Abstract
T cells create vast amounts of diversity in their T cell receptor (TCR) genes, enabling individual clones to recognize specific peptide-MHC ligands. Here we combine TCR sequencing and assay for transposase-accessible chromatin analysis at the single-cell level to provide information on the TCR specificity and epigenomic state of individual T cells. Using this approach, termed Transcript-indexed ATAC-seq (T-ATAC-seq), we identify epigenomic signatures in immortalized leukemic T cells, primary human T cells from healthy volunteers, and primary leukemic T cells from patient samples. In healthy peripheral blood CD4+ T cells, we identify cis and trans regulators of naive and memory T cell states and find substantial heterogeneity in surface marker-defined T cell populations. In patients with cutaneous T cell lymphoma, T-ATAC-seq enabled identification of leukemic and non-leukemic regulatory pathways in T cells from the same individual, separating signals arising from the malignant clone from background T cell noise. Thus, T-ATAC-seq is a new tool that enables analysis of epigenomic landscapes in clonal T cells and should be valuable for studies of T cell malignancy, immunity, and immunotherapy.
Introduction
T lymphocytes recognize self- and foreign antigens and are the central drivers of regulatory and effector immune responses. Each T cell expresses a T cell receptor (TCR), which recognizes antigens in the context of major histocompatibility complex (MHC) molecules displayed on the surface of antigen-presenting or pathogen-infected cells. The major TCR species is composed of α- and β-subunits that are encoded by genes that are somatically-recombined by V(D)J recombination, which produces a diverse repertoire of antigen-reactive T cells, with up to a possible 1014 unique heterodimers in each individual1. As a result of antigen-specific or malignant clonal expansion, the TCR also serves as a faithful identifier of its clonal origin, as T cells expressing identical TCRαβ pairs must almost invariably arise from a common cellular ancestor. The specific pairing of TCRαβ from one cell is necessary to recapitulate its antigen specificity and is critical for weaponizing or disarming an immune response for immunotherapy. Therefore, identification of TCRαβ sequences is critical to understanding the identity of single T cells, and methods which pair TCRαβ sequence with cell and activation states may uncover clonal gene regulatory pathways missed by ensemble measurements.
Recent advances in genome sequencing technologies have enabled single-cell gene expression and epigenetic measurements and have revealed variability in immune cell development and responsiveness2–5. Our groups recently developed methods to efficiently amplify and sequence both TCRα and β chains from single T cells6, and to measure epigenetic changes genome-wide in single cells. The latter method, termed single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq), enables measurement of regulatory DNA elements by direct transposition of sequencing adaptors into regions of accessible chromatin7–9. Unlike methods to measure the transcriptome in single cells, scATAC-seq identifies cell-to-cell variation in cis regulatory elements and trans factors that drive epigenetic cell states. Moreover, analysis of single-cell epigenomic profiles can be used to reveal significant variability within cell surface marker-defined populations and the existence of cell states obscured by ensemble measurements10.
Here we combine these two methodologies to produce a method that can allow one to study both the epigenetic landscape and T cell specificity simultaneously at the single-cell level. This two-way analysis may facilitate discovery of antigens driving a certain T cell fate, or conversely, cis and trans regulators driving the expansion of a T cell clone. We refer to this as transcript-indexed ATAC-seq (T-ATAC-seq). The T-ATAC-seq experimental pipeline integrates scATAC-seq with targeted TCR-seq in the same single cell, followed by high-throughput sequencing and computational integration of both datasets. To demonstrate the performance and utility of T-ATAC-seq, we performed this method on 1,344 human T cells sorted using standard subset-specific cell surface markers and integrated the analysis of regulatory landscapes with TCR identity. T-ATAC-seq in peripheral blood CD4+ T cells from healthy volunteers revealed epigenomic signatures and single-cell variability of naive and memory CD4+ T cells. Importantly, unbiased single-cell analysis identified divergent chromatin states within cell surface marker-defined T cell subtypes. We extended the use of this method to clinical samples from patients with T cell leukemia. T-ATAC-seq enabled the identification of cancer clone-specific epigenomic signatures, which were not apparent from ensemble measurements. These data demonstrate the utility of T-ATAC-seq as a new tool for single-cell epigenomic characterization of T cells in both research and clinical applications.
Results
Performance of T-ATAC-seq in human immortalized T cells
We implemented T-ATAC-seq using an automated microfluidic platform (C1; Fluidigm, Fig. 1a and Supplementary Fig. 1a). For this approach, single cells were first individually captured on the Integrated Fluidics Circuit (IFC) in single-cell chambers and then subjected to cell lysis and DNA transposition with the prokaryotic Tn5 enzyme loaded with sequencing adapters. After transposition of accessible chromatin, Tn5 was released from DNA fragments and TCR RNA within each chamber was subjected to reverse transcription (RT) using primers targeting TCRα and TCRβ constant regions. Immediately after RT, 5′ ends of ATAC-seq fragments were extended and all chamber contents were amplified by PCR. TCR fragments were amplified using primers targeting TCRα and TCRβ constant and variable regions. Single-cell libraries were then collected and TCR or ATAC amplicons were further amplified with cell-identifying barcoded primers, pooled, and sequenced on a high-throughput sequencing instrument.
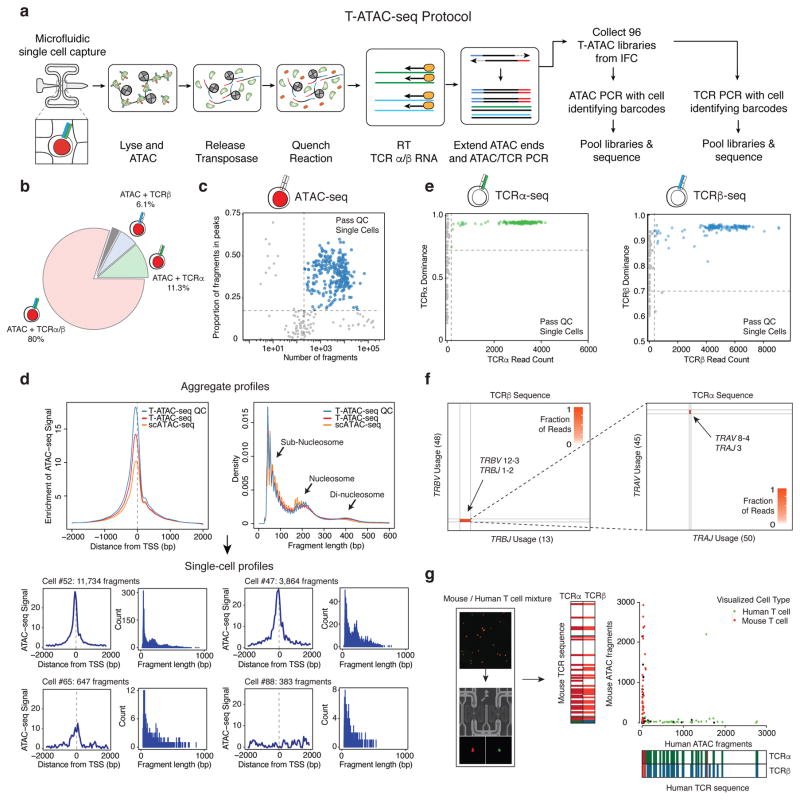
Figure 1. T-ATAC-seq generates open chromatin and TCR profiles in single T cells.
(a) Outline of the T-ATAC-seq protocol. Squares indicate individual microfluidic chambers in the IFC. T cells are individually captured and sequentially subjected to ATAC-seq (chambers 1–3), reverse transcription of TCRα and TCRβ chain transcripts, and amplification of ATAC-seq and TCR-seq amplicons, in nanoliter-scale reaction volumes. Single-cell libraries are then amplified with cell-identifying barcodes and analyzed by high-throughput sequencing. (b) Pie chart indicating overlap of TCR-seq and ATAC-seq data from single Jurkat cells (231 single cells from 3 independent experiments) that passed quality control filters. Shown are the proportion of cells generating ATAC-seq profiles in which TCRα or TCRβ sequence was also obtained. The gray bar indicates the portion of cells in which ATAC-seq data was obtained, but TCRα or TCRβ data was not (2.6%). (c) T-ATAC-seq data quality control filters. Shown are the number of unique ATAC-seq nuclear fragments in each single Jurkat cell compared to the percentage of fragments in ATAC-seq peaks derived from ensemble Jurkat ATAC-seq profiles. (d) Aggregate (top) and single-cell (bottom) T-ATAC-seq profile characteristics. Shown are enrichments of ATAC-seq Tn5 insertions around transcription start sites (TSS) and the nucleosomal periodicity of ATAC-seq fragment lengths. Aggregate profiles obtained from all T-ATAC-seq single cells, T-ATAC-seq single cells passing quality control filters (QC), and scATAC-seq cells are shown. Fragment length indicates the genomic distance between two Tn5 insertion sites, as determined by paired-end sequencing of ATAC fragments. Density indicates the fraction of fragments with the indicated length. The cell # indicates the position of each individual cell in the IFC, and the associated fragment number indicates the number of unique nuclear fragments obtained in that cell. Count indicates the number of fragments for each fragment length. (e) Quality control filters for TCRα (left) and TCRβ (right) sequences. Shown are TCRα or TCRβ paired-end sequencing read counts for each single cell compared to TCR dominance of the top clone for each cell. TCR dominance is quantified as the fraction of reads that support the most prevalent TCR clone by sequence identity6. Dashed lines represent quality control filters of 100 reads and 70% dominance for Jurkat cells. (f) Heat maps showing TCRα or TCRβ rearrangements identified in Jurkat cells. Each axis represents all possible genes within the indicated TCR locus. The labeled genes indicate the sequences identified using T-ATAC-seq. (g) Mouse/human T cell mixing experiment. Shown are visualized cells in the IFC (left), unique nuclear ATAC-seq fragments aligning to the mouse or human genome, and TCR-seq clones identified when compared to mouse or human references (right). In the IFC, human T cells are labeled in green and mouse T cells are labeled in red.
