Abstract
Efforts to develop drugs for Alzheimer’s disease (AD) have shown promise in animal studies, only to fail in human trials, suggesting a pressing need to study AD in human model systems. Using human neurons derived from induced pluripotent stem cells carrying the major genetic risk factor apolipoprotein E4 (apoE4), we demonstrate that apoE4 neurons have higher levels of tau phosphorylation unrelated to their increased Aβ production and displayed GABAergic neuron degeneration. ApoE4 increased Aβ production in human, but not in mouse, neurons. Converting apoE4 to apoE3 by gene editing rescued these phenotypes, indicating the specific effects of apoE4. Neurons lacking apoE behaved like those expressing apoE3, and introducing apoE4 expression recapitulated the pathological phenotypes, suggesting a gain of toxic effects from apoE4. Treating apoE4 neurons with a small-molecule structure corrector ameliorated the detrimental effects, providing a proof of concept that correcting the pathogenic conformation of apoE4 is a viable therapeutic approach for apoE4-related AD.
INTRODUCTION
The complexity and multifactorial etiology of Alzheimer’s disease (AD) pose unique challenges for studying its pathogenesis and developing therapies. Efforts to target AD-related pathways have shown promise in animal studies, only to fail in human trials1,2. There is a pressing need to identify novel mechanisms and therapeutic targets for AD using human model systems, such as human neurons.
AD is pathologically characterized by the formation of intracellular neurofibrillary tangles (NFTs), comprised of hyperphosphorylated tau protein, and extracellular amyloid plaques, comprised of amyloid-beta (Aβ) peptides1. Apolipoprotein (apo) E4, the major genetic risk factor for AD3,4, is found to be associated with increases in both pathologies1. In general, apoE4 increases AD risk and lowers the age of onset in a gene-dose dependent manner5. Remarkably, the lifetime risk estimate of developing AD by age 85 is ~65% in people with two copies of the apoE4 allele but only ~10% in people with two copies of the apoE3 allele6. This striking difference highlights the importance of apoE4 in the pathogenesis of AD.
Human apoE3 and apoE4 differ from each another only at one amino acid residue at the position 112. ApoE3, the common isoform, has Cys-112, whereas apoE4 has Arg-1121,7,8. Structurally, apoE has two domains: the amino-terminal domain and the carboxyl-terminal domain, which contain the receptor-binding region and the lipid-binding region, respectively. The two domains are linked by a structurally flexible hinge region. Interaction between the carboxyl- and amino-terminal domains, called domain interaction, is a unique biophysical property of apoE41,7,8. In apoE4, domain interaction occurs as a result of a salt bridge formation between Arg-61 and Glu-255 due to the effect of Arg-112. This interaction occurs to a much less extent in apoE3 because the side chain of Arg-61 adopts a different conformation owing to Cys-112, resulting in a less accessible side chain conformation for a salt bridge formation with Glu-2551,7,8. Domain interaction has been suggested to be a molecular basis for apoE4’s detrimental effects in AD pathogenesis, and consequently has been pursued as a drug target to identify small molecule structure correctors capable of converting apoE4 to apoE3 both structurally and functionally1,7,8.
Studies in animal models and postmortem human tissues have provided key insights into the pathogenesis of AD1,2,9. However, mouse models of AD do not recapitulate many AD features, and postmortem human brain tissues have characteristics of end-stage disease that may not be present at earlier stages1,2,9. Until recently, studies of the cellular and molecular mechanisms of AD have been hindered by the lack of access to live human neurons. Now, induced pluripotent stem cells (iPSCs) derived from human somatic cells with AD-linked mutations or polymorphisms, together with gene-editing techniques, are promising in vitro models for studying disease pathogenesis in relevant cell types, including human neurons10–15.
Here, we analyzed AD-related phenotypes of cultured neurons derived from human iPSC (hiPSC) lines of different apoE genotypes, including gene-edited isogenic and apoE-deficient lines. We also tested the effects of gene editing to convert apoE4 into apoE3 and of a small-molecule structure corrector to render apoE4 apoE3-like. Our data demonstrate that apoE4 induces AD-related pathological phenotypes, due to a gain of toxic effects, specifically in human neurons, which can be dramatically ameliorated by a small-molecule apoE4 structure corrector.
RESULTS
Generating hiPSC lines from apoE3 and apoE4 homozygotes
hiPSC lines were generated from apoE3/3 and apoE4/4 subjects (Supplementary Table 1) as described16,17. All hiPSC lines were morphologically similar to embryonic stem (ES) cells (Supplementary Fig. 1b) and expressed ES cell markers, such as Nanog, Sox2, TRA-1-60, and TRA-1-81 (Supplementary Fig. 1c–e). DNA sequencing confirmed the apoE genotypes of all hiPSC lines, and chromosomal analysis revealed normal karyotypes (Supplementary Fig. 1f). After injection into immunodeficient mice, all hiPSC lines formed teratomas, confirming their pluripotency16,17. Three apoE3/3-hiPSC lines (E3/3-A, E3/3-B, and E3/3-C) and three apoE4/4-hiPSC lines (E4/4-A, E4/4-B, E4/4-C), each derived from a subject with the corresponding apoE genotype, were fully characterized and used in the current study (Supplementary Table 1). All six of these hiPSC lines developed well into neural stem cells expressing Sox2, nestin, Pax6, and FoxG1 (Supplementary Fig. 1g–i) and then into mature neurons that had neuronal morphology (Supplementary Fig. 1j) and expressed neuronal markers, Tuj1 and MAP2 (Supplementary Fig. 1k,l). Quantification shows that 90 ± 1.5% (mean ± SEM, n = 12 randomly collected images from three independent experiments with total of 326 cells counted) of the cells are positive for neuronal marker, MAP2, indicating the high purity of neuronal culture.
Human apoE4/4 neurons produce less full-length apoE and more apoE fragments than human apoE3/3 neurons
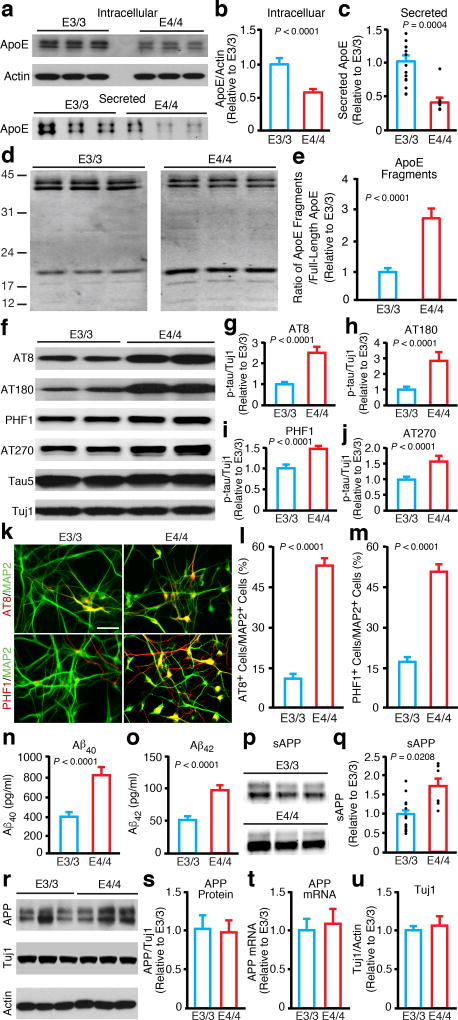
Western blot analyses of neuronal lysates and culture medium revealed that human apoE4/4 neurons produce ~35% less full-length intracellular apoE and secrete ~60% less full-length apoE into the medium than human apoE3/3 neurons, both in individual lines (Supplementary Fig. 2a) and as shown by mean values (Fig. 1a–c). Consequently, the ratio of intracellular apoE to secreted apoE was over 20% higher for apoE4 than apoE3, suggesting that apoE4 tends to be retained inside neurons. The ratio of apoE fragments to full-length apoE in neuronal lysates was also significantly higher in apoE4/4 neurons (Fig. 1d,e and Supplementary Fig. 2b). The major apoE fragments were ~12–20 kDa (Fig. 1d), similar in size to those in brains of neuron-specific apoE4 transgenic mice18 and AD patients with an apoE4 genotype19. ApoE4 fragments were undetectable in the culture medium, suggesting that the fragments tend to accumulate inside neurons.
Figure 1. Human apoE4/4 neurons generate more apoE fragments, have higher p-tau levels, and produce more Aβ than human apoE3/3 neurons.
