Abstract
The choroid plays a critical role in maintaining the portions of the eye responsible for vision. Specific alterations in the choroid have been associated with several disease states, including age-related macular degeneration (AMD), central serous choroiretinopathy, retinitis pigmentosa and diabetes. In addition, choroid thickness measures have been shown as a predictive biomarker for treatment response and visual function. Where several approaches currently exist for segmenting the choroid in optical coherence tomography (OCT) images of healthy retina, very few are capable of addressing images with retinal pathology. The difficulty is due to existing methods relying on first detecting the retinal boundaries before performing the choroidal segmentation. Performance suffers when these boundaries are disrupted or suffer large morphological changes due to disease, and cannot be found accurately. In this work, we show that a learning based approach using convolutional neural networks can allow for the detection and segmentation of the choroid without the prerequisite delineation of the retinal layers. This avoids the need to model and delineate unpredictable pathological changes in the retina due to disease. Experimental validation was performed using 62 manually delineated choroid segmentations of retinal enhanced depth OCT images from patients with AMD. Our results show segmentation accuracy that surpasses those reported by state of the art approaches on healthy retinal images, and overall high values in images with pathology, which are difficult to address by existing methods without pathology specific heuristics.
Keywords: Segmentation, Deep Learning, Convolution Neural Network, Retina, EDI-OCT
1 Introduction
The choroid is the vascular layer located between the retina and sclera in the eye. It plays the vital role of providing nutrients and maintaining the portions of the eye responsible for vision. Specific alterations in the choroid have been associated with several disease states, including age-related macular degeneration (AMD) [1], retinitis pigmentosa [2], Stargardts disease, diabetes [3], sarcoidosis, and Vogt-Koyanagi-Harada syndrome. In addition, choroid thickness measures have been shown as a predictive biomarker for treatment response [4] and visual function [5].
Since the introduction of enhanced depth imaging optical coherence tomography (EDI-OCT) [6], there has been an explosion of interest in studying the choroid in vivo. This has included not only the disease processes previously associated with choroidal abnormalities, but also exploring the role of the choroid in ocular conditions as diverse as myopia and angle closure glaucoma. This quick (<1 second per scan), non-contact, safe, and inexpensive imaging modality has become readily accepted by both clinicians and patients.
To date, most assessments of EDI-OCT images have involved manual evaluation of the borders of the choroid and subjective judgments of choroidal vessel caliber (large, small, dilated, attenuated) and stromal density, which has proven difficult and time consuming. Fully or semi-automated systems are needed to make it possible to use in direct patient care and in clinical studies involving large numbers of patients examined serially over time. While several techniques [7–16] have been designed for the segmentation and measurement of the choroid in OCT, existing methods are primarily focused on images with normal (or normal-appearing) retinal structures with limited pathology.
Current approaches begin by first segmenting the retinal layers using methods such as graph-cuts [8, 10], dynamic programming [7, 9], gradient-based edge detection [11, 15], and active contours [16]. After locating the retinal layers, the choroid is found using an intensity based approach within a sub-region of the image, typically defined by the Bruchs Membrane (BM) located in the first step. Proposed approaches for segmenting the choroid in this sub-region include thresholding and region growing [8], multi-scale filtering with probabilistic estimation [9], constructing statistical [7], morphological models [10], and texture or gradient analysis [15, 16].
In the presence of pathology, existing approaches become less accurate and robust due to changes to the retinal morphology caused by disease. Very few methods have been presented that addresses automated choroid segmentation in the presence of pathology. Notably, [7, 9] proposed addressing the problem by making the retinal boundary segmentation more robust. Their approach first uses image derivative information and edge orientation to prevent unrealistic boundary jumps and shifts. A convex hull is then fit to the estimated boundaries to detect possible detachment of the retina. The method is shown to be robust to many cases of retinal pathology, such as drusen and retinal detachment. However, the algorithm still relies on the initial delineation of the retinal pigment epithelium, which the authors locate using the most hyper-intense boundary in the image. The method will be unreliable in cases where this assumption is violated due to artifacts or pathology. Also relevant, [17] presents an outer retinal-subretinal layer segmentation approach that addresses pathology by augmenting their graph search algorithm with a specialized fluid detection algorithm that is able to determine abnormal fluid-filled structures in the layer.
