SUMMARY
During development, progenitors progress through transition states. The cardiac epicardium contains progenitors of essential non-cardiomyocytes. The Hippo pathway, a kinase cascade that inhibits the Yap transcriptional co-factor, controls organ size in developing hearts. Here, we investigated Hippo kinases, Lats1 and Lats2 in epicardial diversification. Epicardial-specific deletion of Lats1/2 was embryonic lethal, and mutant embryos had defective coronary vasculature remodeling. Single cell RNA-sequencing revealed that Lats1/2 mutant cells failed to activate fibroblast differentiation, but remained in an intermediate cell state with both epicardial and fibroblast characteristics. Lats1/2 mutant cells displayed an arrested developmental trajectory with persistence of epicardial markers and expanded expression of Yap targets Dhrs3, an inhibitor of retinoic acid synthesis, and Dpp4, a protease that modulates extracellular matrix composition (ECM). Genetic and pharmacologic manipulation revealed that Yap inhibits fibroblast differentiation, prolonging a subepicardial-like cell state, and promotes expression of matricellular factors, such as Dpp4, that define ECM characteristics.
eTOC blurb
The epicardium, the outermost cell layer of the heart, contains progenitors that contribute to non-cardiomyocytes. How epicardial progenitors transition to a mature cell type is unknown. Xiao et al. demonstrated that Hippo kinases Lats1/2 promote epicardial-fibroblast transition which is essential for maintaining proper extracellular milieu and coronary vessel development.
INTRODUCTION
The epicardium, cells covering the outer layer of the heart, originates from the extra-cardiac proepicardium. The proepicardium is compartmentalized into populations that give rise to cardiac endothelium and mesenchymal cells: fibroblasts and smooth muscle (Katz et al., 2012; Acharya et al., 2012). At mouse embryonic day (E)9.5, proepicardial cells attach to myocardium, spread as a continuous epithelial sheet, and form a single cell layer covering the entire myocardium. The epicardium expresses a number of important genes including signaling molecules such as Retinaldehyde dehydrogenase 2 (Raldh2), and genes encoding transcription factors: Wilms tumor 1 (Wt1), transcription factor 21 (Tcf21), T-box18 (Tbx18), and multiple C/EBP transcription factor family members (Acharya et al., 2012; Cai et al., 2008; Guadix et al., 2011; Huang et al., 2012; Zhou et al., 2008).
A subset of epicardial cells delaminate, undergo EMT and generate epicardial-derived cells (EPDCs). In mammals, EPDCs emanating from Wt1-expressing lineage primarily give rise to vascular smooth muscle cells and fibroblasts, two supporting cell types that are important for coronary vascular and myocardial development. EPDCs first populate the subepicardial space between epicardium and myocardium, forming the subepicardial mesenchyme, and invade the myocardium where they differentiate into supporting cells of the heart (Wessels and Perez-Pomares, 2004).
Although adult epicardial cells are quiescent, they are activated upon injury and contribute to the repair process by expressing developmental programs in the injured adult heart (Zhou et al., 2011; Huang et al., 2012; Lepilina et al., 2006). Activated adult epicardium is a source of pro-inflammatory signals after myocardial infarction. In the adult heart C/EBP factors activate pro-inflammatory signals after injury whereas Yap and Taz, two Hippo-pathway effectors, promote anti-inflammatory response to injury (Huang et al., 2012; Ramjee et al., 2017).
The Hippo signaling pathway, an organ size control pathway, inhibits cell proliferation and promotes apoptosis (Halder and Johnson, 2011). Hippo pathway components include ste-20 family kinases Mst1 and Mst2 (Mst1/2), which complex with scaffold adaptor protein Salvador (Sav) to phosphorylate the nuclear dbf2-related (NDR) family kinases Lats1 and Lats2 (Lats1/2). Yap and Taz, Hippo pathway effectors, are transcriptional co-factors that are substrates for Lats1/2 kinases. Upon phosphorylation by Lats1/2 kinases, Yap and Taz are excluded from the nucleus and transcriptional activity is inhibited. Removing Hippo pathway components in embryonic or adult myocardium releases the downstream effector Yap from Hippo-dependent suppression, promoting cardiomyocyte (CM) proliferation and tissue regeneration (Heallen et al., 2013; Heallen et al., 2011; Morikawa et al., 2015).
We used single cell transcriptomics to investigate Lats1/2 function in epicardial progenitor cell diversification. A high-throughput single cell (sc) RNA-sequence (seq) platform, Drop-seq, was adopted to characterize E13.5 and E14.5 cardiac cellular composition and heterogeneity in Lats1/2 deficient and control hearts (Macosko et al., 2015). Our data revealed that Lats1/2 activity is required for EPDC progression from a transient subepicardial mesenchyme to fully differentiated cardiac fibroblasts and provide insight into mechanisms coordinating fibroblast development with coronary vascular remodeling in heart development.
RESULTS
Epicardial deletion of Lats1/2 results in defective coronary vessel development
We deleted Lats1/2 in E11.5 epicardium using the Wt1CreERT2 allele (Zhou et al., 2008). Lats1/2 conditional knock out (CKO) embryos failed to survive past E15.5 (Fig. S1A). Lats1/2 CKO E14.5 hearts appeared normal (Fig. S1B,C), but E15.5 mutant hearts were smaller, with less compacted myocardium (Fig. 1A, Fig. S1B). Lats1/2 CKO embryos also displayed skin hemorrhages, as well as, herniated livers and intestines (Fig. S1D–F).
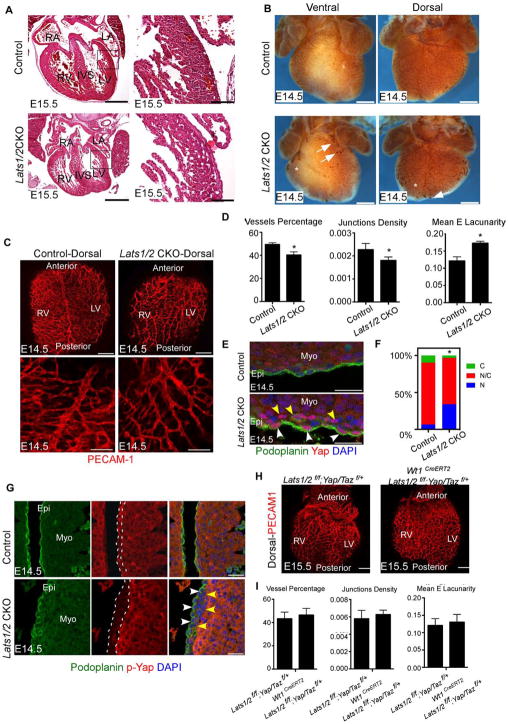
Figure 1. Lats1/2 deficiency results in defective heart development. See also Figure S1 and Figure S2.
(A) E15.5 histology showed reduced compacted myocardium in Lats1/2 CKO. Panels on right are high magnification views of boxed area in panels on left. (B) Coronary vessels visualized by Pecam-1 at E14.5. Lats1/2 CKO had decreased vessel coverage (asterisks) and blood islands (arrows) on ventral and lateral heart. (C) Pecam-1 IF. (D) Quantitation of vasculature in Fig. 1C. (E) Podoplanin labels epicardium and Lats1/2 CKO hearts had increased nuclear Yap in epicardium (white arrowheads) and subepicardium (yellow arrowheads). (F) Quantification of Yap subcellular localization. (G) Lats1/2 CKO had decreased p-Yap in epicardium (white arrows) and subepicardium (yellow arrows). (H–I) Lats1/2 CKO hearts with reduced Yap/Taz were normal at E15.5. Scale bar: A left panels 400μm; right panels 80μm; B 500μm; C upper panels 200μm, bottom panels 100μm, E 25μm, G 50μm, H 200 μm. Data: means ± SD. *P<0.1: Mann-Whitney U test. LA: left atrium; RV: right ventricle; LV: left ventricle; IVS: interventricular septum. RV: right ventricle; LV: left ventricle; N: Nuclear Yap; N/C Nuclear/Cytoplasmic Yap; C: Cytoplasmic Yap; Epi: epicardium; Myo: myocardium.
E14.5 to E15.5 is a critical stage for coronary vessel morphogenesis when the vascular plexus remodels to meet increased oxygen demands (Red-Horse et al., 2010; Viragh and Challice, 1981). Pecam-1 immunostaining in E14.5 Lats1/2 CKO hearts revealed reduced vessel coverage and density with blood island-like structures (Fig. 1B). Pecam-1 immunofluorescence (IF) staining with confocal microscopy and automated quantification revealed dorsal vasculature had decreased branching and reduced vessel coverage with fewer junctions and increased lacunarity (Fig. 1C,D). As controls, we injected tamoxifen to Wt1CreERT2/+ embryos and Cre negative littermates and we injected vehicle (peanut oil) to Wt1CreERT2; Lats1/2 f/f and Cre negative littermates. Coronary vessel development in controls was normal (Fig. S2A,B).
We examined Yap sub-cellular localization and Yap phosphorylation (p-Yap) as a readout of Lats kinase activity. Yap localization in Lats1/2 CKO hearts, detected by total Yap and Podoplanin IF, revealed increased nuclear Yap in both epicardium and subepicardium (Fig. 1E,F). IF revealed decreased p-Yap in Lats1/2 CKO epicardium and subepicardium but no change in CMs since we inactivated Lats 1/2 in the epicardial lineage (Fig. 1G). Podoplanin, restricted to the epicardium in control embryos, was also expressed in Lats1/2 CKO subepicardium suggesting that EMT occurred prior to repression of the epicardial program (Fig. 1G).
Recent work indicated that epicardial deletion of Yap and Taz led to defective EMT (Singh et al., 2016). In situ hybridization with EMT markers revealed that Snai2 was elevated in Lats1/2 CKO hearts, while Twist1 was unchanged (Fig. S2C,D). Tgfβ-signaling that promotes epicardial EMT (Sridurongrit et al., 2008) was elevated in Lats1/2 CKO epicardium as determined by increased nuclear p-Smad2/3, a readout of Tgfβ-signaling (Fig. S2E,F).