(a) Western blot analysis of full-length apoE in lysates (intracellular) or medium (secreted) of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. (b) Quantification of full-length apoE in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 23 biologically independent samples from E3/3 (n=9 from apoE3/3-A; n=8 from apoE3/3-B; n=6 from apoE3/3-C), n = 20 biologically independent samples from E4/4 (n=6 from apoE4/4-A; n=6 from apoE4/4-B; n=8 from apoE4/4-C). (c) Quantification of full-length apoE in culture medium of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 15 biologically independent samples from E3/3 (n=6 from apoE3/3-A; n=6 from apoE3/3-B; n=3 from apoE3/3-C), n = 9 biologically independent samples from E4/4 (n=3 from apoE4/4-A; n=3 from apoE4/4-B; n=3 from apoE4/4-C). (d) Western blot analysis of full-length apoE and apoE fragments in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. (e) Quantification of apoE fragments in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 13 biologically independent samples from E3/3 (n=3 from apoE3/3-A; n=5 from apoE3/3-B; n=5 from apoE3/3-C), n = 14 biologically independent samples from E4/4 (n=3 from apoE4/4-A; n=5 from apoE4/4-B; n=6 from apoE4/4-C). (f) Western blot analysis of p-tau with AT8, AT180, PHF1, and AT270 monoclonal antibodies in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. (g) Quantification of p-tau (AT8) in neuronal lysates. Values are normalized to E3/3. n = 31 biologically independent samples from E3/3 and n = 25 biologically independent samples from E4/4. (h) Quantification of p-tau (AT180) in neuronal lysates. Values are normalized to E3/3. n = 22 biologically independent samples from E3/3 (n=7 from apoE3/3-A; n=7 from apoE3/3-B; n=8 from apoE3/3-C), n = 18 biologically independent samples from E4/4 (n=6 from apoE4/4-A; n=4 from apoE4/4-B; n=8 from apoE4/4-C). (i) Quantification of p-tau (PHF1) in neuronal lysates. Values are normalized to E3/3. n = 17 biologically independent samples from E3/3 (n=4 from apoE3/3-A; n=6 from apoE3/3-B; n=7 from apoE3/3-C), n = 25 biologically independent samples from E4/4 (n=8 from apoE4/4-A; n=8 from apoE4/4-B; n=9 from apoE4/4-C). (j) Quantification of p-tau (AT270) in neuronal lysates. Values are normalized to E3/3. n = 23 biologically independent samples from E3/3 (n=10 from apoE3/3-A; n=10 from apoE3/3-B; n=3 from apoE3/3-C), n = 17 biologically independent samples from E4/4 (n=10 from apoE4/4-A; n=4 from apoE4/4-B; n=3 from apoE4/4-C). (k) Immunostaining for MAP2 and p-tau (AT8 and PHF1) in neuronal cultures derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Scale bar, 50 µm. (l, m) Quantification of the percentage of MAP2-positive neurons that are also positive for p-tau (AT8 and PHF1) in neuronal cultures derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. n = 12 (E3/3: n=12 fields with total of 594 MAP2+ neurons counted for AT8; n=12 fields with total of 945 MAP2+ neurons counted for PHF1), n = 12 (E4/4: n=12 fields with total of 526 MAP2+ neurons counted for AT8; n=12 fields with total of 1030 MAP2+ neurons counted for PHF1). (n) Aβ40 levels in culture medium of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. n = 23 biologically independent samples from E3/3 (n=4 from apoE3/3-A; n=7 from apoE3/3-B; n=12 from apoE3/3-C), n = 21 biologically independent samples from E4/4 (n=5 from apoE4/4-A; n=4 from apoE4/4-B; n=12 from apoE4/4-C). (o) Aβ42 levels in culture medium of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. n = 23 biologically independent samples from E3/3 (n=4 from apoE3/3-A; n=7 from apoE3/3-B; n=12 from apoE3/3-C), n = 21 biologically independent samples from E4/4 (n=5 from apoE4/4-A; n=4 from apoE4/4-B; n=12 from apoE4/4-C). (p) Western blot analysis of sAPP in culture medium of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. (q) Quantification of sAPP in culture medium of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 15 biologically independent samples from E3/3 (n=6 from apoE3/3-A; n=6 from apoE3/3-B; n=3 from apoE3/3-C), n = 6 biologically independent samples from E4/4 (n=2 from apoE4/4-A; n=2 from apoE4/4-B; n=2 from apoE4/4-C). (r) Western blot analysis of APP with 22C11 monoclonal antibody in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. (s) Quantification of APP, as determined by 22C11 monoclonal antibody, in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 11 biologically independent samples from E3/3 (n=4 from apoE3/3-A; n=4 from apoE3/3-B; n=3 from apoE3/3-C), n = 19 biologically independent samples from E4/4 (n=3 from apoE4/4-A; n=8 from apoE4/4-B; n=8 from apoE4/4-C). (t) Quantification of APP mRNA levels by qRT-PCR in neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 12 biologically independent samples from E3/3 (n=4 from apoE3/3-A; n=4 from apoE3/3-B; n=4 from apoE3/3-C), n = 12 biologically independent samples from E4/4 (n=4 from apoE4/4-A; n=4 from apoE4/4-B; n=4 from apoE4/4-C). (u) Tuj1/actin ratios in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 11 biologically independent samples from E3/3 (n=4 from apoE3/3-A; n=4 from apoE3/3-B; n=3 from apoE3/3-C), n = 19 biologically independent samples from E4/4 (n=3 from apoE4/4-A; n=8 from apoE4/4-B; n=8 from apoE4/4-C). All values are expressed as mean ± SEM. Differences between groups were determined with the unpaired two-sided t test.
Phosphorylated tau (p-tau) levels are higher in human apoE4/4 neurons than human apoE3/3 neurons
Western blot analysis with p-tau-specific antibodies (AT8, AT180, PHF1, and AT270) revealed significantly higher p-tau levels in apoE4/4 neurons than apoE3/3 neurons, both in individual lines (Supplementary Fig. 2c–f) and as shown by mean values (Fig. 1f–j). Double immunostaining with anti-p-tau (AT8 or PHF1) and anti-MAP2 antibodies consistently showed that significantly higher percentages of apoE4/4 neurons were also positive for p-tau (Fig. 1k–m). Furthermore, p-tau accumulated mostly in the soma and dendrites of apoE4/4 neurons, suggesting that apoE4 induces both tau phosphorylation and mislocalization of p-tau in human neurons, a pathological hallmark of p-tau in AD brains1,20,21.
Human apoE4/4 neurons produce more Aβ than human apoE3/3 neurons
The levels of Aβ peptides in neuronal culture media were measured by an ELISA10,22. ApoE4/4 neurons secreted >2-fold more Aβ40 and Aβ42 into the culture medium than apoE3/3 neurons, both in individual lines (Supplementary Fig. 2g,h) and as shown by mean values (Fig. 1n,o). The apoE4/4 neurons also secreted greater amounts of soluble APP (sAPP) into the medium (Fig. 1p,q), suggesting increased processing of APP. Interestingly, the full-length APP protein (Fig. 1r,s) and mRNA (Fig. 1t) levels were similar in apoE4/4 and apoE3/3 neurons, as determined by western blot and qRT-PCR analysis respectively, suggesting that apoE4 does not alter APP expression in human neurons. Importantly, the ratios of Tuj1 to actin were similar in apoE3/3 and apoE4/4 neuronal cultures (Fig. 1r,u), indicative of comparable neuronal populations.
Since it has been reported that in human apoE3 and apoE4 knock-in (KI) mice expressing human APP with mutations that cause familial AD, apoE4 appears to reduce Aβ clearance and stimulate Aβ deposition in the brain without affecting Aβ production23, we wondered whether the difference in apoE4’s effect on Aβ production between the current human neuron study and the previously reported mouse data reflects species difference in apoE regulation of Aβ metabolism. To address this question, we also generated mouse iPSC (miPSC) lines from human apoE3-KI and apoE4-KI mouse fibroblasts, differentiated them into neurons in culture for 7–8 weeks, and measured Aβ secretion from the miPSC-derived neurons in culture. Clearly, there were no significant differences in both Aβ40 and Aβ42 levels in the culture medium of neurons derived from apoE3/3-miPSC and apoE4/4-miPSC lines (Supplementary Fig. 3). These data indicate the species difference in apoE isoform regulation of Aβ metabolism—apoE4 stimulates Aβ production in human neurons but not in mouse neurons.
Increased p-tau levels in apoE4/4 neurons are independent of high levels of Aβ
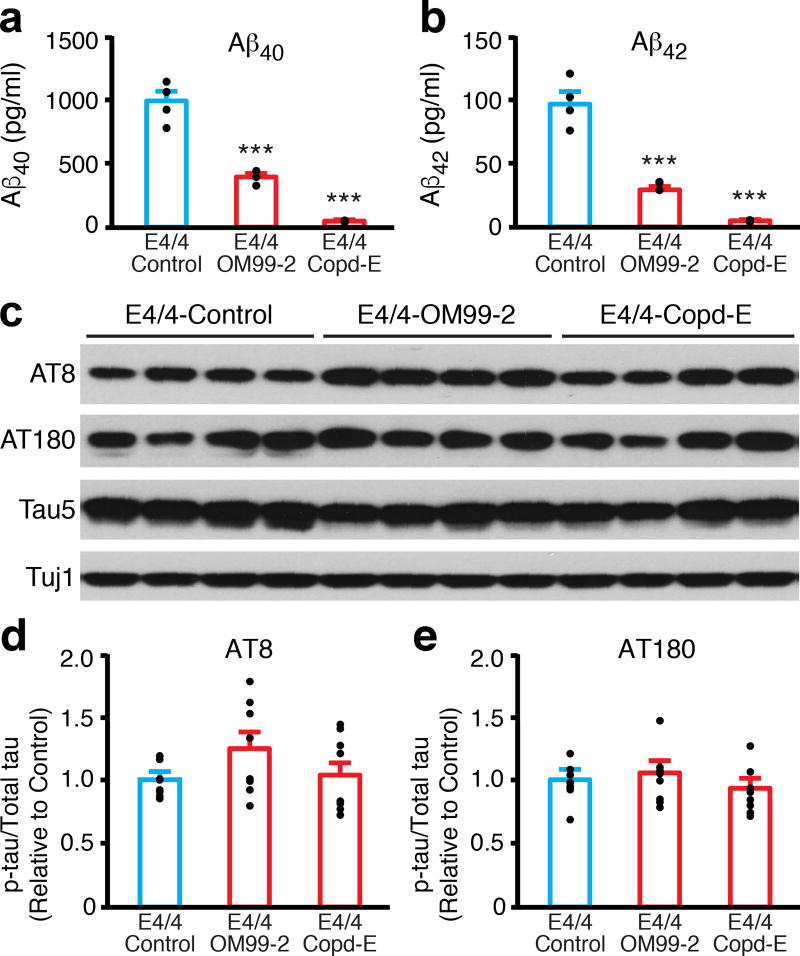
It has been suggested that Aβ accumulation could increase phosphorylation and accumulation of tau24,25. To assess the potential causal relationship between increased Aβ production and increased p-tau levels, we treated apoE4/4 neurons with an inhibitor of β-secretase (OM99-2) or γ-secretase (compound E) for 1 week10. Aβ40 and Aβ42 levels in the culture medium decreased by 60–75% (OM99-2) and >95% (compound E) (Fig. 2a,b). Strikingly, despite these drastic reductions of Aβ levels, p-tau levels were unaffected in apoE4/4 neurons, as shown by western blot analysis with monoclonal antibodies AT8 and AT180 (Fig. 2c–e).
Figure 2. Increased p-tau levels in apoE4/4 neurons are independent of higher Aβ production.
(a) Aβ40 levels in culture medium of apoE4/4 neurons treated with vehicle (control), a β-secretase inhibitor (OM99-2, 750 nM), or a γ-secretase inhibitor (compound E, Copd-E, 200 nM) for 1 week. n = 4 (control), n = 3 (OM99-2), and n = 3 (Copd-E) biologically independent samples. (b) Aβ42 levels in culture medium of apoE4/4 neurons treated with vehicle (control), a β-secretase inhibitor (OM99-2), or a γ-secretase inhibitor (Copd-E) for 1 week. n = 4 (control), n = 3 (OM99-2), and n = 3 (Copd-E) biologically independent samples. (c) Western blot analysis of p-tau with AT8 and AT180 monoclonal antibodies in lysates of apoE4/4 neurons treated with vehicle (control), a β-secretase inhibitor (OM99-2), or a γ-secretase inhibitor (Copd-E) for 1 week. (d) Quantification of p-tau (AT8) in lysates of apoE4/4 neurons treated with vehicle (control), a β-secretase inhibitor (OM99-2), or a γ-secretase inhibitor (Copd-E) for 1 week. Values are normalized to control. n = 8 (control), n = 8 (OM99-2), and n = 8 (Copd-E) biologically independent samples. (e) Quantification of p-tau (AT180) in lysates of apoE4/4 neurons treated with vehicle (control), β-secretase inhibitor (OM99-2), or γ-secretase inhibitor (Copd-E) for 1 week. Values are normalized to control. n = 8 (control), n = 8 (OM99-2), and n = 8 (Copd-E) biologically independent samples. All values are expressed as mean ± SEM. Differences among groups were determined with one-way ANOVA followed with Tukey’s multiple comparison test. ***p < 0.001 versus E4/4-Control.
