Abstract
The Golgi apparatus in plant cells consists of a large number of independent Golgi stack/trans-Golgi network/Golgi matrix units that appear to be randomly distributed throughout the cytoplasm. To study the dynamic behavior of these Golgi units in living plant cells, we have cloned a cDNA from soybean (Glycine max), GmMan1, encoding the resident Golgi protein α-1,2 mannosidase I. The predicted protein of approximately 65 kD shows similarity of general structure and sequence (45% identity) to class I animal and fungal α-1,2 mannosidases. Expression of a GmMan1::green fluorescent protein fusion construct in tobacco (Nicotiana tabacum) Bright Yellow 2 suspension-cultured cells revealed the presence of several hundred to thousands of fluorescent spots. Immuno-electron microscopy demonstrates that these spots correspond to individual Golgi stacks and that the fusion protein is largely confined to the cis-side of the stacks. In living cells, the stacks carry out stop-and-go movements, oscillating rapidly between directed movement and random “wiggling.” Directed movement (maximal velocity 4.2 μm/s) is related to cytoplasmic streaming, occurs along straight trajectories, and is dependent upon intact actin microfilaments and myosin motors, since treatment with cytochalasin D or butanedione monoxime blocks the streaming motion. In contrast, microtubule-disrupting drugs appear to have a small but reproducible stimulatory effect on streaming behavior. We present a model that postulates that the stop-and-go motion of Golgi-trans-Golgi network units is regulated by “stop signals” produced by endoplasmic reticulum export sites and locally expanding cell wall domains to optimize endoplasmic reticulum to Golgi and Golgi to cell wall trafficking.
The Golgi apparatus of plant cells consists of a large number of small, independent stack-trans-Golgi network (TGN) units that are distributed throughout the cytoplasm (Driouich and Staehelin, 1997; Andreeva et al., 1998b; Dupree and Sherrier, 1998). This seemingly random organization within plant cells is in striking contrast to the highly ordered Golgi complex in animal cells (Rambourg and Clermont, 1997). Presumably, these different forms of spatial organization reflect different mechanisms for controlling the localization of Golgi membranes. Similarly, the distinct distributions found in the two organisms likely impose different requirements on the transport of vesicles to and from the Golgi. However, while many of the molecules underlying Golgi positioning and functioning have been identified in animal systems (Barlow, 1998; Lowe and Kreis, 1998), very little is known about these processes for the plant Golgi (Andreeva et al., 1998a).
The juxtanuclear position of the Golgi apparatus in animal cells depends on the presence of an intact microtubule (MT) cytoskeleton (Burkhardt, 1998). Golgi membranes assemble around the centrosome in close proximity to the nucleus, where the cis-cisternae are anchored to the minus ends of MTs (Infante et al., 1999). Vesicular transport from the peripheral endoplasmic reticulum (ER) to this central Golgi complex occurs in a targeted fashion along MTs (Presley et al., 1997; Scales et al., 1997), and post-Golgi transport also often follows MT tracks into the periphery of the cell (Hirschberg et al., 1998; Toomre et al., 1999). In contrast, the Golgi stack-TGN units of angiosperms are dispersed throughout the cytoplasm (Robinson and Kristen, 1982). This distribution conceivably reduces the distance that transport vesicles have to travel, both from ER export sites to the Golgi and from the Golgi to the plasma membrane or vacuole. However, it is unclear whether this traffic is directed, as is the case in animal cells, or whether it occurs by passive diffusion or cytoplasmic streaming (Williamson, 1993).
Cytoplasmic streaming is a process found in many plant cells that moves large quantities of cytoplasm (including organelles) around the cell. This phenomenon is most pronounced in larger, highly vacuolated cells and is generally assumed to allow for efficient mixing and distribution of solutes. Cytoplasmic streaming has been studied extensively in internodal cells of characean algae (Kuroda, 1990; Shimmen and Yokota, 1994). It usually is driven by the acto-myosin system, although instances of microtubule-based organelle movement have been described (e.g. Mizukami and Wada, 1981; Mineyuki and Furuya, 1986). Studies of streaming events in the past have relied on visualization of moving organelles with phase-contrast or Nomarski microscopy. The nature of the organelles was therefore mostly unknown. The participation of Golgi vesicles in cytoplasmic streaming was inferred based only on the effects of inhibitor studies (e.g. Mollenhauer and Morré, 1976).
It also has been proposed that entire Golgi stacks might participate in the streaming motion (Staehelin and Moore, 1995). This postulated movement of Golgi stacks has recently been demonstrated in tobacco (Nicotiana clevelandii) leaf epidermal cells by means of green fluorescent protein (GFP) transiently expressed in the Golgi as a fusion with the targeting domain of mammalian sialyltransferase (Boevink et al., 1998). Golgi stack movement may also provide the mechanism responsible for their dispersed distribution throughout the cytoplasm. At the same time, this movement imposes additional constraints on the possible mechanisms for transport to and from the Golgi.
To investigate these Golgi dynamics in greater detail, we set out to develop a plant-gene-based Golgi marker in a stably transformed cell line. Such a system would allow quantitative analysis of this Golgi movement in great detail, and also enable us to study Golgi dynamics in dividing cells and to carry out novel biochemical fractionation studies. In this report we describe the isolation of a cDNA from soybean (Glycine max) encoding an α-1,2 mannosidase I, GmMan1, the first enzyme of the N-linked oligosaccharide pathway cloned from plants. Fusion of this protein to GFP when expressed in stably transformed tobacco Bright Yellow 2 (BY-2) cells is localized to the cis-Golgi. In living cells, Golgi stacks show characteristic saltatory movements throughout the cytoplasm. Inhibitor treatments demonstrate that this movement is dependent on the acto-myosin system, and that MTs can limit Golgi mobility in a subset of cells.
MATERIALS AND METHODS
Cloning of GmMan1
A partial soybean (Glycine max) α-1,2 mannosidase I clone was generated using degenerate oligonucleotides corresponding to conserved regions 1 and 3 as described previously (Herscovics et al., 1994). The clone (kindly provided by A. Herscovics, McGill University) was sequenced, and gene-specific primers to this partial soybean sequence (MannoN: 5′-TGGTTTTATGARTAYTTGYTGAAA, MannoC: 5′-ATACTTCAGCGTCTCCGCAAG) were designed (marked by thin underlines in Fig. 1). These primers were used in a PCR-based screen of a soybean cDNA library (kindly provided by J. Mullet, Texas A&M University, College Station). Plates of plaques were top-laid with SM medium (Sambrook et al., 1989), which was then tested by PCR for the presence of inserts that could be amplified with MannoN and MannoC. Positive pools were then repeatedly plated at lower plaque densities until a PCR product could be detected on a plate of about 100 plaques. A clone of approximately 2.6 kb was then isolated by plaque-lift screening.
Figure 1.
Predicted amino acid sequence of GmMan1 and its alignment with selected other α-1,2 mannosidase I protein sequences. The sequences and their accession nos. are: soybean, AF126550 (this report); mouse1b, U03458 (Herscovics et al., 1994); Drosophila, X82641 (Kerscher et al., 1995); and yeast, M63598 (Camirand et al., 1991). Homologous residues are highlighted by shading; residues identical in soybean and other sequences are boxed. The solid bar highlights the predicted membrane-spanning domain of GmMan1. The predicted catalytic domain is bracketed by arrows. Dots indicate conserved cysteyl residues that form a disulfide bridge (Lipari and Herscovics, 1996). Conserved acidic residues required for enzymatic activity are highlighted with asterisks (Lipari and Herscovics, 1999). The primers used for cDNA library screening were designed against the underlined sequences.
Sequence Analysis
Multiple sequence alignments were initially performed with the PileUp program from the Genetics Computer Group (Madison, WI) and subsequently optimized manually. Prediction of transmembrane domains was done with TMpred (http://www.ch.embnet.org/software/TMPRED_form.html; Hofmann and Stoffel, 1993). Coiled-coil prediction was carried out with the COILS program (http://www.ch.embnet.org/software/COILS_form.html) using the algorithm of Lupas et al. (1991).
