Abstract
Rationale
Increasing evidence indicates the presence of long noncoding RNAs (lncRNAs) in various cell types. Airn is an imprinting gene transcribed from the paternal chromosome. It is in antisense orientation to the imprinted, but maternally-derived, Igf2r gene, on which Airn exerts its regulation in cis. Although Airn is highly expressed in the heart, functions aside from imprinting remain unknown.
Objective
Here, we studied the functions of Airn in the heart, especially cardiomyocytes.
Methods and Results
Silencing of Airn via siRNAs augmented cell death, vulnerability to cellular stress, and reduced cell migration. To find the cause of such phenotypes, the potential binding partners of Airn were identified via RNA pull-down followed by mass spectrometry, which indicated Igf2bp2 and Rpa1 as potential binding partners. Further experiments showed that Airn binds to Igf2bp2 to control the translation of a number of genes. Moreover, silencing of Airn caused less binding of Igf2bp2 to other mRNAs and reduced translation of Igf2bp2 protein.
Conclusions
Our study uncovers a new function of Airn and demonstrates that Airn is important for the physiology of cardiomyocytes.
Keywords: lncRNA, cardiomyocytes, gene expression, noncoding RNA, transcriptome
Subject Terms: Basic Science Research, Developmental Biology, Functional Genomics, Gene Expression and Regulation, Ischemia
INTRODUCTION
Most of the human genome is transcribed; however, only a minor fraction is translated into proteins. Many of these untranslated RNAs are classified as noncoding RNAs (ncRNAs), including those longer than 200 nucleotides (nt) called “long ncRNAs (lncRNAs)”. To date, numerous lncRNAs being identified in the heart; however, few of them have been functionally studied.1, 2 It is speculated that lncRNAs exert various functions to control the development, homeostasis, and pathophysiological processes in the heart. Thus, lncRNAs might be a key to unravel the regulatory networks controlling cardiomyocyte differentiation and function.
Airn (“antisense Igf2r RNA”, also known as “Air”) is an imprinted gene transcribed from the paternal chromosome.3, 4 In the human and mouse genomes, Airn is located in antisense orientation to the imprinted, but maternally-derived, “insulin-like growth factor 2 receptor (Igf2r)” gene. Airn can regulate nearby protein-coding genes Igf2r, Slc22a2, and Slc22a3 in cis.5–7 In murine hearts, there is no expression difference between male and female mice.8 Although Airn is capped, polyadenylated, a majority of Airn transcripts evade co-transcriptional splicing to give rise to a mature 118 kb transcript, which resides in the nucleus but is highly unstable, whereas the spliced Airn isoforms are as stable as other mRNAs (i.e., Igf2r) and are exported into the cytoplasm.4 Although the function of Airn as an imprinting gene is known, up until now, the functions of Airn isoforms are largely unknown, especially in the heart. Here, we uncover functional roles of Airn isoforms in cardiomyocytes using the murine cardiomyocytic HL-1 cell line.
METHODS
All data and materials have been made publicly available at the Gene Expression Omnibus and can be accessed at GSE87223. An extended methods section is available in the Online Data.
HL-1 cells were cultured according to the original publication.9 Primer and siRNA sequences are provided in Online Table I. Microarray experiment was performed as previously done.10–12 Heteroscedastic two-tail Student’s t-test was applied to calculate a p-value.
Adult male mice were subjected to non-reperfused, coronary artery occlusion to induce heart failure13 and in accordance with the University of Louisville Animal Care and Use Committee.
RESULTS
Airn isoforms are differentially expressed in tissues and cardiomyocytes
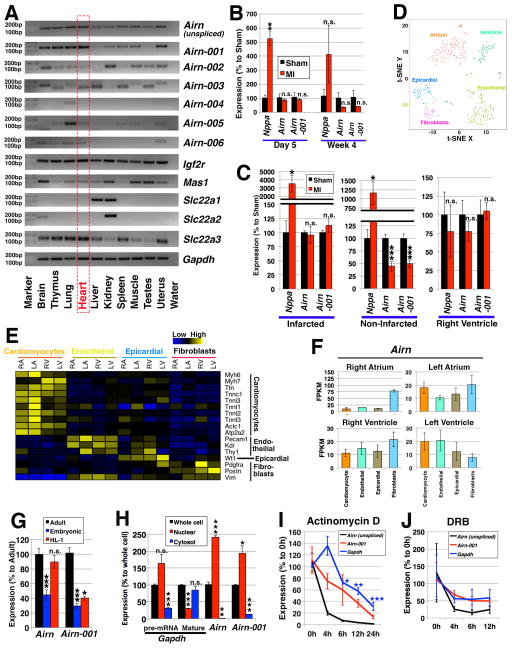
To study Airn isoforms (Online Figure I), we compared expression profiles among 10 murine tissues using a primer pair specifically targeting each isoform (Figure 1A). Of all the isoforms, Airn-001 expression was most similar to the unspliced form of Airn, which is the most studied and is involved in imprinting of nearby protein-coding genes. Other isoforms (Airn-002, -003 and -006) showed high expression in certain tissues, such as brain and kidney, brain and liver, and lung, respectively, which is consistent with previous reports14, 15 (Figure 1A). Based on these results and the high expression of Airn in the heart,4 we focused on unspliced Airn and Airn-001 in this study.