To assess the performance of this method, we carried out T-ATAC-seq in 288 single human Jurkat leukemia cells (Supplementary Fig. 1b). Combined ATAC-seq and TCR-seq (either TCRα or TCRβ) profiles were obtained in 93.9% of captured live cells, and 80% of live cells produced ATAC-seq and paired TCRα and TCRβ sequence (Fig. 1b). Next, we evaluated the quantity and quality of the scATAC-seq data. Microfluidic chambers that produced low quality data (corresponding to empty chambers or dead cell captures) were excluded from further analysis using cut-offs for unique nuclear fragment number and fraction of fragments in accessible chromatin sites, as previously described (Supplementary Fig. 1c and d; Methods)8–11. Chambers passing filter yielded an average of 8.5 × 103 fragments mapping to the nuclear genome, and approximately 38% of fragments were within peaks present in ensemble Jurkat ATAC-seq profiles (Fig. 1c). Single-cell ATAC-seq data recapitulated several characteristics of ensemble ATAC-seq, including fragment-length periodicity and enrichment of fragments at transcription start sites (TSS; Fig. 1d and Supplementary Fig. 1c). Importantly, T-ATAC-seq data quality in single cells were similar to those derived from scATAC-seq alone (Fig. 1d), demonstrating that incorporating targeted RT and PCR of the TCR transcripts did not impact the quality of ATAC-seq data.
We next assessed the performance of T-ATAC-seq in obtaining TCRα and TCRβ sequences from single cells. T-ATAC-seq TCR primers were designed to amplify the complementarity-determining region 3 (CDR3) in the TCRα and TCRβ loci. TCR sequence quality was assessed by TCR sequence read number and single-cell clonal dominance, as previously described6, and only chambers generating high quality TCR sequence were included in downstream analyses. On average, we obtained 2.7 × 103 reads for TCRα and 4.2 × 103 for TCRβ in chambers that passed quality control filters (Fig. 1e and Supplementary Fig. 1d and e). In chambers that produced either ATAC-seq or TCR-seq reads, we obtained TCRα sequence in 89.9% (249/277) cells and TCRβ sequence in 79.1% (219/277) cells, resulting in paired TCRα and TCRβ sequence in 71% of cells (196/277) (Supplementary Fig. 1f). These efficiencies are similar to previous techniques which obtained TCR sequence from single cells6,12,13. TCR sequences in all cells passing filter correctly identified the Jurkat TCR heterodimer as TRBV12–3-TRBJ1–2 and TRAV8-4-TRAJ3 (Fig. 1f). Finally, species mixing experiments using mouse cells (58αβ−/− cells transduced with a mouse TCR, labeled with calcein red) and human T cells (Jurkat, labeled with calcein green) confirmed that T-ATAC-seq correctly paired cells visualized on the microfluidic chip with species-specific open chromatin, TCRα, and TCRβ sequences (Fig. 1g). Human ATAC-seq fragments were always paired with human TCRs, and mouse ATAC-seq fragments with mouse TCRs, with the exception of 1 doublet out of 94 cells. In summary, T-ATAC-seq efficiently and accurately pairs TCRα and TCRβ sequence identity with chromatin accessibility in single T cells.
Single-cell epigenomic analysis using T-ATAC-seq
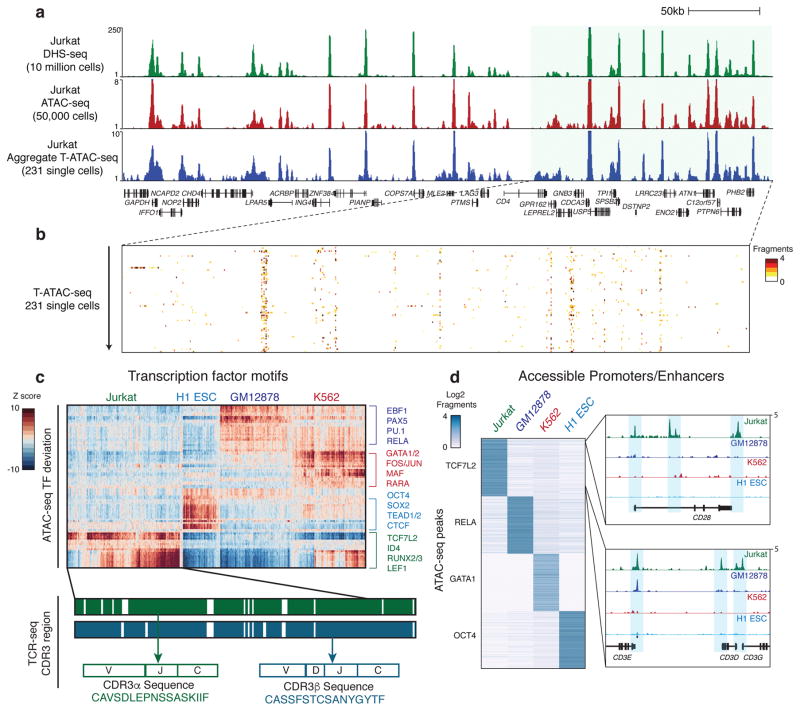
Single-cell epigenomic data can be assessed at the level of (1) regulatory DNA elements or (2) transcription factor (TF) activity across many loci, computed from observed/expected fragments in TF binding sites in each single cell, as previously described8,11,14. T-ATAC-seq performed comparably to scATAC-seq in both measurements. For (1), aggregate T-ATAC-seq profiles from 231 single cells closely reproduced population measurements profiled by DNase I hypersensitivity sequencing (DHS-seq) and ensemble ATAC-seq generated from 107 or 5 × 104 cells, respectively (Fig. 2a)11. Single-cell profiles were strongly enriched for fragments within open chromatin sites present in ensemble profiles (Fig. 2b). For (2), TF motif activity in Jurkat cells identified using T-ATAC-seq or scATAC-seq yielded similar profiles (Supplementary Fig. 2a). Jurkat cells showed high accessibility at DNA regions that contained motifs for T-cell factor (TCF)/lymphoid enhancer-binding factor (LEF) family members, including TCF7L2 and LEF1, and runt-related TF family members RUNX2 and 3, compared to single-cell profiles from H1 embryonic stem cells (ESC), GM12878 B lymphoblastoid cells, and K562 myeloid leukemia cells (Fig 2c and Supplementary Fig. 2b and c). It is important to note that TF motif enrichments (henceforth referred to as TF deviation scores) reflect the activity of all TFs with similar DNA-binding motifs, rather than any particular TF. Therefore, high deviation scores of TCF7L2 in Jurkat cells may reflect the function of additional TCF family members, such as TCF1, which has previously been shown to function in early T cell progenitors to establish T cell fate15. Similarly, high RUNX2/3 deviation scores also encompass RUNX1 activity, as seen in early T cell development16. Differential analysis of ATAC-seq peaks that contained binding sites for each TF identified cell type-specific accessible sites. For example, accessible regions in Jurkat cells containing TCF7L2 motifs included promoters and enhancers for the T cell-specific genes CD28 and CD3D, E, and G (Fig. 2d). Finally, we determined how many single cells were required to reliably recapitulate ensemble ATAC-seq measurements. Strikingly, TF deviation scores were highly accurate even in individual cells when compared to scores derived from ensemble ATAC-seq data (Spearman rank: Rho=0.957, p<0.01; Supplementary Fig. 2d). In contrast, accurately quantifying individual open chromatin sites required the aggregation of approximately 50 single cells in order to reflect population peak profiles (Spearman rank: Rho=0.5, p<0.01; Supplementary Fig. 2e). Therefore, our strategy to assess epigenomic signatures using T-ATAC-seq data was to first characterize cells using TF deviation scores, and then to calculate accessibility differences at individual sites when single cells could be aggregated by their shared immunophenotype or TCRαβ sequence.
Figure 2. T-ATAC-seq identifies epigenomic signatures of clonal Jurkat T cells.
(a) Genome tracks showing a comparison of aggregate single-cell T-ATAC-seq profiles to ensemble ATAC-seq and DHS-seq profiles. (b) Zoom-in of the indicated genome track in (a), showing accessibility profiles for single Jurkat cells. Each pixel represents a 200bp region. (c) Heat map of TF deviation z-scores in single Jurkat cells (231 cells, 3 independent experiments) obtained using T-ATAC-seq compared to previously published profiles from H1 ESC (84 cells), GM12878 (159 cells), and K562 cells (258 cells) obtained using scATAC-seq8. The presence or absence of paired TCRα or TCRβ is indicated by green and blue bars. Amino acid sequences represent the identified CDR3 region, which spans V, (D), J, and C genes. (d) Left, heat map showing ATAC-seq fragment counts in peaks (rows) containing the indicated motifs from aggregated single cells. Cell types analyzed (aggregated from single-cell profiles) are indicated above each column. Right, genome tracks for aggregated single-cell ATAC-seq data. Jurkat-specific peaks in the CD28 and CD3E, D, and G locus are shown.
