Abstract
Mitochondrial ATP synthase comprises a membrane embedded Fo motor that rotates to drive ATP synthesis in the F1 subunit. We used single-particle cryo-EM to obtain structures of the full complex in a lipid bilayer in the absence or presence of the inhibitor oligomycin, at 3.6 Å and 3.8 Å resolution, respectively. To limit conformational heterogeneity, we locked the rotor in a single conformation by fusing the F6 subunit of the stator with the δ-subunit of the rotor. Assembly of the enzyme with the F6-δ fusion caused a twisting of the rotor and a 9° rotation of the Fo c10-ring in the direction of ATP synthesis, relative to the structure of isolated Fo. Our cryo-EM structures show how F1 and Fo are coupled, give insight into the proton translocation pathway and show how oligomycin blocks ATP synthesis.
The mitochondrial ATP synthase is composed of two distinct molecular motors, F1 and Fo (Fig. 1 and Movie 1). F1 includes three catalytic subunits around a central rotor that rotates to effect ATP synthesis. Fo is a transmembrane proton turbine that includes the c10-ring, which rotates in steps of 36° as it moves protons from the cytosol to the matrix space, down an electrochemical gradient (1). The c10-ring in Fo is physically coupled to the rotor in F1 and therefore proton translocation drives the synthesis of ATP. F1 is held in place by a peripheral stator that prevents the rotation of the body of F1 and restricts the wobbling of its central rotor relative to the c10-ring. Relative to the stator, the F1 rotor can be in 3 distinct positions (rotomers) while a revolution of the c10-ring involves 10 discrete steps. This combination of 120° steps of the rotor and 36° steps of the c10-ring results in multiple conformations of the ATP synthase during the reaction cycle. Further, the ATP synthase can form homo-dimers (2–4), which further increases the number of possible conformations of the enzyme complex, making the analysis of the reaction mechanism on a molecular level challenging. Here, we have employed a genetic method to restrict the number of conformations allowing us to study the monomeric form of the yeast ATP synthase. We reconstituted the monomeric enzyme complex into a lipid bilayer formed in nanodiscs to enable structural analysis under near native conditions.
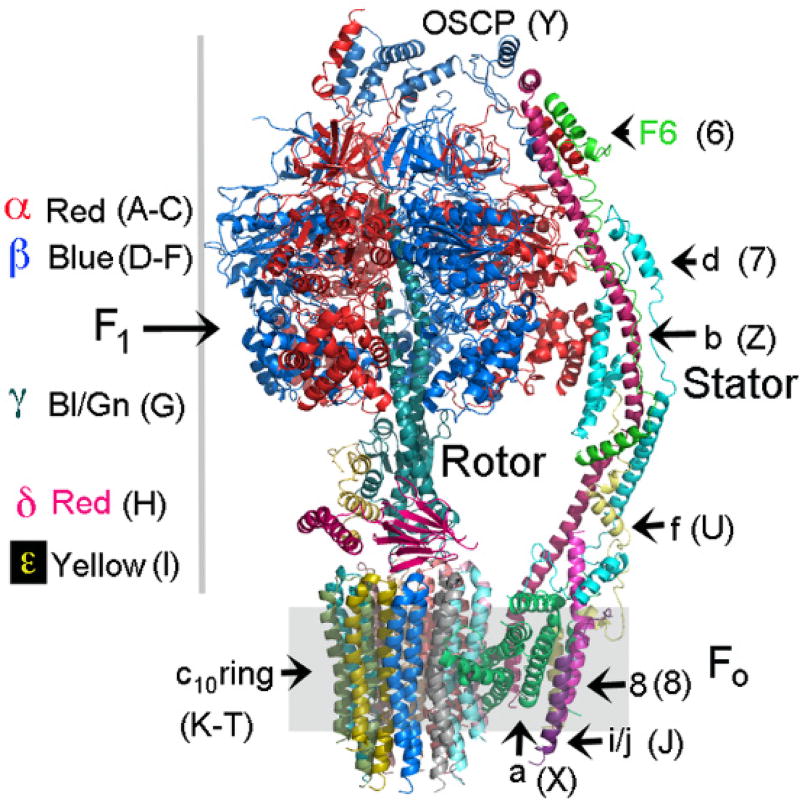
Fig. 1. Subunit composition and architecture of the monomeric yeast ATP synthase.
The subunits are in a variety of colors to allow identification. The chain number or letter used in the pdb is shown in the brackets. There are 3 α- (red) and 3 β-subunits (blue) that with the γ-, δ-, and ε-subunits form F1. The rotor is comprised of the γ-, δ- and ε-subunits of F1. The core of Fo is comprised of the a-subunit and the c10-ring. The subunit of the stator are as shown.
To restrict the number of conformations, we genetically fused subunit F6 of the stator, to the δ-subunit of the rotor. The subunits were linked by T4 lysozyme giving a final fusion of H2N-F6-lysozyme-δ (fig. S1). The mitochondrial leader peptide from the β-subunit (ATP2) was added to direct the import of the protein into the mitochondria and the expression was controlled with the ATP2 transcriptional elements. The plasmid containing this fusion was integrated into the genome in a strain that was deleted in the genes encoding F6 (ATP14) and the δ-subunit (ATP16). The ATP2 gene was also deleted in the yeast strain, but this was complemented by integration of a vector into the genome containing the β-subunit with a His6-tag on the amino terminus allowing rapid purification of the enzyme.