ApoE4 causes GABAergic neuron degeneration/loss in hiPSC-derived neuronal culture
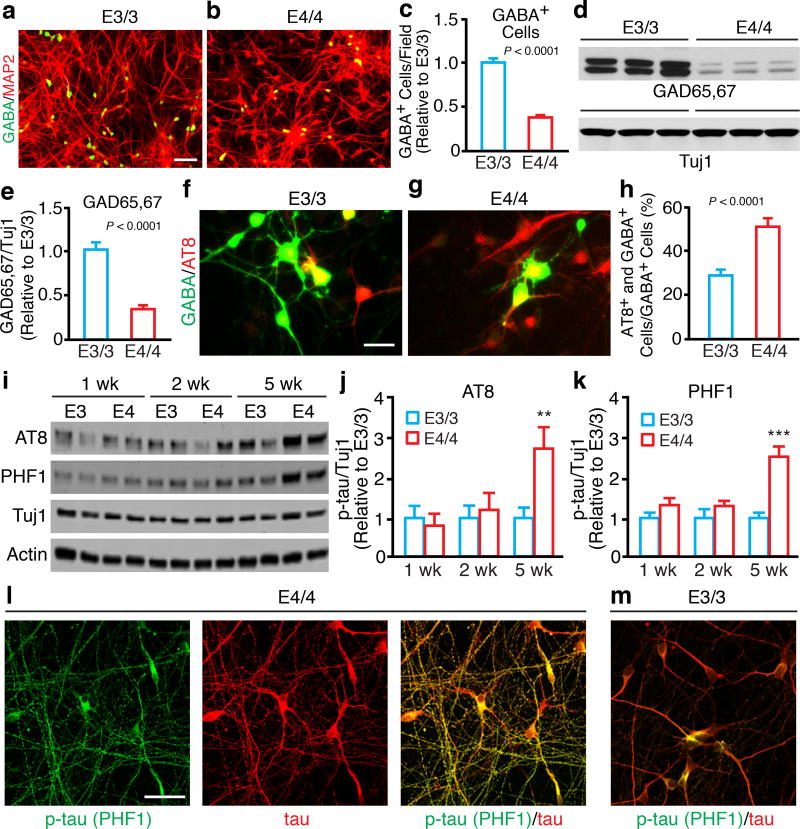
Immunofluorescent staining revealed significantly fewer GABA-positive neurons derived from apoE4/4-hiPSCs than from apoE3/3-hiPSCs, both in individual lines (Supplementary Fig. 2i) and as shown by mean values (Fig. 3a–c). Levels of GAD65/67 (GABAergic neuron marker) were lower in neuronal cultures derived from apoE4/4-hiPSCs than in those derived from apoE3/3-hiPSCs, both in individual lines (Supplementary Fig. 2j) and as shown by mean values (Fig. 3d,e). Immunostaining revealed no significant difference in the numbers of GABA-positive neurons at early time points (within 4 weeks) in culture between apoE4/4-hiPSCs and apoE3/3-hiPSCs (relative to apoE3/3 as 1 ± 0.11, mean ± SEM, n = 5 fields with total of 570 GABA+ neurons counted, apoE4/4 is 0.98 ± 0.14, mean ± SEM, n = 8 fields with total of 894 GABA+ neurons counted), suggesting that the detrimental effect of apoE4 on GABAergic neurons in culture is not a developmental effect. The decrease in GABAergic neuron numbers occurred during the late stage of neuronal differentiation of apoE4/4-hiPSCs, suggesting that apoE4 induces GABAergic neurodegeneration/loss. Importantly, apoE3/3-hiPSCs and apoE4/4-hiPSCs had similar efficiencies in generating Tbr1-positive glutamatergic neurons and TH-positive dopaminergic neurons in culture (Supplementary Fig. 4), suggesting a specific detrimental effect of apoE4 on GABAergic neurons. Furthermore, ~60% of GABAergic neurons derived from apoE4/4-hiPSCs, but only ~30% of those derived from apoE3/3-hiPSCs, were immunopositive for p-tau (Fig. 3f–h).
Figure 3. ApoE4 causes GABAergic neuron degeneration/loss in hiPSC-derived neuronal culture.
(a, b) Double immunostaining for GABA and MAP2 in neuronal cultures derived from apoE3/3-hiPSCs (a) and apoE4/4-hiPSCs (b). Scale bar, 100 µm. (c) Quantification of GABA-positive cells per field in neuronal cultures derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 36 fields from E3/3 (n=16 from apoE3/3-A; n=12 from apoE3/3-B; n=8 from apoE3/3-C, with total of 9325 GABA+ neurons counted), n = 32 fields from E4/4 (n=12 from apoE4/4-A; n=12 from apoE4/4-B; n=8 from apoE4/4-C, with total of 3433 GABA+ neurons counted). (d) Western blot analysis of GAD65/67 in lysates of neurons derived from individual lines of apoE3/3-hiPSCs and apoE4/4-hiPSCs. (e) Quantification of the ratios of GAD65/67 to Tuj1 in lysates of neurons derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. Values are normalized to E3/3. n = 54 biologically independent samples from E3/3 (n=20 from apoE3/3-A; n=25 from apoE3/3-B; n=9 from apoE3/3-C), n =35 biologically independent samples from E4/4 (n=14 from apoE4/4-A; n=12 from apoE4/4-B; n=9 from apoE4/4-C). (f, g) Double immunostaining for GABA and AT8 (p-tau) in neuronal cultures derived from apoE3/3-hiPSCs (f) and apoE4/4-hiPSCs (g). Scale bar, 25 µm. (h) Quantification of the percentage of GABA-positive cells that are also positive for AT8 (p-tau) in neuronal cultures derived from apoE3/3-hiPSCs and apoE4/4-hiPSCs. n = 12 fields from E3/3 (n=4 from apoE3/3-A; n=4 from apoE3/3-B; n=4 from apoE3/3-C, with total of 352 GABA+ neurons counted), n = 12 fields from E4/4 (n=4 from apoE4/4-A; n=4 from apoE4/4-B; n=4 from apoE4/4-C, with total of 144 GABA+ neurons counted). (i) Western blot analysis of p-tau (AT8 and PHF1) in lysates of pure GABAergic neurons generated from apoE3/3-hiPSCs and apoE4/4-hiPSCs. (j) Quantification of p-tau (AT8) in GABAergic neuron lysates. Values are normalized to E3/3. n = 10 biologically independent samples from E3/3 (n=5 from E3/3-A; n=5 from E3/3-B), n = 10 biologically independent samples from E4/4 (n=5 from E4/4-A; n=5 from E4/4-B). (k) Quantification of p-tau (PHF1) in GABAergic neuron lysates. Values are normalized to E3/3. n = 10 biologically independent samples from E3/3 (n=5 from E3/3-A; n=5 from E3/3-B), n = 10 biologically independent samples from E4/4 (n=5 from E4/4-A; n=5 from E4/4-B). (l, m) Double immunostaining for p-tau (PHF1) and total tau in GABAergic neurons generated from apoE4/4-hiPSCs (l) and apoE3/3-hiPSCs (m). The experiments were repeated independently for over three times with similar results. Scale bar, 25µm. All values are expressed as mean ± SEM. Differences between groups were determined with the unpaired two-sided t test in c, e, and h. Differences among groups were determined by two-way ANOVA followed with Sidak’s multiple comparisons test in j and k. **p < 0.01; ***p < 0.001.
To further determine the detrimental effects of apoE4 on GABAergic interneurons, we adapted a protocol of differentiating hiPSCs to medial ganglionic eminence (MGE) cells, which are GABAergic progenitors, and then to mature GABAergic interneurons in culture26–28. The purity of MGE cells and GABAergic interneurons were 96.03 ± 1.25% and 93.15 ± 0.99 (mean ± SEM), respectively, as determined by anti-NKX2.1 (a MGE cell marker) and anti-GABA immunostaining and flow cytometry analysis (Supplementary Fig. 5a–d). Anti-p-tau western blot (AT8 and PHF1) revealed significantly increased p-tau levels in apoE4/4-GABAergic neurons compared to apoE3/3-GABAergic neurons at 5 weeks of culture, while there were no significant differences at 1 and 2 weeks of culture (Fig. 3i–k). Furthermore, immunostaining with p-tau-specific antibodies (AT8 and PHF1) also revealed that 70–100% more GABAergic neurons derived from apoE4/4-MGE cells than from apoE3/3-MGE cells were immunopositive for p-tau (Supplementary Fig. 5e,f). Importantly, p-tau accumulated in degenerating (beading) axons, as determined by anti-p-tau (PHF1) and anti-total tau double immunostaining, in apoE4/4-GABAergic neurons, but not in apoE3/3-GABAergic neurons at 5 weeks of culture (Fig. 3l,m). As in AD brains, p-tau also mislocalized to dendrites of apoE4/4-GABAergic neurons, but not much in apoE3/3-GABAergic neurons, as revealed by anti-p-tau (PHF1) and anti-MAP2 double immunostaining (Supplementary Fig. 5h,i). Quantification of Caspase-3 and GABA double positive cells suggested significantly more apoptotic cell death of apoE4/4-GABAergic neurons than apoE3/3-GABAergic neurons (Supplementary Fig. 5g). Taken together, these data demonstrate clearly that apoE4 induces human GABAergic neuron degeneration/death in culture.
Gene editing reveals the specificity of apoE4 in eliciting AD-related pathologies in human apoE4/4 neurons
To determine whether the AD-related pathologies in apoE4/4 neurons are specifically induced by apoE4, we used gene editing to generate an isogenic apoE3/3-hiPSC line from the apoE4/4-hiPSC-A line (Supplementary Fig. 6a)29. As in the parental line, the isogenic apoE3/3-hiPSC line (iE3/3) had normal expression of pluripotency genes (Supplementary Fig. 6b,c) and a normal karyotype (Supplementary Fig. 6d), and it generated normal neural stem cells, which expressed Sox2, nestin, and FoxG1 (Supplementary Fig. 6e,f) and developed into mature neurons (Supplementary Fig. 6g). Importantly, DNA sequencing analyses revealed no off-target effects of the gene-editing at least on some major AD-related genes (Supplementary Table 2).