Figure 1. Airn and its isoforms.
(A) Expression profiling of Airn and its isoforms in mouse tissues. (B, C) Expression in myocardial infarction (MI). (B) The time-course data of 5 days and 4 weeks post surgery comparing “Sham” control and “MI” mice. n=3 mice for sham (day 5) and n=4 mice for all the other conditions. The expressions were normalized to those of sham samples using Gapdh as internal control. (C) Expression in different regions of hearts 4 weeks after the operation. One heart was divided into 3 regions: “Infarcted”; “Non-Infarcted” (i.e., border and remote regions); and “Right Ventricle”. n=5 mice. The expressions were normalized to those of sham samples using Rn18s as internal control. (D-F) Expression profiling of single-cell RNA-seq data of E10.5 mouse hearts. (D) 5 clusters of cells were defined: cardiomyocytes from atrium or ventricle, endothelial cells, epicardial cells, and fibroblasts. (E) Expressions of marker genes. (F) Expression patterns of the Airn gene. (G) Expression of unspliced Airn (“Airn”) and Airn-001 in “adult” cardiomyocytes compared to those of “embryonic” cardiomyocytes and “HL-1” cells. n=4 samples for adult cardiomyocytes and n=3 samples for others. (H) Subcellular localization in HL-1 cells. As controls, primer pairs for Gapdh of its pre-mRNA (targeting the intron between its exon 2 and 3) and mature RNA (targeting exons 4 and 5) were used. Statistical calculations were made against the expressions in the whole cell. n=3. (I, J) Stability of mRNAs upon the treatment with (I) actinomycin D at 4, 6, 12 and 24 hours; and (J) DRB at 4, 6 and 12 hours. n=3. The expressions were normalized using those of Rn18s. *, ** and *** represent p<0.05, 0.01 and 0.005, respectively. “n.s.” represents “not statistically significant”.
Previous studies show that many lncRNAs were dysregulated following myocardial infarction.1, 2 To identify changes in expression of unspliced Airn and Airn-001, we used hearts from mice subjected to non-reperfused myocardial infarction and from sham-operated mice. When whole hearts were compared, there is a slight tendency of down-regulation for both unspliced Airn and Airn-001 in the infarcted hearts compared to the control 4 weeks post operation (Figure 1B). When the expression patterns of unspliced Airn and Airn-001 were examined spatially, their expressions were down-regulated in the non-infarcted regions of infarcted hearts compared to the similar regions in the sham-operated hearts, whereas no change was detected in the infarct region, which consists of mainly dead cardiomyocytes (Figure 1C; Online Figure II).
Although the above expression profiling is informative, there remains a question of whether Airn is specifically expressed in cardiomyoyctes or not. To this end, we analyzed the published single-cell RNA-seq data of embryonic hearts (Figure 1D, E).16 In all compartments of the heart, Airn expression was recorded, which showed slightly higher expression in the left compared to right atrium and ventricle (Figure 1F). When cell types are compared, in the atrium, Airn is preferentially expressed in fibroblasts, whereas in the left ventricle, the expression of Airn is higher in cardiomyocytes compared to cell types.
