 | 66 | 20.01 |
| C6 | F | 39 | 157 | 67 | 24.59 |
| C7 | F | 29 | 132 | 62 | 24.14 |
| C8 | F | 47 | 140 | 65.5 | 22.94 |
| C9 | F | 59 | 117 | 60 | 22.85 |
| C10 | F | 41 | 110 | 65 | 18.30 |
| C11 | M | 68 | 220 | 76 | 26.78 |
| C12 | M | 63 | 190 | 71 | 26.50 |
| C13 | M | 56 | 235 | 78 | 27.15 |
| C14 | M | 43 | 165 | 70 | 23.67 |
| C15 | M | 49 | 186 | 75 | 23.25 |
| C16 | M | 23 | 165 | 67 | 25.84 |
| Patients | Ketamine response | Age (years) | Weight (lb) | Height (cm) | BMI | |
|---|---|---|---|---|---|---|
| P1 | NA | F | 40 | 128 | 65 | 21.30 |
| P2 | NA | F | 65 | 164 | 67 | 25.68 |
| P3 | NA | F | 47 | 233 | 65 | 38.77 |
| P4 | NA | F | 63 | 102 | 58 | 21.32 |
| P5 | NA | F | 33 | 142 | 62 | 25.97 |
| P6 | NA | F | 31 | 170 | 62 | 31.09 |
| P7 | NA | F | 35 | 230 | 66 | 37.12 |
| P8 | NA | F | 41 | 155 | 67 | 24.27 |
| P9 | NA | F | 32 | 165 | 64 | 28.32 |
| P10 | NA | F | 54 | 212 | 71 | 29.56 |
| P11 | NA | F | 27 | 128 | 62 | 23.41 |
| P12 | NA | F | 51 | 120 | 62 | 21.95 |
| P13 | NA | F | 58 | 272 | 64 | 46.68 |
| P14 | NA | F | 60 | 165 | 69 | 24.36 |
| P15 | NA | F | 38 | 147 | 60 | 28.71 |
| P16 | NA | F | 48 | 140 | 65 | 23.29 |
| P17 | NA | F | 49 | 175 | 66 | 28.24 |
| P18 | NA | F | 28 | 102 | 61 | 19.27 |
| P19 | NA | F | 44 | 99 | 64 | 16.99 |
| P20 | NA | F | 23 | 163 | 61 | 30.80 |
| P21 | NA | F | 38 | 160 | 64 | 27.46 |
| P22 | NA | F | 68 | 154 | 63 | 27.28 |
| P23 | NA | F | 58 | 191 | ||
| P24 | NA | F | 43 | 145 | 68 | 22.04 |
| P25 | NA | F | 57 | 155 | 63 | 27.45 |
| P26 | NA | F | 33 | 166 | 68 | 25.24 |
| P27 | NA | F | 45 | 160 | 61 | 30.23 |
| P28 | Responder | F | 51 | 200 | 64 | 34.33 |
| P29 | Responder | F | 49 | 298 | 67 | 46.67 |
| P30 | Responder | F | 51 | 176 | 68.5 | 26.37 |
| P31 | Responder | F | 39 | 194 | 67 | 30.38 |
| P32 | Responder | F | 43 | 260 | 63 | 46.05 |
| P33 | Responder | F | 65 | 165 | 61 | 31.17 |
| P34 | Responder | F | 49 | 173 | 63 | 30.64 |
| P35 | Poor responder | F | 43 | 175 | 63 | 31.00 |
| P36 | Poor responder | F | 69 | 152 | 61 | 28.72 |
| P37 | Poor responder | F | 35 | 150 | 66 | 24.21 |
| P38 | Poor responder | F | 45 | 160 | 61 | 30.23 |
| P39 | Poor responder | F | 56 | 145 | 63 | 25.68 |
| P40 | Poor responder | F | 33 | 166 | 68 | 25.24 |
Abbbreviation: CRPS, complex regional pain syndrome.