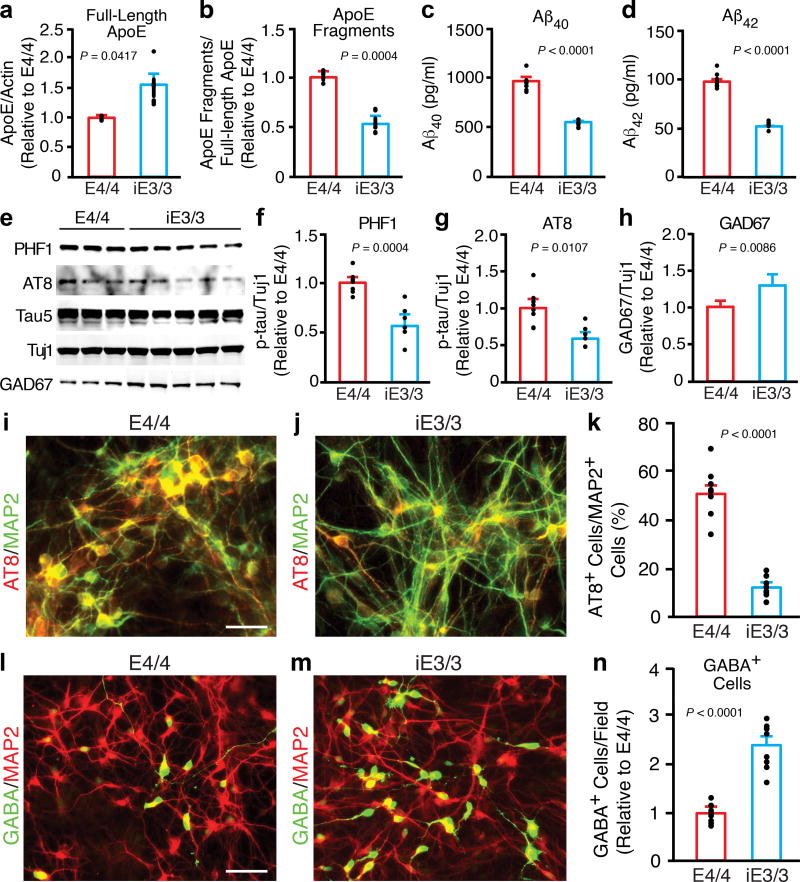
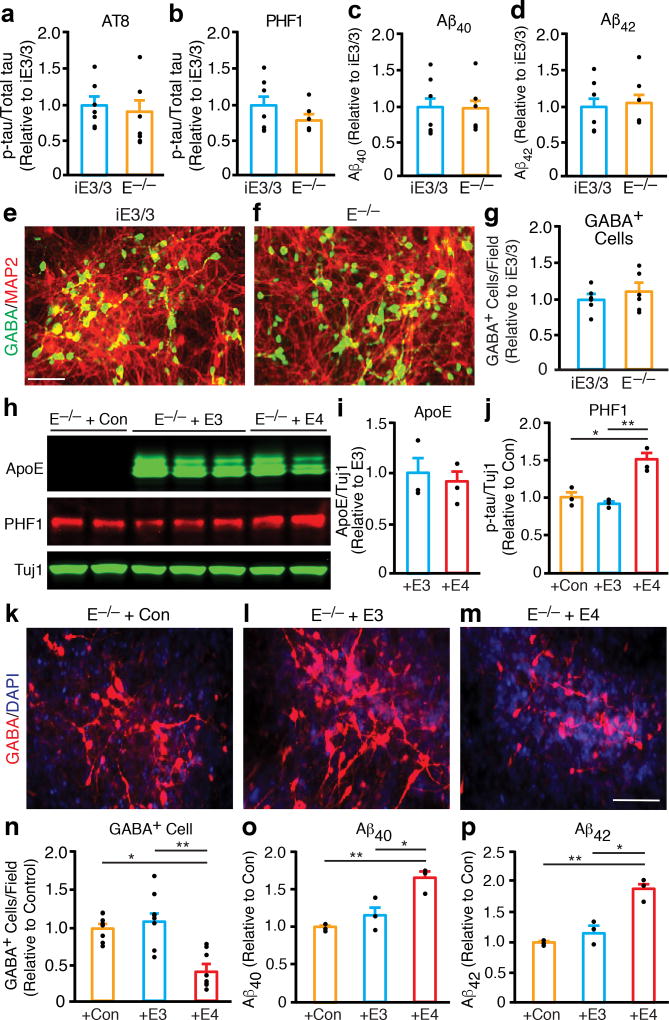
Next, we assessed AD-related pathologies in neurons derived from the isogenic apoE3/3-hiPSC line. The conversion from apoE4 to apoE3 had striking phenotypic effects. It increased the levels of full-length apoE (Fig. 4a), decreased the level of apoE fragments in neuronal lysates (Fig. 4b) and the secretion of Aβ40 and Aβ42 into the culture medium (Fig. 4c,d). P-tau levels decreased (Fig. 4e–g), and there were fewer p-tau-positive neurons (Fig. 4i–k and Supplementary Fig. 7a–c). The conversion also rescued the degeneration/loss of GABAergic neurons in apoE4/4-hiPSC neuronal cultures, as shown by significant increases in GAD67 levels (Fig. 4e,h) and in the number of GABA-positive neurons (Fig. 4l–n). The isogenic apoE3/3-hiPSC cultures also had significantly fewer p-tau-positive GABAergic neurons (Supplementary Fig. 7d–i) than parental apoE4/4-hiPSC cultures. Likewise, GABAergic neurons generated from the isogenic apoE3/3-hiPSC-derived MGE cells also had decreased p-tau levels and axonal degeneration compared to GABAergic neurons derived from the parental apoE4/4-hiPSC-derived MGE cells (Supplementary Fig. 8). In sum, conversion of apoE4 to apoE3 by gene editing abolished the detrimental effects of apoE4 and resulted in cellular phenotypes similar to those of apoE3, strongly suggesting that the AD-related pathologies in apoE4/4-neurons are induced specifically by apoE4.
Figure 4. AD-related pathologies in human apoE4/4 neurons are specifically induced by apoE4.
(a) Quantification of the full-length apoE in lysates of neurons derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). Values are normalized to E4/4. n = 3 (E4/4) and n = 6 (iE3/3) biologically independent samples. (b) Quantification of apoE fragments in lysates of neurons derived from apoE4/4-hiPSCs and the isogenic apoE3/3-hiPSCs (iE3/3). Values are normalized to E4/4. n = 3 (E4/4) and n = 6 (iE3/3) biologically independent samples. (c) Aβ40 levels in culture medium of neurons derived from apoE4/4-hiPSCs and the isogenic apoE3/3-hiPSCs (iE3/3). n = 8 (E4/4) and n = 6 (iE3/3) biologically independent samples. (d) Aβ42 levels in culture medium of neurons derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). n = 8 (E4/4) and n = 6 (iE3/3) biologically independent samples. (e) Western blot analysis for p-tau (PHF1 and AT8) and GAD67 in lysates of neurons derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). (f, g) Quantification of p-tau levels determined with monoclonal antibodies PHF1 (f) and AT8 (g) in lysates of neurons derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). Values are normalized to E4/4. n = 8 (E4/4) and n = 6 (iE3/3) biologically independent samples. (h) Quantification of GAD67 levels in lysates of neurons derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). Values are normalized to E4/4. n = 14 (E4/4) and n = 20 (iE3/3) biologically independent samples. (i, j) Double immunostaining for AT8 and MAP2 in neurons derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). Scale bar, 50 µm. (k) Quantification of the percentage of MAP2-positive cells that are also positive for AT8 (p-tau) in neuronal cultures derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). n = 8 fields from E4/4 with total of 1533 MAP2+ neurons counted, n = 8 fields from iE3/3 with total of 1691 MAP2+ neurons counted. (l, m) Double immunostaining for GABA and MAP2 in neurons derived from apoE4/4-hiPSCs and isogenic apoE3/3-hiPSCs (iE3/3). Scale bar, 50 µm. (n) Quantification of GABA-positive cells per field in neuronal cultures derived from apoE4/4-hiPSCs and the isogenic apoE3/3-hiPSCs (iE3/3). Values are normalized to E4/4 group. n = 8 fields from E4/4 with total of 149 GABA+ neurons counted), n = 8 fields from iE3/3 with total of 359 GABA+ neurons counted). All values are expressed as mean ± SEM. Differences between groups were determined with the unpaired two-sided t test.
Reintroducing different apoE isoforms into apoE-deficient human neurons reveals a gain of toxic effects from apoE4 in hiPSC-derived apoE-null neurons
To determine whether apoE4 induces a loss of function or a gain of toxic effects in human neurons, we also generated a hiPSC line from a subject with apoE deficiency (apoE−/−)30. The apoE−/− hiPSC line had ES cell–like morphology (Supplementary Fig. 9a,b), expressed pluripotency marker genes (Supplementary Fig. 9c–f), and generated neural stem cells (Supplementary Fig. 9g,h) and neurons (Supplementary Fig. 9i). Interestingly, the p-tau levels (Fig. 5a,b and Supplementary Fig. 10a) and the pattern of p-tau immunostaining (Supplementary Fig. 10b–d) in these neurons were similar to those in neurons derived from the isogenic apoE3/3-hiPSCs. ApoE−/− hiPSC-derived neurons and the isogenic apoE3/3-hiPSC-derived neurons secreted similar levels of Aβ40 and Aβ42 into the medium (Fig. 5c,d). Furthermore, apoE−/− hiPSCs and the isogenic apoE3/3-hiPSCs generated similar numbers of GABA-positive neurons (Fig. 5e–g). Thus, the phenotypes of apoE-null neurons were similar to those of apoE3/3 neurons.
Figure 5. ApoE4 confers a gain of toxic effects in hiPSC-derived neurons.
(a, b) Quantification of p-tau levels determined with monoclonal antibodies AT8 (a) and PHF1 (b) in lysates of neurons derived from isogenic apoE3/3-hiPSCs (iE3/3) and apoE−/− hiPSCs (E−/−). Values are normalized to iE3/3. n = 7 (iE3/3) and n = 7 (E−/−) biologically independent samples. (c) Aβ40 levels in culture medium of neurons derived from isogenic apoE3/3-hiPSCs (iE3/3) and apoE−/− hiPSCs (E−/−). Values are normalized to iE3/3. n = 7 (iE3/3) and n = 7 (E−/−) biologically independent samples. (d) Aβ42 levels in culture medium of neurons derived from isogenic apoE3/3-hiPSCs (iE3/3) and apoE−/− hiPSCs (E−/−). Values are normalized to iE3/3. n = 7 (iE3/3) and n = 7 (E−/−) biologically independent samples. (e, f) Immunostaining for MAP2 and GABA in neuronal cultures derived from isogenic apoE3/3-hiPSCs (iE3/3) (e) and apoE−/− hiPSCs (E−/−) (f). Scale bar is 50 µm. (g) Quantification of GABA-positive cells per field in neuronal cultures derived from isogenic apoE3/3-hiPSCs (iE3/3) and apoE−/− hiPSCs (E−/−). Values are normalized to iE3/3. n = 6 fields from iE3/3 with total of 3520 GABA+ neurons counted, n = 6 fields from E−/− with total of 3897 GABA+ neurons counted. (h) Western blot analyses of apoE expression and p-tau (PHF1) levels in lysates of apoE-null neurons transfected with a control lentiviral vector (+Con), a lentiviral apoE3 construct (+E3), or a lentiviral apoE4 construct (+E4). (i) Quantification of the full-length apoE in lysates of apoE-null neurons transfected with a lentiviral apoE3 construct (+E3), or a lentiviral apoE4 construct (+E4). Values are normalized to +E3. n = 3 (+E3) and n = 3 (+E4) biologically independent samples. (j) Quantification of p-tau (PHF1) levels in lysates of apoE-null neurons transfected with a control lentiviral vector (+Con), a lentiviral apoE3 construct (+E3), or a lentiviral apoE4 construct (+E4). Values are normalized to +Con. n = 3 (+Con), n = 3 (+E3), and n = 3 (+E4) biologically independent samples. (k–m) Anti-GABA immunostaining of apoE-null neurons transfected with a control lentiviral vector (+Con), a lentiviral apoE3 construct (+E3), or a lentiviral apoE4 construct (+E4). Scale bar, 50 µm. (n) Quantification of GABA-positive cells per field in apoE-null neuronal cultures transfected with a control lentiviral vector (+Con), a lentiviral apoE3 construct (+E3), or a lentiviral apoE4 construct (+E4). Values are normalized to +Con. n = 8 fields from +Con with total of 1104 GABA+ neurons counted, n = 8 fields from +E3 with total 1234 of GABA+ neurons counted, n = 8 fields from +E4 with total of 637 GABA+ neurons counted. (o) Aβ40 levels in culture medium of apoE-null neurons transfected with a control lentiviral vector (+Con), a lentiviral apoE3 construct (+E3), or a lentiviral apoE4 construct (+E4). Values are normalized to +Con. n = 3 (+Con), n = 3 (+E3), and n = 3 (+E4) biologically independent samples. (p) Aβ42 levels in culture medium of apoE-null neurons transfected with a control lentiviral vector (+Con), a lentiviral apoE3 construct (+E3), or a lentiviral apoE4 construct (+E4). Values are normalized to +Con. n = 3 (+Con), n = 3 (+E3), and n = 3 (+E4) biologically independent samples. All values are expressed as mean ± SEM. Differences between groups were determined with the unpaired two-sided t test in a–d, g, and i. Differences among groups were determined with one-way ANOVA followed with Tukey’s multiple comparison test in j and n–p. *p < 0.05; **p < 0.01.