In this work, we present an approach where the detection and segmentation of the choroid and choroidal vessels are performed without the prerequisite delineation of the retinal layers. Instead we train a convolution neural network to directly identify the interior and exterior boundaries of the choroid in the image. This approach utilizes the fact that 1.) the choroid is often not noticeably affected by retinal pathology, and 2.) the morphology and texture of the choroid are distinct from the retina and retinal pathology. This allows us to directly identify the choroid regardless of the state of the retina in the image. Our approach aims to avoid needing to model and delineate the often unpredictable pathological changes in the retina due to disease.
2 Method
Our method consists of two primary steps. First, a convolution neural network (CNN) is trained to find the pixel-wise probability maps for the interior (Bruch’s membrane) and exterior (choroid-sclera interface) boundaries of the choroid. Once these probability maps are found we use seam carving [18] to estimate the two boundaries in the image and produce the final segmentation of the choroid.
2.1 Generating Edge Probability Maps Using CNN
For our CNN architecture, we use the SegNet design presented in [19]. The network consists of an encoder network, a corresponding decoder network and a pixel-wise classification layer. The SegNet architecture was designed specifically for pixel-wise classification of images where boundary delineation is vital. One of its key features is the ability to reduce the loss of spatial resolution due to max-pooling and sub-sampling by storing the max-pooling indices in the encoder and using it in the respective decoder to upsample the input feature map. This also has the advantage of avoiding the need to learn to upsample, reducing the overall number of parameters.
For our task, we use 6 layers of encoder and 6 layers of decoder, with (8, 16, 16, 32, 32, 64) features in each encoder layer and the reverse order in the decoder layers. A 5-by-5 convolution kernel is used throughout the network. An element-wise rectified linear non-linearity (ReLU) max(0, x) is applied to every layer except the final layer, which uses a softmax function to produce a probability output. Stochastic gradient descent is used for training the network, with a momentum of 0.9 and a fixed learning rate of 0.0001. We used a constant scale factor of 2 in the X and Y directions. We found that the network in general converges within 40 epochs.
Training and Testing
Two different SegNets are trained for our method, one to locate the Bruch’s membrane and the second to locate the choroid-sclera interface. Each network is trained on full EDI-OCT images with manually delineations of the respective boundaries, where each pixel in the training image is marked as either boundary or background. To increase the number of training examples, small random affine perturbations are used to displace each training images. This provides additional examples that can account for anatomical variability in the eye that may not be fully represented in our data. Given an unseen image, both SegNet are applied to the image to produce two probability maps representing the likely location of the two boundaries in the images.
2.2 Seam Carving
To convert the edge probability maps into a binary segmentation of the choroid, we use a technique known as seam carving [18]. Given an image I with dimension n × m, the goal of seam carving is to find a path of connected pixel that fully traverses across the image. For our task, we are interested in finding the horizontal seam,
| (1) |
where y is the mapping y: [1,…, m] ⇒ [1, …, n]. This formulation ensures that the seam only has a single pixel in each column and each pixel in the seam is 8-connected to the pixels in the adjacent columns. To find this seam, we define the cost function
| (2) |
where e is an energy function. For our purpose, e(I(sj)) is the value of the probability map at location sj. Given this energy function, the optimal seam can be found by using dynamic programming as described in [18].
The optimal seams found for our two probability maps are used as the interior and exterior boundaries of the choroid, and the pixels between the boundaries are completely filled to provide the full choroidal segmentation. This approach is advantageous over simply thresholding the probability maps, because it allows us to enforce a specific topology, connectivity, and smoothness to the segmentation.
3 Evaluation and Results
3.1 Data
62 EDI-OCT retinal images from 32 patients diagnosed with age related macular degeneration (AMD) were used for the evaluation of our method. Two images were acquired from each patient, the first image was of a retina with dry (atrophic) AMD from one eye, and the second image was of a retina with wet (exudative) AMD from the other eye. Each EDI-OCT image was a 2D cross-sectional slice centered on the fovea, with approximate dimensions of 1150 by 700 pixels. For each image, a manual segmentation of the choroid was performed by a trained rater using ITK Snap [20]. These manual segmentations serve as the ground truth used in our evaluation.