To determine if Yap function was required for Lats1/2 CKO phenotype, we genetically reduced endogenous Yap and Taz in Lats1/2 CKO embryos by generating Wt1CreERT2; Lats1/2f/f; Yap/Taz f/+ embryos and induced Cre activity at E11.5. The Wt1CreERT2; Lats1/2f/f; Yap/Taz f/+ embryos were viable at E15.5 without major coronary vasculature defects indicating that Lats1/2 kinases are required for normal coronary vessel development by restricting Yap activity (Fig. 1H,I).
Unbiased single-cell transcriptomics of E13.5 and E14.5 embryonic hearts
We used Drop-seq to profile cardiac tissue from control and Lats1/2 CKO E13.5 and E14.5 embryos, the stages preceding the Lats1/2 CKO cardiac phenotype. Graph based clustering was performed on significant principle components and visualized results through non-linear dimensional reduction algorithm, t-Distributed Stochastic Neighbor Embedding (tSNE) (Maaten and Hinton, 2008; Macosko et al., 2015). We acquired 18,757 cells in total across two time points and two genotypes that was reduced to 18,166 single cells, after subtracting red blood cells and platelets (cluster 16), in 27 distinct clusters (Fig. 2A–C). Differential expression analysis on spatially proximal clusters revealed transcriptionally well-defined clusters. Clusters without transcriptional distinctions were merged and classified based on expression of known markers (Fig. 2C–F).
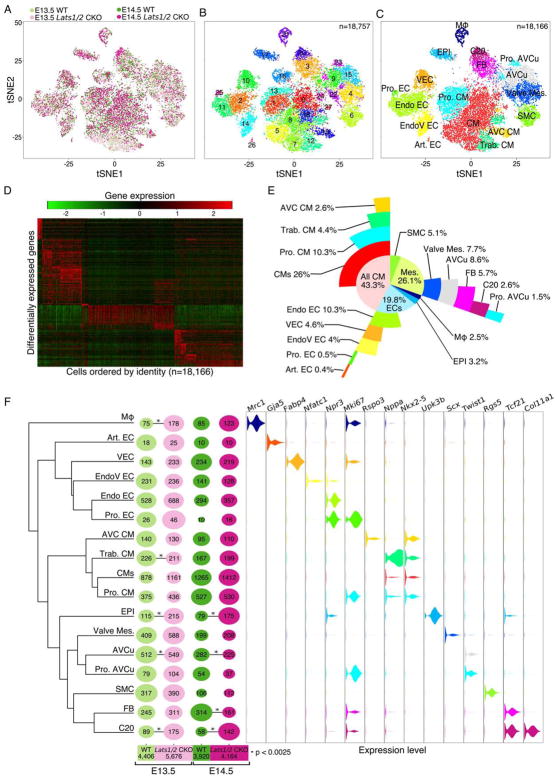
Figure 2. Single cell RNA-seq of embryonic cardiac tissue. See also Figure S3.
(A) two-dimensional tSNE representation of 18,757 single-cell transcriptomes colored by experimental group. (B) tSNE visualization of graph-based clustering carried out on control and Lats1/2 CKO. (C) Classification and merging of clusters shown in Figure 2B. Red blood cells and platelets, cluster 16, are removed. (D) Differentially expressed genes (rows) across final 18,166 cells (columns). (E) Sunburst plot of E13.5–E14.5 cardiac cell composition. Cell classes are colored according to Figure 2C. (F) Lats1/2 CKO and control single-cell composition for individual clusters. Phylogenetic tree of each identified cell type based on 3,217 expressed genes with high dispersion (> 2 standard deviations above average dispersion) (left). Dot plot showing relative proportion of cells belonging to each genotype at E13.5 and E14.5 time points colored according to Figure 2A. Dot size represents percentage of cells within each identity class. Numbers on dots are cell numbers in corresponding cluster and numbers below each genotype are cell numbers analyzed in each group (middle, Chi-squared analysis, * p-value < 0.0025). Violin plot of gene expression for representative markers of each categorical class colored according to Figure 2C (right). Mφ Macrophages; Art. EC, arterial endothelial cells; VEC, vascular endothelial cells; EndoV EC, endocardial valve cells; Endo EC, endocardial cells; Pro. EC, proliferating dndocardial cells; AVC CMs, atrioventricular canal CMs; Trab. CM, trabecular CMs; CMs, standard CMs; Pro. CM, proliferating CMs; EPI, epicardial cells; Valve Mes., valve Mesenchymal cells; AVCu, atrioventricular cushion mesenchymal cells; Pro. AVCu, proliferating atrioventricular cushion mesenchymal cells; SMC, smooth muscle cells; FB, fibroblasts; C20, cluster number 20.
Transcriptional characteristics of cardiomyocytes, endothelial cells, and valve development
CMs, endothelial cells, smooth muscle cells, mesenchymal cells, macrophages, and epicardial cells had cluster sizes that ranged from 63 to 4,716 cells (Fig. 2E,F). We evaluated non-epicardial-derived lineages, specifically CMs and endothelial cells. Analysis of E13.5–E14.5 heart tissue, excluding the atria, revealed a cellular composition that was distinct from adult murine cardiac tissue that is made up of approximately 31% CMs and 43% endothelial cells (Pinto et al., 2016). Our results indicated that E13.5–E14.5 murine heart contains 43% CMs and only 19% endothelial cells (Fig. 2E).
CMs and endothelial cells were heterogeneous and were further sub-categorized (Fig. 2E,F). The two rarest CM populations were atrioventricular canal (AVC) and trabecular CMs. AVC CMs were identified based on a molecular signature that included the Wnt-pathway gene Rspo3 and the Bmp-pathway gene Bmp2 (Fig. 2F, Fig. S3A,B) (Cambier et al., 2014; Ma et al., 2005). In addition to Rspo3 and Bmp2, we detected other AVC CM markers, including Pitx2, Shox2, Wisp1, Tbx2, Tbx3, Tbx5, and Bmp7 (Campione et al., 2001; Habets et al., 2002) (Fig. S3B). Trabecular CMs were identified by a signature that included Nppa and Gja5 (Fig. 2F, Fig. S3B) (Jensen et al., 2012). Further clustering revealed several known and novel CM markers, as well as, proliferating CM signatures (Fig. S3B) (Li et al., 2016).
Endothelial cell subcategories included vascular endothelial cells that were distinguishable by Fabp4 and Apln expression (Fig. S3C) (He et al., 2014; Liu et al., 2015). Arterial endothelial cells were identified by markers, including Gja5, Fbln2, Fbln5, and Sox17 (Fig. 2F and Fig. S3C) (Liu et al., 2015). Endocardial cells were characterized by high levels of Npr3 (Zhang et al., 2016) in addition to markers not previously associated with endocardium including Ednrb, Adgrg6, Plvap, and Smoc1 (Fig. 2F and Fig. S3C). In valvular endocardium, we discovered three distinct clusters of Nfatc1-positive cells (EndoV1-EndoV3) revealing an unappreciated heterogeneity (Fig. S3D). The EndoV3 cluster displayed high expression of endocardial-to-mesenchymal transition (EndoMT) associated genes Enpp2, Prox1, and Fzd10 (Fig. S3C–F) suggesting that dynamic phenotypic changes that characterize EndoMT may add to valvular endocardial heterogeneity (Gong et al., 2014; Lu et al., 2012; Shaul et al., 2014).
Our data make predictions about Wnt-signaling between tissues during valve development. Endo V1 and V2 valvular endothelial cells express Wnt genes Wnt4, Wnt9b, and Wnt16 while the gene encoding Wnt receptor Fzd10 is expressed predominantly in EndoV3 endothelial cells suggesting paracrine signaling between valvular endothelial cells that may promote EndoMT (Fig. S3E,F) (Liebner et al., 2004) (Bosada et al., 2016). Valve mesenchyme expresses Sfrp2, a Wnt inhibitor (Cruciat and Niehrs, 2013), suggesting a mechanism to downregulate Wnt in mesenchymal cell maturation (Fig. S3E,F). We noted heterogeneous expression across valve endocardial clusters of aquaporin (AQPs) genes (e.g. Aqp8 and Aqp1) that encode water channel proteins and control cellular osmotic balance. AQPs have not been implicated in valve development (Fig. S3 E,F) (Rutkovskiy et al., 2013).
An epicardial-derived population composed primarily of Lats1/2 deficient cells
We compared relative proportions of cell types in control and Lats1/2 CKO hearts (Fig. 2F). Although we saw differences in a few non-epicardial-derived cell types, such as macrophages and trabeculated myocardium, we focused on the epicardial lineage. Two clusters showed statistically significant enrichment in Lats1/2 CKO hearts compared to control (chi-squared test, p < 0.0025). One of these was the epicardial cluster, which suggests increased proliferation and self-renewal or defective differentiation of Lats1/2 CKO epicardium. The second predominantly Lats 1/2 mutant cluster, designated Cluster 20 (C20), possessed a gene signature intermediate between that of fibroblasts and epicardium. C20 cells expressed Tcf21, a marker of epicardial cells and resting fibroblasts and Col11a1 (Fig. 2F) (Acharya et al., 2012). Cells that had a transcriptional signature similar to C20 cells were also observed in control hearts at lower frequency than Lats1/2 CKO hearts and IF data revealed that these cells, localized to the subepicardial space, represent subepicardial mesenchyme (see below). Because IF experiments uncovered important differences in protein expression between control and Lats 1/2 CKO subepicardial cells (see below), we will refer to the subepicardial-like cells in Lats 1/2 CKO hearts as C20 cells and in control as subepicardial mesenchyme cells. We also observed a reduction in cardiac fibroblasts in E14.5 Lats1/2 CKO hearts, suggesting a defect in epicardial to fibroblast transition (Fig. 2F).