We performed single-particle cryo-EM analysis on the nanodisc-reconstituted ATP synthase with and without the inhibitor oligomycin. Three preparations of nanodiscs (5) were used differing in their lipid content (Methods). Overall, the cryo-EM density which gave rise to the model of the F1Fo ATP synthase was more complete under the conditions with oligomycin than without, but the structures were similar. Thus we will confine the discussion of the overall structure to the structure with bound oligomycin. To allow analysis of the proton translocation mechanism, the refinement was focused on Fo, which improved resolution of this region but also resulted in poorer resolution of the other parts of the enzyme. For the Fo regions we compare the structures with and without bound oligomycin.
Overall structure of the ATP synthase
The cryo-EM structures of the yeast ATP synthase in nanodisc were determined to overall resolutions of 3.6 Å and 3.8 Å for the entire enzyme complex without and with oligomycin, respectively (see Fig. 2, Table 1, and figs. S1 to S5). The EM density for Fo is weaker and of lower resolution than that of F1, likely owing to flexibility between these domains. Hence, Fo-focused 3D classification and refinement improved the resolution and map quality. In the presence of oligomycin, the flexibility of the ATP synthase was reduced resulting in better EM density for Fo and the stator, and a greater number of residues in Fo with well-defined side chain densities. The resolution varied between subunits and within subunits. Most residues in the F1 subunits display excellent side chain density, while the stator and Fo subunits showed varying resolutions. Many good side chain densities could be seen for most of the components present in the cryo-EM structures, including the central stalk and c10-ring. The secondary structural elements are also well resolved for the stator components. We were able to trace 27 chains in the density (Fig. 3A, Movie 2, and Table 2). The only chains that we did not see are those involved in dimerization of the ATP synthase (as this is the monomer form). The density for the T4 lysozyme molecule used to fuse F6 with δ is seen in one of the cryo-EM maps from 3D classification (fig. S3C). Low-resolution density for the lipid nanodisc clearly delineates the membrane embedded regions of Fo (Fig. 2).
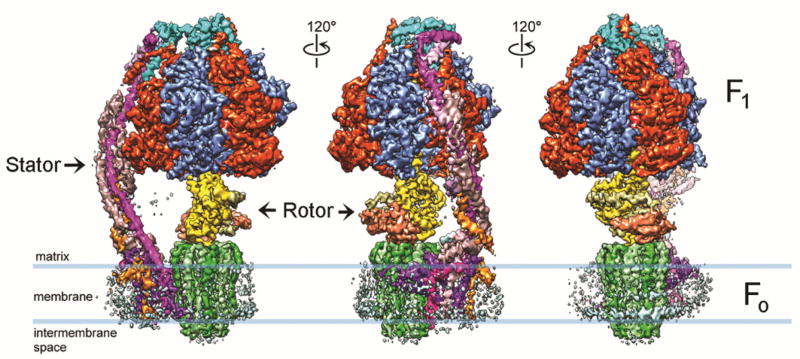
Fig. 2. Cryo-EM 3D reconstruction of nanodisc-embedded yeast ATP synthase bound with oligomycin.
Three side views of the 3D reconstruction filtered to 3.8 Å resolution and rotated by 120° around the y-axis. All subunits are uniquely colored. Blue lines indicate the membrane boundaries as observed by the EM density of the lipid nanodisc.
Table 1.
Statistics of the cryo-EM structures presented in this study.
| Category | ATP synthase | ATP synthase with oligomycin | ||
|---|---|---|---|---|
| Cryo-EM data collection and processing | ||||
| Electron microscope | Polara | Polara | ||
| Voltage (kV) | 300 | 300 | ||
| Electron dose (e−/Å2) | 41 | 41 | ||
| Physical pixel (Å) | 1.23 | 1.23 | ||
| Number of movies | 5,935 | 2,896 | ||
| Number of particles | 541,568 | 346,399 | ||
| F1Fo | Fo | F1Fo | Fo | |
| Number of particles for final map | 160,937 | 109,206 | 104,280 | 104,280 |
| Resolution (Å) | 3.6 | 4.1 | 3.8 | 4.2 |
| Map B-factor (Å2) | −100 | −180 | −100 | −200 |
| Model refinement | ||||
| Number of protein residues | 5094 | 1224 | 5094 | 1224 |