3.2 Results
We evaluated our algorithm on each image using 5-fold cross-validation, where 80% of the images (50 image) were randomly selected and used for training, and the remaining 20% were used to testing. This process was repeated until all images were segmented. The same parameters as described in Sec. 2.1 were used for each SegNet network used in the evaluation. Fig. 1 shows several examples of the automated choroidal boundaries detected by our method. We compared our automated results against the manual ground truth by calculating the average distance error of the Bruch’s membrane (evaluted as the average absolute difference between the manual and automated boundary location in each A-scan), and the overall Dice overlap coefficient [21],
| (3) |
between the automated and manual segmentations.
Fig. 1.
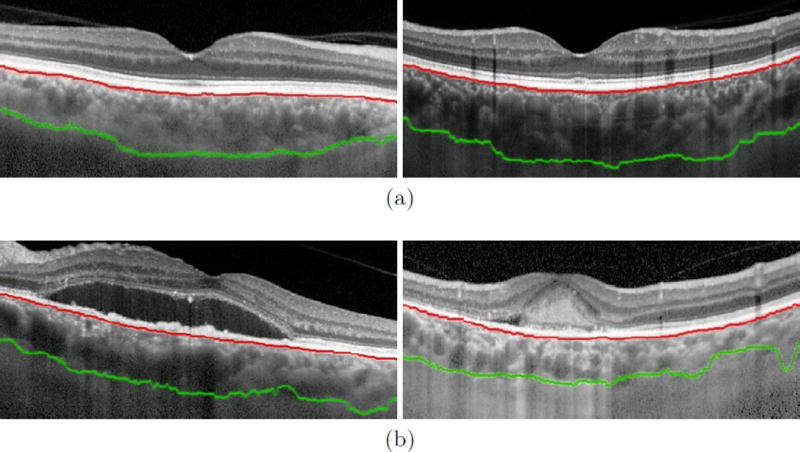
Examples of the interior (Bruch’s membrane, shown in red) and exterior (choroid-sclera interface, shown in green) boundaries of the choroid found by the proposed method on retinal image of eyes diagnosed with (a) dry and (b) wet AMD.
For a baseline comparison, we also performed the same evaluation using a graph cuts based segmentation approach, which have been shown [22, 23] to produce highly accurate segmentations of retinal layers in OCT images of healthy subjects. However, for our comparison, we adapted the approach to find the interior and exterior boundaries of the choroid in our 2D EDI-OCT images. No pathology-specific correction step was used to improving the BM surface in the wet AMD cases [17], but rather a single graph cut optimization with two surfaces was performed. Table 1 shows the average (and standard deviation) of each algorithm’s performance over the dry and wet AMD cases separately and over the full dataset.
Table 1.
The average (standard deviation) of the Dice overlap coefficient and average Bruch’s membrane (BM) boundary distance error (in pixels) of the choroid segmentation, evaluated between each algorithm and the manual ground truth. Each metric was first evaluated over the dry and wet AMD cases separately, and then on the combined overall dataset.
| Dice Overlap | BM Error in Pixels | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Dry | Wet | Overall | Dry | Wet | Overall | |
| Graph Cuts | 0.77(0.11) | 0.66(0.16) | 0.71(0.15) | 5.58(2.48) | 8.55(7.76) | 7.09(6.00) |
| SegNet+Seam Carving | 0.85(0.07) | 0.81(0.10) | 0.83(0.09) | 4.19(2.02) | 7.25(6.92) | 5.72(5.36) |
4 Discussion
From our results we see that, overall, our approach produced segmentations of the choroid that aligned well with the manual delineations. On average our algorithm produced a Dice overlap coefficient of 0.82 (±0.10) relative to manual segmentation. This is comparable to existing literature [8], which reports an average Dice overlap of 0.78 (±0.08) for automated segmentation of the choroid in healthy retinal. While the comparison is indirect, the result is promising considering that our dataset consisted of 2D images with retina pathology, many of which contained large intensity and morphological changes. Conversely, our images are EDI-OCT, which may have allowed the choroidal structures to be more easily segmented than standard OCT.