Construction of GmMan1::GFP Fusion Protein
The gene encoding a modified GFP (HBT-SGFP-TYG-nos in pUC18, obtained from J. Sheen, Massachusetts General Hospital) was subcloned with BamHI and EcoRI into pBluescript (Stratagene, La Jolla, CA) to obtain an in-frame XbaI site at its 5′ end. This was then spliced to an internal XbaI site in GmMan1, thus removing the C-terminal 11 amino acids of the mannosidase-coding region. To drive expression in plant cells a modified cauliflower mosaic virus 35S promoter (with a dual-enhancer element) generated by PCR from a plasmid (pZEV Ω, J. Oliver and K. Danna, University of Colorado, Boulder) was used. This expression cassette was then inserted into the SacI and KpnI sites of pBIN20 (Hennegan and Danna, 1998) and transformed into Agrobacterium tumefaciens strain LBA4404 to yield strain BP37. Transformation of tobacco (Nicotiana tabacum) Bright Yellow 2 (BY-2) suspension-cultured cells (3 d after subculture) was achieved by co-cultivation with strain BP37 for 2 d at 27°C. Cells were then transferred onto selective medium (BY-2 medium plus 500 μg/mL carbenicillin and 100 μg/mL kanamycin).
Growth Conditions
Transformed cells were grown in a modified Linsmaier and Skoog medium (Nagata et al., 1982) with constant shaking (120 rpm) at 27°C in the dark. Cells were subcultured weekly into fresh medium at a dilution of 1:50. Cells were harvested by low-speed centrifugation at 500g for 2 min 6 to 8 d after subculturing and immediately used for experiments.
Electron Microscopy
Transformed BY-2 cells were high-pressure-frozen/freeze-substituted and embedded for transmission electron microscopy, as described in Samuels et al. (1995). For immunogold detection, the following modifications of the standard protocol were applied. After staining with osmium, samples were embedded in LR White. Sections (90 nm) of the samples were cut and placed on formvar-coated 300 mesh nickel grids. After a 20-min exposure to saturated sodium metaperiodate, the grids were washed briefly and blocked with 5% (w/v) nonfat milk in phosphate-buffered saline containing 0.1% (w/v) Tween (PBST). The sections were then exposed to the primary anti-GFP antibody (kindly provided by J. Kahana, Harvard University) for 2 h. Following a thorough rinse with PBST, the sections were labeled with 15 nm of goat anti-rabbit IgG secondary antibody (British BioCell International, Cardiff, UK) for 1 h. The final rinse was with PBST followed by water. The grids were then post-stained in 2% aqueous uranyl acetate and Reynold's lead citrate. Sections were observed on an electron microscope (model CM10, Philips, Eindhoven, The Netherlands).
Fluorescence Microscopy
Cells were observed using the standard fluorescein isothiocyanate filter set. Confocal images were obtained on a Sarastro MultiProbe system (Molecular Dynamics, Sunnyvale, CA) using a ×100 objective (Nikon, Tokyo). Conventional fluorescence microscopy was on an Axioscope microscope (Zeiss, Jena, Germany) with a ×100 objective and a Nikon Eclipse microscope with a ×60 objective (for video capture). Videos of streaming Golgi stacks were captured with a color CCD camera (Optronics, Goleta, CA) at an exposure setting of 1/8 s. Video frames were recorded on a PowerMacintosh computer equipped with a graphics digitizer board (RasterOps) at a rate of 10 frames per s. Individual frames in 1-s intervals were imported into the public domain NIH Image program (developed at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image), optically enhanced, and used for determination of x/y coordinates of individual stacks.
To quantify the streaming behavior of individual stacks, the “streaming coefficient” was calculated according to the formula:
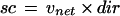 |
1 |
where sc is the streaming coefficient, vnet is the net velocity, and dir is a directionality factor. The net velocity is calculated as:
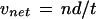 |
2 |
where nd is the net displacement during the observation period (i.e. the distance between the first and last position of the tracing), and t is time (i.e. the duration of the observation period). The directionality factor dir is defined as:
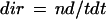 |
3 |
where tdt is the total distance traveled during the observation period. The streaming coefficient for straight trajectories therefore equals the average velocity of the stack. For curved trajectories, the streaming coefficient roughly equals the average velocity in the preferred direction of movement, corrected by the directionality factor. Inclusion of this factor ensures that stacks displaying a high degree of random motion will have a low streaming coefficient, even in the presence of substantial amounts of drift.
Drug Treatments
Drugs (from Sigma, St. Louis) were prepared as a 1,000-fold concentrated stock solution in DMSO and stored at −20°C, except for 2,3-butanedione monoxime (BDM), which was freshly dissolved in BY-2 medium just prior to the experiment. Drug treatments were performed for 15 to 45 min. Controls containing an equivalent concentration of DMSO (0.1%, w/v) did not show any response to the solvent.
RESULTS
Cloning of Soybean α-1,2 Mannosidase I
The enzyme α-1,2 mannosidase I belongs to the class I α-mannosidases, a family of enzymes which remove α-1,2-mannosyl residues from the high Man N-linked oligosaccharides that are synthesized in the ER (Moremen et al., 1994). These are the first modification reactions to occur in the Golgi. A number of class I α-1,2 mannosidases have been cloned from animal and fungal organisms (Herscovics, 1999b), but no enzymes of the N-linked oligosaccharide pathway have been cloned from plants. Degenerate primers based on α-1,2-mannosidase sequences from mouse and yeast (Herscovics et al., 1994) have previously been used to amplify cDNA fragments from other species, among them soybean (A. Herscovics, personal communication). Specific primers were designed based on the termini of this partial soybean clone (target sequences are underlined in Fig. 1) and used in a PCR-based screen of cDNA libraries from soybean, Arabidopsis, and tobacco. Using this approach, a clone was isolated from the soybean library with an insert of approximately 2.5 kb, which could serve as a template for PCR with our gene-specific primers and also hybridized with the original gene fragment from degenerate PCR on a DNA gel blot (data not shown).
Sequencing of the cDNA clone revealed an open reading frame of 1,734 bp, encoding a hypothetical protein of 578 amino acids and a calculated molecular mass of 65,345 D (Fig. 1, GenBank accession no. AF126550). Sequence analysis predicts a single, very short transmembrane domain (amino acids 30… 45 [black line in Fig. 1]) with the N terminus of the protein on the cytoplasmic face of the membrane. This type II orientation is typical of Golgi proteins and is also found in all other known α-1,2-mannosidases (Herscovics, 1999a, 1999b). The lumenal domain consists of a putative stalk region (46… 101), the catalytic domain (102… 548), and a C-terminal tail (549… 578). The stalk region has a high probability of forming a coiled-coil structure. The stalk and tail show no sequence similarity to other known mannosidases.
In contrast, the putative catalytic domain is approximately 55% similar and 45% identical to corresponding parts of α-1,2-mannosidases from either mouse or yeast and also contains several features predicted to be important for enzymatic activity of α-1,2-mannosidases. In particular, two conserved Cys residues that form a required disulfide bond in yeast (Lipari and Herscovics, 1996) are present in the predicted soybean protein (C387 and C420; bullets in Fig. 1). Several carboxyl residues shown by site-directed mutagenesis to be crucial for enzymatic activity (Lipari and Herscovics, 1999) are also conserved (asterisks in Fig. 1). Based on these extensive similarities, we predict that the isolated soybean cDNA encodes a class I α-1,2-mannosidase (EC 3.2.1.113; glycosyl hydrolase family 47, Henrissat and Bairoch, 1996) and propose to call it GmMan1.