| Number of side chains | 5061 | 1198 | 5061 | 1199 |
| Number of atoms | 38814 | 9179 | 38814 | 9434 |
| Geometric parameters (r.m.s.d.) | ||||
| Bond length (Å) | 0.007 | 0.008 | 0.009 | 0.003 |
| Bond angle | 1.026 | 1.309 | 1.119 | 1.354 |
| Ramachandran statistics | ||||
| Residues favored (%) | 93.87 | 92.95 | 92.18 | 92.95 |
| Residues allowed (%) | 5.93 | 6.59 | 7.62 | 7.05 |
| Residues disallowed (%) | 0.20 | 0.46 | 0.20 | 0 |
| Rotamer outliers (%) | 0.15 | 3.23 | 0.36 | 0.21 |
| MolProbity Score | 1.89 | 2.57 | 1.91 | 2.56 |
| EMRinger Score | 1.61 | 0.94 | 1.55 | 0.91 |
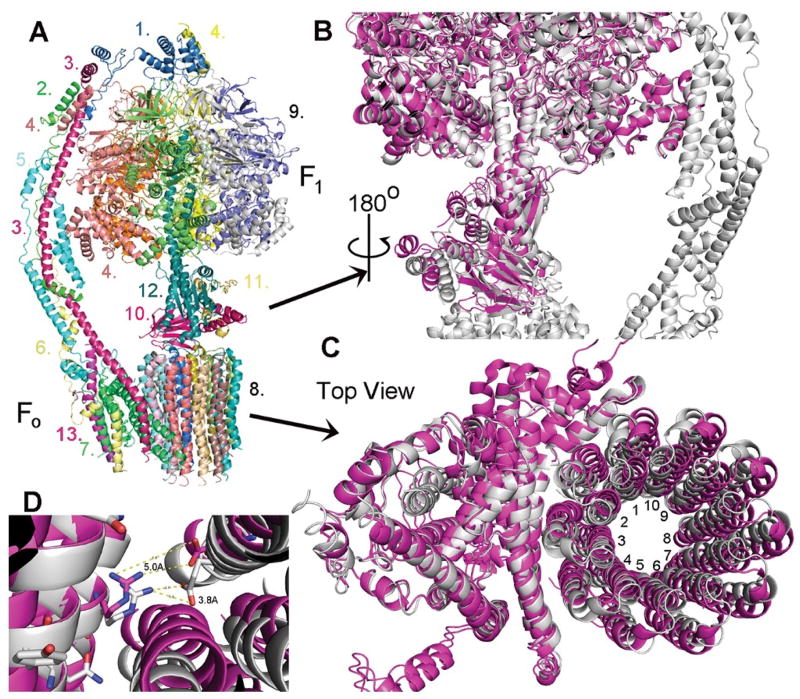
Fig. 3. Overall structure of the yeast ATP synthase.
(A) Molecular model of the F1Fo ATP synthase based on the cryo-EM density map. The subunits are color coded and labeled as: 1. OSCP, 2. F6 3. b-subunit, 4. α-subunit, 5. d-subunit, 6. f-subunit, 7. a-subunit, 8. c10-subunit ring, 9. α3β3 core, 10. δ-subunit, 11. ε-subunit, 12. γ-subunit, 13, i/j-subunit. (B) Superimposition of the crystal structure of the yeast F1 domain (magenta) onto the cryo-EM structure of yeast F1Fo ATP synthase (grey). Three rotor subunits (γ-, δ-, ε-subunits) are displaced by a twisting in the counter clockwise direction. (C) Superimposition of the cryo-EM structure of yeast Fo (magenta, in the absence of F1) on the current structure of F1Fo (grey). The c10-ring is rotated by about 9° in the counter clockwise direction (the direction of ATP synthesis). (D) Relative position of aArg176 and the nearest cGlu59 in the structure of yeast Fo (magenta) and F1Fo (grey). The distance between the side chains of aArg176 and cGlu59 is reduced from about 5.0 Å to about 3.8 Å with the rotation of the c10-ring by 9°.
Table 2.
Summary of subunit composition in this model, chains names, and residues in the model. The DP, TP, and E sites in F1 are composed of chains C and D, B and F, and A and E, respectively.
| Subunit | Alias | Residues | MW (K Da) | Role | Sector | Chain | Modeled |
|---|---|---|---|---|---|---|---|
| α | 510 | 54.9 | catalytic | F1 | A, B, C | 4/6–510 | |
| β | 478 | 51.1 | catalytic | F1 | D, E, F | 6/7–478 | |
| γ | 278 | 30.6 | catalytic | F1 | G | 1–278 | |
| δ | 138 | 14.6 | catalytic | F1 | H | 7–138 | |
| ε | 61 | 6.1 | catalytic | F1 | I | 1–59 | |
| OSCP | sub 5 | 195 | 20.9 | structural | stator | Y | 7–172 |
| sub a | sub 6 | 249 | 25.1 | H+ trans | Fo | X | 26–249 |
| sub b | sub 4 | 209 | 23.3 | Dual | Fo | Z | 53–207 |
| sub c | sub 9 | 76 | 7.76 | H+ trans | Fo | K-T | 1–75/76 |
| sub d | sub 7 | 173 | 19.7 | structural | stator | 7 | 3–173 |
| sub f | 95 | 10.6 | Dual | Fo | U | 1–85 | |
| sub i | sub j | 59 | 6.7 | H+ trans | Fo | J | 1–37 |
| F6 | 92 | 10.4 | structural | stator | 6 | 4–92 | |
| sub 8 | A6L | 48 | 5.8 | structural | stator | 8 | 7–48 |
There are 3 catalytic sites in F1, each of which cycles between 3 states. In the initial structure of the bovine F1 ATPase, the sites were occupied with ADP, ATP, and one site was empty and thus they were named DP, TP, and E (Table 2) (6). The asymmetry of the catalytic sites is determined by the relative position of the γ-subunit within F1. In our cryo-EM structures, the rotor is in a single orientation and the asymmetry of the catalytic sites is conserved. In this orientation, the stator bridges Fo with F1 along the non-catalytic site formed by chains B (αTP) and E (βDP). There is contact between subunit d of the stator with αTP (Chain B).
