Abstract
Research during the past decade witnessed the discovery of [4Fe–4S] clusters in several members of the eukaryotic DNA replication machinery. The presence of clusters was confirmed by UV–visible absorption, electron paramagnetic resonance spectroscopy, and metal analysis for primase and the B-family DNA polymerases δ and ζ. The crystal structure of primase revealed that the [4Fe–4S] cluster is buried inside the protein and fulfills a structural role. Although [4Fe–4S] clusters are firmly established in the C-terminal domains of catalytic subunits of DNA polymerases δ and ζ, no structures are currently available and their precise roles have not been ascertained. The [4Fe–4S] clusters in the polymerases and primase play a structural role ensuring proper protein folding and stability. In DNA polymerases δ and ζ, they can potentially play regulatory role by sensing hurdles during DNA replication and assisting with DNA polymerase switches by oscillation between oxidized-reduced states.
1. INTRODUCTION
The story of the discovery of iron–sulfur clusters in eukaryotic DNA polymerases (Pols) begins with a genetic study of pol3–13 mutations involving mutations of one cysteine (Cys) of the second metal-binding motif of yeast Polδ to an alanine. The pol3–13 mutation was found to be synthetically lethal with mutations in the genes MMS19, NBP35, TAH18, and DRE2 that encode for proteins enigmatic at that time (Chanet & Heude, 2003). We now know these are components of the cytosolic iron–sulfur protein assembly machinery (Netz, Mascarenhas, Stehling, Pierik, & Lill, 2014). The connection of iron–sulfur clusters with yeast and human DNA Pols was made 8 years later (Baranovskiy et al., 2012; Netz et al., 2011). During this time, the large subunit of yeast and human DNA primase was excitingly discovered to possess a [4Fe–4S] cluster (Klinge, Hirst, Maman, Krude, & Pellegrini, 2007; Weiner et al., 2007). Now iron–sulfur clusters have been documented or suspected at various locations characteristic for each B-family Pol (Fig. 1A). These clusters play vital, though not completely understood, roles in Pol transactions at the replication fork (Burgers & Kunkel, 2017). We review here what is known, what inferences can be made, and provide protocols for Pol purification and iron content analysis.
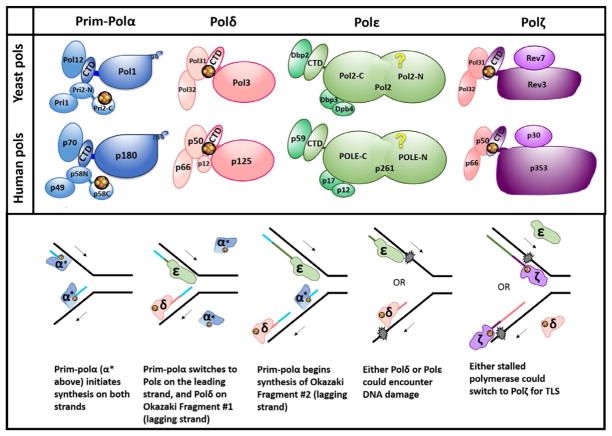
Fig. 1.
B-family DNA polymerases at the replication fork. (A) Subunit structure of yeast and human B-family DNA polymerases. Groups of yellow and orange spheres represent [4Fe–4S] clusters. Only those clusters supported by multiple studies (see Burgers & Kunkel, 2017) are depicted here; others have been proposed but have not been verified yet. These include clusters in CTDs of Polα and Polε. Names of subunits for human Pols corresponding to their molecular mass are shown; the names of the subunits are as follows (from larger to smaller): Polα: POLA1, POLA2, PRIM2, PRIM1; Polδ: POLD1–POLD4; Polε: POLE, POLE2, POLE3, POLE4; Polζ: REV3L, hREV7 (or MAD2L2), POLD3, POLD4. See detailed nomenclature in (Baranovskiy, Gu, et al., 2017). (B) DNA replication under normal conditions and in the presence of DNA damage. Iron–sulfur clusters are shown as in (A). Arrows indicate the direction of DNA synthesis on the respective strand (leading strand top, lagging strand bottom). Newly synthesized DNA is color coded to match the Pol that synthesized that stretch. Blue represents both the primer laid by primase and the DNA laid by Polα. Pol colors match panel (A). For concision, the word “pol” has been omitted to leave only the greek letters to denote Pols, and α* refers to pol α (dark blue portion) plus primase (light blue portion). Recent evidence has suggested that Polδ may also play a role in leading strand synthesis under certain conditions (Burgers & Kunkel, 2017), but this is not shown here.
2. IRON–SULFUR CLUSTERS IN DNA POLYMERASES AND PRIMASE
2.1 DNA Polymerases at the Replication Fork
The basis of heredity is accurate copying of genomic information. Because all DNA Pols synthesize DNA in 5′ →3′ direction, the mechanics of replication of the two DNA strands differ. The leading strand is synthesized continuously in the direction of the moving replication fork, and the lagging strand is synthesized in the opposite direction in ~150-base pair segments called Okazaki fragments (Fig. 1B) (Smith & Whitehouse, 2012). Bulk chromosomal replication is accomplished primarily by the B-family Pols. In eukaryotes, this family is comprised of multisubunit Pols α, δ, ε, and ζ (Fig. 1A). These Pols harbor divergent catalytic domains and assortments of accessory subunits, making each Pol specialized in its role in DNA replication. The C-terminal domains of Pol catalytic subunits (CTDs) contain two metal-binding sites (MBS1 and MBS2) previously thought to contain exclusively Zn2+, as suggested by bioinformatics analysis of B-family Pols (Tahirov, Makarova, Rogozin, Pavlov, & Koonin, 2009) and a crystal structure of the CTD–B-subunit complex of yeast Polα (Klinge, Nunez-Ramirez, Llorca, & Pellegrini, 2009). In 2011, an exciting discovery suggested that MBS2, which participates in interaction with the conserved B-subunit, contains a [4Fe–4S] cluster (Netz et al., 2011). The presence of iron–sulfur clusters in MBS2 of Polα and Polε (Netz et al., 2011) has not been independently confirmed but rather opposed (Baranovskiy, Gu, et al., 2017; Baranovskiy et al., 2012; Klinge et al., 2009; Suwa et al., 2015), as discussed later.