From Table 1 we see that on average our approach produced segmentations with higher Dice overlap and lower BM boundary errors than the graph cuts based approach. However, it would be fair to note that the graph cuts algorithm was adapted from a design developed for 3D OCT volumes of healthy subjects. Thus, the lack of 3D context in our data may explain the lower performance. Additionally, as noted above, the graph cut approach can be augmented with a specific correction technique to handle drusen, cysts and RPE detachment [17], which we have not included. This further contributes to its lower performance in the wet AMD cases. In contrast, our approach is robust to pathology and can be readily deployed even in a single 2D B-scan.
Qualitatively, our proposed algorithm is very robust to retinal pathology (as shown in Fig. 1(b)). This satisfies one of the primary goals of our approach, which was to design an algorithm that can detect the choroid independent of the state of the retina. One disadvantage of using a learning based approach to address this problem is that the success of the algorithm is predicated on the diversity of the training images. The training set must cover the wide range of appearances that the choroid can take in the image. For example, given our limited training set consisting mostly of normal appearing choroids, we can expect the algorithm to fail in the presence of choroidal pathology or imaging artifacts in the choroid. Likewise images acquired using different systems may also pose a problem if the imaging characteristics are sufficiently different.
5 Conclusion
We have introduced an automated approach for segmenting the choroid in EDI-OCT images with retinal pathology. Our results showed high performance relative to manual segmentations, and does not require prerequisite retinal layer segmentations which are necessary for most existing algorithm. There are several direction that this work can be extended. One potential area of exploration is the use of CNN to directly segment the choroid vasculature instead of locating the choroidal edges. This can potentially allow for more accurate and robust segmentation by avoiding the need to detect the thin structure represented by the edges. In addition, 3D information from adjacent (non-EDI) OCT B-scans can potentially be incorporated into the segmentation to allow for more contextual information in the segmentation.
References
- 1.Chung SE, Kang SW, Lee JH, Kim YT. Choroidal thickness in polypoidal choroidal vasculopathy and exudative age-related macular degeneration. Ophthalmology. 2011;118(5):840–845. doi: 10.1016/j.ophtha.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Dhoot DS, Huo S, Yuan A, Xu D, Srivistava S, Ehlers JP, Traboulsi E, Kaiser PK. Evaluation of choroidal thickness in retinitis pigmentosa using enhanced depth imaging optical coherence tomography. British Journal of Ophthalmology. 2013;97(1):66–69. doi: 10.1136/bjophthalmol-2012-301917. [DOI] [PubMed] [Google Scholar]
- 3.Esmaeelpour M, Brunner S, Ansari-Shahrezaei S, Nemetz S, Považay B, Ka-jic V, Drexler W, Binder S. Choroidal Thinning in Diabetes Type 1 Detected by 3-Dimensional 1060 nm Optical Coherence Tomography. Investigative Ophthalmology & Visual Science. 2012;53(11):6803–6809. doi: 10.1167/iovs.12-10314. [DOI] [PubMed] [Google Scholar]
- 4.Kang HM, Kwon HJ, Yi JH, Lee CS, Lee SC. Subfoveal choroidal thickness as a potential predictor of visual outcome and treatment response after intravitreal ranibizumab injections for typical exudative age-related macular degeneration. American Journal of Ophthalmology. 2014;157(5):1013–1021. doi: 10.1016/j.ajo.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Moutray T, Alarbi M, Mahon G, Stevenson M, Chakravarthy U. Relationships between clinical measures of visual function, fluorescein angiographic and optical coherence tomography features in patients with subfoveal choroidal neovascularisation. British Journal of Ophthalmology. 2008;92(3):361–364. doi: 10.1136/bjo.2007.123976. [DOI] [PubMed] [Google Scholar]
- 6.Spaide RF, Koizumi H, Pozonni MC. Enhanced depth imaging spectral-domain optical coherence tomography. American Journal of Ophthalmology. 2008;146(4):496–500. doi: 10.1016/j.ajo.2008.05.032. [DOI] [PubMed] [Google Scholar]