The F1 domain
The conformation of the F1 domain in the cryo-EM structures is nearly identical to that observed in the crystal structures of yeast F1 (7) or yeast F1-c10 (8, 9) which also revealed the 3 distinct states of the catalytic sites, DP, TP, and E. When we superimpose yeast F1 onto the structure of yeast F1Fo, we see that the rotor formed by γ-, δ-, and ε-subunits is twisted by about 9° in the direction of the rotation that leads to ATP synthesis (Fig. 3B). The twisting starts at about γ-Ile271, which is in direct contact with the collar formed by the α- and β-subunits (6). At the bottom of the rotor, this twisting results in a displacement of about 5.6 Å in residues in the δ-subunit. This displacement continues into the c10-ring and causes a commensurate rotation of the c10-ring relative to a-subunit (Fig. 3C). While we believe that the constraint of the central stalk with the stator can be a mimic for twisting of the central stalk during the reaction cycle, this is a first approximation and has not been proven.
The stator domain
The stator (peripheral stalk) is composed of subunits OSCP, b, d, F6, f, i/j, and 8. Subunits OSCP, b, d, f, and F6, primarily serve as structural components of the stator. However, subunits b, d, f, i/j, and 8 are either embedded or attached to the membrane, are part of the complex purified as Fo, and may have a role in proton movement in addition to a structural role in forming the stator. Subunits b and d are similar to their homologs in the crystal structure of bovine subunits F6, b, and d (PDB, 2CLY) (10), however yeast F6 displays differences from bovine F6 including an additional helix (residues 76–91) at the C terminus, which adds additional interactions with the b- and d-subunits. Modeling of the domains of stator subunits that are part of Fo and embedded or bound to the membrane was aided by the high resolution cryo-EM structure of yeast Fo (PDB, 6B2Z) (11) and we did not see any major differences as compared to the reported structure.
The OSCP subunit anchors the stator at the top of F1. As partially seen in the structure of the enzyme from Pichia angusta (12), the N terminus of the three α-subunits interact to stabilize the binding of OSCP to the top of F1 (fig. S7). The helix in αTP (Chain B) formed from BLys4 to BAsn26 interacts with helices from F6 and the b-subunit (fig. S7, region I). Further the 3 helix bundle formed by residues B462-B509 interact with 2 helices of the d-chain from residues 3–48 and a cluster of residues in OSCP ranging from 160–168, but also, residue 122 (Region I). Residues in a random coil from αDP (Chain C, Gln6 to Ser22) run under and interact with OSCP (fig. S7, region III). The helix formed from residues Lys4 to Lys 19 in αE (Chain A) interacts with a helix and turn in OSCP formed by residues Thr46 to His78 (fig. S7, region IV). Thus, the N terminus of each α-subunit makes unique contributions to the stabilization of the anchoring of OSCP to the top of F1.
The Fo domain
Fo is embedded in the inner membrane of the mitochondria. Our cryo-EM map showed clear density for the lipid bilayer in the nanodiscs (Figs. 1 and 4A). The cryo-EM density indicates that the bilayer spans about 37 Å, approximately from Pro49 to Phe74 on the outer helices of the c-subunits. Phe74 is the last residue in helix 2 (residues 42–74) and is considerably shorter than helix 1 (residues 1–40).
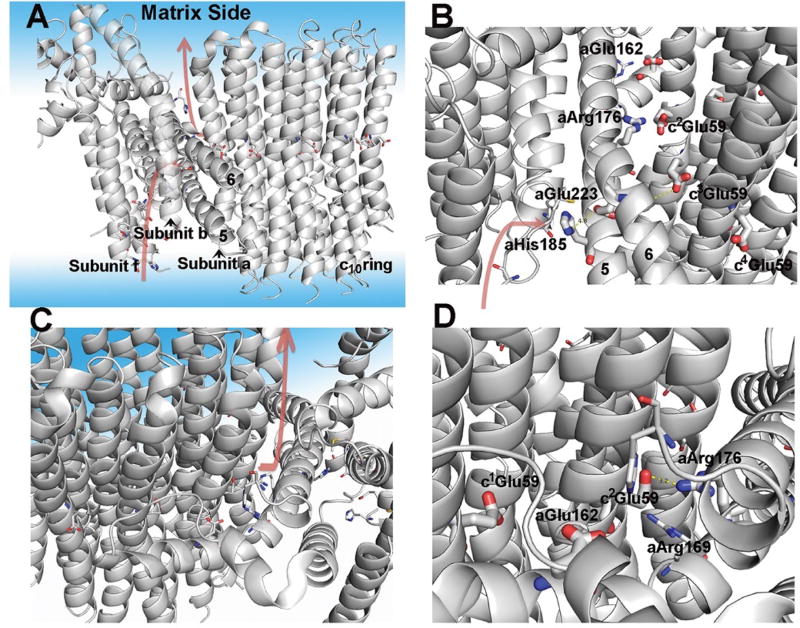
Fig. 4. Model of Fo and the proton pathways.