To further test the potential gain of toxicity conferred by apoE4, we transfected apoE−/− hiPSC-derived neurons with lentiviral cDNA constructs encoding apoE3 or apoE4. The transfected neurons expressed similar protein levels of apoE3 and apoE4 (Fig. 5h,i); however, those expressing apoE4 had higher p-tau levels (Fig. 5h,j) and fewer GABA-positive neurons (Fig. 5m,n) and produced more Aβ40 and Aβ42 (Fig. 5o,p) than neurons expressing apoE3 or controls (Fig. 5j–l, 5n–p). Interestingly, treating apoE−/− hiPSC-derived neurons with purified recombinant human apoE3 or apoE4 did not significantly alter Aβ production (Supplementary Fig. 10e,f), suggesting that the effect of apoE4 on Aβ production depends on the endogenous expression of apoE. Together, these findings strongly suggest that apoE4 confers a gain of toxic effects in hiPSC-derived neurons.
The gain of toxic effects of apoE4 is neuron specific
Although astrocytes are the major source of apoE production in brains1,7, neurons can also produce apoE, especially under stress or in response to injuries1,7,31–33. We confirmed that hiPSC-derived neurons, including glutamatergic and GABAergic neurons, did express apoE in culture (likely represents a “stress” condition) as determined by anti-apoE and anti-neuronal marker double immunostaining (Supplementary Fig. 11). Since our neuronal culture is highly pure, with negligible astrocytes (Supplementary Fig. 12a,c,e), the neuronal apoE is unlikely taken up from astrocyte-secreted pool.
To further determine whether human astrocyte-derived apoE4 also displays detrimental effects in hiPSC-derived neurons, we differentiated hiPSC with different apoE genotypes into mature astrocytes (Supplementary Fig. 12b,d,e) and collected the conditioned media (ACM) with secreted apoE3 or apoE4. The apoE−/− hiPSC-derived neurons in culture were treated for 1 week with the ACM containing 0.35 nM (12 ng/ml) or 1.47 nM (50 ng/ml) of apoE. These concentrations of apoE were similar (0.35 nM) to or 3–4-fold higher (1.47 nM) than those in brain interstitial fluid (ISF) of mice34, which largely reflect the pool of astrocyte-secreted apoE in brains. Clearly, apoE4-ACM treatment at both concentrations did not significantly affect the levels of p-tau (AT8 and PHF1), GAD67 (GABAergic neuron marker), and Aβ40 and Aβ42 production/secretion in apoE−/− hiPSC-derived neuron culture (Supplementary Fig. 13). Thus, human astrocyte-derived apoE4 does not confer detrimental effects in human neurons (at least for tau phosphorylation, GABAergic neuron degeneration, and Aβ production), supporting the conclusion that the gain of toxic effects of apoE4 is neuron specific.
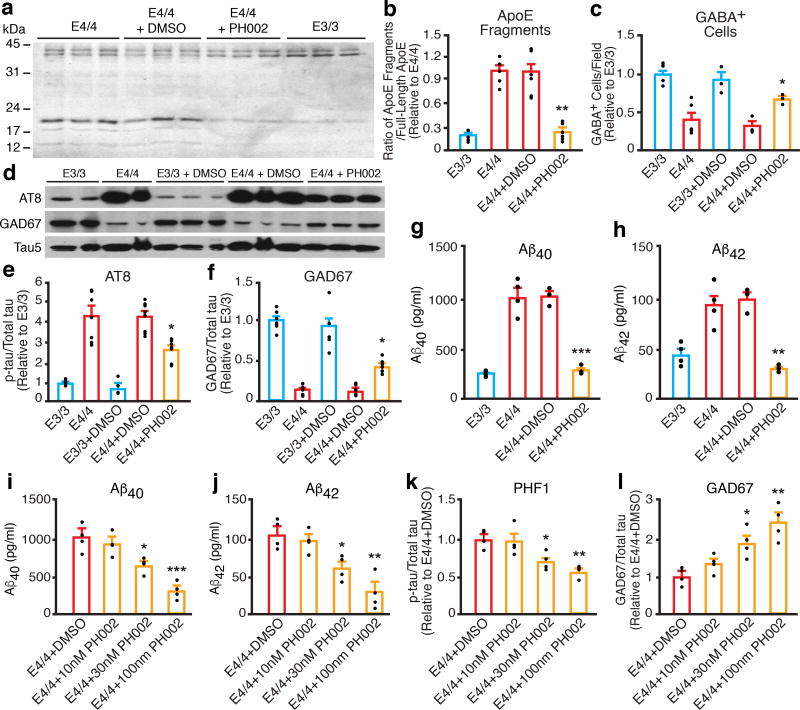
Detrimental effects of apoE4 in hiPSC-derived neurons are ameliorated by a small-molecule structure corrector
We previously reported apoE4 domain interaction (Supplementary Fig. 14a)1,35 and identified small-molecule structure correctors that render apoE4 apoE3-like both structurally and functionally (Supplementary Fig. 14b)36–38. In apoE4/4-hiPSC-derived neurons, treatment with one of the structure correctors, PH002 (Supplementary Fig. 14b)36–38, significantly decreased apoE4 fragment levels (Fig. 6a,b), increased GABAergic neuron numbers (Fig. 6c) and GAD67 levels (Fig. 6d,f), reduced p-tau levels (Fig. 6d,e), and decreased Aβ40 and Aβ42 production/secretion (Fig. 6g,h) in a dose-dependent manner (Fig. 6i–l). Thus, the detrimental effects of apoE4 in hiPSC-derived neurons can be ameliorated by a small molecule apoE4 structure corrector. Importantly, treatment of apoE−/− hiPSC-derived neurons with different concentrations of PH002 did not show any significant effects on Aβ production/secretion, p-tau levels, or GAD67 levels (Supplementary Fig. 14c–f), indicating that the efficacy of PH002 depends on the presence of apoE4.
Figure 6. The gain of toxic effects of apoE4 in hiPSC-derived neurons can be ameliorated by a small-molecule structure corrector.
(a) Western blot analysis of apoE fragment levels in lysates of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with dimethyl sulfoxide (DMSO) or the small-molecule structure corrector PH002. (b) Quantification of apoE fragment levels in lysates of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with DMSO or PH002. Values are normalized to E4/4. n = 5 (E3/3), n = 5 (E4/4), n = 6 (E4/4+DMSO), and n = 6 (E4/4+PH002) biologically independent samples. (c) Quantification of GABA-positive cells per field in cultures of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with DMSO or PH002. Values are normalized to E3/3. n = 6 fields from E3/3 with total of 3071 GABA+ neurons counted) n = 6 fields from E4/4 with total of 1223 GABA+ neurons counted), n = 3 fields from E3/3+DMSO with total of 1358 GABA+ neurons counted), n = 3fields from E4/4+DMSO with total of 511 GABA+ neurons counted, n = 3 fields from E4/4+PH002 with total of 1077 GABA+ neurons counted. (d) Western blot analyses of p-tau (AT8) and GAD67 in lysates of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with DMSO or PH002. (e) Quantification of p-tau (AT8) levels in lysates of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with DMSO or PH002. Values are normalized to E3/3. n = 6 (E3/3), n = 7 (E4/4), n = 5 (E3/3+DMSO), n = 8 (E4/4+DMSO), and n = 8 (E4/4+PH002) biologically independent samples. (f) Quantification of GAD67 levels in lysates of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with DMSO or PH002. Values are normalized to E3/3. n = 7 (E3/3), n = 7 (E4/4), n = 5 (E3/3+DMSO), n = 8 (E4/4+DMSO), and n = 6 (E4/4+PH002) biologically independent samples. (g) Aβ40 levels in culture medium of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with DMSO or PH002. n = 4 (E3/3), n = 4 (E4/4), n = 3 (E4/4+DMSO), and n = 3 (E4/4+PH002) biologically independent samples. (h) Aβ42 levels in culture medium of apoE3/3 neurons, apoE4/4 neurons, and apoE4/4 neurons treated with DMSO or PH002. n = 4 (E3/3), n = 4 (E4/4), n = 3 (E4/4+DMSO), and n = 3 (E4/4+PH002) biologically independent samples. (i–l) Dose effects of PH002 treatment on Aβ40 (i), Aβ42 (j), p-tau (PHF1) (k), and GAD67 (l) levels in culture medium or cell lysates of apoE4/4 neurons. DMSO treatment was used as a control. n = 4 biologically independent samples for each treatment group. All values are expressed as mean ± SEM. Differences among groups were determined with one-way ANOVA followed with Tukey’s multiple comparison test. *p < 0.05; **p < 0.01; ***p < 0.001 versus E4/4 or E4/4+DMSO in b, c, and e–h. *p < 0.05; **p < 0.01; ***p < 0.001 versus E4/4+DMSO in i–l.
DISCUSSION
In this study, we assessed the phenotypes of cultured neurons derived from multiple hiPSC lines of different apoE genotypes, including gene-edited isogenic and apoE-deficient lines. We found that apoE4 specifically induces AD-related pathologies, including the increase in apoE fragmentation, tau phosphorylation, and Aβ production, as well as the degeneration/loss of GABAergic neurons. Importantly, these AD-related pathologies were associated with a neuron-specific gain of toxic effects of apoE4 and were ameliorated by a small-molecule structure corrector that renders apoE4 structurally and functionally similar to apoE336–38.
Lower levels of apoE4 than apoE3 have been reported in human apoE4 and apoE3 knock-in mouse brains39 and in samples of human brain and cerebrospinal fluid1. Indeed, we found that cultured human neurons have ~35% less intracellular apoE4 than apoE3 and secrete ~60% less apoE4 than apoE3. Consequently, the ratio of intracellular apoE to secreted apoE was over 20% higher in apoE4/4 neurons, suggesting that apoE4 tends to be retained inside neurons. These findings might reflect a prolonged retention time of apoE4 in the endoplasmic reticulum and the Golgi apparatus, as reported in cultured rat neuronal cells37. ApoE fragments accumulate in the brains of neuron-specific apoE4 transgenic mice and AD patients carrying apoE417,18. However, our study shows for the first time in cultured human neurons that apoE4 is more susceptible than apoE3 to proteolytic cleavage, generating more neurotoxic fragments.