To study the function of Airn in cardiomyocytes, we employed HL-1 cells.9 Because HL-1 is a cell line, we first quantified the expressions of unspliced Airn and Airn-001 in HL-1 cells by comparing to embryonic and adult cardiomyocytes, which indicated the comparable level of expression for unspliced Airn to adult cardiomyocytes than embryonic ones (Figure 1G). In the case of Airn-001, its expression is lower in HL-1 cells but higher than that of embryonic cardiomyocytes. Next, the subcellular localization of Airn-001 was investigated, which showed its preferential localization in the nucleus, similar to its unspliced form (Figure 1H). Because a previous study showed that Airn isoforms are more stable than the unspliced form,4 we investigated the stability of Airn-001 in comparison to its unspliced form by suppressing RNA synthesis by actinomycin D (Figure 1I) and 5,6-Dichloro-1-β-D-ribofuranosyl benzimidazole (DRB) (Figure 1J). As result, Airn-001 is as stable as the protein-coding gene Gapdh, whereas unspliced Airn is rapidly degraded.
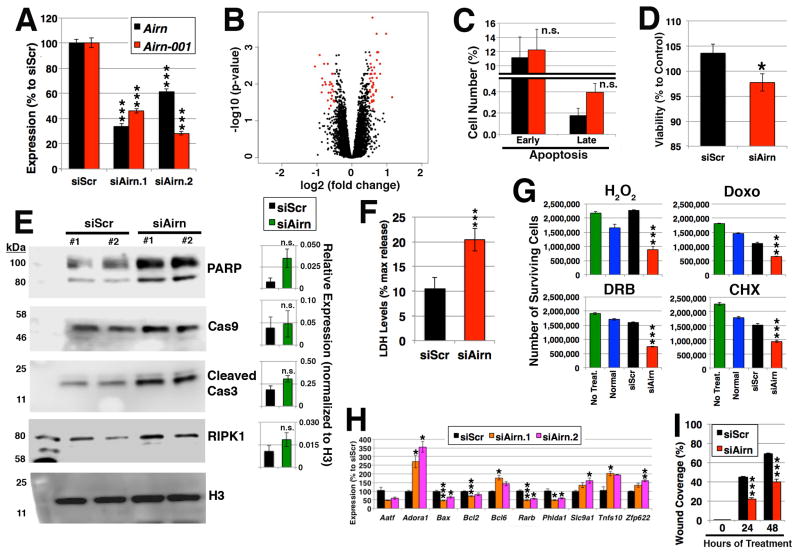
Next, Airn was knocked down in HL-1 cells with siRNAs (Figure 2A) and evaluated by microarrays (Figure 2B). When a threshold of 1.4-fold and p<0.05 were applied, 45 up- and 29 down-regulated genes were identified (Online Table II), which includes genes whose Gene Ontology terms associated with apoptosis (Online Table III). To confirm the molecular profiling, assays to quantify cell death were conducted via Annexin V staining using flow cytometry (Figure 2C) and reduction of 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) (Figure 2D). Silencing Airn showed slightly increased apoptosis, which was associated with caspase activation as detected by Western blotting (Figure 2E). Furthermore, necrosis was significantly induced upon silencing of Airn, which was determined by lactate dehydrogenase (LDH) release (Figure 2F). In line with these observations, when the cells were challenged with hydrogen peroxide (H2O2), doxorubicin (Doxo), DRB, and cycloheximide (CHX), fewer cells survived upon silencing of Airn compared to controls (Figure 2G). Interestingly, similar data were obtained for silencing of highly abundant lncRNAs Malat1 and Neat1 (Online Figure III). These results are in line with changes in expression of apoptotic genes after silencing of Airn compared to the control (Figure 2H). These data suggest that Airn is important for cell survival. Given that migration of cardiomyocytes is important for the proper development of the heart and regeneration upon injury in mice and zebrafish,17, 18 cell migration was measured via scratch assay, which showed reduced migration upon silencing of Airn (Figure 2I; Online Figure IV). Taken together, these results indicate that silencing of Airn affects the physiology of cardiomyocytes.
Figure 2. Silencing of Airn-001.