We used iterative clustering to investigate the transcriptional distinctions between cell populations in more depth. We included vascular endothelial cells (VECs) in our analysis given the Lats 1/2 CKO defective coronary vessel phenotype (Fig. 3A–C). We identified an additional fibroblast-like sub-type, FB2, expressing multiple pericyte markers including Rgs5, Cspg4 (Ng2), Kcnj8, and Pdgfrβ (Fig. 3C and Fig. S4A) (Armulik et al., 2011). Pericytes are precursors of epicardial-derived smooth muscle (Volz et al., 2015). Importantly, the C20 and subepicardial mesenchyme transcriptional signature resembled that of fibroblasts with epicardial features, but was distinct from Fabp4 expressing VECs (Fig. 3C,D). C20 cells and subepicardial mesenchyme expressed a subset of fibroblast markers such as Col1a1 and Spon2, but were deficient for mature fibroblast markers, such as Postn and Sox9 (Fig. 3C,D). C20 and subepicardial mesenchyme also expressed epicardial genes such as Wt1 and Aldh1a2, Dpp4, Smoc2, and Alcam, but failed to express Upk3a and Upk3b that mark mesothelium (Fig. 3C,D) (Rudat et al., 2014). Other genes, not normally expressed in epicardium or fibroblasts, were uniquely enriched in C20 and subepicardial mesenchyme, such as Ephb2 and Vgll3, indicating that C20 and subepicardial mesenchyme have a distinctive signature, while sharing similarity with epicardium and cardiac fibroblasts. The subclustering also revealed that C20 and subepicardial mesenchyme were homogenous and represented an intermediate population between epicardium and fibroblasts (Fig. 3B). Together, the Drop-seq data showed accumulation of epicardial and C20 cells in Lats 1/2 CKO hearts, with a concomitant reduction in differentiated cardiac fibroblasts suggesting that Lats1/2 are required for progression of the normal developmental transition from epicardium to fibroblasts.
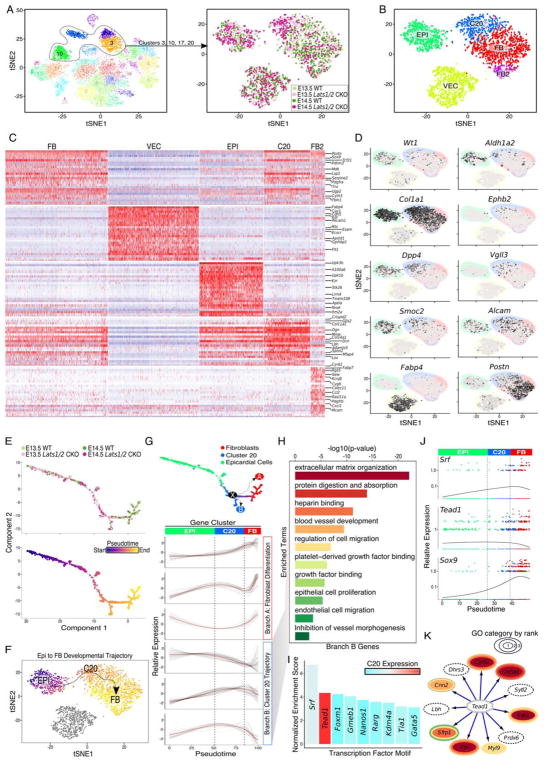
Figure 3. Fibroblast-specified Lats1/2 CKO epicardial cells are arrested along a common differentiation trajectory. See also Figure S3 and Figure S4.
(A) Subsetting and clustering of epicardial and epicardial-derived cells. (Right) View of experimental identity for each single-cell, colored as in Figure 2A. (B) tSNE plot of Lats1/2 CKO enriched clusters. Epicardial cells (EPI), fibroblasts (FB), cluster number 20 (C20), vascular endothelial cells (VEC), and fibroblast cluster 2 (FB2) which resemble pericytes. (C) Heatmap of differentially expressed genes from FB, VEC, EPI, C20, and FB2 clusters. Rows are genes and columns represent single-cells. Red indicates high relative expression. (D) Gene expression projected across tSNE. Black indicates high gene expression and cluster identities and boundaries are set in background and colored according to Figure 3B. (E) Pseudotime trajectory of epicardial cells, fibroblasts, and C20. Cells are colored by experimental groups across epicardial to fibroblast differentiation axis (top). Pseudotime score is assigned to individual cells. Cells with a dark color and bright color represent start and end of pseudotime respectively (bottom). (F) tSNE from Figure 2B is colored by individual cells pseudotime placement and arrow represents epicardium to fibroblast developmental trajectory shown in Fig. 3E. (G) The Monocle2 minimum spanning tree is the underlying black line, and critical node is denoted as ‘X’ with dashed arrows representing the bifurcation of Branch A (fibroblast) and Branch B (C20) (top panel). Hierarchical clustering of genes with branch-specific trends in gene expression across pseudotime. (top three: gene expression pattern enriched in Branch A; bottom three: gene expression pattern enriched in Branch B) (bottom panel). (H) GO analysis of genes identified from branch B, C20-specific, in Figure 3G. Color represents each GO term. (I) Motif enrichment of Branch B, C20, specific genes. Expression of each transcription factor is in percent of cells above threshold in C20. Red indicates high expression. (J) Expression and cell density plots of key transcriptional regulators across pseudotime. Points are each colored by cluster identity from Figure 3B, and 3G. (K) Tead1 target gene visualization and annotation. GO categories assigned and color coordinated based on Figure 3H.
Pseudo-time analysis revealed defective epicardial cell transitions in Lats 1/2 CKO hearts
To investigate epicardial cell developmental trajectory, we used Monocle2 to order cells along a developmental axis progressing from epicardial cells to cardiac fibroblasts (Qiu et al., 2017). Importantly, pseudo-time ordering matched closely with graph-based clustering results, especially when projected across tSNE (Fig. 3E (top panel), F). The left most portion of the epicardial cluster represents the most primitive cellular state and the bottom right section the most differentiated cellular state (Fig. 3E, bottom panel). C20, and a few control subepicardial mesenchyme cells, were isolated centrally along this differentiation axis supporting the notion that C20 is a transition state intermediate between epicardial progenitors and differentiated fibroblasts (Fig. 3G, top panel).
Cells diverging from the Monocle Minimum Spanning Tree (MST) were captured as alternative trajectories by their connection to the full MST path through nodes that represent developmental junctions where cell-fate decisions are made (Trapnell et al., 2014). We identified a node (Fig. 3G, top panel, Node X) proximal to final bifurcation of cardiac fibroblasts (Fig. 3G, top panel, Branch A) and C20 population (Fig. 3G, top panel, Branch B) revealing two distinct EPDC differentiation paths. The “Branch A” trace, composed primarily of control cells (Fig. 3E, top panel), revealed normal EPDC to fibroblast differentiation trajectory and cellular transition from epicardial cells to mature fibroblasts. In contrast, the “Branch B” trace, comprised predominantly of Lats1/2 CKO mutant C20 cells (Fig. 3E top panel), revealed a cell type that was intermediate between epicardial cells and fibroblasts, indicating that most Lats1/2 CKO cells failed to progress to fully differentiated cardiac fibroblasts.
To identify genes expressed in the Node X bifurcation, we examined gene expression patterns across pseudotime and uncovered several co-varying expression patterns. The first expression pattern, characteristic of Branch A fibroblast differentiation, had a higher expression level in differentiated fibroblasts (Fig. 3G bottom panel, top three tracks-Branch A). The second expression pattern was typical of C20 population found in Branch B (Fig. 3G bottom panel, bottom three tracks-Branch B). Gene ontology (GO) analysis on Branch B genes revealed GO terms including extracellular matrix (ECM) and blood vessel development, suggesting that genes expressed by C20 cells contributed to the coronary vessel remodeling defects observed in Lats1/2 CKO hearts (Fig. 3H).
The enriched GO term, ECM, included collagen genes and genes that are essential for collagen processing including SerpinH1 and Plod2 that have also been implicated in human osteogenesis imperfecta syndromes (Table S1) (Ito and Nagata, 2017; Puig-Hervas et al., 2012). In addition, genes encoding growth factors, such as Bmp4 and Edn1 that modulate ECM composition in multiple contexts were also in the Branch B category (Hathaway et al., 2015; Salazar et al., 2016) (Table S1 and Fig. 3K). It has been shown that collagen density regulates proper angiogenesis by modulating the balance between endothelial cell migration and proliferation. An excessive amount of collagen suppresses vessel sprouting (Shamloo et al., 2016). Other ECM genes in Branch B, such as Cyr61 and Ptn (Table S1 and Fig. 3K), are matricellular proteins that promote endothelial cell proliferation (Fang et al., 1992; Hanna et al., 2009; Hinkel et al., 2014). Dpp4, encoding a membrane bound protease, is another ECM gene that is also in “Protein digestion and absorption” GO category (Table S1). Dpp4 proteolyzes both ECM and matrix embedded growth factors to modulate endothelial cell migration (Ghersi et al., 2006). Taken together, Hippo inhibits a gene program that controls multiple aspects of ECM composition, which affects vessel development including endothelial cell proliferation, migration and vessel branching.
Cluster 20-enriched genes are direct Yap-Tead targets
To determine whether C20 and subepicardial mesenchyme expressed genes were direct Yap-Tead targets, we performed unbiased transcription factor DNA-binding motif enrichment analysis across a 20 kb region centered on Transcription Start Site (TSS) of each Branch B gene. Srf and Tead elements were most enriched motifs in Branch B (Fig. 3I). While Tead was highly expressed in C20 and subepicardial mesenchyme, Srf was lowly expressed, suggesting that most Branch B genes were directly regulated by Yap-Tead (Fig. 3I). Tead1 expression was enriched throughout C20 and subepicardial mesenchyme and its expression level decreased moving across pseudotime from C20 and subepicardial mesenchyme toward differentiated fibroblasts (Fig. 3J). Conversely, Sox9 was low in epicardial cells and C20 and subepicardial mesenchyme but high in cardiac fibroblasts (Fig. 3J). We identified Tead1 motif enriched Branch B genes, and performed GO analysis specifically on these Yap-Tead target genes. Enriched GO terms among Branch B Yap-Tead direct target genes were similar to total Branch B enriched GO terms defined above and included ECM organization, regulation of cell migration, and blood vessel development (Fig. 3H,K). Other direct Yap-Tead target genes in C20 and subepicardial mesenchyme were not included in a specific GO category such as Dhrs3, encoding an enzyme that reduces retinoic acid levels (Fig. 3K) (Billings et al., 2013). Other Yap-Tead targets expressed in C20 and subepicardial mesenchyme included Sfrp1, encoding a Wnt antagonist, Lbh, implicated in human congenital heart disease, and Cnn2 encoding an actin binding protein that regulates directed cell migration (McKean et al., 2016; Nusse and Clevers, 2017; Ulmer et al., 2013).