T-ATAC-seq identifies single-cell regulatory signatures in primary CD4+ T cells
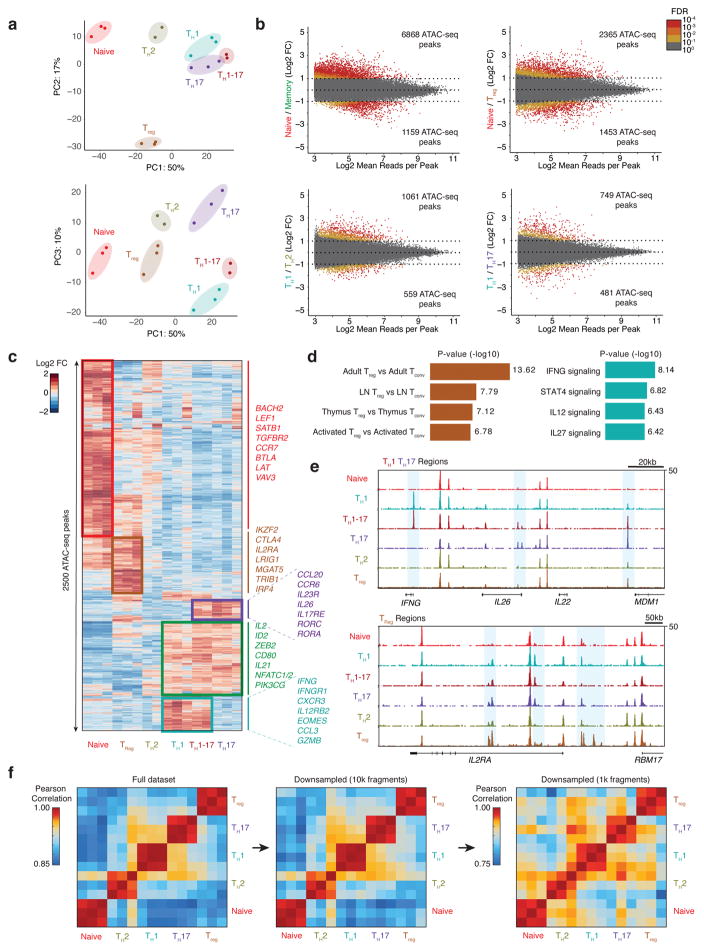
In order to build a comparison dataset for T-ATAC-seq profiles in primary cells and to establish T cell subset-specific chromatin landscape benchmarks, we generated ensemble ATAC-seq profiles from cell surface marker-defined CD4+ naive and memory T cell subtypes17. Peripheral blood CD4+ T cell were obtained from two healthy subjects (3 total replicates), and T cell subsets were isolated by FACS and immediately subjected to ATAC-seq. We profiled naive T cells (CD4+CD45RA+CD25−CD127hi), regulatory T cells (Treg; CD4+CD25+CD127low), T helper 1 cells (TH1; CD4+CD45RA−CD25−CD127hiCXCR3+CCR6−CXCR5−), T helper 17 cells (TH17; CD4+CD45RA−CD25−CD127hiCXCR3−CCR6+CXCR5−), T helper 1-17 cells (TH1-17; CD4+CD45RA−CD25−CD127hiCXCR3+CCR6+CXCR5−), and T helper 2 cells (TH2; CD4+CD45RA−CD25−CD127hiCXCR3−CCR6−CXCR5−) (Supplementary Fig. 3a and b)17. Analysis of ensemble ATAC-seq profiles by principal component analysis (PCA) showed distinct chromatin states for each T cell subset; PC1 distinguished naive and memory T cell subtypes, PC2 distinguished Treg cells from all other subtypes, and PC3 distinguished TH1 and TH17 subtypes (Fig. 3a). Analysis of differential ATAC-seq peaks showed that a large shift in chromatin accessibility accompanied the differentiation of naive T cells to memory T cells, with the majority of differential peaks (6,868 sites) closing in memory cells (Fig. 3b). In contrast, there were relatively fewer differences between T helper subtypes, and cell type-specific open chromatin sites were mainly at functional gene promoters and distal elements (Fig. 3b–e). For example, Treg cells showed increased accessibility at the promoter and upstream elements in the IL2RA locus, consistent with this gene’s critical function in this cell type (Fig. 3c,d)18. Similarly, TH1 and TH1-17 cells showed increased accessibility at the IFNG locus, and TH1-17 and TH17 cells showed increased accessibility at the IL26 and IL22 loci, consistent with the functions of these molecules in T cell-mediated inflammation (Fig. 3e)19,20. Importantly, all naive and memory T cell subtypes could be distinguished from one another when downsampled to the fragment density equivalent to that which is obtained by single-cell T-ATAC-seq data (1 × 103 – 1 × 104 nuclear fragments; Fig. 3f), suggesting that variability in T cell phenotypes could be determined with single-cell measurements.
Figure 3. Epigenomic landscape of ensemble human CD4+ T cell subtypes.
(a) PCA of ensemble ATAC-seq profiles from CD4+ T cell subtypes using the top 2500 variable ATAC-seq peaks (as defined by variance rank of log2 variance-stabilized read counts; n=3, 3 independent experiments). Percentages indicate percent of variance explained by each PC. (b) Differential ATAC-seq peaks for the indicated T cell subtypes. Memory T cell signatures reflect the average accessibility in TH1, TH2, TH17, and TH1-17 cells. (c) Heat map showing clusters for top 2500 varying ATAC-seq peaks. Colors indicate log2 fold-change of reads in each peak compared to the mean across all T cell types. (d) Left, MSigDB Immunologic Signatures of Treg-specific ATAC-seq peaks as obtained from GREAT analysis. Right, MSigDB Pathway Signatures of TH1-specific ATAC-seq peaks as obtained from GREAT analysis (Binomial test, n=3, 3 independent experiments). (e) Ensemble ATAC-seq data genome tracks for the indicated T cell subtypes. Highlighted regions show cell type-specific ATAC-seq peaks. (f) Pearson correlation of PC scores of ensemble ATAC-seq profiles and of ensemble ATAC-seq profiles after downsampling each profile to 10,000 or 1,000 fragments. Downsampling was performed by randomly selecting 10,000 or 1,000 nuclear fragments in each ensemble ATAC-seq .bam file. Heat maps demonstrate that CD4+ T cell subtype profiles can be distinguished from one another using the full dataset or profiles with a fraction of the fragments, as expected in single-cell libraries (16 ATAC-seq profiles obtained from 3 independent experiments).
We next performed T-ATAC-seq in primary human peripheral blood CD4+ T cells (Fig. 3a). We sorted naive T cells (as above), memory T cells that contained all helper phenotypes (CD4+CD45RA−CD25−CD127hi), and memory TH17 cells (CD4+CD45RA−CD25−CD127hiCCR6+) from two healthy individuals, and subjected each population to T-ATAC-seq. Single-cell profiles were filtered using quality controls as described above for immortalized cells. Briefly, single primary T cells displayed high quality ATAC-seq reads; cells passing the filter yielded an average of 2.4 × 103 fragments mapping to the nuclear genome, and an average of 73% of fragments were within peaks derived from ensemble primary T cell ATAC-seq profiles (Supplementary Fig. 4a). Single-cell ATAC-seq data showed enrichment of fragments at TSSs and nucleosomal periodicity of fragment lengths similar to ensemble profiles (Supplementary Fig. 4a). Similarly, TCR sequencing data remained robust in captured single cells, generating on average 1.1 × 103 reads for TCRα sequences and 4.3 × 102 reads for TCRβ sequences (Supplementary Fig. 4b and c).
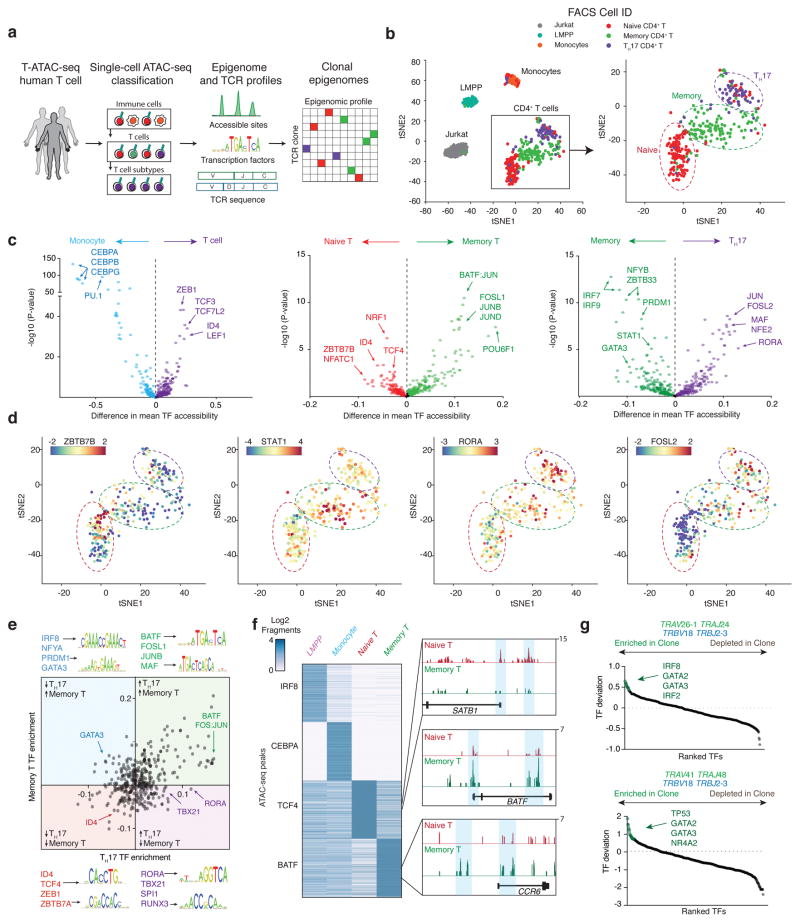
We first analyzed single-cell ATAC-seq profiles using a computational pipeline that integrated reference ensemble ATAC-seq data from T cells (this study) and other hematopoietic cell types10 in order to phenotype individual cells (Fig. 4a). Using a previously described approach to train principal components (PCs) on ensemble ATAC-seq data and project single-cell profiles onto that PC space10, single cells were compared against all ensemble profiles to remove contaminating non-T cells that remained post-sorting (cells sorted to >95% purity). Indeed, while the majority of single-cell profiles showed highest epigenomic correlation with ensemble T cell profiles compared to other cell types, 11/185 naive T cells, 2/134 memory T cells, and 4/148 TH17 cells, showed higher similarity with other immune cell types, particularly with CD4+ monocytes, and were excluded from further analysis (Supplementary Fig. 4d and e). Epigenomic profiles of the remaining T cells (450 cells) were then compared against Jurkat cells (231 cells) and previously published single-cell epigenomic profiles of blood monocytes (92 cells) and lymphoid-primed multipotent progenitor cells (LMPP; 89 cells)9. t-distributed stochastic neighbor embedding (t-SNE) projection21 of single-cell epigenome profiles revealed clustering of single cells largely according to cell type, with primary T cells clustering separately from Jurkat cells, monocytes, and LMPPs (Fig. 4b). Strikingly, T cell profiles generated a continuous spectrum of epigenomic states, rather than distinct subpopulations of naive and memory phenotypes, suggesting significant regulatory variability within cell surface marker-defined sub-populations. In particular, previous studies using high-resolution cell surface marker staining and functional analysis identified significant heterogeneity within the CD45RA+ naive T cell population, including the presence of recent thymic emigrants, ‘super-naive’ cells, early-memory and differentiated cells, and memory stem-like cells22–28. Indeed, single-cell naive T cell chromatin accessibility profiles also showed a spectrum of cell states, including a small population of naive cells present in both individuals (20/174 naive cells, 11.5%) that clustered closely with memory and TH17 cells (Fig. 4b).