(A) Overall model of Fo with subunits a, b, c, and f displayed. The aqueous phase is also displayed with a light blue coloring. The postulated proton pathways for the entry from the intermembrane space and exit to the matrix are shown with the arrows. The protonation pathway during ATP synthesis is a path formed by subunits f, b, and a with the final course formed by helices 5 and 6 of a-subunit. (B) Side view of the entry pathway for protons during ATP synthesis with key residues indicated (see text for discussion). (C) Top side view of proton pathway for proton exit to the matrix. (D) View of proton pathway from the exit site showing helices 5 and 6 of a-subunit and the c-ring.
The a-subunit is adjacent to the c10-ring and contributes to form the proton conduction pathway from the cytosolic side of the mitochondria to protonation sites in the c10-ring and from these sites to the matrix space (13–15). The protonation and deprotonation events at the interface between the a-subunit and the c10-ring couple the translocation of protons to the rotation of the c10-ring and thus effect ATP synthesis. We obtained the near-atomic resolution Fo maps for the enzyme without bound oligomycin and with oligomycin, and the structural models from these maps are nearly identical. This suggests that oligomycin binding does not alter the overall structure of Fo.
The structure of the c10-ring is nearly identical to that observed in a crystal structure of the isolated ring (16). (We have numbered the c-subunits relative to the a-subunit with c1Glu59 corresponding to the first c-subunit with interactions with the a-subunit. The c-subunits are numbered sequentially in the direction of rotation during ATP synthesis. cf, Fig. 3C) Glu59 is the proton acceptor and donor that is responsible for net proton movement during the catalytic cycle. The side chain of Glu59 has been shown to be in one of two conformations: a closed conformation that was suggested to represent the protonated form when in the membrane, and an “open” conformation that was proposed to be present only at the a/c interface and represented the state in which protonation and deprotonation occurs (16, 17). We see 5 instances where Glu59 is in the “open” conformation in the membrane phase suggesting that the “closed” conformation is not a necessary conformation of the protonated side-chain in the membrane phase (fig. S8).
The structure of dimeric yeast Fo was solved by cryo-EM after stripping off F1 using NaBr (11). This structure likely represents the ground state, as it is free from strain caused by assembly of the F1Fo ATP synthase complex. We superimposed the Fo region from the ground state with the Fo region from our native structures using the a- and b-subunits as the anchor (Fig. 3C). This analysis indicates that the c10-ring of the native structure, relative to the a-subunit, is rotated about 9° in the direction of ATP synthesis, as compared to the ground state. This is consistent with the rotation that we observed in the F1 rotor, when comparing our structure to that of F1 alone.
Arg176 of a-subunit is the only residue that is strictly conserved (13) (fig. S9). It has been proposed that Arg176 prevents the short circuiting of protons in the proton pathway (18) as well as acting as a positive pole for attraction of the charged Glu59 (14). In the isolated Fo structure (11) (which we are assigning as the “ground state” structure), atoms in the corresponding side chains of Arg176 and the closest Glu59 (c2Glu59) are separated by 5–7.5 Å, while in this structure, the side-chains move closer to within 3.8 Å (Fig. 3D). Extrapolation on the movement of the c10-ring in the direction of ATP synthesis would predict an even closer configuration of Arg176 and Glu59, thereby creating a potential contributing force for rotation of the c10-ring. This attractive force will be much reduced once the ionized cGlu59 is protonated.
The proton pathway
During ATP synthesis, protons transit from the cytosolic side of the mitochondrial membrane to protonate the c-subunit Glu59 closest to Arg176 (c2Glu59), thereby releasing this interaction. Protons must be released from c1Glu59 into the matrix space (Fig. 4A), although not necessarily in concert with protonation, enabling this side-chain to engage Arg176 following a 36° step. The hydrophilic spaces that provide pathways for protons from the cytosol and then to the matrix space, allowing protonation and later deprotonation of cGlu59, are referred to as half-channels. Figure 4 and Movie 3 show the putative” half-channels” from the cytoplasm and to the matrix. Their location and identification is consistent with recent reports (11, 19–21). The matrix side half-channel is most obvious at the a/c interface, where the side chains of a number of residues appear to form a hydrophilic cavity that extends from c1Glu59 to the surface of the membrane phase (Fig. 4, A, C, and D). aGlu162 is an important, but not essential, residue in this pathway (13). The matrix half-channel extends up to the point where we have determined to be the edge of the membrane. In contrast, the cytoplasmic half-channel is formed by residues in the f-, b-, and a-subunits. The proton conduction pathway appears to include aHis185 and aGlu223 (Fig. 4, A and B). The proton conduction pathway proceeds between helices 5 and 6 of the a-subunit. aHis185 and aGlu223 are interchanged in the enzymes from other species, but remain in comparable positions (fig. S9). The protonation of cGlu59 likely occurs in the transition from c2Glu59 to c3Glu59 and in doing so, eliminates any charge interaction between aArg176 and cGlu59.