Localization of a GmMan1::GFP Fusion to the cis-Golgi in Tobacco Cells
Some α-1,2 mannosidases are localized to the Golgi (Herscovics et al., 1994; Lal et al., 1994), although other members of the family were found in the ER (e.g. Roth et al., 1990; Burke et al., 1996). The localization of GmMan1 was tested by creating an in-frame fusion to a modified GFP (GmMan1::GFP) and expressing it in stably transformed tobacco suspension-cultured cells (BY-2, Nagata et al., 1982). Most of the recovered cell lines displayed a punctate pattern of GFP fluorescence, as would be expected from the dispersed organization of Golgi stacks in plant cells (Fig. 2). Some cell lines showed an additional reticulate fluorescence that resembled the fluorescence seen in cells expressing ER-targeted GFP (data not shown).
Figure 2.
Distribution of GmMan1::GFP in living tobacco BY-2 suspension-cultured cells. A, Single optical section from a confocal microscope through the middle of a group of cells. B, Optical section through the cortical cytoplasm of a single cell. Brightly fluorescing spots can be found throughout the cytoplasm in both cortical regions and transvacuolar strands but not the vacuole (V) or the nucleus (N). The insets (×3 enlargement) demonstrate that some spots appear as short lines (arrowheads), whereas others resemble discs (arrows). C, Conventional epifluorescence picture of cortical cytoplasm. Several Golgi stacks appear as ring-like structures (arrow). Some of the other stacks changed in appearance from lines to rings over time (arrowhead; compare video sequence at http://www.plantphysiol.org/cgi/content/full/121/4/1127/DC1).
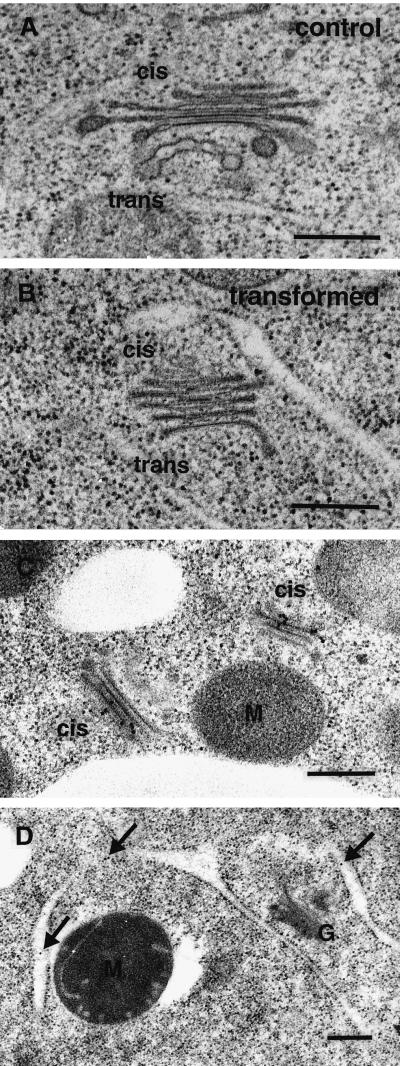
Immuno-electron microscopy on high-pressure frozen/freeze-substituted samples with antibodies against the GFP part of the fusion protein was performed to determine whether the punctate fluorescence does indeed highlight individual Golgi stacks. As shown in Figure 3C, gold label accumulated predominantly over the cis and medial cisternae of stacks, with very little staining in trans and TGN cisternae. In some cells, weak labeling of ER (Fig. 3D) and multivesicular bodies could also be observed. These localizations could mark fusion proteins in transit to the Golgi or targeted for degradation, respectively. Therefore, the GmMan1::GFP fusion construct is predominantly targeted to the cis-Golgi in BY-2 cells. The fluorescent spots shown in Figure 2 thus represent Golgi stacks and can be used to investigate Golgi dynamics in living cells. The overall morphology of Golgi stacks is not altered by overexpression of the fusion protein (compare Fig. 3, A and B). However, we frequently noticed an increase in staining intensity of the cisternal membranes on the cis side of the stack (Fig. 3B). In addition, Golgi stacks in the transgenic lines had a slight reduction of diameter (0.636 versus 0.803 μm, n = 33), which was accompanied by an marginal decrease in average number of cisternae per stack (5.0 versus 5.4, n = 33).
Figure 3.
Ultrastructure of Golgi stacks in untransformed (A) and transformed (B) BY-2 cells and immunogold-localization of the GmMan1::GFP fusion protein (C and D). A, Thin section image of single Golgi stack in high-pressure-frozen/freeze-substituted control cell showing normal appearance of Golgi stacks in BY-2 cells. B, In transformed cells, the cisternae generally have a normal appearance, except for stronger staining of the cis-cisternae. As in control cells, intercisternal elements are present between the trans-cisternae. C, Localization of GmMan1::GFP, as detected by antibodies against the GFP protein, is mostly restricted to the cis-side of the Golgi stack. D, In some cells, weak, non-Golgi labeling was observed over the ER (arrows). M, Mitochondrion; G, Golgi stack. Bar, 0.2 μm.
As illustrated in Figure 2, BY-2 cells contain several hundred individual Golgi stacks. The stacks were more or less evenly distributed throughout the cytoplasm in both the cortical region underlying the plasma membrane and in cytoplasmic strands that traverse the large central vacuole. Careful examination of individual fluorescent spots revealed that they either appeared as discs (arrows in Fig. 2) or as short lines (arrowheads in Fig. 2). The length of the lines was identical to the diameter of the discs and uniform throughout the cell. In living cells, we could observe individual spots change from one shape to the other. These data suggest that the fluorescently labeled Golgi cisternae can be seen in both face-on views, when they appear as discs, and from the side, when they appear as lines. In a few cells we could also observe ring-like fluorescent structures (arrow in Fig. 2C). The diameter of these rings was approximately 1 μm, i.e. similar to Golgi stacks (Fig. 2, A and B). Occasionally these rings could change into short lines of the same length (arrowhead in Fig. 2C), suggesting that the rings represent a face-on view of Golgi stacks in which the fusion protein is restricted to the rim region of the cisternae.
Analysis of Golgi Stack Movement in Living Cells
Observation of living BY-2 cells expressing the GmMan1::GFP fusion protein demonstrated that Golgi stacks can participate in cytoplasmic streaming and move throughout the cell (a video of Golgi stack movement can be viewed at http://www.plantphysiol.org/cgi/content/full/121/4/1127/DC1). This movement was most pronounced in transvacuolar strands and in certain regions of the cortical cytoplasm. Golgi stacks that were not participating in directed movement displayed random “wiggling” reminiscent of Brownian motion. Those stacks that appeared to stream showed saltatory movement in which episodes of rapid movement alternated with periods of relative inactivity and wiggling. Golgi stacks that followed the same track occasionally paused at the same position along the track, suggesting that directed movement is inhibited at these sites. Within a given culture, only about two-thirds of the cells displayed “active streaming” of Golgi stacks. In the remaining cells, the stacks exhibited only wiggling motions (Table II). The percentage of active cells depended on the status of the culture, with younger cultures, which typically contained smaller cells, being less active in terms of their Golgi streaming. Most of the observations of streaming cells were therefore conducted on 7- to 8-d-old cultures. Within a given population, larger cells tended to have more active streaming, whereas small, round cells were mostly inactive.
Table II.
Fraction of cells with active streaming in a population (%)
| Experiment | 3.3 μm Nocodazole | Control | Ratio |
|---|---|---|---|
| 1 | 81 | 74 | 1.09 |
| 2 | 83 | 82 | 1.01 |
| 3 | 84 | 84 | 1.00 |
| 4 | 77 | 64 | 1.20 |
| 5 | 73 | 64 | 1.14 |
| 6 | 78 | 57 | 1.37 |
| 7 | 68 | 60 | 1.13 |
| 8 | 81 | 78 | 1.04 |
| 9 | 53 | 46 | 1.15 |
| 10 | 61 | 56 | 1.09 |
| Mean ± sd | 74 ± 11 | 67 ± 13 | 1.12 ± 0.11 |
Aliquots of a suspension cultures were treated with 3.3 μm nocodazole or 0.1 % (w/v) DMSO (control). After about a 45-min incubation, 100 cells per treatment were classified as displaying active streaming or not. Ten independent experiments were performed. While the average fraction of cells displaying active streaming was not significantly changed by the drug treatment, a consistent small increase was observed in most experiments (P = 0.004, paired t test).