In humans and transgenic mice, apoE4 is associated with increased Aβ accumulation and amyloid plaque formation in the brain1,40. In apoE3 and apoE4 knock-in mice expressing human APP with mutations that cause familial AD, apoE4 appears to reduce Aβ clearance and stimulate Aβ deposition, especially during the initial seeding stage of plaque formation41,42, in the brain without affecting Aβ production23. We found that the endogenously expressed apoE4 significantly stimulates Aβ production in cultured human neurons, likely by enhancing APP processing. Thus, apoE4’s effects on Aβ production are species specific. Importantly, conversion of apoE4 to apoE3 by gene editing significantly lowered Aβ production, consistent with a specific effect of apoE4. This observation highlights the phenotypic differences between mouse and human cellular models of AD. Those differences must be recognized to understand why so many AD drugs in development failed in human clinical trials, even though they worked beautifully in mouse models of AD, and to develop drugs that are more likely effective in AD patients.
It has recently been reported that apoE4 enhances APP expression, leading to increased Aβ production, in a pure excitatory human neuron culture system43. However, we did not observe a similar effect of apoE4 on APP expression in a mixed human neuronal culture system. This could be due to the following differences between the two experimental systems. First, the induced excitatory neurons were generated by artificially expressing a transcription factor Ngn2 in hiPSCs, without inhibitory neurons in the culture43. As reported in previous in vitro and in vivo studies, Aβ production is regulated by neuronal activity—increasing neuronal activity stimulates Aβ production and secretion44,45. Thus, in this pure excitatory neuronal culture, in which there is no inhibitory neurons to control the activity, Aβ production pathway and its regulation could be altered in a way that might not be physiologically relevant. In contrast, in the mixed neuronal culture, the presence of inhibitory neurons balances neuronal activities, as seen under a physiological condition, making Aβ production more physiologically controlled. Second, in the pure excitatory neuron study, conditioned media containing 294 nM (10 µg/ml) of apoE3 or apoE4 were collected from transfected HEK293 cells and were then used to treat pure excitatory neurons in culture43. This apoE concentration is >800-folds higher than the physiological concentrations of apoE (~0.35 nM) in mouse brain ISF34. In addition, it has been reported that HEK293 cell-secreted apoE differs from astrocytes-secreted apoE, including their poor lipidation status, self-aggregation, and not binding well with the LDL receptor46. In our study, however, we used astrocyte-secreted apoE3 or apoE4 at concentrations (0.35 or 1.47 nM) similar to those in mouse brain ISF34.
ApoE4 has been associated with AD-related tau pathologies in both humans and transgenic and gene-targeted mice1,7,17,18. The current study shows that apoE4 in fact increases p-tau levels in cultured human neurons. ApoE4/4 neurons in culture were positive for multiple p-tau-specific monoclonal antibodies (e.g, AT8, AT180, AT270, and PHF1), suggesting hyperphosphorylation of tau. Furthermore, p-tau in apoE4/4 human neurons was largely mislocalized to the soma and dendrites, as seen in AD brains. Again, conversion of apoE4 to apoE3 by gene editing significantly lowered p-tau levels, consistent with a specific effect of apoE4. It has been suggested that Aβ accumulation leads to increase in phosphorylation and accumulation of tau24,25. However, we showed that treatment of apoE4/4 human neurons with a β-secretase or a γ-secretase inhibitor drastically reduced Aβ40 and Aβ42 levels without altering p-tau levels at all. These findings strongly suggest that apoE4 induces p-tau accumulation in an Aβ-independent manner in human neurons at least in culture. In line with this conclusion, a recent study suggests that apoE4 also has an Aβ-independent and gain-of-toxic effect on tau-mediated neurodegeneration in a tauopathy model of mice expressing mutant human tau47.
ApoE4 also had a detrimental effect on GABAergic neurons in hiPSC-derived neuronal cultures. This was especially true in relatively pure GABAergic neuronal cultures derived from hiPSCs and is consistent with observations in human apoE4 knock-in mouse brains48–51. Hyperphosphorylated tau was accumulated in axons of GABAergic neurons, leading to axonal degeneration, and also mislocalized to neuronal dendrites, as seen in AD brains. These effects, too, were apoE4-specific, as they were abolished by conversion of apoE4 to apoE3. GABAergic interneuron loss and neuronal hyperactivity due to loss of inhibitory tone have been observed in apoE4 knock-in mice48,49,52. Importantly, GABAergic interneuron deficits in apoE4 knock-in mice correlate with spatial learning and memory impairment48,49, and deletion of the apoE4 gene in GABAergic interneurons in LoxP-floxed apoE4 knock-in mice restores normal learning and memory and rescues apoE4-induced impairment of hippocampal network activity53,54. Thus, apoE4-induced GABAergic interneuron degeneration/dysfunction could contribute significantly to dementia in AD patients.
A long-standing question has been whether the detrimental effects of apoE4 in AD pathogenesis reflect a loss of normal function or a gain of toxic effects1,55. Studies in animals suggest both possibilities1,55, and it has been difficult to address this question in humans. Our hiPSC-derived neuron model of apoE deficiency and the complementary experiments of reintroducing apoE4 into the apoE-null human neurons strongly argue for a gain of toxic effects of apoE4 in AD pathogenesis, at least for its effects on tau phosphorylation, Aβ production, and GABAergic neuron degeneration. This conclusion is in line with the report of an apoE-deficient patient that detailed neurocognitive studies failed to demonstrate any defects30. This information has critical implications for developing drugs targeting apoE4: reducing apoE4 expression might be an attractive strategy.
Lastly, our findings in hiPSC-derived neurons provide a proof of concept that correcting the pathogenic conformation of apoE4 is a viable therapeutic approach for apoE4-related AD1,8,36–38. Treating apoE4/4 hiPSC-derived neurons with the apoE4 structure corrector PH00236–38 ameliorated the detrimental effects of apoE4 on tau phosphorylation, Aβ production, and GABAergic neuron degeneration in a dose-dependent manner. These findings warrant further development of apoE4 structure correctors and, ultimately, testing in clinical trials.
ONLINE METHODS
Generating and Characterizing hiPSC and miPSC Lines with Different ApoE Genotypes
hiPSC lines were generated as described16,17 from skin fibroblasts of subjects with different apoE genotypes or apoE deficiency (Supplementary Table S1). miPSC lines were generated from fibroblasts derived from human apoE3-KI and apoE4-KI mice using a similar protocol16,17. The hiPSCs were maintained under feeder-free condition in mTeSR1 medium or on irradiated SNL feeder cells in hES cell medium consisting of Dulbecco’s modified Eagle’s medium /F12 with 20% Knockout Serum Replacement, 2 mM glutamine, 0.1 mM nonessential amino acids (all from Life Technologies), 4 ng/ml recombinant human basic fibroblast growth factor (bFGF) (PeproTech), and 0.1 mM 2-mercaptoethanol56. hiPSCs were routinely passaged at 1:2–1:4 by brief treatment with Accutase (Millipore) and scraping. The hiPSC protocol was approved by the Committee on Human Research at the University of California, San Francisco.
Generating and Characterizing Isogenic ApoE3/3 hiPSC Lines by Zinc Finger Nuclease (ZFN)–mediated Gene Editing
To generate isogenic apoE3/3-hiPSC lines, we dissociated the parental apoE4/4 hiPSCs with Accutase (Millipore) and placed 2×106 cells in nucleofection solution (Lonza) containing 4 µg of the APOE-ZFN mRNA pair together with 5 µg of apoE3 donor DNA. A specific pair of six-finger ZFNs engineered to target a region around the Arg-112 of apoE4 allele was designed and prepared by Sigma. The ZFNs bound (uppercase) and cut (lowercase) the following sequence on the APOE gene: CGGGCACGGCTGTCCAAGgagctGCAGGCGGCGCAGGCCCG. The mixture was transferred to a cuvette and nucleofected with Nucleofector II (Lonza) at Nucleofector setting A-23 and the Human Stem Cell Nucleofector Kit I, as reported29. Nucleofected cells were quickly transferred to 500 µl of prewarmed (37°C) hES cell medium, incubated at 37°C for 5 min, and plated at 1×106 cells per well in an irradiated SNL-coated six-well plate containing hES medium supplemented with 10 µM ROCK inhibitor (ROCKi) (Tocris). To maximize viability and efficiency, the entire procedure was completed within 20 min.
Transfection efficiency was assessed by nucleofecting 5 µg of pmaxGFP vector into another set of cells to monitor GFP expression. For clonal isolation of modified hiPSCs after nucleofection, cultures were dissociated into single cells with Accutase and plated at 1 cell/well in 96-well plates coated with irradiated SNL feeders. Cloned hiPSCs were maintained in hES medium supplemented with 10 µM ROCKi for 1 week. Single colonies in 96-well plates were passaged into 48-well plates and expanded in 24-well plates. Cells were then harvested for isolation of genomic DNA with a GenElute Mammalian Genomic DNA Miniprep Kit (Sigma), following the manufacturer’s instructions. Genomic DNA (250 ng) was amplified by PCR with Phusion High Fidelity DNA Polymerase (Thermo Fisher Scientific) and 0.5 µM forward and reverse primers flanking the ZFN binding sequence on APOE to yield a 326-bp fragment. The primer sequences were 5´-CTCGGCCGCAGGGCGCTGATGGA-3´ (forward) and 5´-GCCCCGGCCTGGTACACTGCCA-3´ (reverse). The PCR products were sequenced to identify the ZFN-introduced apoE4 to apoE3 conversion. The converted apoE3 isogenic clones were further confirmed by genomic sequencing of the APOE allele covering the donor region and upstream and downstream regions. The hiPSC lines were karyotyped by Cell Line Genetics.
For immunostaining, hiPSCs cultured in 24-well plates were fixed with 4% paraformaldehyde and stained with primary antibodies against Nanog (ab21624, Abcam), human nuclei (MAB1281, EMD Millipore), Sox2 (sc-17320, Santa Cruz Biotechnology), Oct-3/4(sc-5279, Santa Cruz Biotechnology), SSEA4 (ab16287, Abcam), TRA-1-81 (MAB4381, EMD Millipore), and TRA-1-60 (MAB4360, EMD Millipore). The secondary antibodies were IgG conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Life Technologies). Nuclei were stained with DAPI. Images were taken with a Leica epifluorescence microscope, a Keyence BZ-9000E fluorescence microscope, or a Leica confocal imaging system.