Polα is present in eukaryotic cells in a tight complex with primase (Prim-Polα or primosome, Fig. 1A). Prim-Polα initiates DNA synthesis by synthesizing a primer at the start of the leading strand and each Okazaki fragment (Fig. 1B). Primase synthesizes a 9-mer RNA primer (Baranovskiy, Babayeva, et al., 2016), and then Polα extends it with dNTPs, making a suitable site for loading Polδ or Polε. Polα synthesis is only fairly accurate, as it possesses no internal proofreading capability. Polα is comprised of two subunits (Fig. 1A): catalytic Pol1, possessing 5′ →3′ Pol activity, and accessory B-subunit, Pol12 (yeast subunit names are used in this section; human nomenclature is shown in Fig. 1A and its legend). Primase is also comprised of two subunits: smaller catalytic Pri1 and larger accessory Pri2. The C-terminal domains of Pri2 and p58 (human) contain [4Fe–4S] clusters (Klinge et al., 2007; Weiner et al., 2007) and play important roles in substrate binding and replication initiation (Baranovskiy & Tahirov, 2017).
Once Prim-Polα has dissociated, synthesis is continued by either Polδ or Polε. Polε is required for early steps of DNA replication and is tightly associated with the CMG (Cdc45, Mcm2–7, and GINS) helicase, which travels along the leading strand and unwinds DNA ahead of the fork. Polε participates in DNA synthesis on the same strand (Burgers & Kunkel, 2017; Georgescu et al., 2017; Yeeles, Janska, Early, & Diffley, 2017). Polε synthesis is highly processive due to its unique P-domain (Hogg et al., 2014) and is highly accurate (Ganai & Johansson, 2016; Pavlov, Shcherbakova, & Rogozin, 2006). Its catalytic subunit possesses both 5′ →3′ Pol activity and 3′ →5′ proofreading exonuclease activity. Polε is comprised of four subunits: Pol2, Dpb2, Dpb3, and Dpb4 (Fig. 1A). Catalytic Pol2 is a fusion of two different ancestral B-family Pols and has three domains: two copies of the polymerase/exonuclease module and the CTD (Tahirov et al., 2009). The N-terminal polymerase/exonuclease domain is catalytically active, while the second one is inactive and plays a vital structural role. It has been proposed that the active polymerase/exonuclease domain of Pol2 contains a [4Fe–4S] cluster at the base of the P-domain that enhances template:primer binding (Jain et al., 2014) (question mark in Fig. 1A). However, the crystal structure shows Zn2+ in that position (Hogg et al., 2014). The CTD of Pol2 binds Dpb2, which tethers Polε to the GINS complex of the replisome (Dmowski, Rudzka, Campbell, Jonczyk, & Fijalkowska, 2017; Zhou et al., 2017). Dpb3 and Dpb4 bind the central part of Pol2 and may participate in DNA binding and chromatin remodeling (Bermudez, Farina, Raghavan, Tappin, & Hurwitz, 2011; Hogg & Johansson, 2012).
Polδ takes over synthesis from Polα to complete each Okazaki fragment (Fig. 1B). It is well accepted that Polδ synthesizes primarily the lagging strand, but increasing evidence supports that it also contributes to leading strand synthesis (Pavlov & Shcherbakova, 2010; Stillman, 2015; Waga & Stillman, 1998; Yeeles et al., 2017; Zhou et al., 2017). Polδ is processive when bound to the accessory factor PCNA. It is also very accurate, possessing 3′ →5′ exonuclease activity. Polδ is comprised of three subunits in budding yeast, but four in most organisms (Fig. 1A). MBS2 of the catalytic subunit Pol3 contains a [4Fe–4S]2+ cluster necessary for binding to Pol31 (Baranovskiy et al., 2012; Netz et al., 2011). Pol31 binds the other accessory subunit, Pol32, which can interact with many replication proteins including PCNA.
The fourth B-family member, Polζ, aids in replication when DNA is damaged or when replisome components are defective (Gan, Wittschieben, Wittschieben, & Wood, 2008; Waisertreiger et al., 2012) (Fig. 1B). DNA damage can stall the replication fork because the active sites of accurate Pols ε and δ cannot accommodate the aberrant, bulky structures. When this occurs, one response is translesion synthesis (TLS). During TLS, a A-, B-, or Y-family Pol incorporates a nucleotide across from the damaged site, leaving an aberrant primer terminus that is difficult to extend. Polζ can insert nucleotides across from some lesions, but is most specialized in extending aberrant termini. In this way, Polζ allows for continued DNA replication, but at the risk of mutation due to promiscuous synthesis by the inserter Pol, coupled with Polζ’s low fidelity and lack of proofreading activity. Polζ is responsible for nearly all mutations caused by DNA-damaging agents or by replication stress (Pavlov et al., 2006). Fully functional Polζ is comprised of four subunits (Fig. 1A); accessory subunits include Rev7 and Pol31/Pol32 (latter pair is shared with Polδ) (Baranovskiy et al., 2012; Makarova & Burgers, 2015). Catalytic Rev3 possesses 5′ →3′ Pol activity, and, like Polδ, coordinates a critical [4Fe–4S] cluster in MBS2 (Baranovskiy et al., 2012; Netz et al., 2011).
Pol functions at the fork are summarized in the lower panel of Fig. 1B. This is a simplistic view; many more proteins are involved in vivo. Pols are sophisticated multisubunit and multidomain complexes whose intrinsic and extrinsic acrobatics ensure a variety of transactions during replication. Iron–sulfur clusters play indispensable, though still enigmatic, roles in the dynamics of Pols at the fork. In addition to structural roles discussed later, it has been speculated that they may play roles in Pol switching (Baranovskiy et al., 2012; Stepchenkova, Tarakhovskaya, Siebler, & Pavlov, 2017; Waisertreiger et al., 2012).
2.2 Iron–Sulfur Cluster in the Large Subunit of DNA Primase
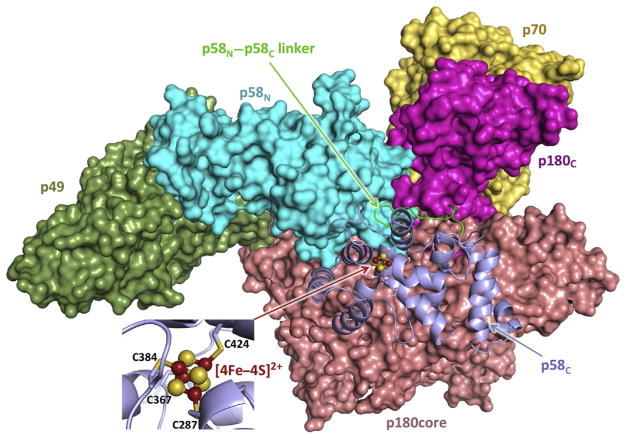
Crystal structures have been reported for full-length human primase alone (Baranovskiy et al., 2015) and for primase within the primosome complex (Baranovskiy, Babayeva, et al., 2016). In the primase structure, the molecule has an extended cashew-like shape. The p58 subunit of primase is folded into two separate domains (Fig. 2): the N-terminal (p58N, residues 1–252) and the C-terminal (p58C, 271–509), connected by a flexible 18-residue linker (253–270). Primase contains three MBS: the Zn2+-binding site of p49 with a proposed structural role, the catalytic site of p49 with two Mg2+ (or Mn2+) ions, and the [4Fe–4S]-binding site of p58C. The latter domain is all-helical and coordinates the iron–sulfur cluster with four conserved cysteines: Cys287, Cys367, Cys384, and Cys424 (Fig. 2). The cluster is fully buried and is in a stable [4Fe–4S]2+ state (Klinge et al., 2007; Weiner et al., 2007). Mutations of iron-coordinating cysteines resulted in disruption of p58C and Pri2C folding, indicating that the cluster plays an indispensable structural role (Klinge et al., 2007; Liu & Huang, 2015; Weiner et al., 2007).