Movement of individual Golgi stacks was quantitated by video microscopy. Cells with bright green spots that showed active streaming were selected for recording over a period of 10 to 75 s with 1/8-s exposure time. The x-y coordinates of individual stacks were extracted in 1-s intervals to obtain tracings reflecting their movements (Fig. 4). This approach allowed for visualization of the relative activity of different regions of the cortical cytoplasm. In Figure 4, the tracings of individual stacks are color-coded according to the level of streaming they displayed over the entire observation period. Streaming stacks are marked with warm colors (yellow–red), whereas tracings of “wiggling” stacks are blue. The streaming level is defined by the “streaming coefficient,” which is calculated as the net velocity of a stack during a certain time interval multiplied by a directionality factor (see “Materials and Methods”). The resulting pattern shows that streaming Golgi stacks follow preferred paths (Fig. 4). These tracks are separated by regions of limited or random movement. It is also evident that large differences in mobility can exist between stacks that are spatially close together.
Figure 4.
Tracing of selected Golgi stacks over 30 s. A, Cortical region in untreated cell. B, Transvacuolar strands in untreated cell. C, Cortical region in cell treated with 40 μm cytochalasin D for 30 min. D, Cortical region in cell treated with 3.3 μm nocodazole for 30 min. The positions of individual Golgi stacks are marked in 1-s intervals. The tracings of the stacks are color coded according to the streaming coefficient of movement over the entire observation period (see text). Tracings in warm colors (red and yellow) represent stacks with a high degree of streaming, tracing in cold colors (blue) represent stacks that are mostly wiggling (compare color scale in C). Note that streaming stacks follow straight, preferred paths. Arrows denote the direction of movement. Cytochalasin D treatment eliminated streaming (C), while nocodazole did not affect directed movement (D). Video sequences can be viewed at http://www.plantphysiol.org/cgi/content/full/121/4/1127/DC1.
The percentage of stacks displaying streaming was higher in cytoplasmic strands than in cortical regions. When stacks were selected at random for tracing analysis (n = 50 per cell, two cells each for cortical regions and transvacuolar strands, respectively), it was found that 15% to 50% of the stacks in cytoplasmic threads had a streaming coefficient greater than 0.2 over the entire observation period, while this fraction was less than 2% for cortical regions (Table I). This difference confirms the observation that large areas of the cortical cytoplasm do not show directed movement (Fig. 4A).
Table I.
Statistics of Golgi stack movement in cortical regions and cytoplasmic strands of two cells per experiment
| Control
|
3.3 μm Nocodazole
|
|||
|---|---|---|---|---|
| Cortical region | Transvacuolar strand | Cortical region | Transvacuolar strand | |
| Average velocity (μm/s) | 0.03–0.46 | 0.11–1.55 | 0.09–1.33 | 0.12–2.37 |
| Maximal velocity (μm/s) | 0.34–2.41 | 0.49–3.85 | 0.34–2.78 | 0.49–4.19 |
| % Fast stacks | 0–6 | 4–42 | 0–7 | 2–73 |
| % Streaming stacks (30 s) | 0, 0 | 14, 47 | 0, 2 | 44, 94 |
Fifty randomly selected stacks in two different cells for either region per treatment (eight cells total) were traced for 30 s. Instantaneous velocities were calculated for every stack at all time intervals. Ranges of maximal and average velocities for all stacks from both cells per region are given. “% Fast stacks” indicates the range of percentages of stacks at any moment that display velocities ≥1 μm/s. “% Streaming stacks” gives the fraction of stacks in the two cells whose movement shows a streaming coefficient ≥ 0.2 over the entire observation period.
The tracing data were used to calculate the average speed of individual stacks as well as instantaneous velocities. As shown in Figure 5, the saltatory movement observed under the microscope is immediately evident in the variations of speed derived from the tracings. Every stack alternated between episodes of rapid movement and periods of relative inactivity. These alterations did not follow any pattern, and Fourier analysis revealed no periodicity (data not shown). The maximal velocity observed for any stack was 4.2 μm/s. Golgi stacks in cortical regions tended to stream more slowly than those in transvacuolar strands (Table I). The variations in instantaneous velocity are also reflected in changes between streaming and wiggling events. Figure 5 exemplifies some of the patterns that could be observed when the streaming coefficient was calculated for 4-s intervals. In particular, some stacks initially displayed a high degree of directional movement (i.e. streaming) that later changed to reduced directionality (i.e. wiggling) (Fig. 5A). Other stacks showed the opposite behavior, with a sudden start of streaming late in the observation period (Fig. 5B). In addition, many stacks showed several reversals of streaming behavior over the 75-s observation period (Fig. 5C). Periods of predominantly translational movement (Fig. 5B, between arrows 2 and 4) could be interrupted by short wiggling events (arrow 3 in Fig. 5B).
Figure 5.
Variation of instantaneous velocity of individual Golgi stacks in control cells over 75 s. Tracing data of three different stacks (A–C) was used to calculate the instantaneous velocity between the marked positions (broken lines). The streaming coefficient was calculated for a 4-s interval centered on the respective time points (solid lines). Note the rapid changes in velocity that do not follow any predictable pattern. Also note that the streaming coefficient of the movement can change over time. Low values for the streaming coefficient indicate wiggling motion; high values indicate streaming motion. Corresponding points on the tracing and the graph are marked with numbered arrows.
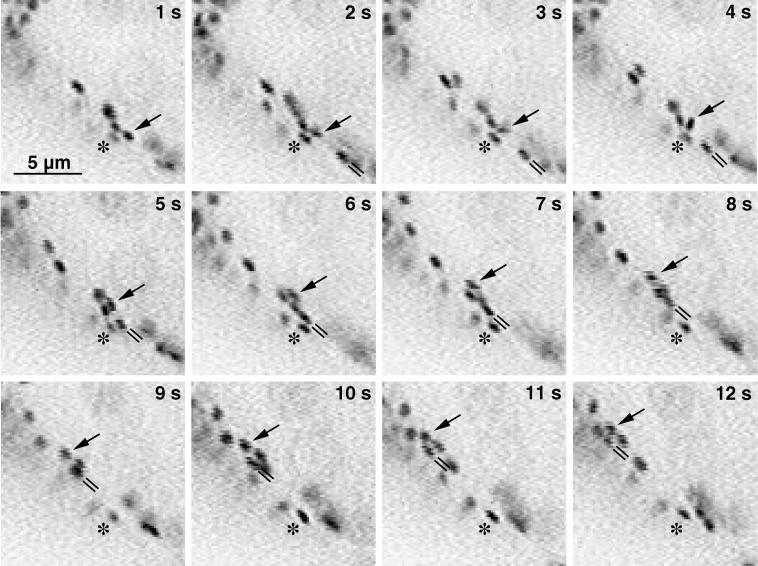
The movement of Golgi stacks showed a high degree of specificity. Rapidly moving stacks were often seen to pass slower stacks in close proximity (e.g. Fig. 6). The differences in instantaneous velocities could be as high as 2.2 μm/s for stacks that were less than 1 μm apart (Fig. 7). When stacks were seen in an edge-on view as they were traveling through the cytoplasm, the edge was usually aligned with the direction of movement and maintained this orientation over the entire streaming event (double lines in Fig. 6).
Figure 6.
Stacks maintain their orientation during streaming events. Sequence of 12 video frames from cortical cytoplasm taken in 1-s intervals. General direction of movement is from lower right to upper left. The stack marked with a double line maintains its orientation from 2 to 8 s and from 10 to 12 s. At 9 s it appears to dive under a slowly moving stack and its orientation cannot be resolved unambiguously. The Golgi stack marked with the arrow first shows a rotational movement (2–4 s), followed by translational movement to the end of the sequence. Asterisks denote a stack that did not move during the entire observation period.
Figure 7.