The C20 cluster resides in subepicardial space
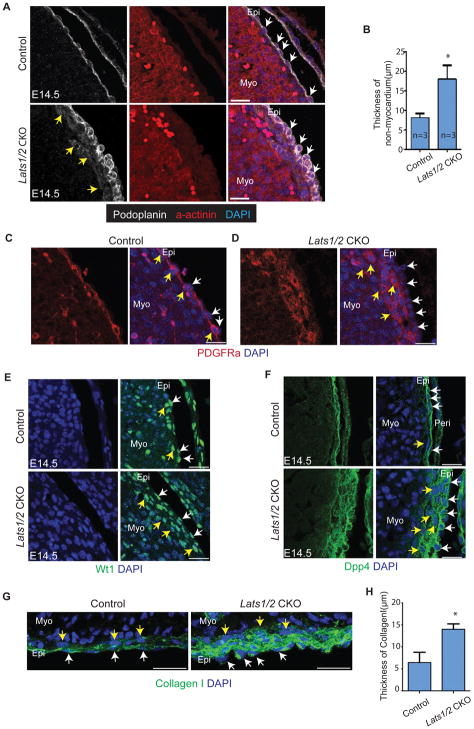
To validate our Drop-seq findings and gain spatial information about C20, we performed IF experiments based on known epicardial markers and newly identified genes expressed in C20. Podoplanin and Keratin expression, markers of the single cell layer epicardium, both expanded into Lats1/2 CKO subepicardium indicating that the Lats1/2 CKO mutant C20 cells still maintained epicardial characteristics that were not observed in control subepicardial mesenchyme (Fig. 4A,B and Fig. S4B) (Acharya et al., 2012). Pdgfr-α, a marker of fibroblasts and subepicardial mesenchyme, was expressed in a single cell layer within E14.5 control subepicardium, while in Lats1/2 CKO embryos Pdgfr-α was expressed in a several layer thick subepicardium containing C20 cells (Fig. 4C,D). Likewise, Wt1, a marker for epicardium and subepicardium, was expressed in Lats1/2 CKO epicardium and C20 cells within the subepicardial space (Fig. 4E) (Rudat and Kispert, 2012). Similarly, Dpp4, Col1a1, and Alcam, expressed in both epicardium and subepicardium in control embryos, were expressed in epicardium and C20 cells in Lats1/2 CKO hearts (Fig. 4F–H and Fig. S4C). Spon2 displayed a different expression pattern by IF (Fig. S4D) compared to its transcriptional expression (Fig. 3C). In Lats 1/2 CKO epicardium, Spon2 was restricted to Lats 1/2 CKO epicardium suggesting posttranscriptional mechanisms inhibit Spon2 protein expression in C20 and subepicardial mesenchyme. Together, the IF data support the conclusion that the C20 population normally resides in subepicardium and that Lats1/2 deficiency results in increased numbers of C20 cells in the subepicardial space.
Figure 4. C20 is located in subepicardium and expands in Lats1/2 CKO hearts. See also Figure S4.
(A–B) Podoplanin labels epicardium (white arrows) and α-actinin labels myocardium. The intervening region is subepicardium and Lats1/2 CKO showed expanded subpicardium weakly stained by Podoplanin (yellow arrows). (C–D) PDGFR-α rarely labels epicardium (white arrows) and extensively labels subepicardium (yellow arrows). Lats1/2 CKO hearts had thickened PDGFR-α positive subepicardium. (E–G) Wt1, Dpp4, and Collagen I labels epicardium (white arrows) and subepicardium (yellow arrows) in Lats1/2 CKO hearts. Lats1/2 CKO hearts had expanded Wt1 positive subepicardium. Scale bar: A,C,E–G, 25μm. In B and H, data are means ± SD. *P<0.1 Mann-Whitney U test.
Lats1/2 epicardial deficiency results in diminished cardiac fibroblast differentiation
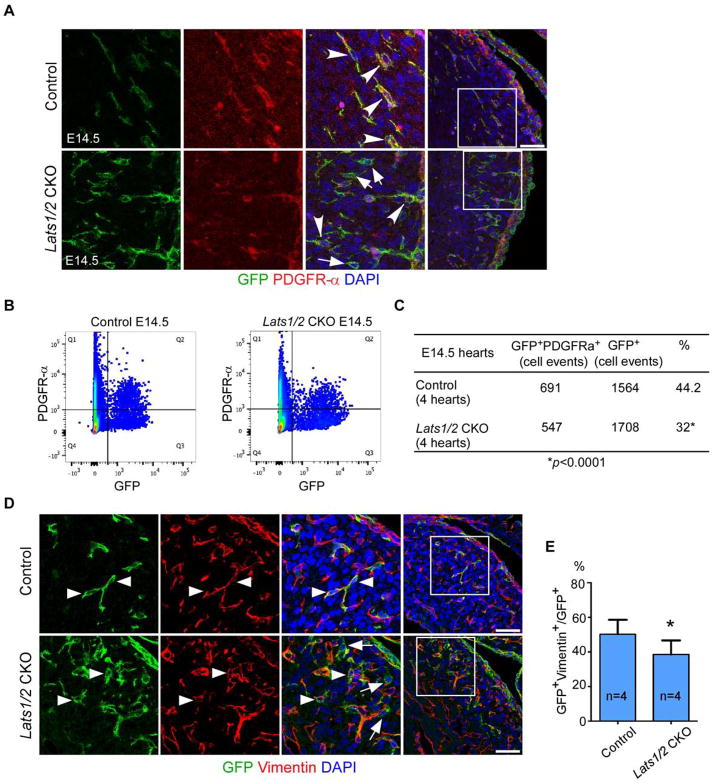
Experiments using Rosa26mTmG reporter to lineage trace epicardial-derived lineages revealed that Lats1/2 CKO hearts showed abundant GFP positive cells in myocardium indicating that EPDC entered the myocardium (Fig. S5A). EdU labeling indicated that Wt1CreERT2 lineage-derived epicardial cells and EPDCs were more proliferative in Lats1/2 CKO embryos than controls (Fig. S5B,C). Proliferation genes were not enriched on the Branch B GO analysis suggesting that proliferation was not the main distinguishing feature between C20 and cardiac fibroblasts (Fig. 3H). Concurrent labeling of the Wt1CreERT2 lineage with Rosa26mTmG and PDGFRα to mark cardiac fibroblasts, revealed by both IF and FACS a reduction in epicardial-derived fibroblasts from 44% to 32% in Lats1/2 CKO mutants compared to controls (Fig. 5A–C). IF with Vimentin, another cardiac fibroblast marker, confirmed the reduction of epicardial-derived fibroblasts (Fig. 5D,E). Together, the lineage tracing data further supported the Drop-seq results, demonstrating that inactivation of Lats1/2 led to defective epicardial differentiation into mature cardiac fibroblasts.
Figure 5. Lats1/2 epicardial deletion leads to reduction of epicardial-derived fibroblast. See also Figure S5.
(A–B) A reduction of epicardial-derived fibroblasts was observed in Lats1/2 CKO hearts. Epicardial-derived fibroblasts are labelled (arrowheads) by GFP and PDGFR-α double positive staining. Other epicardial-derived lineages are indicated (arrows) by GFP single positive staining. Panels on left are a higher magnification view of boxed area in panels on right. (B–C) FACS analysis quantification of percentage of epicardial derived fibroblasts (*P<0.0001: Chi-square test). (D–E) Fibroblast marker Vimentin showed decreased epicardial derived fibroblasts in Lats1/2 CKO hearts. (epicardial derived fibroblasts: arrowheads, other epicardial derived lineages: arrows). Panels on left are higher magnification of boxed area in panels on right. Data are means ± SD. *P<0.1, Mann-Whitney U test. Scale bar: 50μm.
We examined coronary artery smooth muscle differentiation. Smooth muscle myosin heavy chain IF in E15.5 Lats1/2 CKO embryos revealed that coronary arteries established an aortic connection in Lats1/2 CKO embryos and were invested with neural crest-derived smooth muscle (Fig. S6A) (Jiang et al., 2000). PDGFR-β IF combined with lineage tracing uncovered a reduction of E14.5 epicardial-derived smooth muscle cell progenitors suggesting that the smooth muscle lineage differentiation from epicardium was diminished in Lats1/2 CKO hearts. The IF data were consistent with reduction in FB2 pericyte smooth muscle progenitors observed in Drop-seq data (Fig. S6B–D) (Volz et al., 2015).
Yap coordinately regulates fibroblast differentiation and coronary vessel remodeling
To investigate in more depth if genes expressed in C20 and subepicardial mesenchyme are direct Yap target genes, we compared embryonic heart H3K27ac data with cardiac DNase-seq (DNase I hypersensitive site (DHS)) data and extracted TEAD motifs from the E14.5 H3K27ac peaks (GSE52386). Several consensus TEAD motifs, Yap binding sites, were identified at enhancer and promoter regions of genes enriched in Branch B, such as Ogn, Spon2, Gpc3, and Alcam (Fig. S7A,B). Consensus Tead motifs were also found in Dpp4 and Dhrs3. We performed Yap ChIP-qPCR at Tead motifs contained within H3K27ac and DHS peaks using the MEC1 epicardial cell line (Li et al., 2011) to determine if Yap directly bound to these loci. In the Dpp4 and Dhrs3 loci, Yap showed specific binding (Fig. S7B,C).
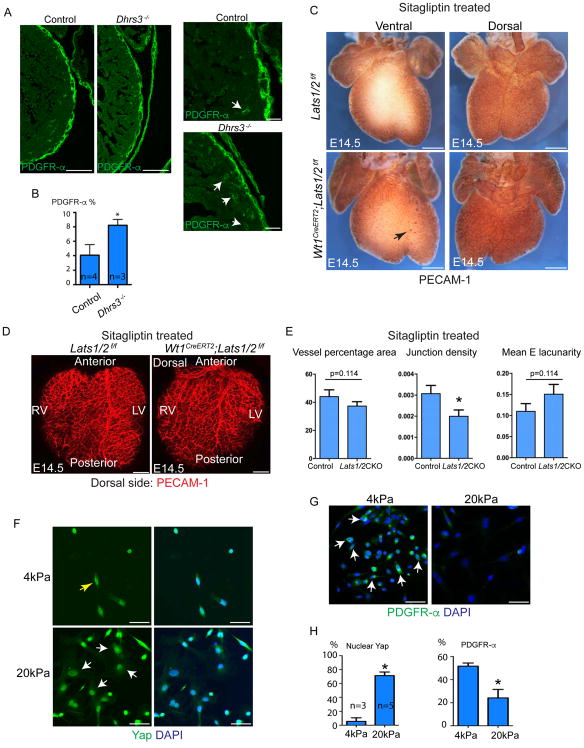
Dhrs3 was up-regulated in Lats 1/2 CKO hearts, which may contribute to impaired fibroblast differentiation by reducing retinoic acid formation and signaling (Billings et al., 2013). Retinoic acid was suggested to play a role in EPDC differentiation into fibroblasts (Braitsch et al., 2012). We used Dhrs3−/− mouse model that had elevated retinoic acid signaling to examine cardiac fibroblast differentiation using PDGFRα+ IF (Billings et al., 2013). We observed more PDGFR-α expressing fibroblasts in Dhrs3−/− myocardium compared with controls supporting the hypothesis that Dhrs3 upregulation, as a downstream Yap target, led to reduction in retinoic acid signaling and promoted fibroblast differentiation arrest in Lat1/2 CKO hearts (Fig. 6A,B).