Possible role of aGlu162 and aGlu223
A requirement for a proton donor/acceptor in the reaction pathway is that the value of the pKa of the donor and acceptor should be around the pH of the medium. The pH of the cytosol is around 7.0 while that of the matrix space during active ATP synthesis is around 8.0. Thus, the pKa of the carboxyl of cGlu59 should be around 7.0 at the a-subunit interface to be an effective proton carrier. If the pKa of the carboxyl was the standard value in water, 5.0, then the carboxyl would rarely be in the protonated state and rotation would be severely impeded during ATP synthesis. While a low pKa would allow the release of the proton to the matrix space during ATP synthesis, it would impede the reversal of the ATP synthase, ATP hydrolysis, which is a fundamental feature of the enzyme. Thus, the pKa of the cGlu59 must be around 7.0 at both the sites for protonation and deprotonation to occur in during ATP synthesis and hydrolysis. While there is a report that the pKa of the c-ring carboxylate is 7.0 (22), these measurements were not made under native conditions nor measured in the intact enzyme. Based on the structure we suggest that aGlu162 and aGlu223 may play a role in shifting the pKa of cGlu59. aGlu162 is highly conserved across species. The corresponding residue in Mycobacteria is aGln, but just one turn away is a pair of Glu residues, thus functionally conserving the role of aGlu162 in the Mycobacteria enzyme (fig. S9). The cGlu59 carboxyl nearest to aGlu162 (c1Glu59) (Fig. 4B) is thus likely to be the proton-releasing site. Rotation of the c10-ring by about 9° places c1Glu59 about 4Å from aGlu162 (fig. S10). On the cytosolic side, aGlu223 is in a dyad interaction with aHis185; this pair is nearly strictly conserved and likely to serve as an intermediate proton-binding site (fig. S9). Rotation of the c10-ring by about 27°, facilitated by the deprotonation of c1Glu59, could bring c2Glu59 close enough to interact with aGlu233 with multiple bridging water molecules. These interactions of aGlu162 and aGlu223 with the side chain of cGlu59 have the potential to shift the pKa of the side chain cGlu59 up to around 7.0 where it needs to be to act as an effective proton transfer group. There is precedence for Glu-Glu interactions and His-Glu interactions to altering the pKa of carboxylates to around 7.0 (23–26). This hypothesis is consistent with biochemical data where replacements at these two positions alter the magnitude of the potential gradient that the enzyme can create with the hydrolysis of ATP (13, 27, 28). Of course, this is still hypothetical and proof will require the structure determination of multiple intermediate states of the reaction cycle.
Inhibition by oligomycin
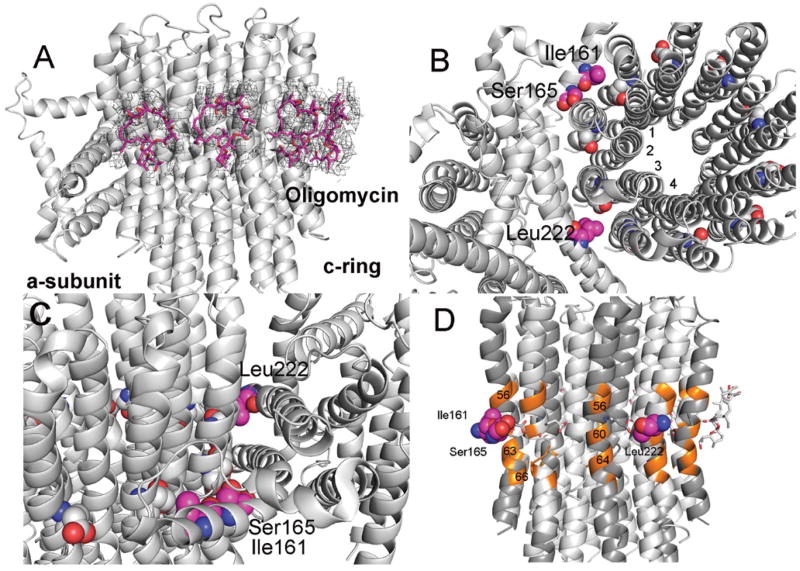
The crystal structure of the isolated yeast c10-ring with bound oligomycin has been determined at 1.9Å (4F4S) (29). Oligomycin was shown to bind to the c10-ring with the inhibitor spanning the outer helices of two adjacent c-subunits and centered over cGlu59. It has been unclear whether oligomycin would bind to the c10-ring in a lipid environment or to alternate sites in the ATP synthase. The cryo-EM analysis here shows densities at 4 sites on the c10-ring (spanning c5c6, c6c7, c7c8, and c8c9) positioned in the membrane phase and at the same position where oligomycin (oligo1-4) was seen to bind in the crystal structure (29) (Fig. 5A). The density map for oligo1-4 suggests that there is some variation in the binding strength at each site. This may reflect slight differences in the conformation of the main and side chains at each binding site. There is also weak density at c4c5, which is at the edge of the a/c interface, but the best fit to the density with an oligomycin molecule (oligo5) places oligomycin in a much different binding mode as compared to those in the other sites (fig. S11). Further, the conformation of oligo5 as modeled is largely different from that observed previously. Likely, the weak density attributed to oligo5 is due in part to surrounding lipid molecules. Thus we believe that this, at best, represents a minor binding mode.
Fig. 5. Inhibition of the ATP synthase by oligomycin.