Movement of several Golgi stacks along one track in cortical cytoplasm of a control cell. A, Positions of individual stacks were marked in 1-s intervals for up to 30 s. Tracing coordinates were transformed so that the abscissa runs along the track, and the ordinate lies perpendicular to it. B, Temporal relationship between stacks that are at the same position along the strand. Note that stacks that are in close proximity (less than 1-μm distance, arrows in A and circles in B) can have drastically different velocities.
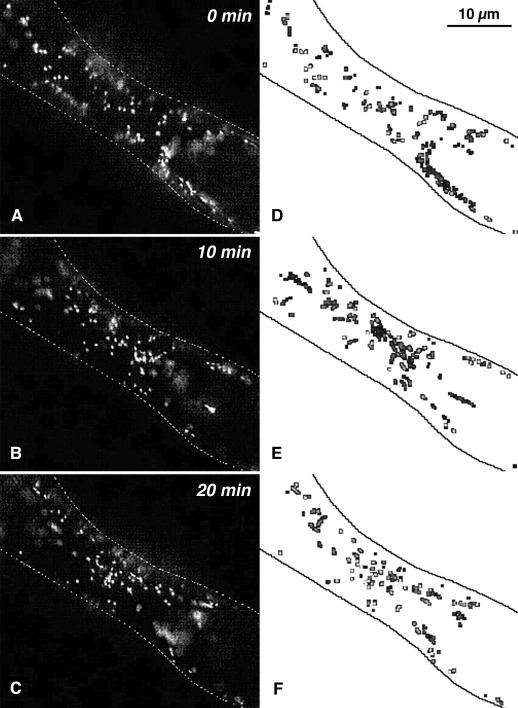
When individual cells were observed for longer periods of time, it was found that the level of activity varied (compare E and F in Fig. 8) and that the regions of the cell with streaming tracks also shifted laterally (compare Fig. 8, D and E). The phenomenon of variable streaming activity was quantified by counting the number of tracks with active streaming in cells that were kept in a perfusion chamber with constant supply of fresh, aerated growth medium (flow rate 0.5 mL/min). Figure 9A shows typical examples for this variability in streaming activity. Cells usually maintained a fairly constant level of streaming activity, which was interrupted by short episodes of reduced activity. Interestingly, sister cells that were connected to each other sometimes displayed parallel fluctuations in activity level (cells A1 and A4 in Fig. 9A), suggesting that a global signal influencing cytoplasmic streaming can travel through plasmodesmata. Most cells also showed a decrease in activity over the course of an experiment (approximately 2 h), with some of them losing streaming activity during the observation period (e.g. cells A1 and A2 in Fig. 9B).
Figure 8.
Shift of regions with active streaming in cortical cytoplasm. Ten-second video sequences of the same cortical region taken in 10-min intervals were analyzed for movement of Golgi stacks. A to C, Single enhanced video images of cortical region in 10-min intervals. D to F, Movement analysis of the video sequences corresponding to A to C, respectively. The positions of Golgi stacks were automatically detected using a peak-finding algorithm. Positions where stacks were detected most of the time (slowly moving and wiggling stacks) are coded in light gray and as hollow spots; positions where stacks were detected only rarely (fast-moving stacks) are coded in black. Video sequences can be viewed at http://www.plantphysiol.org/cgi/content/full/ 121/4/1127/DC1.
Figure 9.
Level of streaming activity of individual cells over time. Individual cells were observed every 5 min, and the number of strands with active streaming in the entire cell were counted. Times are given in minutes from the start of perfusion. In some cases, strands could not be classified unambiguously, since only single stacks showed directed movement or the velocity was slow. A, Activity levels in control cells. ▪, A1; □, A4; ▴, B1; ▵, B2. B, Activity levels prior, during, and after treatment with 30 mm butanedione monoxime. ▾, A1; ▿, A2; ▵, B1; ●, C1; ▪, D1. C, Activity levels prior, during, and after treatment with 10 μm propyzamide. ○, A1; ●, A2; ▪, B1; ▴, C1; ▵, C3. Shaded areas indicate the duration of drug treatments. Cells coded with the same letter are sister cells that share a cell wall. Differences in basal activity are usually related to cell size, with smaller cells showing less Golgi streaming.
Molecular Basis of Golgi Movement
To determine the molecular basis for the observed movement, the streaming behavior of Golgi stacks was investigated after treatment with different drugs that disrupt specific components of the cytoskeleton. Golgi stacks in cells that had been treated with 40 μm cytochalasin D for 30 min to disrupt actin filaments were still wiggling, but showed essentially no translational movement (see tracings in Fig. 4C). Occasionally, single stacks were observed that followed a clear trajectory (not shown). We attribute these few streaming stacks to residual actin filaments that were not affected by the drug. Latrunculin A, another actin-filament-disrupting drug, also inhibited movement of Golgi stacks. At the concentration used (0.1 μm), latrunculin A appeared more potent in that no moving stacks were observed (data not shown). Movement was also reversibly inhibited by 30 mm BDM, an inhibitor of the myosin ATPase (Herrmann et al., 1992; Fig. 9B). All drug effects could be reversed by washing with growth medium; however, most cells were not able to recover fully. It is unclear whether this reduced level of streaming activity reflects normal decrease in cell viability during prolonged observation (see above) or some irreversible component of the drug effect. While the inhibition of directed movement by actin-disrupting drugs was usually accompanied by a breakdown of transvacuolar strands, BDM did not show such an effect (not shown).
A number of microtubule-disrupting drugs was tested, namely nocodazole (at 3.3 μm), colchicine (at 250 μm), and propyzamide (at 6 μm). All had similar effects on the streaming behavior. Here we report data mostly from the nocodazole experiments. Casual observation of nocodazole-treated cells under the microscope revealed that microtubule-disrupting drugs did not inhibit streaming. Instead we noticed an apparent slight increase in streaming activity over control cells. The percentage of cells with actively streaming stacks was increased to a small extent (Table II). This difference, albeit small, is statistically significant when the results from matched drug and control treatments are compared (paired t test, P < 0.004).
While the number of cells with active streaming was increased by microtubule-disrupting drugs, no consistent effect on the activity levels of individual cells could be observed (Fig. 9C), although some cells seemed to show an increase in streaming activity during the drug treatment (e.g. cell A2 in Fig. 9C). Analysis of tracing data for four nocodazole-treated cells (two cortical regions, two thread regions, 50 stacks per cell) showed no significant differences from control cells (Table I). Some of the parameters analyzed appeared elevated in drug-treated cells (e.g. percent streaming stacks in the transvacuolar strands of one of the cells; Table I), but the high degree of variability for the control cells precludes any firm conclusions.
From these experiments it can be concluded that the directed movement of Golgi stacks in plant cells requires intact actin filaments and is probably propelled by myosin motors. Microtubules do not appear to have an effect on movement, except for a subset of cells in which they seem to limit streaming.
DISCUSSION
The Plant α-1,2 Mannosidase Is Homologous to Corresponding Animal and Fungal Enzymes
We have isolated a cDNA from soybean, GmMan1, encoding a protein with a high degree of similarity to animal and fungal class I α-1,2 mannosidases. While the final confirmation of this assignment has to await demonstration of the appropriate enzymatic activity, it appears that the strong conservation of key features found in other mannosidases supports the conclusion that the isolated message encodes this enzymatic function in soybean (Fig. 1). To our knowledge, this is the first report of an enzyme of the N-linked oligosaccharide processing pathway cloned from plants. However, an Arabidopsis expressed sequence tag with 80% similarity to GmMan1 has been reported previously (GenBank accession no. W43154).
GmMan1 shows the typical type II orientation of other Golgi membrane proteins, with a short cytoplasmic tail at its amino terminus followed by a single transmembrane domain. This region, as well as the following putative stalk domain, show little similarity to other α-1,2 mannosidases at the sequence level. In contrast, the catalytic domain is 45% identical in amino acid sequence to either mouse or yeast α-1,2 mannosidase and contains all residues that were shown in other species to be essential for enzymatic function (Fig. 1). This striking distribution of sequence conservation is also found between the α-1,2-mannosidase homologs of other species (Herscovics, 1999a). The soybean cDNA described in this report therefore displays all the hallmarks of known α-1,2 mannosidases.