Figure 4. Single-cell epigenomic and TCR profiling of human CD4+ T cells.
(a) Outline for T-ATAC-seq analysis in primary human T cells. Single cells are first sequentially classified to major blood lineages, and then to T cell subsets, by similarity to ensemble reference ATAC-seq profiles. T-ATAC-seq data from classified single T cells are then analyzed for accessibility at regulatory DNA elements and TF activity using ATAC-seq data, and for TCR sequence identity. Finally, integrative analysis of both data types is performed to identify epigenomic signatures in T cell clones. (b) t-SNE projection of naive and memory T cells (T-ATAC-seq, 320 cells, 6 independent experiments), Jurkat T cells (T-ATAC-seq, 145 cells, 3 independent experiments), Monocytes (scATAC-seq, 71 cells), and Lymphoid-primed multipotent progenitors (LMPP; scATAC-seq, 86 cells). See Methods for generation of tSNE plots from high-quality single-cell libraries. (c) TF bias-corrected deviation enrichments (chromVAR) in aggregated single-cell populations. TF enrichments are calculated as the difference in mean TF motif accessibility between two populations of single cells. Shown are enrichments for all T cells compared to monocytes (left), memory T cells compared to naive T cells (classified according to t-SNE clustering; middle), and TH17 cells compared to memory T cells (right). P-values were calculated using a two-tailed t-test. (d) t-SNE projection of single T cells colored by ZBTB7B (enriched in naive cells), STAT1 (enriched in memory cells), RORA (enriched in TH17 cells), and FOSL2 (enriched in TH17 cells) motif accessibility TF z-scores. Scale bars indicate the range of TF z-scores. (e) Mean bias-corrected deviations ranked for difference in aggregated TH17 cells vs aggregated naive cells (x-axis) and aggregated memory (non-TH17) cells vs aggregated naive cells (y-axis). Shown are TF motifs for selected factors in each quadrant. For example, BATF motifs show increased accessibility in memory T cells and TH17 cells. In contrast, RORA motifs show increased accessibility in TH17 cells, but not in memory T cells. (f) Left, heat map showing ATAC-seq fragment counts in peaks (rows) containing the indicated motifs from aggregated single cells. Cell types analyzed are indicated above each column. Right, aggregated single-cell genome tracks for naive T cell- and memory T cell-specific peaks in the SATB1, BATF, and CCR6 loci. (g) TCR clone and CDR3 sequences for two memory T cell clones and associated TF deviation enrichments in clonal cells vs non-clonal memory T cells. Clone #1 is shown in the top panel, and clone #2 is shown in the bottom panel. Epigenomic profiles from each clone were aggregated and compared against an aggregate profile from all non-clonal memory T cells.
We next measured TF deviation scores and variation in single cells and aggregated by cell type. In aggregate, all T cells exhibited high deviations in TCF/LEF family members, compared to monocytes, suggesting that these factors direct T cell lineage specification through changes in chromatin accessibility (Fig. 4c)29. In contrast, monocytes exhibited high activity of CCAAT/enhancer-binding protein (CEBP) family members and PU.1. A comparison of naive cells and memory cells identified a large shift in epigenomic profile from high activity of T cell specification TFs in naive T cells, including TCF family factors and zinc finger and BTB domain containing 7B (ZBTB7B), to T cell activation TFs in memory cells, including the activator protein-1 (AP-1) factors FOS, JUN, and basic leucine zipper ATF-like (BATF; Fig. 4c). Finally, comparison of memory T cells and TH17 cells showed high activities for STAT, GATA, and IRF factors in memory cells, and AP-1, MAF, RUNX, and RAR-related orphan receptor (ROR) factors in TH17 cells, consistent with the critical roles of these TFs in memory and TH17 cells, respectively30–38 (Fig. 4c). Cell type-specific TFs identified in aggregated single-cell profiles were remarkably similar to profiles obtained from ensemble measurements in 500 times more cells. Ensemble naive T cell profiles showed similar enrichments of accessibility at TCF/LEF family members, and ZBTB7B, while memory cells demonstrated high deviations in AP-1 factors (Supplementary Fig. 5a and b). Similarly, TH17 cells showed high activities for ROR, AP-1, and RUNX factors, compared to all other memory T cell types (Supplementary Fig. 5a–c). Finally, an examination of TH1, TH2, and Treg cells identified TF signatures associated that aligned well with previously identified master regulators in each lineage, including TBX21 (T-BET) and Eomesodermin (EOMES) in TH1 cells, GATA3 in TH2 cells, and FOXP3 in Treg cells (Supplementary Fig. 5a–c).
We next integrated information from ensemble profiles and cell surface marker staining to visualize epigenomic variability in these canonical populations. As observed in the t-SNE projections, CD45RA+ naive T cells displayed significant TF heterogeneity that could be broken into at least three sub-clusters that spanned the continuum of naive to memory cell differentiation. The majority of naive cells (132/174, 75.9%) were present in the first cluster of ‘true-naive’ cells, and demonstrated high TF deviation scores for ensemble naive T cell TFs, including ZBTB7B, and low scores for ensemble memory cell TFs (Fig. 4d). A second cluster of ‘early-differentiating’ naive cells (22/174, 12.6%) showed lower deviation scores of naive cell TFs and higher scores for memory cell TFs, including AP-1, IRF, and STAT factors, albeit lower than true memory cells (Fig. 4d and Supplementary Fig. 6a and b). Finally, a small minority of naive cells existed in a differentiated state (20/174, 11.5%) with high AP-1 and RUNX activity (Fig. 4d and Supplementary Fig. 6a and b). Extensive variability was also observed in sorted memory T cells, with variation in known T helper phenotypes as expected, and a small fraction of cells clustering closely with naive T cells, suggesting an early differentiated memory state (Fig. 4d). The observed TF variability in T cell subtypes was greater than expected in background ATAC-seq peaks matched for GC bias, peak height, and transposition rate, and variability was not driven by single cells with low quality ATAC-seq data, such as low fragment numbers (Supplementary Fig. 6b and c).
Comparing all three populations of T cells revealed two categories of factors; (1) factors involved in general memory or naive T cell differentiation, and (2) factors specific to T helper cell subtypes (Fig. 4e). Surprisingly, relatively few TFs were enriched in the latter category, suggesting that large-scale changes occur during transition from naive to memory phenotypes, which dominate the epigenomic landscape, while subtype-specific changes are comparatively fewer and controlled by specific factors (Fig. 4e). This principle was also supported by an unbiased analysis of TF modules, in which we correlated TF activity across single cells (Supplementary Fig. 6d). We found several TF programs corresponding to subset-specific functions, and that these TFs functioned in concert with a common memory program (Supplementary Fig. 6d). Interestingly, modules encompassing TH1 and TH2 phenotypes could be observed in this analysis, even though these populations were not specifically enriched by cell sorting, demonstrating that this information could be derived de novo from single-cell profiles. Finally, differential analysis of ATAC-seq peaks that contained binding sites for cell state-specific TFs identified cell type-specific cis-regulatory elements, including SATB1 locus elements in naive T cells and BATF and CCR6 locus elements in memory T cells.
We next integrated TCR sequencing results with single-cell epigenomic profiles in these healthy individuals. We identified two clonal populations within the memory population in one individual with a history of atopy, which could be identified by common expression of TRBV18 TRBJ2–3, suggesting that they may have expanded to shared antigens (Fig. 4g). Interestingly, neither clonotype was present in the sampled naive cells from the same individual. Analysis of epigenomic signatures in these cells revealed common high TF deviation scores for GATA factors, consistent with a TH2 phenotype (Fig. 4g). In summary, these data demonstrate that T-ATAC-seq can effectively capture ensemble epigenomic measurements while at the same time preserving single-cell regulatory and TCR information.