(A) Model of oligomycin bound to the Fo. The electron density (shown at 4.5σ) fitted with a model of an oligomycin molecule (magenta) at 4 super-imposable positions on the c10-ring. (B) Positions of residues in a-subunit at which replacements can confer resistance to oligomycin. The three residues, Ile161, Ser165, and Leu222, in a-subunit are positioned at the interface between subunits c1c2, and c3c4 and directly in line with c-Glu59 (Fig. 5, C and D). This position is precisely where oligomycin binds when the site is exposed to the membrane bilayer. (C) Side view of the residues in a-subunit. (D) Positions of residues in c-subunits that interact with oligomycin. The c-subunit residues colored orange (Ala56, Ala60, Leu63, Phe64) are a subset of residues that provide binding interactions with oligomycin (29). (Three of the c-chains are shaded darker grey.)
To test the hypothesis that oligomycin can bind in a stable mode to the c10-ring in a membrane environment, we evaluated the free-energy of formation of this complex using all-atom molecular dynamics simulations (fig. S12). This binding free-energy can be defined as ΔGb = −kBT ln N + ΔGint + ΔGr + ΔGt, where N is the number of available binding sites on the c10-ring, ΔGint is the free-energy difference between the associated and dissociated complex, and ΔGr and ΔGt denote the free-energy penalties due to the loss of rotational and translational entropy of the inhibitor upon binding, respectively. The values calculated from the simulation data are (detailed in Methods): ΔGint = −6.6 kcal/mol, ΔGr = +1.6 kcal/mol and ΔGt = +4.4 kcal/mol, where the latter assumes that the mole-fraction of oligomycin in the membrane is 0.01. For N = 7 (i.e., the number of c-subunits exposed to the membrane), the resulting value of the binding free energy, ΔGb = −1.7 kcal/mol, implies that oligomycin is about 20 times more likely to be bound to the c10-ring than free in the lipid bilayer. (Note that ΔGt would be reduced for higher oligomycin densities and thus binding would be more favorable. For a mole-fraction of 0.05, ΔGt is +3.5 kcal/mol and ΔGb would range from −2.7 kcal/mol for N = 7 to −1.5 kcal/mol for N = 1, which translate into binding probabilities ranging from 90:1 to 10:1.) This energetic analysis hence supports the conclusion that oligomycin binds to the c10-ring subunits exposed to the lipid bilayer. All available data therefore suggests that oligomycin inhibits the enzyme by first binding to the c10-ring and thus impairing its rotation against subunit-a. Likely, the c-subunit with bound oligomycin is sterically prevented from entering the a-c interface, but if it did, it would be unable to either release the bound proton or accept a proton during the catalytic cycle.
There are 3 sites reported in the a-subunit where mutations cause yeast to be resistant to oligomycin. The mutations conferring resistance are: Ile161Met, Ser165Thr/Cys/Tyr, and Leu222Phe (30). The sites are mapped onto the model for Fo (Fig. 5, B to D). This poses a paradox in that these mutations map to locations that are separated by as much as 61 residues along a single α-helix and distant from the binding sites mapped on the c10-ring. However, the resistance forming residues all face the c10-ring and are positioned almost at the center of the region where oligomycin is known to bind, though not at those specific sites. These bulky replacements in a-subunit will disrupt the c10-ring at the point of their interaction but in doing so, also disturb the interaction between the neighboring 2 c-subunits. We propose that the disruption in the conformation of the c10-ring, possibly just on the surface, is propagated through all c-subunits thereby allowing a stable c10-ring and altering the conformation of all oligomycin binding sites. As such, the conformation of the wild type c-subunit (represented here as cOs) is converted to a conformation that is resistant to oligomycin, cOR. However, for a c10-ring to form, each subunit must have identical contacts between the c10-ring and thus c1ORc9OS gets converted to c10OR (where the subscript represents the number of c-subunits), in presence of the oligomycin resistance mutation in a-subunit. This suggests that the conformation of the c10-ring surface is plastic in that it can assume new stable forms that allow it to function. This represents a mechanism that differs from allosteric interactions as these are normally thought of in relationship to enzyme regulation.
Materials and methods
Yeast strains and plasmids
Yeast wild type, USY006 (Matα, ade2-1, his3-11,15, leu2-1, trp1-1, ura3-52, atp2::LEU2, pRS304-ATP2H6::TRP1) and yeast with F6δ fusion, DMY741 (Mata, ade2-1, his3-11,15, trp1-1, ura3-52, can1R100, atp2::loxP, atp7::loxP, atp1::KANR, F6δ::LEU2, ATP2-H6::TRP1, ATP1::URA3) were used throughout. pMSP1E3D1 (31) was a gift from Stephen Sligar (Addgene plasmid # 20066) and was transformed into BL21 (DE3) Gold (Agilent, Santa Clara, CA). Plasmid pRK792 (32) (to express TEV protease) was a gift from David Waugh (Addgene plasmid # 8830). The yeast was grown in a 60l fermentor in semi-synthetic media with the glucose concentration controlled to about 0.2% (33). Bacteria were grown in LB media.