The high degree of conservation in the catalytic domain is contrasted by the near complete absence of sequence similarity at the N terminus of the protein. This part of the protein is most likely responsible for correct localization of the mannosidase (for review, see Colley, 1997). The lack of sequence conservation led to the formulation of two models that explain the targeting/retention of Golgi proteins in terms of structural features. According to one model, Golgi proteins with their typically short transmembrane helices preferentially partition into the thinner membranes of the early secretory pathway (Pelham and Munro, 1993). The other model proposes a “kin-recognition” mechanism, where resident proteins of the same compartment can physically interact and thereby maintain their specific localization (Nilsson et al., 1993). The sequence of GmMan1 is compatible with both models. The predicted membrane-spanning domain is unusually short, only 16 amino acids. At the same time, the stalk region has a high probability of forming a coiled-coil structure, which is often indicative of protein-protein interactions. Interestingly, two recent reports describe proper targeting of mammalian sialyltransferase to the trans-Golgi in plant cells (Boevink et al., 1998; Wee et al., 1998), suggesting that the targeting mechanism of Golgi oligosaccharide processing enzymes could be a common structural motif conserved between animals and plants.
GmMan1::GFP Fusion Can Be Used as an in Vivo Marker for cis/Medial Golgi Cisternae
The fusion protein of GmMan1 to GFP is localized to the cis side of plant Golgi stacks of suspension-cultured tobacco BY-2 cells (Fig. 3) and can therefore act as a marker of Golgi stack localization in living plant cells. The Golgi stacks of BY-2 cells appear to be randomly distributed throughout the cytoplasm. This is consistent with predictions based on electron micrographs (Robinson and Kristen, 1982) and observations made in a number of other systems. For example, similar patterns of fluorescent dots have been observed in chemically fixed maize root cells stained with the monoclonal antibody JIM84 (Satiat-Jeunemaitre and Hawes, 1992), which recognizes Golgi- and plasma membrane-specific Lewis a type epitopes (Fitchette-Lainé et al., 1997), as well as in transgenic Arabidopsis cells expressing an epitope-tagged sialyltransferase (Wee et al., 1998) and in tobacco leaf epidermal cells transiently expressing GFP fusions of sialyltransferase and the KDEL receptor protein AtERD2 (Boevink et al., 1998).
Occasionally, we could observe cells that had a green fluorescent ER in addition to Golgi stacks (data not shown). It is not clear whether this reflected a bottleneck in the export from the ER or an increased rate of retrograde transport from the “saturated” Golgi. A few cells also contained Golgi-sized fluorescent structures that appeared as small rings (Fig. 2C). Similar structures have been observed when a GFP-tagged sialyltransferase was transiently expressed in tobacco leaf epidermis cells; the images were interpreted as edge-on views of highly curved stacks (Boevink et al., 1998). The observation that the rings can change into lines with a length similar to that of the ring diameter (compare video sequence at http://www.plantphysiol.org/cgi/content/full/121/4/1127/DC1), however, suggests that they represent face-on views of Golgi stacks in which the fusion protein is concentrated in the rims of the cisternae. This distribution could be caused by conditions that lead to an osmotic collapse of the cisternal lumen and the exclusion of the bulky fusion proteins from the central region of the cisternae where the membranes became appressed. This interpretation could also explain the distribution of the sialyltransferase-GFP fusion construct, since the trans-cisternae in cryo-fixed and freeze-substituted cells typically exhibit a collapsed central domain with minimal intracisternal volume (Staehelin et al., 1990).
The Saltatory Movement of Plant Golgi Stacks Is Acto-Myosin Based
The Golgi stacks of BY-2 cells usually alternate between episodes of random “wiggling” motion reminiscent of Brownian motion and directed movement along linear tracks. The wiggling periods can last from seconds to minutes and during such periods individual stacks can “drift” several micrometers without participating in any directed translocation events. In contrast, the stacks displaying directed movement appear to follow the same tracks as cytoplasmic streaming (Williamson, 1993). This was confirmed by staining mitochondria with a rhodamine-labeled dye (MitoTracker, Molecular Probes) and observing the movement of the two organelles in the same cell. Streaming Golgi stacks and mitochondria typically traveled along the same paths and also showed qualitatively similar stop-and-go movement (data not shown). Individual Golgi stacks followed more or less straight trajectories, often with several stacks following the same track (Fig. 7). Progress along these tracks was intermittent, so individual stacks showed stop-and-go movement (Figs. 5 and 7). Interestingly, stacks moving along the same track often paused at the same position, suggesting that these sites can inhibit movement (see below).
To determine the molecular basis for this saltatory motion, BY-2 cells were treated with a number of inhibitors that interfere with normal functioning of cytoskeletal elements. Drugs that disrupt the actin cytoskeleton (cytochalasin D, latrunculin A) also stopped the streaming of Golgi stacks (Fig. 4C). “Wiggling” motion usually was not affected. Cytochalasin D used at 40 μm often did not lead to a complete block of directed movement, and in some instances produced localized circular movements. We assume that this drug concentration did not disrupt all actin filaments and that the few remaining filaments allowed Golgi stack movement to occur. Streaming of Golgi stacks therefore requires intact actin microfilaments. A similar conclusion was reached by Boevink et al. (1998). Interestingly, these authors found a tight co-localization of actin filaments and tubular ER, suggesting that the movement of Golgi stacks occurs in close association with the ER. This is consistent with the finding of Lichtscheidl et al. (1990) that actin filament bundles in the epidermal cells of Drosera tentacles often co-localize with tubular ER cisternae. In our transformed BY-2 cells this ER-Golgi streaming relationship was less evident.
The involvement of myosin motors in this movement was tested by applying the myosin inhibitor BDM, which has been shown to inhibit the ATPase function on isolated skeletal muscle myosin II (Herrmann et al., 1992). Movement of Golgi stacks could be stopped by adding 30 mm BDM to the perfusion medium of the cells (Fig. 9B), suggesting that myosin provides the motive force for the observed motion. This conclusion is consistent both with observations that have implicated myosin as the motor for cytoplasmic streaming (Shimmen and Yokota, 1994) and with reports in which the translocation of organelles in pollen tubes has been linked to myosin (Kohno and Shimmen, 1988). We assume that the movement of plant Golgi stacks occurs by active translocation along actin filaments as opposed to passive drift in a general cytoplasmic stream. This interpretation is based on, among others, on the high degree of specificity observed in the movement of individual stacks (see e.g. Fig. 6). This implies that Golgi movement is mediated by a specific myosin, presumably associated with the Golgi-matrix that surrounds the individual Golgi stack-TGN units.
Microtubule Disruption Does Not Inhibit Golgi Saltatory Movements
In animal cells, the localization of the Golgi complex and the movement of transport carriers to and from the Golgi depend on the presence of intact microtubules (Presley et al., 1997; Scales et al., 1997; Burkhardt, 1998; Hirschberg et al., 1998; Toomre et al., 1999). Therefore, we tested the role of microtubules in directed movement of plant Golgi stacks. Treatment of BY-2 tobacco cells with 3.3 μm nocodazole, 250 μm colchicine, or 6 μm propyzamide did not inhibit streaming (Figs. 4D and 9C; data not shown), indicating that microtubules are not required for the movement of Golgi stacks. Quite unexpectedly, we noticed an apparent increase in streaming activity during the drug treatments. Such a stimulatory effect of microtubule-disrupting drugs on cytoplasmic streaming has to our knowledge not been described before.