Fig. 2.
Overall view of the human primosome. p58C with an adjacent linker and the rest of the molecule are represented as cartoon and surface, respectively. [4Fe–4S] is represented as ball-sticks, with iron and sulfur colored red and yellow, respectively. The inset shows close-up view of [4Fe–4S]2+ coordination by four conservative Cys of p58C. Coordinates of the human primosome (PDB ID 5EXR) were used to draw this structure. All structural figures are prepared using the PyMOL Molecular Graphics System (version 1.8, Schrödinger, LLC).
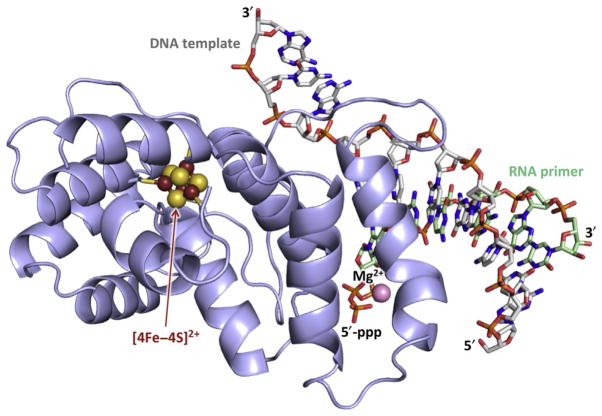
Biochemical studies of substrate binding by human primase revealed that p58C is responsible for the bulk of primase interactions with the template: primer (Baranovskiy, Zhang, et al., 2016). In particular, the 5′-triphosphate of theRNAprimer, which is retained from the initiating nucleotide, and the 3′-overhang of the DNA template interact with p58C during all stages of primer synthesis. During primer synthesis, p58C stays bound to the junction at the 5′-end of the primer, while p49 moves with the growing 3′-end. Confirmation for such a model of primase–substrate interactions was provided by X-ray crystallography (Fig. 3) (Baranovskiy, Babayeva, et al., 2016). The iron–sulfur cluster is located 10Å from the single-stranded DNA-binding surface and 18Å from the double-stranded DNA-binding surface. This structure, together with structures of human primosome and primase with incoming NTP, provided the basis for models of the primosome during initiation, elongation, and termination of primer synthesis and the switch of the template:primer to Polα (Baranovskiy, Babayeva, et al., 2016; Baranovskiy & Tahirov, 2017; Kilkenny, Longo, Perera, & Pellegrini, 2013). The formation of the tight p58C/template:primer complex ensures: (a) the synthesis of a predominantly 9-mer RNA primer by primase; (b) additional verification of primer length after the switch from p49 to Polα, preventing dNTP addition to RNA primers shorter than 9-mer; and (c) coordination of primase and Polα catalytic activities, since primase is unable to initiate new primer synthesis until dissociation of p58C from the template:primer. Recent work proposed the role of redox switching and charge transfer in initiation and termination of RNA synthesis by primase (O’Brien et al., 2017). This idea still awaits rigorous testing because the proposed charge transfer channel in p58C may not exist in the natural protein (Baranovskiy, Babayeva, et al., 2017; Pellegrini, 2017). It is also unclear how the charge transfer is involved in initiation of RNA synthesis on single-stranded DNA.
Fig. 3.
Structure of p58C–DNA:RNA complex. DNA template and RNA primer with 5′-triphosphate are shown as sticks with carbons colored gray and green, respectively. Oxygens, nitrogens, and phosphorus are colored red, blue, and orange, respectively. Mg2+ ion coordinated by 5′-triphosphate is shown as pink sphere with scaling 0.5. The figure was drawn using the coordinates of the p58C/DNA:RNA complex (PDB ID 5F0Q).
2.3 Iron–Sulfur Clusters in B-Family DNA Polymerases
Critical analysis of current biochemical and structural data indicates that only two B-family DNA Pols, δ and ζ, contain the [4Fe–4S] cluster in MBS2 (Baranovskiy et al., 2012; Baranovskiy & Tahirov, 2017; Netz et al., 2011). Iron content in the human CTD–B-subunit (CTD–B) complexes of Polδ and Polζ was about three ions/protein (Baranovskiy et al., 2012), while in purified yeast Polδ it was approximately four (Netz et al., 2011). Variation could be due to different estimates of protein concentration and/or to higher stability of the cluster in intact Pol complexes. Several factors may lead to reduced iron content: oxidation of the [4Fe–4S]2+ cluster to [3Fe–4S]1+, complete disruption of the cluster in a fraction of molecules, and/or incomplete extraction of iron during measurement. Stoichiometry of iron binding below four is quite common after purification of proteins containing [4Fe–4S]2+ clusters; for example, the iron/protein ratio was around three for both p58C and Pri2C, where the iron–sulfur cluster is considered to be stable at aerobic conditions (Klinge et al., 2007; Weiner et al., 2007). Only a small fraction of iron–sulfur clusters in the yeast Polδ sample (<10%) was oxidized upon treatment with ferricyanide (Netz et al., 2011), which might be due to low accessibility of the cluster, its low sensitivity to this reagent, or its high stability, as seen for other cluster-containing enzymes (Fuss, Tsai, Ishida, & Tainer, 2015; Lukianova & David, 2005).