Figure 6. Hippo/Yap signaling controls fibroblast differentiation and coronary vessel patterning. See also Figure S7.
(A) Cardiac fibroblast labelled with PDGFR-α in control and Dhrs3−/− hearts at E14.5. Dhrs3−/− exhibited more fibroblasts in myocardium (arrows). Right two panels are high magnification of left two panels. (B) Quantification of Figure 6A. (C) Vasculature of Sitagliptin treated E14.5 embryonic hearts were visualized by Pecam-1. Ventral side of Lats1/2 CKO hearts treated with Sitagliptin exhibited some blood islands (arrows), but dorsal side showed comparable vessel coverage. Vessel phenotype was partially suppressed compared with Fig. 1B. (D) Pecam-IF of Sitagliptin treated E14.5 heart. Vessel phenotype was partially suppressed compared with Fig. 1C. (E) Quantification of vessel pattern in Fig. 6D. Junction density was reduced in Lats1/2 CKO hearts treated with Sitagliptin, but vessel percentage and Mean E lacunarity have no difference between Lats1/2 CKO and controls. (F) Primary epicardial cells growing on 4kPa and 20kPa stiffness hydrogel showing cell shape and Yap localization. (G) PDGFR-α expression in primary epicardial cells on different stiffness.(H) Quantification of F, G. Scale bar: A, 50μm, C, 400μm, D, 200μm, F–G, 50μm. In B, E and I, data are means ± SD. Statistics: Mann-Whitney U test.
We treated Lats1/2 CKO embryos and controls with the Dpp4 inhibitor Sitagliptin (Choy and Lam, 2007). Pecam IF and automated quantification revealed that Sitaglipin treatment partially suppressed the disorganized coronary vessel in Lats1/2 CKO hearts (Fig. 6C–E). While some blood islands still appeared on the ventral side of Sitaglipin treated Lats1/2 CKO hearts, the dorsal side lacked blood islands (Fig. 6C). Moreover, the dorsal side vessel coverage phenotype was also suppressed in Sitaglipin treated Lats1/2 CKO hearts when compared to untreated Lats1/2 CKO hearts (compare Fig. 1B,C to Fig. 6C,D). Pecam-1 IF also revealed that defects of vessel percentage area and mean lacunarity were suppressed by Dpp4 inhibition (Fig. 6D,E), supporting our model that Lat1/2 CKO coronary vessel defects were partially caused by elevated Dpp4 activity.
Mechanical signaling regulates Hippo activity in epicardial cells
Mechanical tension is an upstream physiologic regulator of Hippo/Yap activity (Dupont et al., 2011). To examine mechanical tension in epicardial differentiation, we cultured Wt1-expressing primary epicardial cells on hydrogels of different stiffness (Fig. S7D). Primary epicardial cells cultured on 4kPa matrix, approximating embryonic heart stiffness (Majkut et al., 2013), exhibited a spindle shape with Yap distributing equally to nucleus and cytoplasm (Fig. 6F). In contrast, epicardial cells grown on a stiffer 20kPa matrix had a flat shape with increased nuclear Yap localization (Fig. 6F). Interestingly, spindle-shape cells strongly expressed fibroblast marker PDGFRα while flat-shape cells were weakly positive for PDGFRα (Fig. 6G,H). These data suggest that mechanical tension is an upstream physiologic signal controlling Hippo/Yap activity in epicardial cells and that increased nuclear Yap, perhaps resulting from elevated mechanical tension, impedes epicardial cell to fibroblast transition.
DISCUSSION
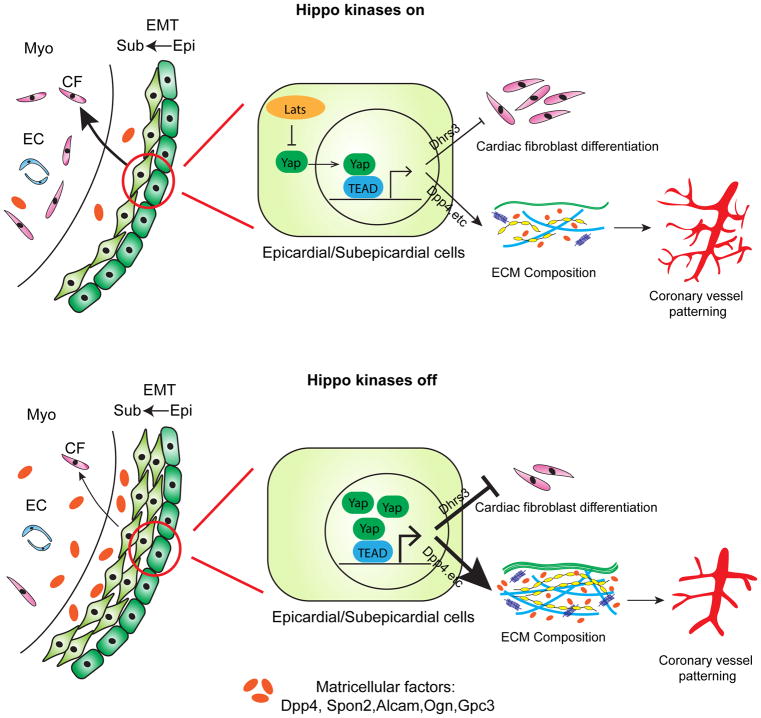
We uncovered an essential role for the Hippo pathway kinases, Lats1/2, in promoting the transition from epicardial progenitors into differentiated cardiac fibroblasts while concurrently controlling ECM composition and vascular remodeling (Fig. 7). Hippo signaling promotes retinoid signaling by inhibiting a negative regulator of retinoid signaling, Dhrs3, enhancing subepicardial mesenchyme to cardiac fibroblast differentiation. Our data suggest that the subepicardial transition state may be responsive to specific physiologic cues, mediated through the Hippo pathway, that modulate genes encoding matricellular factors such as Dpp4 that control ECM characteristics and vascular remodeling (Fig. 7). Our data represent the first in vivo characterization of a transition state at the single cell level in the heart.
Figure 7. Model of Lats kinase regulation of subepicardial cell differentiation and coronary vessel patterning.
In presence of Lats (Hippo kinases on), epicardial cells undergo EMT and transform into subepicardial mesenchyme. Subepicardial mesenchymal cells migrate and differentiate into cardiac fibroblasts (upper left). In absence of Lats (Hippo kinases off), epicardial cells undergo EMT and transform into subepicardium. Differentiation from subepicardium to cardiac fibroblasts is impaired (bottom left). In epicardial and subepicardial cells (middle and right), Lats1/2 kinases restrict nuclear Yap that is required for proper Dhrs3 activity. Nuclear Yap controls ECM composition and coronary patterning by matricellular factor regulation.
Lats1/2 regulate epicardial progenitor differentiation into cardiac fibroblasts
We discovered an autonomous function for Hippo signaling in promoting differentiation of cardiac fibroblasts. Lats1/2 inhibit Dhrs3, a Yap target gene and a negative modulator of retinoic acid signaling, as subepicardial cells differentiate into cardiac fibroblasts. Our data are consistent with previous findings showing that retinoid signaling is required for cardiac fibroblast differentiation and epicardial EMT (Braitsch et al., 2012; Wang et al., 2018). We found that Dhrs3 mutant embryos had more cardiac fibroblasts. Little is know about the interaction of retinoic acid signaling with the Hippo pathway. In an in vitro model of neural crest development, in contrast to our findings Yap synergizes with retinoids to promote the neural crest phenotype suggesting context dependent modifiers of Yap and retinoid interactions (Hindley et al., 2016).
Cluster 20, a cell intermediate between epicardium and cardiac fibroblasts
In Lats1/2 CKO hearts, we discovered an expansion of C20, a cell type with characteristics of both epicardium and primitive cardiac fibroblasts. Importantly, Drop-seq revealed that C20 was found in both wild type and mutant hearts, indicating that C20 is a transition cell, related to subepicardial mesenchyme, that exists during normal epicardial diversification. Furthermore, IF validation revealed that C20 was located in the subepicardial space supporting the hypothesis that C20 is closely related to subepicardial cells. IF experiments also revealed that cells in the Lats 1/2 CKO subepicardial space, containing C20 cells, expressed Podoplanin and Keratin that are restricted epicardial markers indicating that C20, while closely related to control subepicardial cells, also had distinct epicardial characteristics (Acharya et al., 2012) (Fig. 4A and Fig. S4B). The distinct Podoplanin and Keratin expression between Lats1/2 CKO C20 cells and control subepicardial cells was not detected by Drop-seq suggesting that Podoplanin and Keratin expression was regulated post transcriptionally or their transcripts were below the level of detection for Drop-seq.
Hippo signaling coordinates vascular development with fibroblast differentiation
An important insight from our study, not previously appreciated, is the non-autonomous connection between cardiac fibroblast differentiation and vascular patterning. Our data suggest that the disorganized coronary vessel patterning in Lats1/2 CKO hearts is due to aberrant signaling from Lats1/2 mutant C20 cells. Importantly, we validated Dpp4 as a direct Yap target and found that Dpp4 inhibition partially suppressed the coronary vessel remodeling defect in Lats1/2 CKO hearts. Cross talk, mediated by growth factors, between epicardium and myocardium is critical for coronary vessel angiogenesis and myocardium growth during heart development (Smart et al., 2011; Zhou et al., 2011). Our data reveal a new interaction between developing fibroblasts that express matricellular factors that modulate ECM composition and control signaling to developing vasculature.