- 7.Kajić V, Esmaeelpour M, Považay B, Marshall D, Rosin PL, Drexler W. Automated choroidal segmentation of 1060 nm OCT in healthy and patho-logic eyes using a statistical model. Biomedical Optics Express. 2012;3(1):86–103. doi: 10.1364/BOE.3.000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Lee K, Niemeijer M, Mullins RF, Sonka M, Abramoff MD. Automated segmentation of the choroid from clinical SD-OCT. Investigative Ophthalmology & Visual Science. 2012;53(12):7510–7519. doi: 10.1167/iovs.12-10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajić V, Esmaeelpour M, Glittenberg C, Kraus MF, Honegger J, Othara R, Binder S, Fujimoto JG, Drexler W. Automated three-dimensional choroidal vessel segmentation of 3D 1060 nm OCT retinal data. Biomedical Optics Express. 2013;4(1):134–150. doi: 10.1364/BOE.4.000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z, Wu X, Ouyang Y, Ouyang Y, Sadda SR. Semiautomated Segmentation of the choroid in spectral-domain optical coherence tomography volume scans. Investigative Ophthalmology & Visual Science. 2013;54(3):1722–1729. doi: 10.1167/iovs.12-10578. [DOI] [PubMed] [Google Scholar]
- 11.Tian J, Marziliano P, Baskaran M, Tun TA, Aung T. Automatic segmentation of the choroid in enhanced depth imaging optical coherence tomography images. Biomedical Optics Express. 2013;4(3):397–411. doi: 10.1364/BOE.4.000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Buitendijk GH, Lee K, Sonka M, Springelkamp H, Hofman A, Vingerling JR, Mullins RF, Klaver CC, Abràmoff MD. Validity of automated choroidal segmentation in SS-OCT and SD-OCT. Investigative Ophthalmology & Visual Science. 2015;56(5):3202–3211. doi: 10.1167/iovs.14-15669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vupparaboina KK, Nizampatnam S, Chhablani J, Richhariya A, Jana S. Automated estimation of choroidal thickness distribution and volume based on OCT images of posterior visual section. Computerized Medical Imaging and Graphics. 2015;46:315–327. doi: 10.1016/j.compmedimag.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Philip AM, Gerendas BS, Zhang L, Faatz H, Podkowinski D, Bogunovic H, Abramoff MD, Hagmann M, Leitner R, Simader C, et al. Choroidal thickness maps from spectral domain and swept source optical coherence tomography: algorithmic versus ground truth annotation. British Journal of Ophthalmology. 2016;0:1–5. doi: 10.1136/bjophthalmol-2015-307985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Q, Fan W, Niu S, Shi J, Shen H, Yuan S. Automated choroid segmentation based on gradual intensity distance in hd-oct images. Optics express. 2015;23(7):8974–8994. doi: 10.1364/OE.23.008974. [DOI] [PubMed] [Google Scholar]
- 16.González-López A, Remeseiro B, Ortega M, Penedo MG, Charlón P. International Conference on Computer Aided Systems Theory. Springer; 2015. A texture-based method for choroid segmentation in retinal edi-oct images; pp. 487–493. [Google Scholar]
- 17.Zhang L, Sonka M, Folk JC, Russell SR, Abramoff MD. Quantifying Disrupted Outer Retinal-Subretinal Layer in SD-OCT Images in Choroidal Neovascularization. Investigative Ophthalmology & Visual Science. 2014 Apr;55:2329–2335. doi: 10.1167/iovs.13-13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avidan S, Shamir A. Seam carving for content-aware image resizing. ACM Transactions on graphics (TOG) 2007;26(3):10. ACM. [Google Scholar]
- 19.Badrinarayanan V, Kendall A, Cipolla R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. arXiv preprint arXiv:1511.00561. 2015 doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 20.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3d active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 22.Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M. Automated 3-d intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE transactions on medical imaging. 2009;28(9):1436–1447. doi: 10.1109/TMI.2009.2016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang A, Carass A, Hauser M, Sotirchos ES, Calabresi PA, Ying HS, Prince JL. Retinal layer segmentation of macular oct images using boundary classification. Biomedical optics express. 2013;4(7):1133–1152. doi: 10.1364/BOE.4.001133. [DOI] [PMC free article] [PubMed] [Google Scholar]