(A) Efficiency of knockdown for two siRNAs against Airn (siAirn.1 (n=6) and siAirn.2 (n=5)) in comparison to the control siRNA (siScr (n=6)). (B) Volcano plot of microarray results. Genes that were selected with threshold values of 1.4-fold and FDR<0.05 are colored in red. n=6 for siScr, n=4 for siAirn.1, and n=4 for siAirn.2. (C, D) Quantification of apoptosis via (C) flow cytometry for Annexin V compared to the total number of cells measured. n=2 (each with three technical replicates); and (D) MTT assay. The data were normalized to the untreated cells. n=3 (each with 24 technical replicates). (E) Western blotting of marker proteins of apoptosis. PARP, Cas9, cleaved Cas3, and RIPK1. Anti-histone H3 antibody was used as loading control. The representative blotting images are shown. The quantification of each corresponding band was normalized to that of H3 and represented as relative expression in each bar graph next to the corresponding blotting image (n=6 samples). (F) LDH assay. n=5 (each with 16 technical replicates). A total of 80 wells were counted. (G) Numbers of surviving cells. n=4 technical replicates. (H) qRT-PCR for apoptotic genes upon silencing of Airn compared to siScr. n=3 samples. (I) Scratch assay. The recovered areas were quantified. n=3. *, ** and *** represent p<0.05, 0.01 and 0.005, respectively. “n.s.” represents “not statistically significant”.
Airn binds to Igf2bp2 and Rpa1
To identify the mechanism by which Airn affects cardiomyocyte survival and function, we employed RNA pull-down experiment by labeling full-length Airn-001 with biotin and mixing it with cellular lysates of HL-1 cells to identify proteins that bind to Airn-001 via mass spectrometry analysis (Figure 3A). Among nine potential binding partners, two proteins (Igf2bp2 and Rpa1) were selected for further validation due to their subcellular localizations and the availability of working antibodies. When Western blotting experiments were performed after RNA pull-down, the binding of Rpa1 was observed for both sense and antisense Airn-001 (Figure 3B), suggesting that Rpa1 is not specifically bound to Airn-001, but nonspecifically binds to RNA. In contrast, Igf2bp2 preferentially binds to sense Airn-001 (Figure 3C). The binding of Igf2bp2 is further confirmed by RNA immunoprecipitation (RIP) followed by RT-PCR. Unspliced Airn and Airn-001 showed preferential binding to Igf2bp2 (Figure 3D), which confirms that Airn binds to Igf2bp2.
Figure 3. Binding partners.
(A) Silver-stained SDS-PAGE gel after RNA pull-down assay. The band specific to the sense sample but missing in the antisense sample was used for mass spectrometry analysis. In addition, the similar region in the antisense sample was analyzed by mass spectrometry. As result, 9 proteins listed were detected only in the sense Airn-001 compared to antisense one (negative control) in two idependent assays. (B, C) Western blotting after RNA pull-down assay for (B) Rpa1; and (C) Igf2bp2 “M” stands for marker; “I” for 5% input; “A” for antisense; and “S” for sense probe for Airn-001. Representative image from 5 independent assays. (D) RIP using anti-Igf2bp2 antibody. The expression values were normalized to those of the input. * and ** represent p<0.05 and 0.01, respectively. n=3.
Igf2bp2 protein binds various mRNA in cardiomyocytes
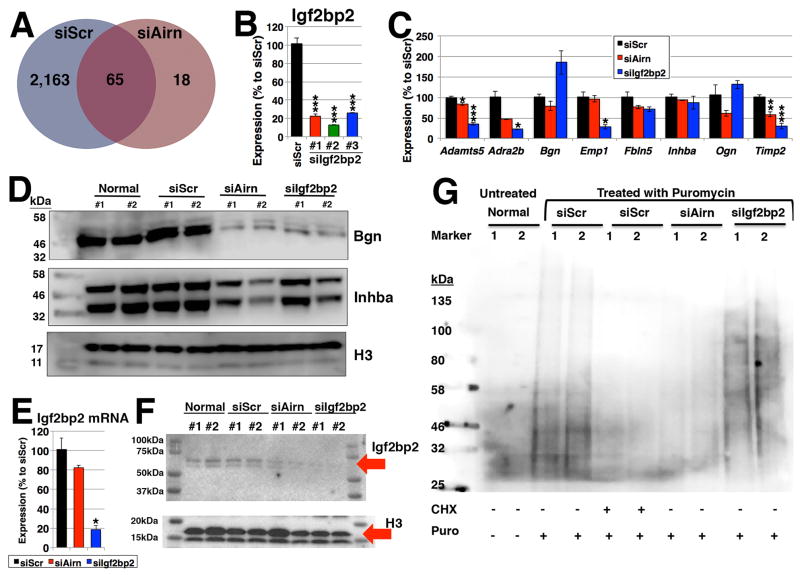
To understand the causal relationship between Airn and targets of Igf2bp2, RIP assay followed by microarray (RIP-chip) was performed. Compared to the control, there were significantly fewer Igf2bp2-bound genes upon silencing of Airn (2,228 and 83 genes, respectively) at the threshold value of 1.5-fold enrichment and p<0.05 over anti-IgG antibody as negative control (Figure 4A, Online Table IV), suggesting that Airn assists the binding of Igf2bp2 to mRNAs.