The cell surface serine protease, Dpp4, and its direct inhibitor Gpc3 regulate multiple, essential signaling events in coronary vascular development (Khurana et al., 2013; Ou et al., 2013). Dpp4 controls the functional activity of chemokines and cytokines that contain Dpp4 proteolytic motifs. In the context of human cord blood, the chemokine Cxcl12 is cleaved by Dpp4 and inhibits the function of uncleaved Cxcl12 (Christopherson et al., 2002). Recent data reveal that Cxcl12, signaling through Cxcr4, promotes endothelial cell migration and coronary plexus pruning and maturation (Cavallero et al., 2015; Harrison et al., 2015). Our Dpp4 inhibitor findings also suggest that a prolonged C20 transition state results in more Dpp4 activity resulting in endothelial remodeling defects.
Mechanical tension determines cardiac fibroblast differentiation by controlling Yap activity
During heart development, organ vascularization is coordinated with overall heart size to meet growing cardiac metabolic needs. Our data suggest that the epicardium and subepicardium are important for coordinating organ vascularity with organ size. In other contexts, it has been shown that in addition to Hippo pathway kinases, mechanical tension acts as another mode of regulation of nuclear Yap activity (Dupont et al., 2011). Our data, using primary epicardial cells, further reveal the importance of mechanical signaling in regulating Yap. A tempting model would be that with heart growth, a gradual tension increase in epicardium and subepicardium may transiently promote nuclear Yap activity and EMT with subepicardial cell proliferation and activation of Yap target genes. In this scenario, Hippo kinase activity may modify the influence of mechanical tension on EMT, proliferation, and differentiation so that the correct number of subepicardial cells and EPDCs are formed.
In addition to new insights into Hippo signaling in epicardial diversification, our data provide important insight into all developing cardiac cell types at E13.5–E14.5, including rare cell types like macrophages and arterial endothelial cells. Along with other recently described data sets, our data will be useful for further investigation of CM and non-CM characteristics in both the developing and adult heart (DeLaughter et al., 2016; Li et al., 2016; Skelly et al., 2018).
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, James F. Martin (jfmartin@bcm.edu).
Experimental Model and Subject Details
Mice strain
Wt1CreERT2 (Zhou et al., 2008), Lats1/2f/f (Heallen et al., 2011), Yap/Taz f/f (Xin et al., 2011), Rosa26mTmG (Muzumdar et al., 2007), Dhrs3−/− (Billings et al., 2013) alleles have been described previously. Mice were on a mixed genetic background of C57BL/6 and 129SV. All animals were maintained in pathogen-free BCM Transgenic Mouse Facility (TMF). All animal protocols and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Baylor College of Medicine in Houston, Texas.
Method Details
Cre activity induction
Tamoxifen was dissolved in peanut oil with 5% ethanol at 10mg/ml. For Wt1CreERT2, 0.1mg/g body weight tamoxifen was administrated to plugged female by intraperitoneal injection at embryonic (E) day 11.5.
Cell preparation and Drop-seq workflow
Atria were removed prior to single cell suspension preparation, droplet generation, cDNA amplification, and library preparation. Following sequencing of Drop-seq libraries, a minimum gene per cell threshold of 500 was set for inclusion into the data set resulting in a final digital gene expression matrix with a median of 1,005 genes per cell and 1,500 Unique Molecular Identifiers (UMI) per cell (Fig. S2A). Batch effects were corrected for and principle components analysis was carried out. Cells were dissociated as previously described (Dyer and Patterson, 2013). Hearts were chopped into several pieces and digested by 1mg/ml collagenase I for 20 min at 37°C. Every 10min, the sample was gently pipetted sample up and down to mechanically dissociate the cells. 10% FBS in DMEM was applied to quench the protease, and samples were passed through a 100μm strainer. Following tissue dissociation cells were diluted to a concentration of 150,000 cells/mL in PBS with 0.1% BSA. Next, Drop-seq was performed according to Macosko et. al (Macosko et al., 2015). Briefly, cells were co-encapsulated into nano-liter sized droplets containing barcoded microparticles and lysis buffer using a Drop-seq specific microfluidics device (custom made by FlowJEM, Toronto, Canada). After droplet breakage, reverse transcription, and exonuclease treatment, total cDNA was amplified, pooled, and purified with Ampure XP beads and then quality control, quantification and size determination was performed using a Fragment analyzer (Advanced Analytical Technologies, Inc.) for quality control, quantification, and size determination. Tagmentation and library preparation was performed with the Illumina Nextera XT kit. Final libraries were triple purified with a successive Ampure XP based protocol, whereby two successive 0.6X (beads to sample ratio) purifications were performed, followed by a final 1X purification step. All libraries were sequenced on an Illumina Nextseq500 instrument.
Drop-seq data analysis
Sequencing data was processed as described in Shekhar et. al. (2016) (Macosko et al., 2015; Shekhar et al., 2016). Briefly, raw fastq files were converted to BAMs with Picard tools (MergeSamFiles) and then used as input for STAR alignment (Dobin et al., 2013), cell barcode correction, and digital gene expression (DGE) matrix generation via the Drop-seq tools software package (available at http://mccarrolllab.com/dropseq/). Next, DGEs from each experiment were merged. We then imported the comprehensive DGE into Seurat (version 1.4.0.5) where normalization was performed according to package default settings (Satija et al., 2015). Batch effects were corrected for with the application of Combat, from the R SVA package (version 3.18.0), and by further regressing out the number of molecules per cell and the percentage of mapped mitochondrial reads with the RegressOut function (Seurat package) (Leek et al., 2012). Next, principle components analysis was performed and significant PCs were used as input for graph-based clustering. Finally, 2-dimesnsional visualization of the multi-dimensional data set was done with tSNE (Maaten and Hinton, 2008). Differential expression of the various clusters was also performed with Seurat, using the likelihood-ratio test for single cell gene expression (McDavid et al., 2013). For iterative clustering, cell barcodes were selected from each cell type and deposited into a new digital expression matrix prior to principle component analysis (PCA), graph based clustering, and tSNE visualization. We evaluated these cell types and performed differential expression analysis (likelihood-ratio test for single cell gene expression (McDavid et al., 2013). For pseudotime analysis, the normalized data from the indicated clusters calculated in Seurat was then passed directly into Monocle2 (Qiu et al., 2017b). The Monocle2 branched expression analysis modeling (BEAM) statistical test was utilized to isolate the branch-specific gene expression patterns (256 genes with qval <0.1). Motif enrichment analysis was carried out with the Cytoscape plug-in iRegulon using the mouse genome’s default parameters (Janky et al., 2014). All gene ontology (GO) analysis was performed with the Metascape tool (http://metascape.org) (Tripathi et al., 2015).
Histology and immunofluorescence
Epicardium-restricted-Lats1/2 mutant embryos (Lats1/2 CKO) were generated by crossing Wt1CreERT2 with Lats1/2f/f or Lats1/2f/f; Rosa26mTmG. Control embryos were generated by crossing Wt1CreERT2 with Rosa26mTmG or Cre negative littermate control. For H&E staining and immunofluorescence staining, hearts were fixed in 4% PFA overnight at 4°C and dehydrated in a serial ethanol, xylene and embedded in paraffin. Sections of 7μm thick sections were stained with H&E for histological analyses (Singhal and Martin, 2015). For some immunofluorescence staining, cryosections were used. Antibodies used for immunofluorescence staining were as follows: GFP (1:200, Abcam ab290, ab6673), phospho-YAP (1:200, Cell signaling technology, 4911), Yap (1:200, Novus NB110-583538), PECAM-1 (1:100–1:200, BD Pharmingen 550274), Vimentin (1:200, Abcam, ab92547), Wt1 (1:200, Abcam ab89901), Podoplanin (1:200, Developmental Studies Hybridoma Bank 8.1.1), α-actinin (1:200, Abcam ab68687), Collagen I (1:200, Abcam ab21286), Dpp4 (1:100, Biolegend H194-112), pan-Keratin (1:200, Abcam ab9377), pSmad2/3 (1:200, Abcam ab52903), PDGFRα (1:100, Cell signaling technology 3174), PDGFRβ (1:100, Cell signaling technology 3169), SM-MHC (1:200, Alfa Aesar BT-562), Spon2 (1:200, Thermo Fisher Scientific PA5-59087), Alcam (1:200, Abcam ab109215). To visualize some antigens, Alexa-647 was employed. When applications required green and red co-staining, sections were pre-treated with 0.3% H2O2 in PBS for 20min at room temperature to quench the endogenous GFP and Tomato signals, which come from the Rosa26mTmG reporter line. In some cases, Tyramide Signal Amplification Systems (1:100, Perkin Elmer) were used to amplify signal. PECAM-1 whole mount staining was performed as previously described (Mukouyama et al., 2012). PECAM-1 (1:100) staining was followed either by anti-rat-HRP (1:200, Life Technologies, 62-9520) and a DAB kit (Vector lab) for color development or anti-rat-Alexa-647 for immunofluorescence staining. The vessel pattern was quantified by Angiotool.
RNA in situ hybridization
The tissues for RNA in-situ hybridization were prepared as described above except adding Diethylpyrocarbonate (DEPC) was added to avoid RNAse contamination. RNA in-situ hybridization was performed by the RNA In Situ Hybridization Core at Baylor College of Medicine. The Twist1 (Ma et al., 2005) and Snai2 (Jiang et al., 1998) probes were previously described.
Gross heart images were captured by Zeiss SteREO Discovery.V12 microscope. Histology and RNA in situ images were captured by Nikon Eclipse 80i microscope. Immunofluorescence images were captured on a Leica TCS SP5 confocal microscope, a Zeiss LSM 780 confocal microscope or Nikon A1-Rs inverted laser scanning microscope.
EdU incorporation assay
To study cell proliferation, pregnant females were injected with 0.08mg/g body weight EdU 2hr before harvesting embryos. Hearts were processed as described above. EdU incorporation was assayed using the Click-it EdU imaging kit.