Careful analysis of a variety of streaming parameters revealed that the enhanced streaming effect of microtubule disruptors is brought about by increasing the percentage of cells displaying saltatory Golgi movement (Table II). Movement of individual stacks was unaffected with respect to average and maximal velocities, maximal accelerations, and changes between streaming and wiggling events. A few cells appeared to have a higher percentage of streaming stacks after drug treatment (Fig. 9C, cell A2; Table I, cell 2 of transvacuolar strands), while the other cells were not affected. It is not known whether the cells with higher activity are examples for the increase in number of cells with streaming Golgi stacks (see Table II), or whether they represent extremes within the normal variability that was also seen in control cells.
It is unclear how disruption of microtubules can trigger cytoplasmic streaming in a subset of cells. One possibility is that cells in a certain phase of the cell cycle have reduced streaming, and that this inhibition could be mediated in part by microtubules. Disruption of the microtubule scaffold would then release the constraints on the Golgi translocation machinery. For example, it is known that cytoplasmic streaming ceases during mitosis (Mineyuki et al., 1984). However, the percentage of cells in mitosis is too small to account for the observed increase in numbers of cells with streaming. Therefore, we speculate that Golgi movement may also be constrained during another stage of the cell cycle. We are currently investigating this possibility in synchronized cells.
Is the Stop-and-Go Movement of Plant Golgi Stacks a Regulated Process? A Hypothesis
The rapid stop-and-go movement of plant Golgi stacks raises the question of how the secretory pathway in plant cells can function efficiently. One of the consequences of the streaming behavior of Golgi stacks is that there is no fixed spatial relationship between the ER and Golgi. The variable spatial orientation of Golgi stacks to nearby ER cisternae has been known to plant cell biologists for over 20 years (for review, see Robinson and Kristen, 1982), but the mechanistic basis for this variability has remained an enigma. This report and that by Boevink et al. (1998) help to explain this apparent random distribution as a steady-state intermediate of continuously moving stacks. At the same time, these findings raise the question of whether vesicle trafficking between the ER and Golgi is based strictly on random encounters, or if it is regulated by mechanisms yet to be discovered.
The observation that in leaf epidermal cells Golgi stacks usually track along the well-defined ER elements underlying the plasma membrane has led to the suggestion that the Golgi stacks act as “vacuum cleaners” that move around to pick up products from the ER (Boevink et al., 1998). However, this movement was predominantly observed along tubular ER strands (Boevink et al., 1998), which may not be very active in protein synthesis. It is also difficult to envision the targeting of ER-derived transport vesicles to cis-Golgi cisternae when the stacks travel at speeds most likely greater than vesicle diffusion rates. In the model depicted in Figure 10, we offer an alternative hypothesis, in which the stop-and-go motion of Golgi stacks is postulated to be regulated to increase the efficiency of ER-to-Golgi transport, as well as the delivery of secretory products to specific cell wall domains.
Figure 10.
Model of regulated stop-and-go movement of Golgi stacks and its relationship to the transport of products through the secretory pathway. A, Golgi stacks with myosin motors attached to the Golgi matrix move along actin filaments. B, Activation of ER exit sites may release a local signal that inhibits movement of Golgi stacks and allows for uptake of ER-to-Golgi transport vesicles. C, Local cell wall expansion or sites of secondary wall thickenings may lead to a signal resulting in stopping of Golgi stack movement and to the release of Golgi-derived secretory vesicles.
Our model postulates that active ER export sites produce a localized signal that leads to the uncoupling of nearby Golgi stacks from the actin tracks and to their pausing in the vicinity of the activated ER export site (Fig. 10B). This would increase the efficiency of ER-to-Golgi (and Golgi-to-ER) trafficking. Upon completion of the transfer, the ER stop signal would be turned off, allowing the stacks to resume their movement. A similar stop signal may be produced by regions where Golgi products are required, such as areas of wall expansion or sites of secondary cell wall thickenings (Fig. 10C). It is well-known that secretion can be directed to specific cell wall sites (Fowler and Quatrano, 1997), and a patch-like distribution of secretory vesicle profiles in freeze-fracture images of plasma membranes of root tip and cultured cells has been reported (Staehelin and Chapman, 1987; Craig and Staehelin, 1988). The gradual shifting of Golgi streaming domains over larger time intervals (Fig. 8) may help to ensure an even deposition of cell wall products over time.
A potential candidate for the postulated stop signal is calcium, since it is known that elevated calcium concentrations can block cytoplasmic streaming (Shimmen and Yokota, 1994) by a calmodulin-mediated inhibition of myosin (Yokota et al., 1999). Such sites of localized inhibition of movement may be recognized by studying the behavior of several stacks within one streaming strand. Indeed, we have found positions along a strand where two or three sequentially arriving stacks stop briefly before moving on (compare videos at http://www.plantphysiol.org/cgi/content/full/121/4/1127/DC1). Of course, this circumstantial evidence is not conclusive, as discontinuities in actin filaments could cause similar effects. We are currently initiating experiments to address this question more directly.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Dr. Annette Herscovics (McGill University, Montreal) for providing the partial soybean clone and insightful comments concerning the manuscript. Work in her laboratory was supported by the National Institutes of Health (grant no. GM31265) and The Medical Research Council of Canada. We also thank Dr. J. Mullet (Texas A&M University) for the soybean cDNA library, Dr. J. Sheen (Massachusetts General Hospital, Boston) for the GFP gene, and Dr. J. Kahana (Harvard University, Cambridge, MA) for the anti-GFP antibodies. Dr. Dan LaFlamme provided valuable advice regarding cloning techniques. We thank Dr. Bradley Olwin for the generous use of his video microscope setup.
Footnotes
This work was supported by the National Institutes of Health (grant no. GM18639) to L.A.S.
LITERATURE CITED
- Andreeva AV, Kutuzov MA, Evans DE, Hawes CR. Proteins involved in membrane transport between the ER and the Golgi apparatus: 21 putative plant homologues revealed by dbEST searching. Cell Biol Int. 1998a;22:145–160. doi: 10.1006/cbir.1998.0235. [DOI] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA, Evans DE, Hawes CR. The structure and function of the Golgi apparatus: a hundred years of questions. J Exp Bot. 1998b;49:1281–1291. [Google Scholar]
- Barlow C. COPII and selective export from the endoplasmic reticulum. Biochim Biophys Acta. 1998;1404:67–76. doi: 10.1016/s0167-4889(98)00047-0. [DOI] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Sant Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Burke J, Lipari F, Igdoura S, Herscovics A. The Saccharomyces cerevisiae processing α1,2-mannosidase is localized in the endoplasmic reticulum, independently of known retrieval motifs. Eur J Cell Biol. 1996;70:298–305. [PubMed] [Google Scholar]
- Burkhardt JK. The role of microtubule-based motor proteins in maintaining the structure and function of the Golgi complex. Biochim Biophys Acta. 1998;1404:113–126. doi: 10.1016/s0167-4889(98)00052-4. [DOI] [PubMed] [Google Scholar]
- Camirand A, Heysen A, Grondin B, Herscovics AA. Glycoprotein biosynthesis in Saccharomyces cerevisiae: isolation and characterization of the gene encoding a specific processing alpha-mannosidase. J Biol Chem. 1991;266:15120–15127. [PubMed] [Google Scholar]