Figure 4. Binding of Igf2bp2.
(A) Venn diagram of Igf2bp2-bound genes upon silencing of Airn. RIP-chip experiments (n=4 samples) were performed using anti-Igf2bp2 antibody and anti-IgG antibody as negative control. The thresholds of 1.5-fold enrichment and FDR<0.05 were applied. (B) Efficiency of Igf2bp2 knockdown. n=3. (C) qRT-PCR. n=3. (D) Western blotting using anti-Bgn and -Inhba antibodies. (E, F) Expression of Igf2bp at the levels of (E) RNA (n=3) and (F) protein (n=2 and representative images from four independent assays). (G) SUnSET. The addition of CHX is used as a negative control. Representative image from 3 independent assays. *, ** and *** represent p<0.05, 0.01 and 0.005, respectively. “n.s.” represents “not statistically significant”.
To further validate these findings, expressions of Igf2bp2-bound mRNAs were quantified. If, indeed, Airn affects the binding of Igf2bp2 to these mRNAs, their gene expression should not change upon silencing of Airn, while the level of their proteins should be reduced. When the expression of the selected genes were quantified, 5 genes (Bgn, Emp1, Fbln5, Inhba and Ogn) among 8 genes tested did not show statistically significant changes upon silencing of Airn at the mRNA level (Figure 4B, C). At the protein level, biglycan (Bgn) and inhibin beta-A (Inhba) were reduced to a similar extent by silencing of Airn or Igf2bp2 (Figure 4D). Of note, although the secretion of Bgn was not affected by silencing of Airn or Igf2bp2, the reduction in the secreted level of Inbha was observed (Online Figure V). These results suggest that their translation is inhibited due to the reduced expression of Airn that is necessary for the functionality of Igf2bp2 protein as a translation regulator.
The reduction of Igf2bp2-bound genes raises a question whether the expression of Igf2bp2 is altered upon silencing of Airn. Although no reduction of Igf2bp2 was observed upon silencing of Airn compared to the control at the mRNA level (Figure 4E), to our surprise, silencing of Airn caused a reduction of Igf2bp2 protein (Figure 4F). To further confirm this finding, surface sensing of translation (SUnSET) assay19 was performed to monitor global protein synthesis. In this assay, puromycin is used to label newly synthesized peptides. Upon silencing of Airn, the amount of puromycin-labelled peptides is similar to the negative control (treated with CHX to block translation) than other conditions (Figure 4G). Based on these results, we conclude that Airn affects the translation (but not transcription) of Igf2bp2, which binds to many mRNAs to control their translation efficiencies.
Discussion
In this study, we uncovered the role of Airn other than imprinting in cardiomyocytes; that is, Airn binds to Igf2bp2 protein and control translation of Igf2bp2 as well as other genes. Compared to protein-coding genes, lncRNAs are less sequence-conserved among species20; however, some well conserved lncRNAs do exist (e.g. MALAT121), especially those that are involved in genomic imprinting (e.g. Meg322, 23). Given that Airn is an imprinting gene and conserved in humans,24 the question is whether a similar function exists in humans. Indeed, human AIRN is highly expressed in the heart (Online Figure VIA). Upon its silencing by siRNA in human embryonic kidney cells 293 (HEK-293) (Online Figure VIB), the survival of cells was reduced when treated with H2O2 (Online Figure IIIC), which indicates the similar phenotype as silencing of mouse Airn. Further investigation is necessary to understand the exact mechanism of reduced survival upon silencing of human AIRN.