MBS2 is important for interactions between the catalytic and B-subunits of all B-family Pols (Johansson & Macneill, 2010). The crystal structures of the CTD–B complexes of human Pols α and ε (Baranovskiy, Gu, et al., 2017; Suwa et al., 2015) show that Zn2+-MBS2 snugly fits a docking site on the B-subunit (Fig. 4A and B). Sulfur atoms of four zinc-coordinating cysteines form the tetrahedron with length of edges from 3.5 to 3.9Å. In comparison, the edges of [4Fe–4S]2+-coordinating tetrahedron in primase vary from 6.5 to 7.0Å. Therefore, coordination of the [4Fe–4S] by MBS2 of Pols α and ε would require the cysteines to move apart by ~3Å, resulting in significant conformational changes affecting the CTD–B interaction. Consistent with that, iron–sulfur clusters were not detected in pure and fully functional samples of human Polα and Polε, nor in their CTD–B complexes (Baranovskiy, Babayeva, et al., 2016; Baranovskiy et al., 2012; Zahurancik, Baranovskiy, Tahirov, & Suo, 2015; Zhang, Baranovskiy, Tahirov, & Pavlov, 2014). Earlier detection of iron–sulfur clusters in partially purified yeast Polα and Polε (Netz et al., 2011) can be explained by the presence of free catalytic subunits with misincorporated clusters in their CTDs, cluster binding by the Polε catalytic core (Jain et al., 2014), and the presence of primase in the Polα sample. Unfortunately, there is no high resolution structural information for Polδ and Polζ CTDs or their CTD–B complexes. Their [4Fe–4S]-binding modules may interact with the B-subunit differently than the Zn2+-MBS2 in Polα and Polε. In support of this, protein fold predictions suggest the absence of β-strands in the [4Fe–4S]-binding module of Polδ and Polζ, and that two cysteines coordinating the cluster are located on the α-helix (Baranovskiy et al., 2012).
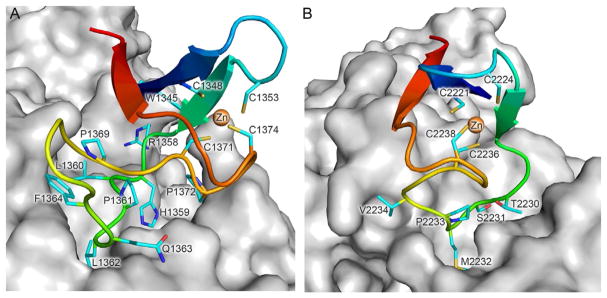
Fig. 4.
Interaction interface of MBS2 of human Polα (A) and Polε (B) with the corresponding B-subunits. MBS2 and the B-subunit are represented as cartoon and surface, respectively. Main chain atoms of MBS2 are colored by a gradient from blue (N-terminus) to red (C-terminus). Side chains of cysteines coordinating Zn2+ as well as the residues participating in interaction with the B-subunit (except those located on β-strands) are shown as sticks and colored cyan for carbons, red for oxygens, and blue for nitrogens. Zn2+ ions are shown as orange spheres with scaling 0.5. The figure was drawn using the coordinates of human Polα and Polε CTD–B complexes (PDB IDs 4Y97 and 5VBN, respectively).
It is worth emphasizing the fixed relative position of two cysteines in MBS2 of Polα: Cys1348 is located on the β-sheet and Cys1353 is located on the short structured loop between the two β-strands (Fig. 4A). This makes [4Fe–4S] cluster coordination difficult and explains why CTD of Polα purified anaerobically was almost iron free, while CTDs of other B-family Pols contained significant amounts of iron and acid-labile sulfide (Netz et al., 2011). In MBS2 of Polε, the relative position of these two Cys is not so well fixed as in Polα (Fig. 4B): there is no proline in the loop between Cys2221 and Cys2224, the β-strand following Cys2224 is shorter, and its residues make four less hydrogen bonds with other residues of MBS2. This structural peculiarity explains the significant level of iron present in Polε CTD samples after purification under aerobic or anaerobic conditions (Baranovskiy et al., 2012; Netz et al., 2011). However, the Polε catalytic subunit (or its CTD) with a misincorporated [4Fe–4S] cluster at MBS2 cannot form a stable complex with the B-subunit, and is efficiently separated during appropriately designed purification (Baranovskiy et al., 2012; Zahurancik et al., 2015) (see Section 4). MBS1 is well structured in both Polα and Polε, precluding [4Fe–4S] coordination.
Intriguingly, coordination of [4Fe–4S] clusters by the CTDs of Polδ and Polζ could contribute to their ability to share the same B-subunit. According to fold predictions, CTDs of human and yeast Pols δ and ζ have very similar secondary structures, especially in the [4Fe–4S]-binding modules (Baranovskiy et al., 2012; Johnson, Prakash, & Prakash, 2012). It is possible that the iron–sulfur cluster integrity and/or changes in its oxidation state affect CTD–B complex stability and determine the mechanism of catalytic subunit switching between Polδ and Polζ during TLS (Baranovskiy et al., 2012; Waisertreiger et al., 2012). For example, oxidation to the [3Fe–4S]1+ state would result in disruption of one of the four bonds between the cluster and the protein, with potential effects on MBS2 folding and its interaction with a B-subunit. In support of this idea, the fourth subunit of human Polδ (p12), proposed to stabilize the complex between the catalytic (p125) and B (p50) subunits (Li et al., 2006), undergoes degradation in response to DNA damage induced by UV, alkylating agents, oxidative, and replication stresses (Lee et al., 2012). The stabilizing role of p12 could be in protecting the iron–sulfur cluster from oxidation. Dissociation and degradation of p12 upon Polδ stalling would trigger dissociation of the catalytic subunit from p50/p66-PCNA/DNA and its replacement by two-subunit Polζ (p353/p30 in humans or Rev3/Rev7 in yeast) (Baranovskiy et al., 2012). In this scenario, the catalytic subunit of Polζ makes a temporary complex with Polδ accessory subunits p50/p66 only during lesion bypass. The alternative model of Polδ to Polζ switching during TLS, where the entire Polδ dissociates from PCNA and is replaced by four-subunit Polζ (p353/p30-p50/p66), is based on successful purification of four-subunit yeast and human Polζ after overexpression (Johnson et al., 2012; Lee, Gregory, & Yang, 2014; Makarova, Stodola, & Burgers, 2012). This model of Pol switch is consistent with the fact that p12, along with p66 and maybe p125, participates in interaction with PCNA (Li et al., 2006); therefore, dissociation of p12 could weaken the Polδ–PCNA complex. The additional role of four-subunit Polζ might be in replacing stalled Polε on the leading strand (Kraszewska, Garbacz, Jonczyk, Fijalkowska, & Jaszczur, 2012) or being a repository of the stabilized form of p353/p30 (with the iron–sulfur cluster protected from oxidation by p50), which replaces p125 on the lagging strand.