- Colley KJ. Golgi localization of glycosyltransferases: more questions than answers. Glycobiology. 1997;7:1–13. doi: 10.1093/glycob/7.1.1-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig S, Staehelin LA. High pressure freezing of intact plant tissues: evaluation and characterization of novel features of the endoplasmatic reticulum and associated membrane systems. Eur J Cell Biol. 1988;46:80–93. [PubMed] [Google Scholar]
- Driouich A, Staehelin LA. The plant Golgi apparatus: structural organization and functional properties. In: Berger EG, Roth J, editors. The Golgi Apparatus. Basel: Birkhäuser Verlag; 1997. pp. 275–301. [Google Scholar]
- Dupree P, Sherrier DJ. The plant Golgi apparatus. Biochim Biophys Acta. 1998;1404:259–270. doi: 10.1016/s0167-4889(98)00061-5. [DOI] [PubMed] [Google Scholar]
- Fitchette-Lainé A-C, Gomord V, Cabanes M, Michalski JC, Saint-Macary M, Foucher B, Cavelier B, Hawes C, Lerouge P, Faye L. N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997;12:1411–1417. doi: 10.1046/j.1365-313x.1997.12061411.x. [DOI] [PubMed] [Google Scholar]
- Fowler JE, Quatrano RE. Plant cell morphogenesis: plasma membrane interactions with the cytoskeleton and cell wall. Annu Rev Cell Dev Biol. 1997;13:697–743. doi: 10.1146/annurev.cellbio.13.1.697. [DOI] [PubMed] [Google Scholar]
- Hennegan KP, Danna KJ. pBIN20: an improved binary vector for Agrobacterium-mediated transformation. Plant Mol Biol Rep. 1998;16:129–131. [Google Scholar]
- Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C, Wray J, Travers F, Barman T. Effect of 2,3-butanedione monoxime on myosin and myofibrillar ATPases: an example of an uncompetitive inhibitor. Biochemistry. 1992;31:12227–12232. doi: 10.1021/bi00163a036. [DOI] [PubMed] [Google Scholar]
- Herscovics A. Glycosidases of the asparagine-linked oligosaccharide processing pathway. In: Pinto BM, editor. Comprehensive Natural Products Chemistry. Ed 3. New York: Elsevier Science Publishing; 1999a. pp. 13–35. [Google Scholar]
- Herscovics A. Processing glycosidases of Saccharomyces cerevisiae. Biochim Biophys Acta. 1999b;1426:275–285. doi: 10.1016/s0304-4165(98)00129-9. [DOI] [PubMed] [Google Scholar]
- Herscovics A, Schneikert J, Athanassiadis A, Moremen KW. Isolation of a mouse Golgi mannosidase cDNA, a member of a gene family conserved from yeast to mammals. J Biol Chem. 1994;269:9864–9871. [PubMed] [Google Scholar]
- Hirschberg K, Miller CM, Ellenberg J, Presley JF, Siggia ED, Phair RD, Lippincott-Schwartz J. Kinetic analysis of secretory protein traffic and characterization of Golgi to plasma membrane transport intermediates in living cells. J Cell Biol. 1998;142:1485–1503. doi: 10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios RM. GMAP-210, a cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J Cell Biol. 1999;145:83–98. doi: 10.1083/jcb.145.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher S, Albert S, Wucherpfennig D, Heisenberg M, Schneuwly S. Molecular and genetic analysis of the Drosophila mas-1 (mannosidase-1) gene which encodes a glycoprotein processing alpha 1,2-mannosidase. Dev Biol. 1995;168:613–626. doi: 10.1006/dbio.1995.1106. [DOI] [PubMed] [Google Scholar]
- Kohno T, Shimmen T. Accelerated sliding of pollen tube organelles along Characeae actin bundles regulated by Ca2+ J Cell Biol. 1988;106:1539–1543. doi: 10.1083/jcb.106.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K. Cytoplasmic streaming in plant cells. Int Rev Cytol. 1990;121:267–307. [Google Scholar]
- Lal A, Schutzbach JS, Foresee WT, Neame PJ, Moremen KW. Isolation and expression of murine and rabbit cDNAs encoding an α1,2-mannosidase involved in the processing of asparagine-linked oligosaccharides. J Biol Chem. 1994;13:9872–9881. [PubMed] [Google Scholar]
- Lichtscheidl IK, Lancelle SA, Hepler PK. Actin-endoplasmic reticulum complexes in Drosera: their structural relationship with the plasmalemma, nucleus, and organelles in cells prepared by high pressure freezing. Protoplasma. 1990;155:116–126. [Google Scholar]
- Lipari F, Herscovics A. Role of the cysteine residues in the α1,2-mannosidase involved in N-glycan biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 1996;271:27615–27622. doi: 10.1074/jbc.271.44.27615. [DOI] [PubMed] [Google Scholar]
- Lipari F, Herscovics A. Calcium binding to the class I α-1,2-mannosidase from Saccharomyces cerevisiae occurs outside the EF hand motif. Biochemistry. 1999;38:1111–1118. doi: 10.1021/bi981643i. [DOI] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. Regulation of membrane traffic in animal cells by COPI. Biochim Biophys Acta. 1998;1404:53–66. doi: 10.1016/s0167-4889(98)00046-9. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mineyuki Y, Furuya M. Involvement of colchicine-sensitive cytoplasmic element in premitotic nuclear positioning of Adiantum protonemata. Protoplasma. 1986;130:83–90. [Google Scholar]
- Mineyuki Y, Takagi M, Furuya M. Changes in organelle movement in the nuclear region during the cell cycle of Adiantum protonemata. Plant Cell Physiol. 1984;25:297–308. [Google Scholar]
- Mizukami M, Wada S. Action spectrum for light-induced chloroplast accumulation in a marine coenocytic alga, Bryopsis plumosa. Plant Cell Physiol. 1981;22:1245–1255. [Google Scholar]
- Mollenhauer HH, Morré DJ. Cytochalasin B, but not colchicine, inhibits migration of secretory vesicles in root tips of maize. Protoplasma. 1976;87:39–48. doi: 10.1007/BF01623957. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Trimble RB, Herscovics A. Glycosidases of the asparagine-linked oligosaccharide processing pathway. Glycobiology. 1994;4:113–125. doi: 10.1093/glycob/4.2.113. [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol. 1982;132:1–30. [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G. Kin recognition: a model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Pelham HRB, Munro S. Sorting of membrane proteins in the secretory pathway. Cell. 1993;75:603–605. doi: 10.1016/0092-8674(93)90479-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJM, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y. Three-dimensional structure of the Golgi apparatus in mammalian cells. In: Berger EG, Roth J, editors. The Golgi Apparatus. Basel: Birkhäuser Verlag; 1997. pp. 37–61. [Google Scholar]
- Robinson DG, Kristen U. Membrane flow via the Golgi apparatus of higher plant cells. Int Rev Cytol. 1982;77:89–127. [Google Scholar]
- Roth J, Brada D, Lackie PM, Schweden J, Bause E. Oligosaccharide trimming Man9-mannosidase is a resident ER protein and exhibits a more restricted and local distribution than glucosidase II. Eur J Cell Biol. 1990;53:131–141. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Samuels AL, Giddings TH, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satiat-Jeunemaitre B, Hawes C. Redistribution of a Golgi glycoprotein in plant cells treated with brefeldin A. J Cell Sci. 1992;103:1153–1166. [Google Scholar]
- Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Shimmen T, Yokota E. Physiological and biochemical aspects of cytoplasmic streaming. Int Rev Cytol. 1994;155:97–139. [Google Scholar]
- Staehelin LA, Chapman RL. Secretion and membrane recycling in plant cells: novel intermediary structures visualized in ultrarapidly frozen sycamore and carrot suspension-culture cells. Planta. 1987;171:43–57. doi: 10.1007/BF00395066. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Giddings TH, Kiss JZ, Sack FD. Macromolecular differentiation of Golgi stacks in root tips of Arabidopsis and Nicotiana seedlings as visualized in high pressure frozen and freeze-substituted samples. Protoplasma. 1990;157:75–91. doi: 10.1007/BF01322640. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Moore I. The plant Golgi apparatus: structure, functional organization and trafficking mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:261–288. [Google Scholar]
- Toomre D, Keller P, White J, Olivo J-C, Simons K. Dual-color visualization of trans-Golgi network to plasma membrane traffic along microtubules in living cells. J Cell Sci. 1999;112:21–33. doi: 10.1242/jcs.112.1.21. [DOI] [PubMed] [Google Scholar]
- Wee EG-T, Sherrier DJ, Prime TA, Dupree P. Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell. 1998;10:1759–1768. doi: 10.1105/tpc.10.10.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RE. Organelle movements. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:181–202. [Google Scholar]
- Yokota E, Muto S, Shimmen T. Inhibitory regulation of higher-plant myosin by Ca2+ ions. Plant Physiol. 1999;119:231–239. doi: 10.1104/pp.119.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.