T-ATAC-seq reveals regulatory signatures in T cell leukemia and host immunity
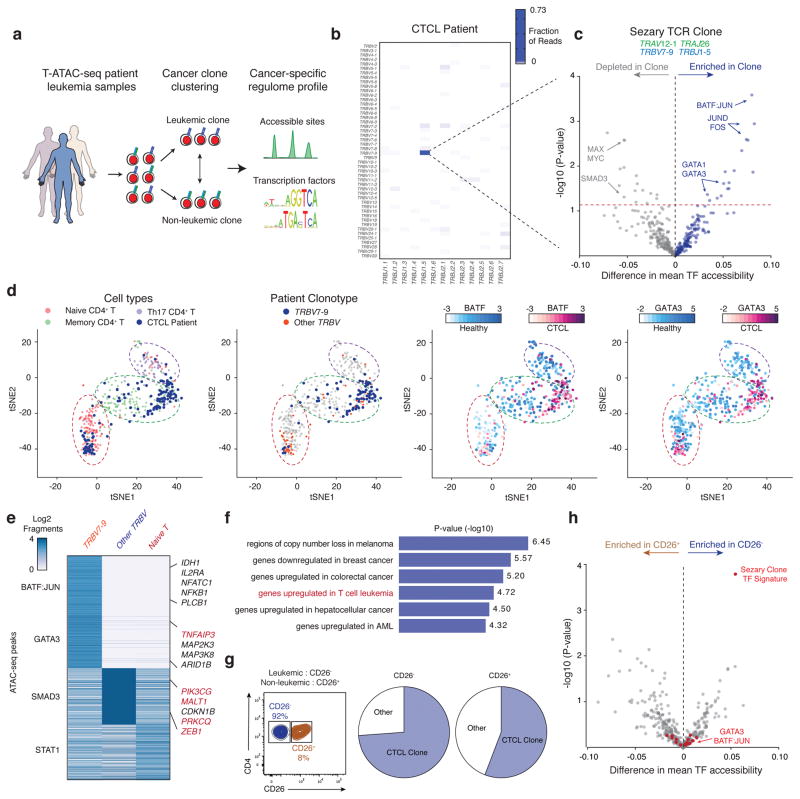
We performed T-ATAC-seq on clinical blood samples from patients with Sézary syndrome, which is a leukemic form of cutaneous T cell lymphoma (CTCL). Identification of cancer cell regulatory signatures can be challenging since only a fraction of circulating CD4+ T cells are malignant, and standard immunophenotypic methods to distinguish healthy and cancer clones are imprecise and not applicable to some patients39,40. These observations have been the basis for the recent development of TCR clonality assays for the identification of malignant T cell expansion and minimal residual disease in clinical CTCL samples41,42. Therefore, we asked whether the integrated analysis of T-ATAC-seq could improve the identification of cancer-specific epigenomic signatures of malignant cells (Fig. 5a). We first isolated CD4+ T cells from a patient with Sézary syndrome and subjected these cells to T-ATAC-seq (3 independent experiments). Strikingly, 73% of all CD4+ T cells (157/215 cells) expressed a single TCRβ sequence TRBV7–9 TRBJ1–5, representing the putative leukemic clone (Fig 5b and Supplementary Fig. 7a). These cells showed TCRβ pairing with TRAV12-1 TRAJ26. We next aggregated all cells according to leukemic or non-leukemic clonotype and compared epigenomic profiles. Leukemic cells showed high TF deviation scores for memory T cell-specific TFs, including BATF, JUN, and FOS, and GATA motifs, including the TH2-specific TF GATA3 (Fig. 5c). These findings are consistent with the long-standing hypothesis, based on cytokine and cell surface marker expression, that Sézary cells represent a malignant counterpart of TH2 memory T cells, which may contribute to disease persistence and pathogenesis43,44. t-SNE projection of single-cell T-ATAC-seq PCA scores revealed that almost all of the memory T cells in this patient were replaced by leukemic TH2 cells, while the non-malignant T cells were predominantly in a naive state. The non-malignant T cell clones in the CTCL patient exhibited strong SMAD3-associated chromatin accessibility, which may reflect an immunosuppressive TGF-beta pathway (Fig. 5c,e). These findings identify a possible cause for systemic immunodeficiency associated with Sézary syndrome, since nearly all memory T cells have been replaced by the leukemic clone (Fig. 5d)45. Interestingly, analysis of individual cis-regulatory changes contributing to the overall shift in TF landscape identified genes that have previously been shown to be recurrently mutated in CTCL and other cancer types (Fig. 5e)46,47. These included genes in T cell survival and activation pathways such as TNFAIP3, PIK3CG, and PRKCQ. Analysis of MSigDB signatures pathways enriched in cis-elements that were more accessible in leukemic cells demonstrated that these elements significantly overlapped with genes that are upregulated in T cell leukemia, as well as in other cancer types (Fig. 5f).
Figure 5. T-ATAC-seq identifies epigenomic signatures of clonal leukemic T cells in patient samples.
(a) Outline for T-ATAC-seq analysis in patient T cell leukemia samples. Single cells are first classified according to TCR sequence identity as leukemic cells or non-leukemic cells. ATAC-seq data from classified single T cells are then analyzed for accessibility at regulatory DNA elements and TF activity. (b) Heat map showing TCRβ rearrangements in peripheral blood samples from a patient with Sezary syndrome (3 independent experiments). (c) TF bias-corrected deviation enrichments in aggregated clonal T cells as compared to all other T cells. Shown is the TCR sequence identified in the putative leukemic T cell clone (top). TF enrichments (bottom) are calculated as the difference in mean TF motif accessibility between aggregated leukemic T cell clone profiles and non-clonal T cell profiles in the same patient. Selected TF motifs enriched or depleted in the T cell clone are indicated. P-values were calculated using a two-tailed t-test. (d) t-SNE projection of healthy naive and memory T cells (320 cells, 6 independent experiments) and patient cells (139 cells, 3 independent experiments) colored by Cell ID (left), clonal vs non-clonal cells (middle left), BATF TF score (middle right), and GATA3 TF score (right). Scale bars indicate range of TF z-scores. (e) Heat map showing ATAC-seq fragment counts in peaks containing the indicated motifs (left). Labels indicate genes associated with differential peaks, including genes previously shown to be mutated in CTCL (red). (f) MSigDB Perturbation Signatures of TRB7–9-specific ATAC-seq peaks as obtained from GREAT analysis (Binomial test, 102 aggregated single cells, 3 independent experiments). (g) Sort strategy for CD26+ and CD26− CD4+ T cells (left), and clonal TCR profiles in each population (right). The lack of CD26 expression has been previously used to distinguish leukemic cells from non-leukemic cells. (h) TF bias-corrected deviation enrichments in aggregated CD26− cells (56 single cells) compared to CD26+ cells (49 single cells). P-values were calculated using a two-tailed t-test. TFs identified above the dashed line in (c) are highlighted in red.
Finally, we asked whether the leukemia-specific signature could be identified using standard immunophenotypic FACS strategies for cancer cells. We sorted CD4+ cells according to their expression of CD26 (also known as dipeptidyl peptidase-4; DPP4), a cell surface protein whose loss of expression is clinically used as a diagnostic tool to identify malignant Sézary cells (Supplementary Fig. 7b)48. Surprisingly, we observed the presence of the CTCL clone in both CD26+ and CD26− cell populations, demonstrating that, at least in a subset of patients, this marker does not accurately identify circulating malignant cells (Fig. 5g)40. Accordingly, aggregating single cells based on their immunophenotype, rather than clonotype, obscured cancer-specific epigenomic signatures, since memory and TH2-specific TFs were not enriched in CD26− cells compared to CD26+ cells (Fig. 5h). T-ATAC-seq of two additional CTCL patients confirmed the superiority of TCR clonotype over CD26 immunophenotype to isolate leukemic clones and their epigenomic signatures (Supplementary Fig. 7c and d). Altogether, this use of T-ATAC-seq in T cell leukemia demonstrates that this method is applicable to clinical blood samples and can be used to separate clonal and non-clonal regulatory pathways in cells from the same individual.
Discussion
The expression of uniquely recombined TCRs on individual T cells is the central driver of immune responsiveness and connects specific antigen recognition to a particular effector function. In addition, since the diversity of possible human T cell receptors is estimated at ~1014, single-cell TCR sequencing can serve as a powerful lineage tracer, either in the context of a normal immune response, or in the context of malignant transformation. Therefore, pairing TCR identity to functional phenotype represents an important strategy to investigate T cell clonal dynamics, phenotypic plasticity, and tumor heterogeneity6,12,13. In this study, we report the technical development and application of T-ATAC-seq to immortalized and primary human T cells. We have found it to be robust and reproducible across T cell types and individuals and to compare favorably with previous technologies capable of assaying single-cell epigenomes. T-ATAC-seq pairs epigenomic data, identifying cis and trans determinants of cell identify, with high-fidelity RNA sequence of TCR loci providing a platform for multi-omic investigation of T cell diversity.
We used ensemble ATAC-seq data and TF binding sites genome-wide as scaffolds to map single-cell chromatin states and developed a step-wise approach to use single-cell chromatin accessibility to phenotype immune cells. Each single cell is sequentially classified to major blood lineages, and then to T cell subsets – a scheme that recapitulates the chromatin landscape during physiologic development. Previous efforts to characterize single-cell epigenomes highlighted the presence of inter- and intra-population variability in cell lines and distinct hematopoietic cell types8,10,14. We demonstrate that this approach may also be informative to distinguish more subtle phenotypes in primary T cells and reveals heterogeneity in T cell populations which can appear similar by cell surface marker profiling. For example, a small fraction of naive CD4+ T cells, characterized by the expression CD45RA, exhibited chromatin states more similar to memory T cells, showing accessibility at genomic sites bound by AP-1 TFs. This observation is supported by previous functional studies that identified a memory T cell population with stem-like properties in the CD45RA+ naive T cell gate27. Similarly, single cells with memory T cell or TH17-defining cell surface markers displayed significant epigenomic heterogeneity, particularly in cell type-specific TFs such as IRF, STAT, and ROR factors. These results suggest that memory T cells may exist in a phenotypic continuum, rather than in distinct quantal chromatin states3. Future studies with more extensive sampling of single T cells in homeostatic and inflammatory conditions could use this approach to define the continuous landscape of single T cell states and variability within cell-surface marker-defined subtypes.
We exploited the ability of T-ATAC-seq to pair TCRs with chromatin state information to identify cancer-associated epigenomic changes in patients with T cell leukemia. The clinical diagnosis of T cell leukemia is based on several factors including clinical presentation, histopathologic findings, and the identification of a clonal T cell population. However, all of these diagnostic findings, including the expansion of T cell clones, are often present in benign inflammatory skin conditions, and it remains a significant challenge to distinguish small populations of malignant cells from benign, but oligoclonal, T cell proliferations42,49. Using T-ATAC-seq, we were able to define epigenomic signatures of clonal cancer cells that are missed by ensemble or standard FACS-based separation methods, demonstrating the promise of this approach. This result has potentially significant clinical applications since recent studies have described distinct epigenomic classifications of CTCL that are associated with differential responses to currently used clinical therapies that target the epigenome, such as histone deacetylase inhibitors50,51. Future studies on larger patient cohorts are needed to establish whether integration of epigenomic information with T cell clonality can (1) improve diagnostic precision compared to standard clinical techniques currently in use, and (2) predict or monitor successful clinical responses to therapies that target the epigenome.