Although its polymorphisms are linked to type 2 diabetes,25 the function of Igf2bp2 (also known as “IMP2”) in cardiomyocytes is unknown. There are studies of Igf2bp2 in related cell types (i.e. myoblasts and skeletal muscle), reporting the following: (i) It is highly expressed in myoblasts, where Igf2bp2 controls cell adhesion and motility via direct binding to the mRNA of Lims2 as well as that of Trim54 for the stabilization of microtubules;26 and (ii) Under the transcriptional control via Hmga2, Igf2bp2 contributes to myoblast proliferation by promoting the translation of various mRNAs, including Myc, Sp1 and Igf1r, but not Igf2.27 Here, we confirmed that Igf2bp2 directly binds to various mRNAs in cardiomyocytes (Figure 4A). Furthermore, through the interaction with Airn and its isoforms, the translation of Igf2bp2 protein itself is controlled (Figure 4E, F). Thus, we conclude that Airn lies in the upstream of Igf2bp2 to control the translation of many mRNAs, which include genes involved in apoptosis directly related to cell survival. Taken together, our study uncovered a novel function of Airn in controlling translation of various protein-coding genes through the regulation of Igf2bp2 protein synthesis in cardiomyocytes.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
The majority of the mammalian genome is transcribed as RNA, but only a minor part corresponds to exons of protein-coding genes.
The number of long noncoding RNAs (the noncoding transcripts longer than 200 nucleotides) is more than protein-coding genes, but their functions are largely unknown.
Airn (“antisense Igf2r RNA”, also known as “Air”) is an imprinted gene transcribed from the paternal chromosome, but besides imprinting, its functions remain unknown.
What New Information Does This Article Contribute?
Silencing of Airn augmented cell death, vulnerability to cellular stress, and reduced cell migration.
Airn binds to the RNA-binding protein Igf2bp2 to control the translation of a number of genes via reducing the translation of Igf2bp2 protein.
Although lncRNAs are present in the heart, their functions remain largely unknown. Of the few predicted functions, binding to macromolecules (e.g., nucleic acids or proteins) is of great interest because lncRNAs may function as molecular scaffolds or switches to activate or to inhibit biological processes. Recent evidence indicates that dysregulation of lncRNAs alters the transcriptome and leads to cardiac dysfunction. Because the number of lncRNAs exceeds that of protein-coding genes, more systematic study of lncRNAs in the heart is needed to uncover the causal relationship between lncRNAs and cardiac health and disease. Here, we show that the lncRNA Airn has a function besides its well known function as an imprinting gene. Our findings on the translational control of a number of genes via the interaction between Airn and Igf2bp2 have a broad translational impact as its manipulation may allow for post-transcriptional control of various protein-coding genes and their protein products. Although a majority of lncRNAs are not conserved across species, Airn is one of the few well defined lncRNAs that are conserved between humans and mice. These findings could significantly advance the understanding of human lncRNAs and their function in the heart.
Acknowledgments
The authors thank Wenjun Jin for technical assistance, the late Prof. Bill Claycomb for HL-1 cells, Dr. Chiara Cencioni for flow cytometry, Nicole Ritter for isolating embryonic cardiomyocytes, and UofL Cardinal Research Cluster for computing resources.
SOURCES OF FUNDING
This study was supported by the Deutsche Forschungsgemeinschaft (UC 67/2-1); LOEWE Center for Cell and Gene Therapy (Hessen, Germany); V.V. Cooke Foundation (Kentucky, U.S.A.); University of Louisville School of Medicine; and the startup funding from the Mansbach Family, the Gheens Foundation and other generous supporters at the University of Louisville. Dr. Jones has been supported by grants from the NIH (R01 HL083320, R01 HL094419, HL131647, P20 GM103492, and P01 HL078825). Dr. Dassanayaka was supported by American Heart Association Predoctoral Fellowship—Great Rivers Affiliate (14PRE19710015).
Nonstandard Abbreviations and Acronyms
- IncRNA
long noncoding RNA
- NGS
next generation sequencing
- NcRNA
noncoding RNA
- Nt
nucleotides
Footnotes
DISCLOSURES
None.