FACS analysis
Cells were dissociated as previously described in the method section of “Cell preparation and Drop-seq workflow”. BV421 Rat Anti-Mouse CD140A antibody (BD Biosciences 562774) was used to detect PDGFR-α and BV421 Rat IgG2α, κ Isotype (BD Biosciences 562602) was used for gating control. Cells were analyzed using BD Biosciences CORP Aria I and BD Biosciences LSRII and images were processed with FlowJo software.
Motif analysis and ChIP-q-PCR
Gene regulatory region information was extracted from GEO database: E14.5 H3K27ac (GSE52386), P0 H3K27ac (GSE52386) and Adult Heart DHS (GSE37074). TEAD motif was analyzed by Homer. Chromatin immunoprecipitation was performed with an anti-Yap antibody (Novus NB110-58358) in the MEC1 epicardial cell line (Li et al., 2011). Three biological replicates were included in each group. The primers used for detecting TEAD binding region at Dhrs3 and Dpp4 are as follows:
Dpp4 site1-Forward 5′- GGAGGAAGATTATGCACAACAAC -3′;
Dpp4 site1-Reserve 5′- TGTGGAGACATGAAAGACTAAGG -3′;
Dpp4 site2-Forward 5′- GGAGCTCATGAATGCCTGATT -3′;
Dpp4 site2-Reserve 5′- CTGCAGAAGAACTGTGCTCTTA -3′;
Dhrs3-Forward 5′- CCTACCCACACAAGACATCAA -3′;
Dhrs3-Reserve 5′- CTCAGGAGATGATCCAACAAGAA -3′.
Primary epicardial cell culture
Embryonic hearts were isolated from embryos from ICR(CD-1) at E11.5. One litter of the hearts were pooled and digested with 1mg/ml Collagenase I (Worthington). The hearts were maintained intact to expose epicardium only to the digestion buffer. The hearts were digested twice with 500μl Collagenase I at 37 degree with agitation (~100rpm). After every 5 min digestion, pipet hearts up and down for about tens times using disposable dropper. Cell suspension was collected and new digestions buffer was added to the tissue. Next, cell suspension was collected in an equal volume of 10%FBS DMEM and kept on ice. Cell suspension with 10% FBS DMEM from two-time digestion were pooled and filtered through 100-micron tissue strainer (Falcon). Spin down cells at 400g for 5min and re-suspend cells in 10% FBS DMEM number for culturing on stiffness hydrogel. About 2000–3000 cells/well were plated in 24 well plate with stiffness hydrogel made on 12mm round coverslide. After one-day culture, cell identity was detected by IF and >90% cells express epicardial cell marker Wt1 (Fig. S7D). To examine the effect of stiffness on epicardial-fibroblast differentiation, cells were continuously cultured in 10% FBS DMEM supplemented with 100ng/ml b-Fgf (R&D Systems) for 10 days and medium was changed everyday.
Stiffness hydrogel preparation
Tunable stiffness hydrogels were prepared as previously described (Tse and Engler, 2010). 12mm round coverslips were used for making hydrogel. Coverslips were pre-treated with 0.1M NaOH and (3-Aminopropyl) trimethoxysilane (Sigma) for 5 and 10 mins respectively. After rinsed with water, coverslips were covered with 0.5% Glutaraldehyde (Sigma) solution for 30 mins. Another same amount of coverslips were coated with Dichlorodimethylsilane (Sigma) and let them air dry. Stiffness was adjusted based on the relative concentration of acrylamide and bis-acrylamide. For 4kPa, 5% acrylamide and 0.15% bis-acrylamide were mixed with ammonium persulfate and TEMED; for 20kPa, 8% acrylamide and 0.264% bis-acrylamide were mixed with ammonium persulfate and TEMED. On the day for cell culture, 1ml of 50mM HEPES Ph8.5 was mixed with 10ul 50mg/ml Sulfo-SANPAH (sulfosuccinimidyl 6-(4′-azido-2′-nitrophenylamino) hexanoate) (Thermo Fisher Scientific) and was applied to hydrogel under 375nm,15W UV light for 5mins twice. Hydrogel were rinsed with 50mM HEPES PH8.5 several times and were freshly coated with type I rat tail collagen (0.5mg/ml in 0.2% acetic acid) for 2 hours. After this, hydrogels were washed three times by sterile PBS and sterilized under UV in cell culture hood for 30min. And hydrogels were ready to use.
Fibroblast detection on hydrogel
After 10-day culture, only the region with less than 50% confluency were studied. Cells were stained with Yap to visualize Yap cellular localization and PDGFR-α for fibroblast.
Sitagliptin treatment
Dpp4 inhibitor Sitagliptin (Sitagliptin phosphate, Sigma, 1612903) was dissolved in saline. 0.01mg/g body weight Sitagliptin was administrated to plugged female by gavage at E11.5, E12.5 and E13.5.
Quantification and Statistical Analysis
Images taken by Nikon A1-Rs inverted laser scanning microscope were process with FIJI software. Contrast and image size of IF images were adjusted with Adobe photoshop CC or GIMP. Images of Drop-seq were produced in R. Flow cytometry graphs were generated by FlowJo. Bar graphs were generated by GraphPad Prism 6. All figures were made in Adobe Illustrator CC 2015 or Canvas X or Inkscape.
Coronary vessel pattern was analyzed by AngioTool. Statistical analyses were performed in SPSS 21.0. Sample size was labelled on the corresponding bar graph, otherwise n=3 in all groups, which represent the number of hearts were analyzed in each experimental group. N number of cell composition in Drop-seq (Fig. 2F) and FACS analysis (Fig. 5C) indicated number of cells were analyzed. For IF images, three fields of views were analyzed in each heart. For quantification of PDGFR-α cells Fig. 6A, percentage of PDGFR-α was calculated by PDGFR-α+ cell number was divided by total cell number in compact myocardium. For quantification of pSmad2/3high in Fig. S2E and Fig. 6F, in each experimental group, epicardial cells from 2–4 different hearts were analyzed and counted as total observation number. Mann-Whitney U test were used for most studies, except the cell composition in Drop-seq (Fig. 2F), FACS analysis (Fig. 5C), and pSmad2/3high cells in epicardium, in which Chi-square were used. The cut-off value for statistical significance were indicated in corresponding figure legend.
Data and Software Availability
The Drop-seq dataset has been deposited in Gene Expression Omnibus (GEO) with accession number GSE100861.
Supplementary Material
Supplemental Table 1. Related to Figure 3. Gene ontology (GO) analysis of the genes enriched in Branch B.
HIGHLIGHTS.
Single-cell transcriptomic analysis of 18,166 cells from E13.5 and E14.5 heart
Characterization of rare cardiac cell types
Lats1/2 are required for subepicardial transition into cardiac fibroblasts
Lats1/2 are required for proper extracellular milieu and coronary vessel formation
Acknowledgments
Supported by Intellectual and Developmental Disability Research Center grant (1U54 HD083092) from Eunice Kennedy Shriver NICHD; Mouse Phenotyping Core at Baylor College of Medicine with funding from NIH (U54 HG006348); grants from NIH (DE 023177, HL 127717, HL 130804, and HL 118761 to J.F.M.; HD 077260 to A.R.M.; F31HL136065 to M.C.H.) and Vivian L. Smith Foundation and MacDonald Research Fund Award 16RDM001 (J.F.M.). AHA 16GRNT31330023 (J.D.W.). Transatlantic Network of Excellence Award LeDucq Foundation Transatlantic Networks of Excellence in Cardiovascular Research 14CVD01: “Defining genomic topology of atrial fibrillation.” (J.F.M.).