It is possible that different mechanisms of Polδ to Polζ switching operate in yeast (Polδ↔Polζ) and humans (p125↔p353/p30). Yeast tetrameric Polζ and trimeric Polδ (naturally missing the fourth subunit) are shown to be stable; therefore, their dissociation during Pol switching might be difficult (Johnson et al., 2012; Makarova et al., 2012). In contrast, our studies with human proteins indicate that the CTD–B complexes of Polδ and Polζ are less stable than complexes of Polα and Polε (unpublished data). In line with that, p50 and p66 were underrepresented in purified human Polζ, indicating that the interaction between p353 and p50 is not strong (Lee et al., 2014). Reduced stability might be an intrinsic property of the CTD–B complexes of human Polδ and Polζ.
3. GENETIC EVIDENCE FOR THE IMPORTANT ROLES OF IRON–SULFUR CLUSTERS IN DNA REPLICATION IN VIVO
The [4Fe–4S] clusters in primase and the CTDs of Polδ and Polζ are coordinated by four Cys residues (Cys1–4, simplified naming according to their order in the polypeptide). However, mutational analysis suggests that not all of them are equally important for the role of any given cluster in vivo.
The [4Fe–4S] cluster in Pri2 has been proposed to contribute to the stability of the Prim-Polα complex and its recruitment to replication start sites (Liu & Huang, 2015). Single mutations affecting Cys1, Cys2, and Cys4 had no effect on the growth of yeast cells, but a strain with C434A substitution (Cys3) was temperature sensitive (Ts), displaying severe growth defects at 30°C and 37°C (normal/permissive is 25–30°C). Notably, substitution of Cys3 with Tyr (C434Y) was lethal to cell growth at 37°C, possibly due to steric interference between the bulky tyrosine and the neighboring residues. Various double mutant combinations (excluding Cys3) also showed growth defects at 30°C and 37°C. The triple mutant (C336A/C417A/C474A) showed severe growth defects and cells permanently arrested in G1 phase, unable to replicate DNA. Mutations leading to C434A, C336A/C417A, and the triple mutant also resulted in decreased Pri2 protein levels at 23°C. Genetic data are generally in agreement with biochemical data with mutant proteins; mutations that cause the Pri2 protein to lose the most iron (signifying degradation/destabilization of the cluster) lead to the most severe phenotypes in vivo in yeast (Klinge et al., 2007).
Single mutations affecting Cys that coordinate the [4Fe–4S] cluster of Pol3 (Polδ) led to a variety of phenotypes (Giot, Chanet, Simon, Facca, & Faye, 1997). The C1059S (Cys2) substitution had no effect on yeast growth; C1069S (Cys3) was Ts, while C1069N was lethal. The C1074S (Cys4) mutant (pol3–13, mentioned first in Section 1) has been more thoroughly characterized. This mutant is Ts at 37°C, reflecting defects in DNA replication. These cells also show hypersensitivity to DNA damaging agents, severely decreased levels of induced mutagenesis (Giot et al., 1997), and, surprisingly, high levels of Polζ-dependent spontaneous mutations (Stepchenkova et al., 2017). This highlights the role of Polδ and its cluster in DNA repair and damage-avoidancemechanisms, potentially through its direct participation or through Pol switches required during TLS.
When the catalytic activity of Polζ is disrupted by mutations affecting the critical aspartate in the active site, almost all induced mutations in yeast vanish (Johnson et al., 2012; Siebler, Lada, Baranovskiy, Tahirov, & Pavlov, 2014). Strains with substitutions in all four cysteines coordinating the [4Fe–4S] of Rev3 behave similarly to the catalytically inactive mutant, showing a drastic decrease in survival after UV and almost complete loss of mutagenesis (Baranovskiy et al., 2012). The C1468S (Cys3) single substitution results in milder responses to UV; no effect was seen in the CAN1 forward mutation assay, but there was a decrease in reversion mutations induced by UV (Johnson et al., 2012). This difference may be because the reversion assay tests for specific base pair substitutions at a particular site, whereas the forward assay tests for a variety of different mutations across the whole CAN1 gene arising by diverse mechanisms. A Cys3/Cys4 double mutant (C1449S/C1473S) showed no binding to Pol31 and an almost complete loss of mutagenesis (Makarova et al., 2012). The complexity of transactions dependent on the iron–sulfur cluster in Polζ is illustrated by a mutant where the whole CTD is deleted, but induced mutagenesis is partially retained (Siebler et al., 2014). Overall, changes of single-Cys residues responsible for [4Fe–4S] binding have varying effects on Pol function, and substitution of more cysteines causes progressively more severe phenotypes.
4. PURIFICATION OF EUKARYOTIC B-FAMILY DNA POLYMERASES
The enzymatic and metal-binding properties of catalytic subunits may significantly differ from those of entire Pol complexes. Therefore, it is important to reduce the level of free catalytic subunits in purified Pol samples to the minimum. This task can be achieved only by placing the affinity tag on the accessory subunit, because it is difficult to remove the excess of catalytic subunit from Pol sample by the ion-exchange or size-exclusion chromatography. Here, we provide the core of a purification scheme applicable to all B-family Pols (yeast and human). Immobilized metal affinity chromatography is used as the first step; a 6×histidine tag (His6) is placed on the N-terminus of the B-subunit. The next important step is chromatography on Heparin HiTrap HP column (GE Healthcare Life Sciences), which allows for the removal of aggregated molecules and excess of the B-subunit. Heparin imitates the sugar-phosphate backbone of DNA substrate and provides tight binding for Pols. The combination of these two chromatography steps usually yields Pol samples with decent purity in 1 day. Higher purity can be achieved by applying additional purification steps, for example, chromatography on MonoS or MonoQ columns (GE Healthcare Life Sciences). Chromatography on the hydroxyapatite column usually does not provide significant improvement in purity, but allows direct loading of protein samples with high salt concentration. Size-exclusion chromatography is preferable as a final polishing step that yields the protein in a storage buffer. Expression of Pol accessory subunits and CTD–B complexes is usually performed in Escherichia coli cell culture at 20°C (the growth media is supplemented with 50 μM FeCl3). Obtaining substantial levels of human Pols or their subcomplexes containing the large catalytic subunits require the employment of more expensive and time-consuming expression in insect cells, which are infected with a recombinant Autographa californica baculovirus. We prefer to use Sf21 cells because of fast duplication, as well as a separate baculovirus for each Pol subunit with the cDNA placed under control of the polyhedrin promoter.