References
- 1.Uchida S, Dimmeler S. Long noncoding rnas in cardiovascular diseases. Circulation research. 2015;116:737–750. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 2.Uchida S, Bolli R. Short and long noncoding rnas regulate the epigenetic status of cells. Antioxidants & redox signaling. 2017 doi: 10.1089/ars.2017.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the igf2r gene depends on an intronic cpg island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 4.Seidl CI, Stricker SH, Barlow DP. The imprinted air ncrna is an atypical rnapii transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zwart R, Sleutels F, Wutz A, Schinkel AH, Barlow DP. Bidirectional action of the igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001;15:2361–2366. doi: 10.1101/gad.206201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcho C, Bevilacqua A, Tremblay KD, Mager J. Tissue-specific regulation of igf2r/airn imprinting during gastrulation. Epigenetics Chromatin. 2015;8:10. doi: 10.1186/s13072-015-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, Pauler FM. Imprinted igf2r silencing depends on continuous airn lncrna expression and is not restricted to a developmental window. Development. 2013;140:1184–1195. doi: 10.1242/dev.088849. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L. A comprehensive mouse transcriptomic bodymap across 17 tissues by rna-seq. Scientific reports. 2017;7:4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr Hl-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gellert P, Ponomareva Y, Braun T, Uchida S. Noncoder: A web interface for exon array-based detection of long noncoding rnas. Nucleic acids research. 2013;41:e20. doi: 10.1093/nar/gks877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gellert P, Teranishi M, Jenniches K, De Gaspari P, John D, Kreymborg K, Braun T, Uchida S. Gene array analyzer: Alternative usage of gene arrays to study alternative splicing events. Nucleic acids research. 2012;40:2414–2425. doi: 10.1093/nar/gkr1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreymborg K, Uchida S, Gellert P, Schneider A, Boettger T, Voswinckel R, Wietelmann A, Szibor M, Weissmann N, Ghofrani AH, Schermuly R, Schranz D, Seeger W, Braun T. Identification of right heart-enriched genes in a murine model of chronic outflow tract obstruction. Journal of molecular and cellular cardiology. 2010;49:598–605. doi: 10.1016/j.yjmcc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Muthusamy S, DeMartino AM, Watson LJ, Brittian KR, Zafir A, Dassanayaka S, Hong KU, Jones SP. Microrna-539 is up-regulated in failing heart, and suppresses o-glcnacase expression. The Journal of biological chemistry. 2014;289:29665–29676. doi: 10.1074/jbc.M114.578682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The air noncoding rna epigenetically silences transcription by targeting g9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 15.Pauler FM, Stricker SH, Warczok KE, Barlow DP. Long-range dnase i hypersensitivity mapping reveals the imprinted igf2r and air promoters share cis-regulatory elements. Genome research. 2005;15:1379–1387. doi: 10.1101/gr.3783805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR, Wu SM. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Developmental cell. 2016;39:491–507. doi: 10.1016/j.devcel.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, Kawakami Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development. 2012;139:4133–4142. doi: 10.1242/dev.079756. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt EK, Clavarino G, Ceppi M, Pierre P. Sunset, a nonradioactive method to monitor protein synthesis. Nature methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 20.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The gencode v7 catalog of human long noncoding rnas: Analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding rna malat1 regulates endothelial cell function and vessel growth. Circulation research. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 22.Boon RA, Hofmann P, Michalik KM, Lozano-Vidal N, Berghauser D, Fischer A, Knau A, Jae N, Schurmann C, Dimmeler S. Long noncoding rna meg3 controls endothelial cell aging and function: Implications for regenerative angiogenesis. Journal of the American College of Cardiology. 2016;68:2589–2591. doi: 10.1016/j.jacc.2016.09.949. [DOI] [PubMed] [Google Scholar]
- 23.Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, Garg A, Remke J, Zimmer K, Batkai S, Thum T. Inhibition of the cardiac fibroblast-enriched lncrna meg3 prevents cardiac fibrosis and diastolic dysfunction. Circulation research. 2017;121:575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
- 24.Yotova IY, Vlatkovic IM, Pauler FM, Warczok KE, Ambros PF, Oshimura M, Theussl HC, Gessler M, Wagner EF, Barlow DP. Identification of the human homolog of the imprinted mouse air noncoding rna. Genomics. 2008;92:464–473. doi: 10.1016/j.ygeno.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christiansen J, Kolte AM, Hansen T, Nielsen FC. Igf2 mrna-binding protein 2: Biological function and putative role in type 2 diabetes. Journal of molecular endocrinology. 2009;43:187–195. doi: 10.1677/JME-09-0016. [DOI] [PubMed] [Google Scholar]
- 26.Boudoukha S, Cuvellier S, Polesskaya A. Role of the rna-binding protein imp-2 in muscle cell motility. Molecular and cellular biology. 2010;30:5710–5725. doi: 10.1128/MCB.00665-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Gilbert JA, Zhang Y, Zhang M, Qiu Q, Ramanujan K, Shavlakadze T, Eash JK, Scaramozza A, Goddeeris MM, Kirsch DG, Campbell KP, Brack AS, Glass DJ. An hmga2-igf2bp2 axis regulates myoblast proliferation and myogenesis. Developmental cell. 2012;23:1176–1188. doi: 10.1016/j.devcel.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.