Acknowledgments
We gratefully acknowledge funding from Drexel University Clinical and Translational Research Institute, Rita Allen Foundation, and National Institutes of Health Grant 1R01NS102836-01A1 to Seena K Ajit. Botros B Shenoda is a recipient of the Fulbright Foreign Student Program fellowship funded by the US Department of State, Bureau of Educational and Cultural Affairs, and Dean’s Fellowship from Drexel University College of Medicine.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mogil JS. Sex differences in pain and pain inhibition: multiple explanations of a controversial phenomenon. Nat Rev Neurosci. 2012;13(12):859–866. doi: 10.1038/nrn3360. [DOI] [PubMed] [Google Scholar]
- 2.Bartley EJ, Fillingim RB. Sex differences in pain: a brief review of clinical and experimental findings. Br J Anaesthesia. 2013;111(1):52–58. doi: 10.1093/bja/aet127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Mos M, de Bruijn AG, Huygen FJ, Dieleman JP, Stricker BH, Sturkenboom MC. The incidence of complex regional pain syndrome: a population-based study. Pain. 2007;129(1–2):12–20. doi: 10.1016/j.pain.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Birklein F, Schlereth T. Complex regional pain syndrome-significant progress in understanding. Pain. 2015;156(Suppl 1):S94–S103. doi: 10.1097/01.j.pain.0000460344.54470.20. [DOI] [PubMed] [Google Scholar]
- 5.Gierthmuhlen J, Binder A, Baron R. Mechanism-based treatment in complex regional pain syndromes. Nat Rev Neurol. 2014;10(9):518–528. doi: 10.1038/nrneurol.2014.140. [DOI] [PubMed] [Google Scholar]
- 6.Schwartzman RJ, Erwin KL, Alexander GM. The natural history of complex regional pain syndrome. Clin J Pain. 2009;25(4):273–280. doi: 10.1097/AJP.0b013e31818ecea5. [DOI] [PubMed] [Google Scholar]
- 7.Douglas SR, Shenoda BB, Qureshi RA, et al. Analgesic response to intravenous Ketamine is linked to a circulating microRNA signature in female patients with complex regional pain syndrome. J Pain. 2015;16(9):814–824. doi: 10.1016/j.jpain.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Orlova IA, Alexander GM, Qureshi RA, et al. MicroRNA modulation in complex regional pain syndrome. J Transl Med. 2011;9(1):195. doi: 10.1186/1479-5876-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 10.Peeters SB, Cotton AM, Brown CJ. Variable escape from X-chromosome inactivation: identifying factors that tip the scales towards expression. Bioessays. 2014;36(8):746–756. doi: 10.1002/bies.201400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang P, Liu R, Zheng Y, et al. MiR-34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res. 2012;318(10):1175–1184. doi: 10.1016/j.yexcr.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Mathe E, Nguyen GH, Funamizu N, et al. Inflammation regulates microRNA expression in cooperation with p53 and nitric oxide. Int J Cancer. 2012;131(3):760–765. doi: 10.1002/ijc.26403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shenoda BB, Alexander GM, Ajit SK. Hsa-miR-34a mediated repression of corticotrophin releasing hormone receptor 1 regulates pro-opiomelanocortin expression in patients with complex regional pain syndrome. J Transl Med. 2016;14:64. doi: 10.1186/s12967-016-0820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription. 2010;1(2):81–84. doi: 10.4161/trns.1.2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen QR, Yu LR, Tsang P, et al. Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR-34a. J Proteome Res. 2011;10(2):479–487. doi: 10.1021/pr1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Stovall DB, Inoue K, Sui G. The oncogenic role of Yin Yang 1. Crit Rev Oncog. 2011;16(3–4):163–197. doi: 10.1615/critrevoncog.v16.i3-4.