More broadly, T-ATAC-seq represents an important technical advance towards achieving an atlas of human cell types and states52 in that it is able to generate genome-wide chromatin accessibility maps, while simultaneously preserving and measuring RNA sequence. T-ATAC-seq may be particularly well-suited for the examination of TF activity and specific enhancer elements underlying cell states compared to existing methods that pair whole transcriptome profiles with TCR sequence in single cells. While we employed an unbiased approach and sequenced all captured cells, which is applicable to settings of significant clonal expansion such as CTCL, the use of T-ATAC-seq to interrogate rare clonal populations may be technically challenging at the current throughput of 96 cells per microfluidic chip. One strategy to address this challenge may be to selectively sequence single-cell epigenomes after identifying TCRs of interest (or vice versa), but further technical improvements focused on increasing throughput of T-ATAC-seq will be critical for the analysis of rare T cell clones. Given the inherent challenges in obtaining large amounts of RNA from T cells compared to other cell types, we believe that this strategy should be easily adaptable to other cell types where RNA is more abundant. In particular, T-ATAC could be adapted to determine RNA sequences of other cell identity-specific transcripts, such as B cell receptors, olfactory receptors, lncRNAs, and cytokines, or perhaps with additional technical development, even to measure whole transcriptomes. Finally, the sequential reaction conditions employed to assay chromatin and RNA sequences from single cells can be easily scaled-up to obtain both types of information from ensemble samples where material is limited, such as rare cell types or clinical samples.
We envision that T-ATAC-seq will be complementary to approaches for unbiased identification of TCR ligands, enabling integration of T cell epigenomic state, TCR sequence, and TCR ligands53,54. The application of this strategy to human diseases such as cancer and autoimmune disease, particularly in the context of immunotherapy, could be invaluable in generating comprehensive profiles of beneficial and harmful T-cell responses; the regulatory networks underlying either response, and the antigens that drive these networks.
Methods
Human subjects
This study was approved by and performed in compliance with the ethical regulations of the Stanford University Administrative Panels on Human Subjects in Medical Research. Written informed consent was obtained from all participants.
Code availability
All custom code used in this work is available upon request.
Cell culture and T cell isolation
Jurkat cells were obtained ATCC (Clone E6-1) and cultured in RPMI- 1640 Medium with 10% FBS and Penicillin/Streptomycin. For single Jurkat cell experiments, cells were sorted into single-cell suspension prior to capture on the C1. Mouse 58αβ-negative hybridoma cells were retrovirally-transduced with a paired TCRαβ sequence, and these cells were used in mouse/human mixing experiments55,56. CD4+ T cells from healthy volunteers or Sezary syndrome patients were enriched from peripheral blood using the RosetteSep Human CD4+ T Cell Enrichment Cocktail (StemCell Technology). For single-cell experiments, CD4+ T helper cells were sorted as naive T cells (CD4+CD25−CD45RA+), memory T cells (CD4+CD25−CD45RA−), or TH17 cells (CD4+CD25−CD45RA−CCR6+CXCR5−). 200,000 cells from two healthy volunteers were sorted into RPMI + 10% FBS, washed, and loaded onto the C1 IFC, as described below. For ensemble ATAC-seq experiments, CD4+ T helper cells were sorted as naive T cells (CD4+CD25−CD45RA+), Treg (CD4+CD25+IL7Rlo), TH1 (CD4+CD25−IL7RhiCD45RA−CXCR3+CCR6−), TH2 (CD4+CD25−IL7RhiCD45RA−CXCR3−CCR6−), TH17 (CD4+CD25−IL7RhiCD45RA−CXCR3−CCR6+), and TH1-17 (CD4+CD25−IL7RhiCD45RA−CXCR3+CCR6+) (Supplementary Fig. 5). 55,000 cells from two healthy volunteers (3 replicates total) were sorted into RPMI + 10% FBS, washed with PBS, and immediately transposed as described below. Post-sort purities of > 95% were confirmed by flow cytometry for all samples.
Antibodies
The following antibodies were used in this study: anti-human CD45RA-PERCPCy5.5 (Clone HI100, Lot# B213966, Cat# 304107, Biolegend), anti-human CD127-Brilliant Violet 510 (Clone A019D5, Lot# B197159, Cat# 351331, Biolegend), anti-human CD4-APC-Cy7 (Clone OKT4, Lot# B207751, Cat# 317417, Biolegend), anti-human CCR6-PE (Clone G034E3, Lot# B203239, Cat# 353409, Biolegend), anti-human CD25-FITC (Clone BC96, Lot# B168869, Cat# 302603, Biolegend), anti-human CXCR3-Brilliant Violet 421 (Clone G025H7, Lot# B206003, Cat# 353715, Biolegend), anti-human CXCR5-AlexaFluor647 (Clone RF8B2, Lot# 5302868, Cat# 558113, BD Pharmingen), anti-human CD26-PE (Clone 2A6, Lot# 4301881, Cat# 12-0269-42, Thermo Fisher), and anti-human CD3E-Pacific Blue (Clone UCHT1, Lot# 4341657, Cat# 558117, BD Biosciences). All antibodies were validated by the manufacturer in human peripheral blood samples, used at a 1:200 dilution, and compared to isotype and no staining control samples.
Ensemble ATAC-seq
Cell isolation and transposase reaction
Cells were isolated and subjected to ATAC-seq as previously described16. Briefly, 55,000 cells were pelleted after sorting and washed once with 100μL PBS. Cell pellets were then resuspended in 50μL lysis buffer (10mM Tris-HCl, pH 7.4, 3mM MgCl2, 10mM NaCl, 0.1% NP-40 (Igepal CA-630)), and immediately centrifuged at 500g for 10 min at 4°C. The nuclei pellets were resuspended in 50μL transposition buffer (25μL 2X TD buffer, 22.5μL dH20, 2.5μL Illumina Tn5 transposase), and incubated at 37°C for 30 min. Transposed DNA was purified with MinElute PCR Purification Kit (Qiagen), and eluted in 10μL EB buffer.
Primary data processing and peak calling
ATAC-seq libraries were prepared as previously described, barcoded, and sequenced on an Illumina Nextseq at the Stanford Functional Genomics Facility. Adapter sequence trimming, mapping to Hg19 using Bowtie2, and PCR duplicate removal using Picard Tools were performed. All samples were merged for peak calling using MACS2. The number of raw reads, Tn5 offset corrected, mapped to the union peak set for each sample was quantified using intersectBed in BedTools. Peak raw counts were normalized using the “CQN” package in R. Peak intensity was defined as the variance stabilized log2 counts using the “DESeq2” package in R. After these steps, an N×M data matrix was obtained where N indicates the number of merged peaks, M indicates the number of samples, and value Di,j indicates the peak intensity of peak i (i=1 to N) in sample j (j=1 to M). Pearson correlation was calculated based on the log2 normalized counts of all the peaks. Unsupervised correlation of the Pearson correlation matrix was performed using Cluster 3.0 and visualized in Java Treeview.
Transcript-indexed single-cell ATAC-seq (T-ATAC-seq)
Step 1. Cell isolation and loading onto the IFC
We adapted the C1 Single-Cell Auto Prep System with its Open App™ program (Fluidigm, Inc.) to perform T-ATAC-seq. Single T cells were captured using the C1 IFC microfluidic chips (small; 5–10micron) and custom-built T-ATAC-seq scripts generated using the C1™ Script Builder Software (scripts available from Fluidigm and upon request). Jurkat cells or peripheral blood T cells were first isolated by FACS sorting and then washed three times in C1 DNA Seq Cell Wash Buffer (Fluidigm). Cells were resuspended in DNA Seq Cell Wash Buffer at a concentration of 300 cells/μL and mixed with C1 Cell Suspension Reagent at a ratio of 3:2. 15μL of this cell mix was loaded onto the IFC. After cell loading, captured cells were visualized by imaging on a Leica CTR 6000 microscope.
Step 2. Microfluidic reactions on the IFC: reagents and conditions
On the C1, cells were subjected sequentially to lysis and transposition, transposase release, MgCl2 quenching, reverse transcription, and PCR, as described (Fig. 1a and Supplementary Fig. 1a), using the custom T-ATAC-seq script “T-ATAC-seq: Sample Prep (1861×, 1862×, 1863×).” For lysis and transposition (in chamber #1), 30μL of Tn5 transposition mix was prepared (22.5μL 2× TD buffer, 2.25μL transposase (Nextera DNA Sample Prep Kit, Illumina), 2.25μL C1 Loading Reagent without salt (Fluidigm), and 0.45μL 10% NP40). For transposase release (in chamber #2), 20μL of Tn5 release buffer mix was prepared (2μL 500mM EDTA, 1μL C1 Loading Reagent without salt, and 17μL 10mM Tris-HCl Buffer, pH 8). For MgCl2 quenching (in chamber #3), 20μL of MgCl2 quenching buffer mix was prepared (18μL 50mM MgCl2, 1μL C1 Loading Reagent without salt, and 1μL 10mM Tris-HCl Buffer, pH 8). For reverse transcription (in chamber #4), 30μL of RT mix was prepared (15.55μL H20, 3.7μL 10× Sensiscript RT buffer (Qiagen), 3.7μL 5mM dNTPs, 1.5μL C1 Loading Reagent without salt (Fluidigm), 1.85μL Sensiscript (Qiagen), and 3.7μL 6μM TCR primer mix (described below). Finally, for ATAC and TCR PCR (in chamber #5), 30μL of PCR mix was prepared (8.62μL H20, 13.4μL 5× Q5 polymerase buffer (NEB), 1.2μL 5mM dNTPs, 1.5μL C1 Loading Reagent without salt, 0.67μL Q5 polymerase (2U/μL; NEB), 0.8μL 25μM non-indexed custom Nextera ATAC-seq PCR primer 1, 0.8μL 25μM non-indexed custom Nextera ATAC-seq primer 2, and 3μL 6μM TCR primer mix). The primer sequences for the non-indexed custom Nextera ATAC-seq primers are listed in Supplementary Table 1 in a prior study8.