4.1 Procedure
All steps are performed on ice or at 4°C unless otherwise stated. The frozen cell pellet (stored at −80°C in 3–6 g aliquots) is quickly thawed by incubation in room temperature water bath for 5 min and placed on ice. Cells are resuspended in nine volumes of bufferN(bN) containing 20 mMTris–HCl, pH 7.9, 200 mM NaCl, 10 mM K-phosphate, pH 7.9, 3% glycerol, 4 mM β-mercaptoethanol, and supplemented with protease inhibitors: 0.5 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich) and 1 μg/mL leupeptin (Acros Organics). After cell disruption by three passes through the EmulsiFlex-C3 homogenizer (Avestin Inc., Canada), the extract is cleared by centrifugation at 45,000×g for 15 min and loaded to Profinity IMAC resin (Bio-Rad, USA) charged with Ni2+ ions. In the case of robust protein expression, 7 mL of resin is optimal for 14 g of cell pellet. The resin is washed with the following buffers: three column volumes (CV) of bN; five CV of bN containing 400 mM NaCl and 40 mM K-phosphate, pH 7.9; and one CV of bN. The bound material is eluted by 10 CV gradient (0–0.2 M) of imidazole–HCl, pH 7.9, in bN. After analysis by electrophoresis in 12% SDS-PAGE, the purest fractions are combined (approximately two CV) and loaded to 5 mL Heparin HiTrap HP column (GE Healthcare Life Sciences). It is imperative to estimate protein amount and load no more than 3 mg of protein per mL of resin. After washing the column with two CV of bN, Pols are eluted by 10 CV gradient bN200–bN900 (subscript indicates NaCl concentration in mM). After purity analysis by SDS-PAGE, the purest fractions are combined (approximately one CV), dialyzed 3 h against 50 volumes of storage buffer: 10 mM Tris–HCl, pH 7.9, 200 mM NaCl (or KCl), 3% glycerol, and 1 mM dithiothreitol, and flash frozen in small aliquots.
4.2 Notes
Purity analysis by electrophoresis may be skipped if the elution place of Pol peak is well known.
According to structural and biochemical data, His-tag on the N-terminus of the B-subunit does not affect the formation of the Pol complex or its activity, but it can potentially affect interaction with other replication factors. In this case, a cleavable affinity tag is preferable, which can be implemented by adding the short recognition sequence for highly specific protease, like human rhinovirus 3C protease (common name of precision protease) or nuclear inclusion protease from tobacco etch virus. Tag cleavage usually requires overnight digestion, which prolongs purification and increases possibility of sample degradation and oxidation.
We found that most of the expressed Pol complexes accumulate in the cytoplasm of insect cells and can be easily extracted by cell pellet resuspension in bN after one freeze–thaw cycle.
Buffer exchange is carried out by placing the Pol sample into the Spectra/Pore 7 dialysis membrane with molecular weight cut off 25 kDa. This membrane is made of regenerated cellulose and chemically treated to minimize the heavy metal and sulfur content. Dialysis is conducted with continuous buffer mix by placing the beaker on a stirrer; beaker contains a stir bar of adequate size and must be covered by Saran to prevent the buffer and sample aeration.
5. ANALYSIS OF IRON CONTENT IN PROTEIN SAMPLES
Protein samples with [4Fe–4S] clusters have a yellow-brown color, which turns darker upon higher concentration. Iron–sulfur clusters show a wide absorbance peak around 400 nm, which is masked by a peak at 280 nm in large proteins. A detailed review of iron–sulfur cluster detection and analysis methods is provided in (Fuss et al., 2015). Most of these methods require expensive equipment and a significant amount of pure protein. Here, we provide a simple protocol for nonheme iron detection, routinely used in our laboratory to quickly analyze freshly purified protein samples. This calorimetric method is based on the reaction between ferrous ions (Fe2+) and the chromogen ferrozine, resulting in a violet-colored complex whose intensity can be measured spectrophotometrically at 560 nm. Ferrozine shows higher sensitivity in comparison to other chromogens used in iron determination, including thiocyanate o-phenanthroline, bathophenanthroline, and 2,4,6-Tris(2-pyridyl)-s-triazine (TPTZ). Ferrozine does not react with ferric ions (Fe3+), so the reducing agent hydroxylamine hydrochloride should be present to have all iron in Fe2+ form. The method described here is based on the protocol provided with the proprietary reagent set for measurement of the total iron-binding capacity of serum (Pointe Scientific, Inc.). Treatment at 37°C in acidic buffer containing surfactant allows for protein denaturation and quantitative iron extraction. Unlike serum, pure DNA Pol samples do not have detectable absorbance at 560 nm, so the original protocol was simplified by excluding absorbance reading before adding the ferrozine to a protein sample treated with an acidic buffer. The color intensity is proportional to the iron concentration in the range of 0–90 μM.
5.1 Procedure
The reaction is set up at room temperature in a 0.6-mL tube; 10 μL of protein sample is diluted with 51 μL of premixed reagent solution containing 50 μL of buffer (220 mM hydroxylamine hydrochloride in acetate buffer, pH 4.5, with surfactant) and 1 μL of iron color reagent (16.7 mM ferrozine in hydroxylamine hydrochloride). After 10 min at 37°C, samples were spun for 3 min at 15,000×g at room temperature to remove denatured protein. Supernatant (60 μL) is used to record the absorbance at 560 nm against a blank solution, using trUView cuvette (Bio-Rad; allows for measuring absorbance in volumes as low as 50 μL) and Nanodrop 2000c (Thermo Scientific). Blank solution and iron standard are prepared and incubated in a similar way, except 10 μL of the iron-free buffer and 90 μM FeCl2 in hydroxylamine hydrochloride, respectively, are added instead of protein. Iron concentration in the protein sample is calculated using the following formula: A560(sample)/A560(standard)×90 μM. Finally, the ratio of iron and protein concentration gives the iron content/protein molecule.
5.2 Notes
For accurate iron determination, protein samples should contain 0.1–0.9 nmol of iron, which corresponds to 25–225 pmol of protein with [4Fe–4S] cluster (corresponding to 1.25–11.25 μg of 50 kDa protein).
Protein concentration determined by different methods can vary. We prefer measuring absorbance at 280 nm and using extinction coefficients calculated with the ProtParam software.
The absorbance reading of the iron standard should be close to 0.4.
Increasing reaction time to 1 h and temperature to 45°C might be useful for iron extraction from highly stable proteins.
The readings are stable for several hours at room temperature after sample preparation.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (NIGMS) Grant GM101167 to T.H.T. and Eppley Institute Pilot grant to Y.I.P. The Eppley Institute’s X-ray Crystallography Core Facility is supported by the Cancer Center Support Grant P30CA036727.