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makhlouf M, Ouimette JF, Oldfield A, Navarro P, Neuillet D, Rougeulle C. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat Commun. 2014;5:4878. doi: 10.1038/ncomms5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34C:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 22.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592(1):8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 23.Dixon-McDougall T, Brown C. The making of a Barr body: the mosaic of factors that eXIST on the mammalian inactive X chromosome. Biochem Cell Biol. 2016;94(1):56–70. doi: 10.1139/bcb-2015-0016. [DOI] [PubMed] [Google Scholar]
- 24.Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database issue):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Liu H, Wang Z, et al. The role of transcription factors Sp1 and YY1 in proximal promoter region in initiation of transcription of the mu opioid receptor gene in human lymphocytes. J Cell Biochem. 2008;104(1):237–250. doi: 10.1002/jcb.21616. [DOI] [PubMed] [Google Scholar]
- 26.Joo M, Wright JG, Hu NN, et al. Yin Yang 1 enhances cyclooxygenase-2 gene expression in macrophages. Am J Physiol Lung Cell Mol Physiol. 2007;292(5):L1219–L1226. doi: 10.1152/ajplung.00474.2006. [DOI] [PubMed] [Google Scholar]
- 27.Schmeier S, MacPherson CR, Essack M, et al. Deciphering the transcriptional circuitry of microRNA genes expressed during human monocytic differentiation. BMC Genom. 2009;10:595. doi: 10.1186/1471-2164-10-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atchison M, Basu A, Zaprazna K, Papasani M. Mechanisms of Yin Yang 1 in oncogenesis: the importance of indirect effects. Crit Rev Oncog. 2011;16(3–4):143–161. doi: 10.1615/critrevoncog.v16.i3-4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Lin X, Williams MA, Hamid Q, Georas SN. Yin-Yang 1 regulates effector cytokine gene expression and T(H)2 immune responses. J Aller Clin Immunol. 2008;122(1):195–201. 201.e1–e5. doi: 10.1016/j.jaci.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorge RE, LaCroix-Fralish ML, Tuttle AH, et al. The Yin and Yang of pain: variability in formalin test nociception and morphine analgesia produced by the Yin Yang 1 transcription factor gene. Genes Brain Behav. 2013;12(4):405–413. doi: 10.1111/gbb.12030. [DOI] [PubMed] [Google Scholar]
- 31.Lin J, He Y, Chen J, Zeng Z, Yang B, Ou Q. A critical role of transcription factor YY1 in rheumatoid arthritis by regulation of interleukin-6. J Autoimmun. 2017;77:67–75. doi: 10.1016/j.jaut.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Hertlein E, Bakkar N, et al. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007;27(12):4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75(7):1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han X, Yang F, Cao H, Liang Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29(7):3054–3064. doi: 10.1096/fj.14-259952. [DOI] [PubMed] [Google Scholar]
- 35.Ren C, Li X, Wang T, et al. Functions and mechanisms of long non-coding RNAs in ovarian cancer. Int J Gynecol Cancer. 2015;25(4):566–569. doi: 10.1097/IGC.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 36.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(7):7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Y, Ma J, Xue Y, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359(1):75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 38.Salvador MA, Wicinski J, Cabaud O, et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin Cancer Res. 2013;19(23):6520–6531. doi: 10.1158/1078-0432.CCR-13-0877. [DOI] [PubMed] [Google Scholar]