Footnotes
DECLARATION OF INTERESTS
Authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Conceptualization/Methodology: Y.X, J.F.M.; Software/data curation: M.C.H., M.Z., T.J.M.; Investigation: Y.X., M.C.H. Y.M. and J.D.W, Resources: S.W., A.R.M; Writing – Original Draft Y.X., M.C.H., Writing –Review & Editing: J.F.M., J.D.W., A.R.M.; Visualization: Y.X., M.C.H.; Supervision, project administration, funding acquisition: J.F.M.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A, Genove G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Billings SE, Pierzchalski K, Butler Tjaden NE, Pang XY, Trainor PA, Kane MA, Moise AR. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 2013;27:4877–4889. doi: 10.1096/fj.13-227967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosada FM, Devasthali V, Jones KA, Stankunas K. Wnt/beta-catenin signaling enables developmental transitions during valvulogenesis. Development. 2016;143:1041–1054. doi: 10.1242/dev.130575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitsch CM, Combs MD, Quaggin SE, Yutzey KE. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol. 2012;368:345–357. doi: 10.1016/j.ydbio.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier L, Plate M, Sucov HM, Pashmforoush M. Nkx2-5 regulates cardiac growth through modulation of Wnt signaling by R-spondin3. Development. 2014;141:2959–2971. doi: 10.1242/dev.103416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, Blum M, Franco D, Moorman AF. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- Cavallero S, Shen H, Yi C, Lien CL, Kumar SR, Sucov HM. CXCL12 Signaling Is Essential for Maturation of the Ventricular Coronary Endothelial Plexus and Establishment of Functional Coronary Circulation. Dev Cell. 2015;33:469–477. doi: 10.1016/j.devcel.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy M, Lam S. Sitagliptin: a novel drug for the treatment of type 2 diabetes. Cardiol Rev. 2007;15:264–271. doi: 10.1097/CRD.0b013e318123f771. [DOI] [PubMed] [Google Scholar]
- Christopherson KW, 2nd, Hangoc G, Broxmeyer HE. Cell surface peptidase CD26/dipeptidylpeptidase IV regulates CXCL12/stromal cell-derived factor-1 alpha-mediated chemotaxis of human cord blood CD34+ progenitor cells. Journal of immunology (Baltimore, Md: 1950) 2002;169:7000–7008. doi: 10.4049/jimmunol.169.12.7000. [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, et al. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell. 2016;39:480–490. doi: 10.1016/j.devcel.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Dyer LA, Patterson C. Isolation of embryonic ventricular endothelial cells. J Vis Exp. 2013 doi: 10.3791/50463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Hartmann N, Chow DT, Riegel AT, Wellstein A. Pleiotrophin stimulates fibroblasts and endothelial and epithelial cells and is expressed in human cancer. J Biol Chem. 1992;267:25889–25897. [PubMed] [Google Scholar]
- Ghersi G, Zhao Q, Salamone M, Yeh Y, Zucker S, Chen WT. The protease complex consisting of dipeptidyl peptidase IV and seprase plays a role in the migration and invasion of human endothelial cells in collagenous matrices. Cancer Res. 2006;66:4652–4661. doi: 10.1158/0008-5472.CAN-05-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Qu S, Lv XB, Liu B, Tan W, Nie Y, Su F, Liu Q, Yao H, Song E. BRMS1L suppresses breast cancer metastasis by inducing epigenetic silence of FZD10. Nature communications. 2014;5:5406. doi: 10.1038/ncomms6406. [DOI] [PubMed] [Google Scholar]
- Guadix JA, Ruiz-Villalba A, Lettice L, Velecela V, Munoz-Chapuli R, Hastie ND, Perez-Pomares JM, Martinez-Estrada OM. Wt1 controls retinoic acid signalling in embryonic epicardium through transcriptional activation of Raldh2. Development. 2011;138:1093–1097. doi: 10.1242/dev.044594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M, Liu H, Amir J, Sun Y, Morris SW, Siddiqui MA, Lau LF, Chaqour B. Mechanical regulation of the proangiogenic factor CCN1/CYR61 gene requires the combined activities of MRTF-A and CREB-binding protein histone acetyltransferase. J Biol Chem. 2009;284:23125–23136. doi: 10.1074/jbc.M109.019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MR, Bussmann J, Huang Y, Zhao L, Osorio A, Burns CG, Burns CE, Sucov HM, Siekmann AF, Lien CL. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev Cell. 2015;33:442–454. doi: 10.1016/j.devcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway CK, Grant R, Hagaman JR, Hiller S, Li F, Xu L, Chang AS, Madden VJ, Bagnell CR, Rojas M, et al. Endothelin-1 critically influences cardiac function via superoxide-MMP9 cascade. Proc Natl Acad Sci U S A. 2015;112:5141–5146. doi: 10.1073/pnas.1504557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Tian X, Zhang H, Wythe JD, Zhou B. Fabp4-CreER lineage tracing reveals two distinctive coronary vascular populations. J Cell Mol Med. 2014;18:2152–2156. doi: 10.1111/jcmm.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, Martin JF. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley CJ, Condurat AL, Menon V, Thomas R, Azmitia LM, Davis JA, Pruszak J. The Hippo pathway member YAP enhances human neural crest cell fate and migration. Scientific reports. 2016;6:23208. doi: 10.1038/srep23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkel R, Trenkwalder T, Petersen B, Husada W, Gesenhues F, Lee S, Hannappel E, Bock-Marquette I, Theisen D, Leitner L, et al. MRTF-A controls vessel growth and maturation by increasing the expression of CCN1 and CCN2. Nature communications. 2014;5:3970. doi: 10.1038/ncomms4970. [DOI] [PubMed] [Google Scholar]
- Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–1603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nagata K. Biology of Hsp47 (Serpin H1), a collagen-specific molecular chaperone. Seminars in cell & developmental biology. 2017;62:142–151. doi: 10.1016/j.semcdb.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. 2014;10:e1003731. doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen B, Boukens BJ, Postma AV, Gunst QD, van den Hoff MJ, Moorman AF, Wang T, Christoffels VM. Identifying the evolutionary building blocks of the cardiac conduction system. PLoS One. 2012;7:e44231. doi: 10.1371/journal.pone.0044231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell. 2012;22:639–650. doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Margamuljana L, Joseph C, Schouteden S, Buckley SM, Verfaillie CM. Glypican-3-mediated inhibition of CD26 by TFPI: a novel mechanism in hematopoietic stem cell homing and maintenance. Blood. 2013;121:2587–2595. doi: 10.1182/blood-2012-09-456715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR, et al. Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev Cell. 2016;39:491–507. doi: 10.1016/j.devcel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Cavallero S, Gu Y, Chen TH, Hughes J, Hassan AB, Bruning JC, Pashmforoush M, Sucov HM. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebner S, Cattelino A, Gallini R, Rudini N, Iurlaro M, Piccolo S, Dejana E. Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. The Journal of cell biology. 2004;166:359–367. doi: 10.1083/jcb.200403050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hu T, He L, Huang X, Tian X, Zhang H, He L, Pu W, Zhang L, Sun H, et al. Genetic targeting of sprouting angiogenesis using Apln-CreER. Nature communications. 2015;6:6020. doi: 10.1038/ncomms7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MH, Huang CC, Pan MR, Chen HH, Hung WC. Prospero homeobox 1 promotes epithelial-mesenchymal transition in colon cancer cells by inhibiting E-cadherin via miR-9. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:6416–6425. doi: 10.1158/1078-0432.CCR-12-0832. [DOI] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9 [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majkut S, Idema T, Swift J, Krieger C, Liu A, Discher DE. Heart-specific stiffening in early embryos parallels matrix and myosin expression to optimize beating. Curr Biol. 2013;23:2434–2439. doi: 10.1016/j.cub.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean DM, Homsy J, Wakimoto H, Patel N, Gorham J, DePalma SR, Ware JS, Zaidi S, Ma W, Patel N, et al. Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nature communications. 2016;7:12824. doi: 10.1038/ncomms12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Zhang M, Heallen T, Leach J, Tao G, Xiao Y, Bai Y, Li W, Willerson JT, Martin JF. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci Signal. 2015;8 doi: 10.1126/scisignal.2005781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukouyama YS, James J, Nam J, Uchida Y. Whole-mount confocal microscopy for vascular branching morphogenesis. Methods in molecular biology. 2012;843:69–78. doi: 10.1007/978-1-61779-523-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Ou X, O’Leary HA, Broxmeyer HE. Implications of DPP4 modification of proteins that regulate stem/progenitor and more mature cell types. Blood. 2013;122:161–169. doi: 10.1182/blood-2013-02-487470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D’Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, et al. Revisiting Cardiac Cellular Composition. Circ Res. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Hervas MT, Temtamy S, Aglan M, Valencia M, Martinez-Glez V, Ballesta-Martinez MJ, Lopez-Gonzalez V, Ashour AM, Amr K, Pulido V, et al. Mutations in PLOD2 cause autosomal-recessive connective tissue disorders within the Bruck syndrome--osteogenesis imperfecta phenotypic spectrum. Human mutation. 2012;33:1444–1449. doi: 10.1002/humu.22133. [DOI] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjee V, Li D, Manderfield LJ, Liu F, Engleka KA, Aghajanian H, Rodell CB, Lu W, Ho V, Wang T, et al. Epicardial YAP/TAZ orchestrate an immunosuppressive response following myocardial infarction. J Clin Invest. 2017;127:899–911. doi: 10.1172/JCI88759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudat C, Grieskamp T, Rohr C, Airik R, Wrede C, Hegermann J, Herrmann BG, Schuster-Gossler K, Kispert A. Upk3b is dispensable for development and integrity of urothelium and mesothelium. PLoS One. 2014;9:e112112. doi: 10.1371/journal.pone.0112112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudat C, Kispert A. Wt1 and epicardial fate mapping. Circ Res. 2012;111:165–169. doi: 10.1161/CIRCRESAHA.112.273946. [DOI] [PubMed] [Google Scholar]
- Rutkovskiy A, Valen G, Vaage J. Cardiac aquaporins. Basic Res Cardiol. 2013;108:393. doi: 10.1007/s00395-013-0393-6. [DOI] [PubMed] [Google Scholar]
- Salazar VS, Gamer LW, Rosen V. BMP signalling in skeletal development, disease and repair. Nature reviews Endocrinology. 2016;12:203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamloo A, Mohammadaliha N, Heilshorn SC, Bauer AL. A comparative study of collagen matrix density effect on dndothelial sprout formation using experimental and computational approaches. Ann Biomed Eng. 2016;44:929–941. doi: 10.1007/s10439-015-1416-2. [DOI] [PubMed] [Google Scholar]
- Shaul YD, Freinkman E, Comb WC, Cantor JR, Tam WL, Thiru P, Kim D, Kanarek N, Pacold ME, Chen WW, et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323. e1330. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Ramesh S, Cibi DM, Yun LS, Li J, Li L, Manderfield LJ, Olson EN, Epstein JA, Singh MK. Hippo Signaling Mediators Yap and Taz Are Required in the Epicardium for Coronary Vasculature Development. Cell Rep. 2016;15:1384–1393. doi: 10.1016/j.celrep.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Martin PT. A role for Galgt1 in skeletal muscle regeneration. Skeletal muscle. 2015;5:3. doi: 10.1186/s13395-014-0028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dubé KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridurongrit S, Larsson J, Schwartz R, Ruiz-Lozano P, Kaartinen V. Signaling via the Tgf-beta type I receptor Alk5 in heart development. Dev Biol. 2008;322:208–218. doi: 10.1016/j.ydbio.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe. 2015;18:723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Current protocols in cell biology/editorial board, Juan S Bonifacino [et al] 2010;Chapter 10(Unit 10):16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- Ulmer B, Hagenlocher C, Schmalholz S, Kurz S, Schweickert A, Kohl A, Roth L, Sela-Donenfeld D, Blum M. Calponin 2 acts as an effector of noncanonical Wnt-mediated cell polarization during neural crest cell migration. Cell Rep. 2013;3:615–621. doi: 10.1016/j.celrep.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- Volz KS, Jacobs AH, Chen HI, Poduri A, McKay AS, Riordan DP, Kofler N, Kitajewski J, Weissman I, Red-Horse K. Pericytes are progenitors for coronary artery smooth muscle. Elife. 2015;4 doi: 10.7554/eLife.10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yu J, Jones JW, Pierzchalski K, Kane MA, Trainor PA, Xavier-Nieto J, Moise AR. Retinoic Acid Signaling Promotes the Cytoskeleton Rearrangement of Embryonic Epicardial Cells. Faseb J. 2018 doi: 10.1096/fj.201701038R. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat Rec A Discov Mol Cell Evol Biol. 2004;276:43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pu W, Li G, Huang X, He L, Tian X, Liu Q, Zhang L, Wu SM, Sucov HM, et al. Endocardium Minimally Contributes to Coronary Endothelium in the Embryonic Ventricular Free Walls. Circ Res. 2016;118:1880–1893. doi: 10.1161/CIRCRESAHA.116.308749. [DOI] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Related to Figure 3. Gene ontology (GO) analysis of the genes enriched in Branch B.