The authors declare no competing financial interests.
ABBREVIATIONS
- CTD
C-terminal domain of Pol catalytic subunits
- CTD–B
complex between CTD and B-subunit
- MBS
metal-binding site
- Pol
polymerase
- TLS
translesion synthesis
- Ts
temperature sensitive
References
- Baranovskiy AG, Babayeva ND, Zhang Y, Blanco L, Pavlov YI, Tahirov TH. Comment on “The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport”. Science. 2017;357(6348):eaan2396. doi: 10.1126/science.aan2396. https://doi.org/10.1126/science.aan2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Babayeva ND, Zhang Y, Gu J, Suwa Y, Pavlov YI, et al. Mechanism of concerted RNA-DNA primer synthesis by the human primosome. The Journal of Biological Chemistry. 2016;291(19):10006–10020. doi: 10.1074/jbc.M116.717405. https://doi.org/10.1074/jbc.M116.717405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Gu J, Babayeva ND, Kurinov I, Pavlov YI, Tahirov TH. Crystal structure of human Pol epsilon B subunit in complex with C-terminal domain of catalytic subunit. The Journal of Biological Chemistry. 2017;292(38):15717–15730. doi: 10.1074/jbc.M117.792705. https://doi.org/10.1074/jbc.M117.792705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Lada AG, Siebler HM, Zhang Y, Pavlov YI, Tahirov TH. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. The Journal of Biological Chemistry. 2012;287(21):17281–17287. doi: 10.1074/jbc.M112.351122. https://doi.org/10.1074/jbc.M112.351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Tahirov TH. Elaborated action of the human primosome. Genes (Basel) 2017;8(2):E62. doi: 10.3390/genes8020062. https://doi.org/10.3390/genes8020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Zhang Y, Suwa Y, Babayeva ND, Gu J, Pavlov YI, et al. Crystal structure of the human primase. The Journal of Biological Chemistry. 2015;290(9):5635–5646. doi: 10.1074/jbc.M114.624742. https://doi.org/10.1074/jbc.M114.624742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Zhang Y, Suwa Y, Gu J, Babayeva ND, Pavlov YI, et al. Insight into the human DNA primase interaction with template-primer. The Journal of Biological Chemistry. 2016;291(9):4793–4802. doi: 10.1074/jbc.M115.704064. https://doi.org/10.1074/jbc.M115.704064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez VP, Farina A, Raghavan V, Tappin I, Hurwitz J. Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. The Journal of Biological Chemistry. 2011;286(33):28963–28977. doi: 10.1074/jbc.M111.256289. https://doi.org/10.1074/jbc.M111.256289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PMJ, Kunkel TA. Eukaryotic DNA replication fork. Annual Review of Biochemistry. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. https://doi.org/10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet R, Heude M. Characterization of mutations that are synthetic lethal with pol3-13, a mutated allele of DNA polymerase delta in Saccharomyces cerevisiae. Current Genetics. 2003;43(5):337–350. doi: 10.1007/s00294-003-0407-2. https://doi.org/10.1007/s00294-003-0407-2. [DOI] [PubMed] [Google Scholar]
- Dmowski M, Rudzka J, Campbell JL, Jonczyk P, Fijalkowska IJ. Mutations in the non-catalytic subunit Dpb2 of DNA polymerase epsilon affect the Nrm1 branch of the DNA replication checkpoint. PLoS Genetics. 2017;13(1):e1006572. doi: 10.1371/journal.pgen.1006572. https://doi.org/10.1371/journal.pgen.1006572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss JO, Tsai CL, Ishida JP, Tainer JA. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochimica et Biophysica Acta. 2015;1853(6):1253–1271. doi: 10.1016/j.bbamcr.2015.01.018. https://doi.org/10.1016/j.bbamcr.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan GN, Wittschieben JP, Wittschieben BO, Wood RD. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Research. 2008;18(1):174–183. doi: 10.1038/cr.2007.117. https://doi.org/10.1038/cr.2007.117. [DOI] [PubMed] [Google Scholar]
- Ganai RA, Johansson E. DNA replication—A matter of fidelity. Molecular Cell. 2016;62(5):745–755. doi: 10.1016/j.molcel.2016.05.003. https://doi.org/10.1016/j.molcel.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, Zhang D, et al. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(5):E697–E706. doi: 10.1073/pnas.1620500114. https://doi.org/10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L, Chanet R, Simon M, Facca C, Faye G. Involvement of the yeast DNA polymerase delta in DNA repair in vivo. Genetics. 1997;146(4):1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Johansson E. DNA polymerase epsilon. Sub-Cellular Biochemistry. 2012;62:237–257. doi: 10.1007/978-94-007-4572-8_13. https://doi.org/10.1007/978-94-007-4572-8_13. [DOI] [PubMed] [Google Scholar]
- Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, Sauer-Eriksson AE, et al. Structural basis for processive DNA synthesis by yeast DNA polymerase epsilon. Nature Structural & Molecular Biology. 2014;21(1):49–55. doi: 10.1038/nsmb.2712. https://doi.org/10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- Jain R, Vanamee ES, Dzikovski BG, Buku A, Johnson RE, Prakash L, et al. An iron-sulfur cluster in the polymerase domain of yeast DNA polymerase epsilon. Journal of Molecular Biology. 2014;426(2):301–308. doi: 10.1016/j.jmb.2013.10.015. https://doi.org/10.1016/j.jmb.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends in Biochemical Sciences. 2010;35(6):339–347. doi: 10.1016/j.tibs.2010.01.004. https://doi.org/10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(31):12455–12460. doi: 10.1073/pnas.1206052109. https://doi.org/10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny ML, Longo MA, Perera RL, Pellegrini L. Structures of human primase reveal design of nucleotide elongation site and mode of Pol alpha tethering. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(40):15961–15966. doi: 10.1073/pnas.1311185110. https://doi.org/10.1073/pnas.1311185110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nature Structural & Molecular Biology. 2007;14(9):875–877. doi: 10.1038/nsmb1288. https://doi.org/10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Nunez-Ramirez R, Llorca O, Pellegrini L. 3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases. The EMBO Journal. 2009;28(13):1978–1987. doi: 10.1038/emboj.2009.150. https://doi.org/10.1038/emboj.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraszewska J, Garbacz M, Jonczyk P, Fijalkowska IJ, Jaszczur M. Defect of Dpb2p, a noncatalytic subunit of DNA polymerase epsilon, promotes error prone replication of undamaged chromosomal DNA in Saccharomyces cerevisiae. Mutation Research. 2012;737(1–2):34–42. doi: 10.1016/j.mrfmmm.2012.06.002. https://doi.org/10.1016/j.mrfmmm.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Lee YS, Gregory MT, Yang W. Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(8):2954–2959. doi: 10.1073/pnas.1324001111. https://doi.org/10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Zhang S, Lin SH, Chea J, Wang X, LeRoy C, et al. Regulation of human DNA polymerase delta in the cellular responses to DNA damage. Environmental and Molecular Mutagenesis. 2012;53(9):683–698. doi: 10.1002/em.21743. https://doi.org/10.1002/em.21743. [DOI] [PubMed] [Google Scholar]