- 39.Zhuang LK, Yang YT, Ma X, et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haupl T, Yahyawi M, Lubke C, et al. Gene expression profiling of rheumatoid arthritis synovial cells treated with antirheumatic drugs. J Biomol Screen. 2007;12(3):328–340. doi: 10.1177/1087057107299261. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y, Li S, Xiao M, Shi Y, Zhao CM. Effects of XIST/miR-137 axis on neuropathic pain by targeting TNFAIP1 in a rat model. J Cell Physiol. 2017;233(5):4307–4316. doi: 10.1002/jcp.26254. [DOI] [PubMed] [Google Scholar]
- 42.Bianchi I, Lleo A, Gershwin ME, Invernizzi P. The X chromosome and immune associated genes. J Autoimmun. 2012;38(2–3):J187–J192. doi: 10.1016/j.jaut.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Invernizzi P, Pasini S, Selmi C, Gershwin ME, Podda M. Female predominance and X chromosome defects in autoimmune diseases. J Autoimmun. 2009;33(1):12–16. doi: 10.1016/j.jaut.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 44.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol. 2010;10(8):594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 45.Vacca M, Della Ragione F, Scalabrì F, D’Esposito M. X inactivation and reactivation in X-linked diseases. Semin Cell Dev Biol. 2016;56:78–87. doi: 10.1016/j.semcdb.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
The demographics of control and CRPS patients
| Controls | Sex | Age (years) | Weight (lb) | Height (cm) | BMI |
|---|---|---|---|---|---|
| C1 | F | 42 | 120 | 63 | 21.25 |
| C2 | F | 37 | 134 | 67 | 20.99 |
| C3 | F | 40 | 240 | 67 | 37.59 |
| C4 | F | 24 | 113 | 62 | 20.67 |
| C5 | F | 34 | 124 | 66 | 20.01 |
| C6 | F | 39 | 157 | 67 | 24.59 |
| C7 | F | 29 | 132 | 62 | 24.14 |
| C8 | F | 47 | 140 | 65.5 | 22.94 |
| C9 | F | 59 | 117 | 60 | 22.85 |
| C10 | F | 41 | 110 | 65 | 18.30 |
| C11 | M | 68 | 220 | 76 | 26.78 |
| C12 | M | 63 | 190 | 71 | 26.50 |
| C13 | M | 56 | 235 | 78 | 27.15 |
| C14 | M | 43 | 165 | 70 | 23.67 |
| C15 | M | 49 | 186 | 75 | 23.25 |
| C16 | M | 23 | 165 | 67 | 25.84 |
| Patients | Ketamine response | Age (years) | Weight (lb) | Height (cm) | BMI | |
|---|---|---|---|---|---|---|
| P1 | NA | F | 40 | 128 | 65 | 21.30 |
| P2 | NA | F | 65 | 164 | 67 | 25.68 |
| P3 | NA | F | 47 | 233 | 65 | 38.77 |
| P4 | NA | F | 63 | 102 | 58 | 21.32 |
| P5 | NA | F | 33 | 142 | 62 | 25.97 |
| P6 | NA | F | 31 | 170 | 62 | 31.09 |
| P7 | NA | F | 35 | 230 | 66 | 37.12 |
| P8 | NA | F | 41 | 155 | 67 | 24.27 |
| P9 | NA | F | 32 | 165 | 64 | 28.32 |
| P10 | NA | F | 54 | 212 | 71 | 29.56 |
| P11 | NA | F | 27 | 128 | 62 | 23.41 |
| P12 | NA | F | 51 | 120 | 62 | 21.95 |
| P13 | NA | F | 58 | 272 | 64 | 46.68 |
| P14 | NA | F | 60 | 165 | 69 | 24.36 |
| P15 | NA | F | 38 | 147 | 60 | 28.71 |
| P16 | NA | F | 48 | 140 | 65 | 23.29 |
| P17 | NA | F | 49 | 175 | 66 | 28.24 |
| P18 | NA | F | 28 | 102 | 61 | 19.27 |
| P19 | NA | F | 44 | 99 | 64 | 16.99 |
| P20 | NA | F | 23 | 163 | 61 | 30.80 |
| P21 | NA | F | 38 | 160 | 64 | 27.46 |
| P22 | NA | F | 68 | 154 | 63 | 27.28 |
| P23 | NA | F | 58 | 191 | ||
| P24 | NA | F | 43 | 145 | 68 | 22.04 |
| P25 | NA | F | 57 | 155 | 63 | 27.45 |
| P26 | NA | F | 33 | 166 | 68 | 25.24 |
| P27 | NA | F | 45 | 160 | 61 | 30.23 |
| P28 | Responder | F | 51 | 200 | 64 | 34.33 |
| P29 | Responder | F | 49 | 298 | 67 | 46.67 |
| P30 | Responder | F | 51 | 176 | 68.5 | 26.37 |
| P31 | Responder | F | 39 | 194 | 67 | 30.38 |
| P32 | Responder | F | 43 | 260 | 63 | 46.05 |
| P33 | Responder | F | 65 | 165 | 61 | 31.17 |
| P34 | Responder | F | 49 | 173 | 63 | 30.64 |
| P35 | Poor responder | F | 43 | 175 | 63 | 31.00 |
| P36 | Poor responder | F | 69 | 152 | 61 | 28.72 |
| P37 | Poor responder | F | 35 | 150 | 66 | 24.21 |
| P38 | Poor responder | F | 45 | 160 | 61 | 30.23 |
| P39 | Poor responder | F | 56 | 145 | 63 | 25.68 |
| P40 | Poor responder | F | 33 | 166 | 68 | 25.24 |
Abbbreviation: CRPS, complex regional pain syndrome.