7μL lysis and transposition mix, 7μL transposase release buffer, 7μL MgCl2 quenching buffer, 24μL RT mix, and 24μL PCR mix were added to the IFC inlets. On the IFC, Tn5 lysis and transposition reaction was carried out for 30 min at 37°C. Next, transposase release was carried out for 30 min at 50°C. MgCl2 quenching buffer was immediately added and chamber contents were immediately incubated with RT mix for 30 min at 50°C. Finally, gap filling and 8 cycles of PCR were performed using the following conditions: 72°C for 5 min and then thermocycling at 94°C for 30s, 62°C for 60s, and 72°C for 60s. The amplified transposed DNA was harvested in a total of 13.5μL C1 Harvest Reagent. Following completion of the on-chip protocol (~4–5hrs), chamber contents were transferred to 96-well PCR plates, mixed, and divided for further amplification of ATAC-seq fragments (5 μl) or TCR-seq fragments (6–7 μl).
Step 3. Amplification of TCR-seq libraries
On-chip PCR: TCR sequence from single cells were obtained by a series of three PCR reactions (phases) as previously described, with slight modifications for implementation on the IFC6,57. The design principles and validation of all TCR primers were described previously6, and primer sequences are listed in Supplementary Table 1 in that study. In order to integrate TCR amplification into the T-ATAC-seq protocol, the RT and first phase PCR was carried out in chambers 4 and 5 of the IFC using the conditions described above. The phase 1 TCR primer mix included multiple Vα and Vβ region primers and Cα and Cβ region primers; each V-region primer was at a concentration of 0.06μM, and each C-region primer was at a concentration of 0.3μM. RT was performed using the Cα and Cβ region primers, and the cDNA was then subjected to 8 cycles of PCR using both Vα and Vβ region primers and Cα and Cβ region primers (simultaneously as ATAC fragments were also being amplified in the same chamber using distinct primers described above).
Off-chip phase 1 PCR: Following completion of the on-chip protocol, 6–7μL of the harvested libraries were further amplified using TCR primers. First, an additional 8 cycles of PCR was done using the following cycling conditions: 95°C 15 min and thermocycling at 94°C for 30s, 62°C for 1 min, and 72°C for 1 min; 72°C 10 min; 4°C.
Off-chip phase 2 PCR: Thereafter, a 1μL aliquot of this final phase 1 product was used as a template for a 12μL phase 2 PCR reaction. The following cycling conditions were used for a 25-cycle phase 2 PCR: 95°C for 15 min and thermocycling at 94°C for 30s, 64°C for 1 min, and 72°C for 1 min; 72°C 5 min; 4°C. For the phase 2 reaction, multiple internally nested TCRVα, TCRVβ, TCRCα and Cβ primers were used (V primers 0.6μM, C primers 0.3μM). The phase 2 primers of TCR V-region contained a common 23-base sequence at the 5′ end to enable further amplification (during the phase 3 reaction) with a common 23-base primer.
Off-chip phase 3 PCR: Finally, 1μL of the final phase 2 PCR product was used as a template for a 14μL phase 3 PCR reaction, which incorporates barcodes and enables sequencing on the Illumina MiSeq platform. For the phase 3 PCR reaction, amplification was performed using a 5′ barcoding primer (0.05μM) containing the common 23-base sequence and a 3′ barcoding primer (0.05μM) containing sequence of a third internally nested Cα and/or Cβ primer, and Illumina paired-end primers (0.5μM each). The following cycling conditions were used for a 25-cycle Phase 3 PCR: 95°C 15 min and thermocycling at 94°C for 30 s, 66°C for 30 s, and 72°C for 1 min; 72°C 5 min; 4°C. The final phase 3 barcoding PCR reactions for TCRα and TCRβ were done separately. For the Phase 3 reaction, 0.5μM of the 3′ Cα barcoding primer and the 3′ Cβ barcoding primer were used. In addition to the common 23-base sequence at the 3′ end (that enables amplification of products from the second reaction) and a common 23-base sequence at the 5′ end (that enables amplification with Illumina paired-end primers), each 5′ barcoding primer contains a unique 5-base barcode that specifies plate and a unique 5-base barcode that specifies row within the plate. In addition to the internally nested TCR C-region sequence and a common 23-base sequence at the 3′ end (that enables amplification with Illumina paired-end primers), each 3′ barcoding primer contains a unique 5-nucleotide barcode that specifies column.
Library purification and sequencing: After the phase 3 PCR reaction, each PCR product should have a unique set of barcodes incorporated that specifies plate, row and column and have Illumina paired-end sequences that enable sequencing on the Illumina MiSeq platform. The PCR products were combined at equal proportion by volume, run on a 1.2% agarose gel, and a band around 350 to 380bp was excised and gel purified using a Qiaquick gel extraction kit (Qiagen). This purified product was then sequenced.
Step 4. Amplification of ATAC-seq libraries
5μL of harvested libraries were amplified in a 50μL PCR reaction for an additional 17 cycles with 1.25μM Nextera dual-index PCR primers8 in 1× NEBnext High-Fidelity PCR Master Mix) using the following PCR conditions: 72°C for 5 min; 98°C for 30s; and thermocycling at 98°C for 10s, 72°C for 30s, and 72°C for 1 min. The PCR products were pooled and purified on a single MinElute PCR purification column (Qiagen). Libraries were quantified using qPCR prior to sequencing.
Data processing of single-cell TCR-seq libraries
TCR sequencing data was analyzed as previously described6,57. Briefly, raw sequencing data were demultiplexed using a custom computational pipeline and primer dimers were removed. All paired-end reads were assembled by finding a consensus of at least 100 bases in the middle of each read. A consensus sequence was obtained for each TCR gene. Because multiple TCR genes might be present in a given well, we established sequence identity cutoffs according to sequence identity distributions in each experiment (generally >80% sequence identity within a given well). The sequence identity cutoff ensures that all sequences derived from the same transcript would be properly assigned, even given a PCR error rate of 1/9,000 bases, and sequencing error rate up to 0.4%. TCR V, D and J segments were assigned by VDJFasta. For downstream analysis, an additional read cut-off of 100 reads was used for each identified TCR sequence. For confirmation of identified TCRβ sequences, select patient samples were also sequenced by immunoSEQ (Adaptive Biotechnologies), according to the Survey protocol.
Data processing of single-cell ATAC-seq libraries
All single-cell ATAC-seq libraries were sequenced using paired-end, dual-index sequencing. Single-cell ATAC-seq data was pre-processed as previously described8. Briefly, adapter sequences were trimmed, mapped to Hg19 using Bowtie2 using the parameter –X2000, and PCR duplicates were removed. Reads mapping to mitochondria or unmapped contigs were also removed and not considered in further analysis. As with ensemble ATAC-seq data, peaks were called with MACS2, and filtered single-cell libraries were required to contain >15% of unique fragments in called peaks and a library size of >500 fragments for most downstream analysis. For t-SNE projections, a further filtering step was performed to only include high-quality libraries that contained >40% of unique fragments in called peaks and a library size of >500 fragments. For example, conclusions regarding primary T cell subsets are derived from 450 single T cells that pass the 15% fragments in peaks cut-off. t-SNE projections show 320 high-quality cells that pass the 40% fragments in peaks cut-off to ensure that all conclusions based on clustering results are also true for high-quality single cell libraries.
We validated that single-cell ATAC-seq libraries did not contain contaminating fragments from TCR libraries in the T-ATAC-seq protocol. First, the phase 1 TCR primer mix used on the IFC (described above) was designed to exclude ATAC-seq Nextera primer binding sites. Therefore, TCR fragments present in the ATAC-seq library would not amplify in library preparation steps or be sequenced. Second, we did not observe TCR library fragments in filtered and aligned ATAC-seq reads. Third, ATAC-seq data derived from T-ATAC-seq in Jurkat cells displayed similar accessibility and TF motif measurements as ATAC-seq data derived from scATAC-seq in Jurkat cells.
PCA and t-SNE clustering
We performed PCA projections of ensemble ATAC-seq and single-cell T-ATAC-seq profiles as previously described10,11. For ensemble ATAC-seq T cell profiles, after removing unmapped contigs, 97,395 peaks were used for further downstream analysis, and PCA analysis was performed on the 2500 peaks exhibiting the highest variance across T cell subtypes (log2 variance-stabilized). For single-cell T-ATAC-seq analysis, we used a reference set of ensemble ATAC-seq profiles encompassing a wide array of hematopoietic cell types that included previously published hematopoietic progenitors and end-stage cell types9,10 as well as CD4+ helper subtypes generated in this study (Supplementary Fig. 4 and Supplementary Fig. 5d). After removing peaks that aligned to annotated promoters, chrX, chrY and unmapped contigs, 455,057 peaks were used for the PCA projection analysis. To normalize ensemble ATAC-seq profiles, we identified 18,858 low variance promoters across all ensemble samples and normalized each sample by the mean fragment counts within the low variance promoters. We subsequently performed PCA on the normalized values aggregated by similar ensemble cell types. To score single cells for each component, we used the weighted coefficients for each peak and PC (determined using PCA-SVD of the ensemble data above) and calculated the product of the weighted PC coefficients by the centered count values for each cell, taking the sum of this value resulted in a matrix of cells by PCs. We then normalized each cell across the PC-scored values using the sum-of-squares. The matrix of cells by PCs, normalized by the sum-of-squares, was used as an input to a MATLAB implementation of t-SNE (https://lvdmaaten.github.io/tsne/). Data was visualized with scHemeR10.
TF deviation and variability scores using ChromVAR
Single-cell ATAC-seq data processing and calculation of TF deviation was performed using chromVAR11. Human TF motifs were obtained from the JASPAR database58 and included many T cell-specific motifs derived from high-throughput SELEX and ChIP-seq experiments59. All analysis was repeated using a curated list of human TF motifs from the cisBP database without substantial differences11,60. JASPAR motif results are presented in all Figures, except Supplementary Figure 5. Briefly, for each TF, ‘raw accessibility deviations’ were computed by subtracting the expected number of ATAC-seq fragments in peaks for a given motif (from the population average) from the observed number of ATAC-seq fragments in peaks for each single cell. For this calculation, either 455,057 hematopoietic peaks (as defined above) or a subset of 114,653 peaks called using only ensemble T cell subsets, monocyte, and LMPP data were used, with similar results. Next, the accessibility deviation value for each cell is subtracted by the mean deviation calculated for sets of ATAC-seq peaks with similar accessibility and GC content (background peak set) to obtain a bias-corrected deviation value, and additionally divided by standard deviation of the deviation calculated for the background peak sets to obtain a Z-score. For TF differences between single cells or aggregate single-cell populations, either bias-corrected deviations or Z-scores are used to identify cell-specific motifs, as indicated in figure legends. Volcano plots were generated by calculating the mean difference in bias-corrected TF deviation score between two aggregate single-cell populations. Significance was tested using a two-tailed Student’s t-test. The variability of a TF motif across single cells was determined by computing the standard deviation of the z-scores across the cells8,11. The expected value of this metric is 1 if the motif is no more variable than the background peak sets for that motif.