- Li H, Xie B, Zhou Y, Rahmeh A, Trusa S, Zhang S, et al. Functional roles of p12, the fourth subunit of human DNA polymerase delta. The Journal of Biological Chemistry. 2006;281(21):14748–14755. doi: 10.1074/jbc.M600322200. https://doi.org/10.1074/jbc.M600322200. [DOI] [PubMed] [Google Scholar]
- Liu L, Huang M. Essential role of the iron-sulfur cluster binding domain of the primase regulatory subunit Pri2 in DNA replication initiation. Protein & Cell. 2015;6(3):194–210. doi: 10.1007/s13238-015-0134-8. https://doi.org/10.1007/s13238-015-0134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Current Opinion in Chemical Biology. 2005;9(2):145–151. doi: 10.1016/j.cbpa.2005.02.006. https://doi.org/10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Makarova AV, Burgers PM. Eukaryotic DNA polymerase zeta. DNA Repair (Amst) 2015;29:47–55. doi: 10.1016/j.dnarep.2015.02.012. https://doi.org/10.1016/j.dnarep.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova AV, Stodola JL, Burgers PM. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Research. 2012;40(22):11618–11626. doi: 10.1093/nar/gks948. https://doi.org/10.1093/nar/gks948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJ, Mascarenhas J, Stehling O, Pierik AJ, Lill R. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends in Cell Biology. 2014;24(5):303–312. doi: 10.1016/j.tcb.2013.11.005. https://doi.org/10.1016/j.tcb.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nature Chemical Biology. 2011;8(1):125–132. doi: 10.1038/nchembio.721. https://doi.org/10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien E, Holt ME, Thompson MK, Salay LE, Ehlinger AC, Chazin WJ, et al. The [4Fe4S] cluster of human DNA primase functions as a redox switch using DNA charge transport. Science. 2017;355(6327):eaag1789. doi: 10.1126/science.aag1789. https://doi.org/10.1126/science.aag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV. DNA polymerases at the eukaryotic fork—20 years later. Mutation Research. 2010;685(1–2):45–53. doi: 10.1016/j.mrfmmm.2009.08.002. https://doi.org/10.1016/j.mrfmmm.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. International Review of Cytology. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. https://doi.org/10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- Pellegrini L. Comment on “The [4Fe4S] cluster of human DNAprimase functions as a redox switch using DNA charge transport”. Science. 2017;357(6348):eaan2954. doi: 10.1126/science.aan2954. https://doi.org/10.1126/science.aan2954. [DOI] [PubMed] [Google Scholar]
- Siebler HM, Lada AG, Baranovskiy AG, Tahirov TH, Pavlov YI. A novel variant of DNA polymerase zeta, Rev3DeltaC, highlights differential regulation of Pol32 as a subunit of polymerase delta versus zeta in Saccharomyces cerevisiae. DNA Repair (Amst) 2014;24:138–149. doi: 10.1016/j.dnarep.2014.04.013. https://doi.org/10.1016/j.dnarep.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483(7390):434–438. doi: 10.1038/nature10895. https://doi.org/10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepchenkova EI, Tarakhovskaya ER, Siebler HM, Pavlov YI. Defect of Fe-S cluster binding by DNA polymerase delta in yeast suppresses UV-induced mutagenesis, but enhances DNA polymerase zeta-dependent spontaneous mutagenesis. DNA Repair (Amst) 2017;49:60–69. doi: 10.1016/j.dnarep.2016.11.004. https://doi.org/10.1016/j.dnarep.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Stillman B. Reconsidering DNA polymerases at the replication fork in eukaryotes. Molecular Cell. 2015;59(2):139–141. doi: 10.1016/j.molcel.2015.07.004. https://doi.org/10.1016/j.molcel.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwa Y, Gu J, Baranovskiy AG, Babayeva ND, Pavlov YI, Tahirov TH. Crystal structure of the human Pol alpha B subunit in complex with the C-terminal domain of the catalytic subunit. The Journal of Biological Chemistry. 2015;290(23):14328–14337. doi: 10.1074/jbc.M115.649954. https://doi.org/10.1074/jbc.M115.649954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Evolution of DNA polymerases: An inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biology Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. https://doi.org/10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNAreplication fork in eukaryotic cells. Annual Review of Biochemistry. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. https://doi.org/10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Waisertreiger IS, Liston VG, Menezes MR, Kim HM, Lobachev KS, Stepchenkova EI, et al. Modulation of mutagenesis in eukaryotes by DNA replication fork dynamics and quality of nucleotide pools. Environmental and Molecular Mutagenesis. 2012;53(9):699–724. doi: 10.1002/em.21735. https://doi.org/10.1002/em.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner BE, Huang H, Dattilo BM, Nilges MJ, Fanning E, Chazin WJ. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. The Journal of Biological Chemistry. 2007;282(46):33444–33451. doi: 10.1074/jbc.M705826200. https://doi.org/10.1074/jbc.M705826200. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Janska A, Early A, Diffley JF. How the eukaryotic replisome achieves rapid and efficient DNA replication. Molecular Cell. 2017;65(1):105–116. doi: 10.1016/j.molcel.2016.11.017. https://doi.org/10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahurancik WJ, Baranovskiy AG, Tahirov TH, Suo Z. Comparison of the kinetic parameters of the truncated catalytic subunit and holoenzyme of human DNA polymerase varepsilon. DNA Repair (Amst) 2015;29:16–22. doi: 10.1016/j.dnarep.2015.01.008. https://doi.org/10.1016/j.dnarep.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Baranovskiy AG, Tahirov TH, Pavlov YI. The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase alpha-primase complex. The Journal of Biological Chemistry. 2014;289(32):22021–22034. doi: 10.1074/jbc.M114.570333. https://doi.org/10.1074/jbc.M114.570333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, Kotecha A, et al. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(16):4141–4146. doi: 10.1073/pnas.1700530114. https://doi.org/10.1073/pnas.1700530114. [DOI] [PMC free article] [PubMed] [Google Scholar]