For the teratoma formation assay, 1×106 cells of each hiPSC line were dissociated with Accutase (Millipore), spun down, and resuspended in 100 µl of PBS. NOD-SCID mice were anesthetized by standard procedures, and the cell suspensions were injected into the right and left thigh muscles (1×106 cells per site) of three mice. The mice were monitored during recovery. Tumors were apparent by about 4 weeks. The tumors were collected, fixed, sectioned, and stained with hematoxylin-eosin to identify teratomas. The protocol was approved by the Animal Care and Use Committees of the Gladstone Institutes and the University of California, San Francisco.
cDNA Sequencing of Alzheimer’s Disease Related Genes
Total RNAs were purified from neurons derived from the parental apoE4/4-hiPSC and the isogenic apoE3/3-hiPSC lines using Qiagen Rneasy mini kit. cDNAs were reverse transcribed from total RNA using oligo dT as primers (SuperScript™ III First-Strand Synthesis System, ThermoFisher Scientific). Using the resultant cDNAs as templates, the coding sequence of APP, MAPT (Tau), BACE1, and PS1 were amplified with corresponding 2–3 pairs of PCR primers. The coding sequence of the NPLOC4 gene was also amplified because it was predicted as a potential off-target gene of the gene editing approach using the program Prognos. All PCR products were then purified and sequenced. The sequencing data were then used to blast search database for DNA sequence alignment.
Neuronal Differentiation of hiPSCs and miPSCs
hiPSCs were differentiated into neurons as described57,58 with modifications. hiPSCs were treated with collagenase IV (Stem Cell Technologies) and grown in suspension as embryoid bodies in hES medium without bFGF. The medium was replaced every day for 5 days. On day 5, embryoid bodies were grown in Neural Induction Medium containing Dulbecco’s modified Eagle’s medium/F12 and Neurobasal Medium (1:1, Thermo Fisher), 1% N2 Supplement (Life Technologies), 1% B27 Supplement (Life Technologies), nonessential amino acids, and 0.5% penicillin/streptomycin (Life Technologies) supplemented with inhibitors of the TGF-β receptor (SB431542, Stemgent; 5 µM) and the bone morphogenetic protein receptor (LDN-193189, Stemgent; 0.25 µM). On day 7, spheres were transferred to wells coated with Matrigel (BD Biosciences) and grown in Neural Progenitor Medium, which consisted of Neural Induction Medium without SB and LDN, but containing 10 ng/ml bFGF (PeproTech), 10 ng/mL epidermal growth factor (EGF) (PeproTech), and 2 µg/ml heparin (Sigma). Attached neuroepithelial cells were fed every other day for 7 days. Neural rosette structures appeared by around day 15, were lifted off the plate, and grown in suspension in Neural Progenitor Medium to form neurospheres. For neuronal differentiation, neurospheres were dissociated with Accutase and plated on culture plates coated with 100 µg/ml poly-l-ornithine (Sigma) and 10 µg/ml laminin (Sigma). Neuronal cultures were maintained in Neural Differentiation Medium consisting of Neural Progenitor Medium, brain-derived neurotrophic factor (10 ng/ml; PeproTech), and glial cell–derived growth factor (10 ng/ml; PeproTech) without bFGF, EGF, or heparin. Neurons were observed after several days and were further differentiated for 30 days or longer. Most of the experiments were performed on the neuronal culture differentiated beyond 8 weeks.
miPSCs were differentiated into neurons using a similar protocol as described above. This culture condition generated 92 ± 3% MAP2+ neurons, including 74 ± 4% vGLUT+ total glutamatergic neurons, 32 ± 4% Tbr1+ subclass of glutamatergic neurons, 13 ± 3% GABA+ GABAergic neurons (for apoE3/3-miPSCs; 5 ± 2% for apoE4/4-miPSCs), and 5 ± 2% TH+ dopaminergic neurons. In this neuronal culture, GFAP+ astrocytes were 3 ± 1%. The percentages of these different cell types generated from miPSCs were very similar to those generated from hiPSCs.
Generation of MGE Cells and Maturation into GABAergic Neurons from hiPSCs
hiPSCs were differentiated into MGE progenitors as previously reported with modifications26–28. On day 1 through day 8 of differentiation, MGE spheres were grown in suspension in KSR medium supplemented with Rock Inhibitor (RI, Tocris Bioscience), the TGF-ß receptor inhibitor SB431542 (SB, Stemgent), the Wnt pathway inhibitor Wnt Antagonist II (IWP-II, Millipore), the Hedgehog pathway activator Smoothened Agonist (SAG, Millipore), and the bone morphogenetic protein receptor inhibitor LDN-193189 (LDN, Stemgent). On day 8, MGE spheres were transferred to KSR media supplemented with FGF-8 (PeproTech), IWP-II, SAG, and LDN. On day 21, MGE-progenitors were dissociated with Accutase and plated on culture plates coated with PLL overnight at 37°C and Laminin for ≥ 1hour at 37°C in neural induction medium containing DMEM/F12 and Neurobasal Medium (1:1, Thermo Fisher), N2 Supplement, B27 Supplement (Life Technologies), NEAA, Glutamax, and penicillin/streptomycin supplemented with Brain-Derived Neurotrophic Factor (BDNF, PeproTech), Glial-Derived Neurotrophic Factor (GDNF, PeproTech), the γ secretase inhibitor DAPT (Tocris), the MEK/ERK pathway inhibitory SU5402 (STEMCELL Technologies), and the MEK inhibitor PD0325901 (Tocris). From day 28 on, maturing GABAergic inhibitory interneurons were cultured in neural induction medium containing BDNF and GDNF for up to 5 weeks.
Flow Cytometry Analysis of hiPSC-derived MGE Progenitors and Mature GABAergic neurons
Flow cytometry analysis was performed at Day 35 of GABAergic interneuron differentiation in order to assess the purity of the cell population. Cells were dissociated and fixed with 4% paraformaldehyde and stained with the following antibodies: NKX2.1 (sc-13040, Santa Cruz Biotechnology), and GABA (A2052, Sigma). The secondary antibody was IgG-conjugated Alexa Fluor 488 (Life Technologies). Live and dead cells were identified using the Zombie UV Fixable Viability Kit (Biolegend). Unstained as well as IgG-conjugated Alexa Fluor 488 (Life Technologies) secondary antibody alone was used as a control and to gate positive populations. Cells were analyzed on a BD LSR II Flow Cytometer (BD Biosciences), and data were interpreted with FlowJo Version 10.
Immunocytochemical Analysis of hiPSC-derived Neuronal Cultures
All neural progenitors, including MGE cells, and further differentiated cells, including MGE cell-derived GABAergic neurons, were fixed in 4% paraformaldehyde and stained with the following primary antibodies: nestin (MAB5326, EMD Millipore), Pax6 (Developmental Studies Hybridoma Bank), FoxG1 (sc-48788, Santa Cruz Biotechnology), MAP2 (MAB3418, AB5622, EMD Millipore), Tuj1 (MAB1637, EMD Millipore; MRB-435P, Covance), apoE (178479, Calbiochem), PHF1 (gift from Peter Davies), AT8 (MN1020, Thermo Fisher Scientific), AT180 (MN1040, Thermo Fisher Scientific), Tau5 (577801, EMD Millipore), total-tau (T6402, Sigma), NKX2.1 (sc-13040, Santa Cruz Biotechnology), GABA (A2052, Sigma), and cleaved Caspase-3 (D3E9, Cell Signaling Technology). The secondary antibodies were IgG-conjugated Alexa Fluor 488 or Alexa Fluor 594 (Life Technologies). Nuclei were stained with DAPI. Images were taken with a Leica epifluorescence microscope, a Keyence BZ-9000E fluorescence microscope, or a Leica confocal imaging system.
Western Blot Analysis of hiPSC-derived Neurons
hiPSC-derived neurons, including MGE cell-derived GABAergic neurons, in culture were washed with PBS and collected in the presence of a high-detergent buffer consisting of 50 mM Tris, 150 mM sodium chloride, 2% Nonidet P-40, 1% sodium deoxycholate, 4% sodium dodecyl sulfate, and supplemented with complete protease inhibitor cocktail (Roche), phosphatase inhibitor cocktail 1 (P2850, Sigma), and phosphatase inhibitor cocktail 2 (P5726, Sigma). The total protein in cell lysates was quantitated with the BCA protein assay kit (#23227, Pierce). In some experiments, the media were also collected for analysis of apoE and soluble APP levels by western blot. The samples were separated by SDS-PAGE on 4−20% Bis-Tris polyacrylamide gels (Life Technologies) or Criterion XT 4–12% Bis-Tris gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad). The membranes were then blocked in Odyssey Blocking Buffer (PBS) (LI-COR) and probed with the following primary antibodies: apoE (178479, Calbiochem), actin (A5060, Sigma), Tuj1 (MAB1637, EMD Millipore; MRB-435P, Covance), GFAP (Z0334, Dako), PHF1 (gift from Peter Davies), AT8 (MN1020, Thermo Fisher Scientific), AT180 (MN1040, Thermo Fisher Scientific), AT270 (MN1050, Thermo Fisher Scientific), Tau-5 (577801, EMD Millipore), soluble APP-β (SIG-39138, Covance), 22C11 (MAB348, EMD Millipore), GAD65/67 (AB1511, EMD Millipore), and GAD67 (MAB5406, EMD Millipore). The secondary antibodies were IgG labeled with IRDye 800 or IRDye 680 (LI-COR). The blotted membranes were scanned with an Odyssey CLx Imaging System (LI-COR). Signals were analyzed with Image Studio Lite 4.0 (LI-COR). Unprocessed original scans of blots are shown in Supplementary Figure 15.
Measuring Aβ40 and Aβ42 in hiPSC- and miPSC-derived Neuron Culture Medium
Medium from neuronal cultures was harvested and stored at –80°C. Cell lysates were collected for cellular protein normalization and analysis of p-tau levels by western blot analysis. Human and mouse Aβ peptides were measured with MSD Human V-PLEX Aβ Peptide Panel 1 (6E10) Kit (K15200E, Meso Scale Discovery) and Thermo Fisher Mouse Aβ Peptide ELISA Kits (KMB3481 and KMB3441, Thermo Fisher Scientific) according to the product manuals. For human Aβ measurement, the plates were read with a Sector Imager 2400, and the data were acquired and analyzed with Discovery Workbench software. For mouse Aβ measurement, the plates were read with a FlexStation-III, and the data were analyzed with Prism-6 software. For experiments with secretase inhibitors, neuronal cultures were treated with a γ-secretase inhibitor (compound E, Copd-E, at a final concentration of 200 nM), a β-secretase inhibitor (OM99-2, final concentration of 750 nM), or dimethyl sulfoxide (vehicle) as described10. All inhibitors were from EMD Millipore.
Astrocyte Differentiation of hiPSCs and Treatment of Neurons from ApoE-deficient hiPSCs with Conditioned Media from Astrocyte Culture
hiPSCs were differentiated into astrocyte progenitors and astrocytes as previously reported with some modifications59. The astrocyte progenitors were generated using the procedure as described for neuronal differentiation of hiPSCs. Instead, these progenitor spheres were kept in suspension for further expansion in the presence of 10 ng/ml bFGF (PeproTech) and 10 ng/mL EGF (PeproTech), and passaged by disaggregating into small clusters with a Pasteur pipette for around 90 days. For astroglial differentiation, progenitor spheres were dissociated with accutase to single cells, and seeded onto the plates coated with PLL and laminin in Neural Differentiation Medium without bFGF and EGF. The astrocyte conditioned media (ACM) were collected from the astrocyte culture, and the apoE levels of these media were measured with Human ApoE (AD2) ELISA kit (Cat # EHAPOE, Thermo Scientific). The amount of ACM for treatment of the neurons derived from apoE-deficient hiPSCs were adjusted to the same apoE concentration (0.35 nM or 1.47 nM), based on apoE levels in the ACM collected from apoE3/3 or apoE4/4 astrocyte culture. After ACM treatment for 1 week, the media and cell lysates of neurons derived from apoE-deficient hiPSCs were collected and further analyzed.