Modification of T-ATAC-seq for additional RNA targets
For method development and RT primer troubleshooting, the T-ATAC-seq protocol can be performed on 1000 cells in Eppendorf tubes with each reaction performed in 1000X volume. Following lysis, transposition, and transposase release, RNA can be reverse-transcribed and subjected to PCR amplification to check RNA quality and quantity for a chosen primer set.
Data availability
All ensemble and single-cell sequencing data are available through the Gene Expression Omnibus (GEO) under accession GSE107817. Two replicates of the ensemble ATAC-seq data for Naïve, TH17, and Treg cells were previously published and are available under GEO accession GSE10149861. In addition, we have generated an open-access interactive web browser, which enables single-cell TCR sequence and ATAC-seq TF deviation exploration (Supplementary Fig. 8; tcr.buenrostrolab.com). This browser includes all single-cell data presented in the study.
A WashU browser session with ensemble T cell subtype ATAC-seq data is available here: http://epigenomegateway.wustl.edu/browser/?genome=hg19&session=N7ew2XJpWK&statusId=293545209
Supplementary Material
Acknowledgments
We thank members of the Chang, Davis, and Greenleaf laboratories, including Y. Shen and K. Qu, for helpful discussions. We thank X. Ji, D. Wagh, and J. Coller at the Stanford Functional Genomics Facility. This work was supported by the Parker Institute for Cancer Immunotherapy (A.T.S., H.Y.C., and M.M.D.), the National Institutes of Health (NIH) P50HG007735 (H.Y.C. and W.J.G.) and 5U19AI057229 (M.M.D.), and the Scleroderma Research Foundation (H.Y.C.). A.T.S. was supported by a Parker Bridge Scholar Award from the Parker Institute for Cancer Immunotherapy and a Cancer Research Institute Irvington Fellowship from the Cancer Research Institute. N.S. was supported by the National Multiple Sclerosis Society Post-Doctoral Fellowship. J.D.B. acknowledges the Broad Institute Fellows and Harvard Society of Fellows programs for funding. M.R.C. was supported by a grant from the Leukemia and Lymphoma Society Career Development Program. W.J.G is a Chan Zuckerberg Biohub investigator. M.M.D. is an investigator of the Howard Hughes Medical Institute. Sequencing was performed by the Stanford Functional Genomics Facility (NIH S10OD018220).
Footnotes
Conflict of Interest
H.Y.C. and W.J.G. are founders of Epinomics and members of its scientific advisory board. H.Y.C. is a founder of Accent Therapeutics and a member of its scientific advisory board. H.Y.C. is a member of the scientific advisory board of Spring Discovery.
Author Contributions
A.T.S., N.S., M.M.D., and H.Y.C. conceived the project. A.T.S, N.S., and J.D.B. performed experiments and analyzed data. B.W. and Y.Q. performed T-ATAC-seq experiments. R.L., J.M.G., M.R.M., and D.G.G. performed ensemble ATAC-seq experiments and analyzed data. W.S.S. performed TCR-seq experiments. Y.W., A.J.R., K.R.P., C.A.L., A.N.S., and M.R.C. analyzed data. M.S.K. and Y.H.K. obtained clinical specimens. H.Y.C, M.M.D, W.J.G., and P.A.K. guided experiments and data analysis. A.T.S, M.M.D., and H.Y.C. wrote the manuscript with input from all authors.
References
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaublomme JT, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul F, et al. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016 doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenrostro JD, et al. Single-cell epigenomics maps the continuous regulatory landscape of human hematopoietic differentiation. bioRxiv. 2017:109843. doi: 10.1101/109843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stubbington MJT, et al. T cell fate and clonality inference from single cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afik S, et al. Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res. 2017;45:e148. doi: 10.1093/nar/gkx615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cusanovich DA, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meller S, et al. TH17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat Immunol. 2015;16:970–979. doi: 10.1038/ni.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 22.Kimmig S, et al. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J Exp Med. 2002;195:789–794. doi: 10.1084/jem.20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 24.Harari A, Vallelian F, Pantaleo G. Phenotypic heterogeneity of antigen-specific CD4 T cells under different conditions of antigen persistence and antigen load. Eur J Immunol. 2004;34:3525–3533. doi: 10.1002/eji.200425324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao C, Davies JD. A peripheral CD4+ T cell precursor for naive, memory, and regulatory T cells. J Exp Med. 2010;207:2883–2894. doi: 10.1084/jem.20100598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song K, et al. Characterization of subsets of CD4+ memory T cells reveals early branched pathways of T cell differentiation in humans. Proc Natl Acad Sci U S A. 2005;102:7916–7921. doi: 10.1073/pnas.0409720102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17:1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiskopf D, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci U S A. 2015;112:E4256–4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014;14:529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 31.Lohoff M, Mak TW. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat Rev Immunol. 2005;5:125–135. doi: 10.1038/nri1552. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 33.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutz S, et al. Transcription factor c-Maf mediates the TGF-β-dependent suppression of IL-22 production in T(H)17 cells. Nat Immunol. 2011;12:1238–1245. doi: 10.1038/ni.2134. [DOI] [PubMed] [Google Scholar]
- 38.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bigler RD, Boselli CM, Foley B, Vonderheid EC. Failure of anti-T-cell receptor V beta antibodies to consistently identify a malignant T-cell clone in Sézary syndrome. Am J Pathol. 1996;149:1477–1483. [PMC free article] [PubMed] [Google Scholar]
- 40.Kelemen K, Guitart J, Kuzel TM, Goolsby CL, Peterson LC. The usefulness of CD26 in flow cytometric analysis of peripheral blood in Sézary syndrome. Am J Clin Pathol. 2008;129:146–156. doi: 10.1309/05GFG3LY3VYCDMEY. [DOI] [PubMed] [Google Scholar]
- 41.Weng W-K, et al. Minimal Residual Disease Monitoring with High-Throughput Sequencing of T Cell Receptors in Cutaneous T Cell Lymphoma. Sci Transl Med. 2013;5:214ra171–214ra171. doi: 10.1126/scitranslmed.3007420. [DOI] [PubMed] [Google Scholar]
- 42.Sufficool KE, et al. T-cell clonality assessment by next-generation sequencing improves detection sensitivity in mycosis fungoides. J Am Acad Dermatol. 2015;73:228–236.e2. doi: 10.1016/j.jaad.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 43.Rook AH, Vowels BR, Jaworsky C, Singh A, Lessin SR. The immunopathogenesis of cutaneous T-cell lymphoma. Abnormal cytokine production by Sézary T cells. Arch Dermatol. 1993;129:486–489. [PubMed] [Google Scholar]
- 44.Vowels BR, et al. Th2 cytokine mRNA expression in skin in cutaneous T-cell lymphoma. J Invest Dermatol. 1994;103:669–673. doi: 10.1111/1523-1747.ep12398454. [DOI] [PubMed] [Google Scholar]
- 45.Krejsgaard T, Odum N, Geisler C, Wasik MA, Woetmann A. Regulatory T cells and immunodeficiency in mycosis fungoides and Sézary syndrome. Leukemia. 2012;26:424–432. doi: 10.1038/leu.2011.237. [DOI] [PubMed] [Google Scholar]
- 46.Ungewickell A, et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47:1056–1060. doi: 10.1038/ng.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi J, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47:1011–1019. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernengo MG, et al. Prognostic factors in Sézary syndrome: a multivariate analysis of clinical, haematological and immunological features. Ann Oncol Off J Eur Soc Med Oncol. 1998;9:857–863. doi: 10.1023/a:1008397323199. [DOI] [PubMed] [Google Scholar]
- 49.Kirsch IR, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. 2015;7:308ra158–308ra158. doi: 10.1126/scitranslmed.aaa9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 51.Qu K, et al. Chromatin Accessibility Landscape of Cutaneous T Cell Lymphoma and Dynamic Response to HDAC Inhibitors. Cancer Cell. 2017;32:27–41.e4. doi: 10.1016/j.ccell.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Regev A, et al. The Human Cell Atlas. bioRxiv. 2017;121202 doi: 10.1101/121202. [DOI] [Google Scholar]
- 53.Birnbaum ME, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newell EW, Davis MM. Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells. Nat Biotechnol. 2014;32:149–157. doi: 10.1038/nbt.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letourneur F, Malissen B. Derivation of a T cell hybridoma variant deprived of functional T cell receptor alpha and beta chain transcripts reveals a nonfunctional alpha-mRNA of BW5147 origin. Eur J Immunol. 1989;19:2269–2274. doi: 10.1002/eji.1830191214. [DOI] [PubMed] [Google Scholar]
- 56.Huse M, et al. Spatial and temporal dynamics of T cell receptor signaling with a photoactivatable agonist. Immunity. 2007;27:76–88. doi: 10.1016/j.immuni.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 57.Glanville J, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathelier A, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mumbach MR, et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet. 2017 doi: 10.1038/ng.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ensemble and single-cell sequencing data are available through the Gene Expression Omnibus (GEO) under accession GSE107817. Two replicates of the ensemble ATAC-seq data for Naïve, TH17, and Treg cells were previously published and are available under GEO accession GSE10149861. In addition, we have generated an open-access interactive web browser, which enables single-cell TCR sequence and ATAC-seq TF deviation exploration (Supplementary Fig. 8; tcr.buenrostrolab.com). This browser includes all single-cell data presented in the study.
A WashU browser session with ensemble T cell subtype ATAC-seq data is available here: http://epigenomegateway.wustl.edu/browser/?genome=hg19&session=N7ew2XJpWK&statusId=293545209