Treatment of hiPSC-derived Neurons with a Small-molecule Structure Corrector
Neuronal cultures derived from hiPSC lines were treated with a small-molecule structure corrector, PH002 (final concentration, 100 nM; prepared in dimethyl sulfoxide) as described36–38. After 7 days of treatment, culture medium was collected for measurement of Aβ40 and Aβ42, and cells were homogenized for western blot analyses. For the dose-effect experiments, different doses of PH002 (i.e., 0 nM, 10 nM, 30 nM, and 100 nM) were used to treat the culture neurons for 1 week.
Treatment of hiPSC-derived Neurons with Recombinant Human ApoE
Neuronal cultures derived from apoE−/− hiPSCs were treated with purified recombinant human apoE3 or apoE4 (final concentration, 220 nM or 7.5 µg/ml). After 2 days of treatment with recombinant human apoE, culture medium was collected for measurement of Aβ40 and Aβ42, and cells were homogenized for protein normalization and western blot analysis.
Statistical Analyses
Power analysis of a pilot cohort (n = 3) showed that a sample size of three would be sufficient to show genotype effects; all groups have n ≥3. Values are expressed as mean ± SEM. All n numbers represent the numbers of biological replicates or fields used for image analysis. The distribution of data was assessed with the Shapiro-Wilk normality test; most of the data were normally distributed. The variances between groups were similar as shown by Bartlett’s test. Statistical significance was calculated with GraphPad Prism (GraphPad Software). Differences between groups were determined with the unpaired two-sided t test. For multiple comparisons, one-way analysis of variance (ANOVA) with Tukey’s post-test was used. P < 0.05 was considered significant. Researchers were not blinded to genotypes during experiments.
Compliance with Relevant Ethical Regulations and Animal Use Guidelines
All experimental and animal protocols and procedures comply with the university and institutional ethical regulations and animal use guidelines.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary information file) and are available at the corresponding author’s lab.
Supplementary Material
Acknowledgments
This work was supported by grants AG048030, AG048017, and AG056305 to Y.H., AG023501 to B.L.M. and Y.H. from the National Institutes of Health and grants RN2-00952 and TRAN1-09394 to Y.H. from the California Institute for Regenerative Medicine and by a gift from the Roddenberry Foundation. C.W. was partially supported by a fellowship from the California Institute for Regenerative Medicine. PHF1 antibody was from Dr. Peter Davies at Albert Einstein College of Medicine. We thank Anna Karydas for assistance with skin biopsies and Stephen Ordway and Theodora Pak for editorial assistance.
Footnotes
AUTHOR CONTRIBUTIONS
C.W. and Y.H. designed and coordinated the study. C.W. did most of the studies and data analyses. R.N. performed all MGE studies and related data analysis. Q.X. and D.J. helped with off-target analysis of gene-editing and apoE lentiviral vectors and performed some western blots. D.W. designed and prepared the apoE3 donor DNA for gene editing. D.W. and S.Y.Y. helped with genetic screening. M.E.B. helped with the cell culture. H.Y. helped with MGE studies and image collection. G.L. helped with miPSC studies. Z.A.M., B.L.M. and M.J.M. provided apoE-null human skin biopsy. C.W. and Y.H. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
Y.H. is a cofounder and scientific advisory board member of E-Scape Bio, Inc. Other authors declare no competing financial interests.
References
- 1.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golde TE, Schneider LS, Koo EH. Anti-Aβ therapeutics in Alzheimer's disease: the need for a paradigm shift. Neuron. 2011;69:203–213. doi: 10.1016/j.neuron.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corder E, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 4.Saunders AM, et al. Association of apolipoprotein E allele e4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 5.Farrer LA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. J. Am. Med. Assoc. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 6.Genin E, et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Mol. Psychiatry. 2011;16:903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y. Aβ-independent roles of apolipoprotein E4 in the pathogenesis of Alzheimer's disease. Trends Mol Med. 2010;16:287–294. doi: 10.1016/j.molmed.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Mahley RW, Huang Y. Apolipoprotein E sets the stage: response to injury triggers neuropathology. Neuron. 2012;76:871–885. doi: 10.1016/j.neuron.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashe KH, Zahs KR. Probing the biology of Alzheimer's disease in mice. Neuron. 2010;66:631–645. doi: 10.1016/j.neuron.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Israel MA, et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yahata N, et al. Anti-Aβ drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer's disease. PLoS ONE. 2011;6:e25788. doi: 10.1371/journal.pone.0025788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yagi T, et al. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 13.Almeida S, et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012;2:789–798. doi: 10.1016/j.celrep.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo T, et al. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, et al. Effect of potent γ-secretase modulator in human neurons derived from multiple presenillin 1-induced pluripotent stem cell mutant carriers. JAMA Neurol. 2014 doi: 10.1001/jamaneurol.2014.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Brecht WJ, et al. Neuron-specific apolipoprotein E4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J. Neurosci. 2004;24:2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris FM, et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc. Natl. Acad. Sci. USA. 2003;100:10966–10971. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoover BR, et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron. 2010;68:1067–1081. doi: 10.1016/j.neuron.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thies E, Mandelkow EM. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J Neurosci. 2007;27:2896–2907. doi: 10.1523/JNEUROSCI.4674-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodruff G, et al. The presenilin-1 ΔE9 mutation results in reduced γ-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep. 2013;5:974–985. doi: 10.1016/j.celrep.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellano JM, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci. Transl. Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busciglio J, Lorenzo A, Yeh J, Yankner BA. beta-amyloid fibrils induce tau phosphorylation and loss of microtubule binding. Neuron. 1995;14:879–888. doi: 10.1016/0896-6273(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 25.Jin M, et al. Soluble amyloid beta-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci USA. 2011;108:5819–5824. doi: 10.1073/pnas.1017033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham M, et al. hPSC-derived maturing GABAergic interneurons ameliorate seizures and abnormal behavior in epileptic mice. Cell Stem Cell. 2014;15:559–573. doi: 10.1016/j.stem.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholas CR, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn S, Kim TG, Kim KS, Chung S. Differentiation of human pluripotent stem cells into Medial Ganglionic Eminence vs. Caudal Ganglionic Eminence cells. Methods. 2016;101:103–112. doi: 10.1016/j.ymeth.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong H, et al. Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells. Stem Cell Reports. 2013;1:226–234. doi: 10.1016/j.stemcr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak AC, et al. Effects of the absence of apolipoprotein e on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol. 2014;71:1228–1236. doi: 10.1001/jamaneurol.2014.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Q, et al. Profile and regulation of apolipoprotein E (apoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the apoE locus. J. Neurosci. 2006;26:4985–4994. doi: 10.1523/JNEUROSCI.5476-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Q, et al. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J. Neurosci. 2008;28:1452–1459. doi: 10.1523/JNEUROSCI.3253-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu P-T, et al. Specific regional transcription of apolipoprotein E in human brain neurons. Am. J. Pathol. 1999;154:601–611. doi: 10.1016/S0002-9440(10)65305-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulrich JD, et al. In vivo measurement of apolipoprotein E from the brain interstitial fluid using microdialysis. Mol Neurodegener. 2013;8:13. doi: 10.1186/1750-1326-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: A causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen HK, et al. Small-molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J. Biol. Chem. 2012;287:5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodbeck J, et al. Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J. Biol. Chem. 2011;286:17217–17226. doi: 10.1074/jbc.M110.217380. J. Biol. Chem. 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahley RW, Huang Y. Small-molecule structure correctors target abnormal protein structure and function: Structure corrector rescue of apolipoprotein E4-associated neuropathology. J. Med. Chem. 2012;55:8997–9008. doi: 10.1021/jm3008618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bien-Ly N, Gillespie AK, Walker D, Yoon SY, Huang Y. Reducing human apolipoprotein E levels attenuates age-dependent Aβ accumulation in mutant human amyloid precursor protein transgenic mice. J. Neurosci. 2012;32:4803–4811. doi: 10.1523/JNEUROSCI.0033-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huynh TV, et al. Age-Dependent Effects of apoE Reduction Using Antisense Oligonucleotides in a Model of β-amyloidosis. Neuron. 2017;96:1013–1023. doi: 10.1016/j.neuron.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu CC, et al. ApoE4 Accelerates Early Seeding of Amyloid Pathology. Neuron. 2017;96:1024–1032. doi: 10.1016/j.neuron.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang YA, Zhou B, Wernig M, Südhof TC. ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Aβ Secretion. Cell. 2017;168:427–441. doi: 10.1016/j.cell.2016.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamenetz F, et al. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- 45.Cirrito JR, et al. Synaptic activity regulates interstitial fluid amyloid-β levels in vivo. Neuron. 2005;48:913–922. doi: 10.1016/j.neuron.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 46.LaDu MJ, et al. Self-Assembly of HEK Cell-Secreted ApoE Particles Resemble ApoE-Enrichment of Lipoproteins as a Ligand for the LDL Receptor-Related Protein. Biochemistry. 2006;45:381–390. doi: 10.1021/bi051765s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, et al. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549:523–527. doi: 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrews-Zwilling Y, et al. Apolipoprotein E4 causes age- and Tau-dependent impairment of GABAergic interneurons, leading to learning and memory deficits in mice. J. Neurosci. 2010;30:13707–13717. doi: 10.1523/JNEUROSCI.4040-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung L, et al. Apolipoprotein E4 causes age- and sex-dependent impairments of hilar GABAergic interneurons and learning and memory deficits in mice. PLoS ONE. 2012;7:e53569. doi: 10.1371/journal.pone.0053569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li G, et al. GABAergic interneuron dysfunction impairs hippocampal neurogenesis in adult apolipoprotein E4 knockin mice. Cell Stem Cell. 2009;5:634–645. doi: 10.1016/j.stem.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong LM, et al. Inhibitory interneuron progenitor transplantation restores normal learning and memory in apoE4 knock-in mice without or with Aβ accumulation. J Neurosci. 2014;34:9506–9515. doi: 10.1523/JNEUROSCI.0693-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nuriel T, et al. Neuronal hyperactivity due to loss of inhibitory tone in APOE4 mice lacking Alzheimer's disease-like pathology. Nat Commun. 2017;8:1464. doi: 10.1038/s41467-017-01444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knoferle J, et al. Apolipoprotein E4 produced in GABAergic interneurons causes learning and memory deficits in mice. J Neurosci. 2014;34:14069–14078. doi: 10.1523/JNEUROSCI.2281-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillespie AK, et al. Apolipoprotein E4 causes age-dependent disruption of slow gamma oscillations during hippocampal sharp-wave ripples. Neuron. 2016;90:740–751. doi: 10.1016/j.neuron.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 57.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibiton of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu BY, Zhang SC. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat. Protoc. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krencik R, Weick JP, Liu Y, Zhang ZJ, Zhang SC. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary information file) and are available at the corresponding author’s lab.