The parent strain for DMY741 was W303-1A (Rodney Rothstein via Alex Tzagoloff, Columbia Univ.). Strain DMY741 was developed by sequentially deleting out the genes (34), ATP1, ATP2, ATP7, and ATP16 and then introducing the genes on plasmids containing ATP1, ATP2 with a His6 tag, and the fusion construct contain subunit F6 (ATP7), T4-lysozyme, and the δ-subunit (ATP16). The fusion construct was made by total synthesis of the DNA (Genscript, Piscataway, NJ). The DNA sequence that encoding T4-lysozyme was codon optimized for expression in yeast and was flanked by 2 NarI sites that added Gly-Ala to the N- and C terminus of lysozyme. The assembled gene was used in a “gap repair” reaction to introduce it into the gene encoding the β-subunit such that it was inserted just behind the codons encoding the mitochondrial leader peptide on the 5′ end and then at the stop codon on the 3′ end of the gene, as described (35). The fusion gene was subcloned into the integrating vector, pRS304 (36) and transformed (37) into the yeast strain.
Biochemistry
We chose to incorporate the yeast ATP synthase into nanodiscs as this is under near native conditions and the oligomycin binding site is at the surface of the c10-ring in the membrane phase.
Yeast ATP synthase was purified essentially as described except a Co-affinity column was substituted for the Ni-affinity column (33). For reconstitution of the ATP synthase into nanodiscs, the enzyme was purified only through the Co-affinity column.
The membrane scaffolding protein (MSP), MSP1E3D1, was expressed in BL21-Gold (DE3) strain (Agilent, Santa Clara, CA) as described (38) with the following changes. After binding to the MSP to the Ni-column, the column was washed with 5 column volumes of WB (40mM Tris-Cl, 0.3M NaCl, 25mM imidazole, 6M guanidinium hydrochloride, 5mM ε-amino caproic acid and 5mM benzamidine HCl, 1mM PMSF, pH 8.0) and then the MSP was eluted with 40mM Tris-Cl, 0.3M NaCl, 0.4M imidazole, 6M guanidinium hydrochloride, 5mM ε-amino caproic acid and 5mM benzamidine HCl, 1 mM PMSF, pH 8.0. The buffer was exchanged with a HiPrep 26/10 desalting column (GE Healthcare, Uppsala, Sweden) equilibrated with 25mM Tris-Cl, 0.15M NaCl, 1mM EDTA, pH 8.0. The MSP was digested with TEV protease at 30°C for 6hrs (TEV/MSP ratio was 1/100 (mg/mg)). The TEV digestion reaction mixture was passed through a HiPrep 26/10 desalting column (GE Healthcare, Uppsala, Sweden) equilibrated with WB containing 75mM imidazole (WBI). The MSP was loaded onto Ni+2- column equilibrated with WBI. The fractions containing protein that did not bind to the column were collected, pooled together, and dialyzed against storage buffer (25mM Tris-Cl, 0.1M NaCl, pH 8.0) at 4°C. The protein concentration of MSP1E3D1, was determined using the extinction coefficient of 26,930 M−1 cm−1 at 280nm (38). MSP1E3D1 was lyophilized and stored at −20°C.
For reconstitution of F1Fo into nanodiscs, we used 3 different lipid preparations. In the case of the structural analysis in the absence of oligomycin, we used nanodiscs prepared form total polar E. coli lipids spiked with ergosterol at a ratio of 3:1 on a molar basis and we also used 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC). For the structure in the presence of oligomycin, we used 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) mixed with cardiolipin (C:18), (CL) at a ratio of 3:1 by weight. The lipids were (obtained from Avanti Polar Lipids, Alabaster, Al) dissolved in chloroform, and stored under argon at −80°C.
The most improved method for reconstitution is reflected in the preparation using POPC:CL. In this procedure, the lipids were added to a glass test tube and the chloroform was removed with a gentle stream of nitrogen. The sample was placed under vacuum for overnight to remove the residual chloroform. The lipids were dissolved in 20mM Tris-Cl, 0.15M NaCl, pH 7.4 with 60mM DDM with the aid of a water bath sonicator. Solubilized lipid was mixed with MSP1E3D1 and F1Fo ATP synthase at a molar ratio of 120:15:2.5 (lipid:MSP:F1Fo) and the mixture was incubated at 30°C for 1hr with constant agitation. To initiate nanodisc assembly, Amberlite XAD-2 (Sigma Aldrich, Milwaukee, WI) beads (0.6 g/ml, final) were added and the mix was incubated at 30°C for 24hrs with constant agitation. Adsorbent beads were added in three batches; in first batch 8% of total beads were added and the reaction was incubated for 6hrs; in the second batch, additional 10% of total beads were added and the reaction mix was incubated for another 16hrs; in the final step of addition, the final 82% of total beads were added and the reaction was incubated for another 2hrs. For reconstitution using E. coli total polar lipids or DMPC, the adsorbent beads were added in 4 batches, 25% of the total beads per hour for a total of 4hrs. The beads were removed and the reaction mixture was purified on a Superose-6 (GE Healthcare, Uppsala, Sweden) size exclusion column equilibrated with 20mM Tris-Cl, 50mM sucrose, 4mM MgSO4, 1mM EDTA, 0.25mM ADP, 0.15M NaCl, 5mM ε-amino caproic acid and 5mM benzamidine HCl, 1mM PMSF, pH 8.0. The buffer was exchanged using a centrifugal column (39) (1ml) using Bio-Gel P-6 (Bio-Rad, Hercules, CA) and buffer containing, 20mM Tris-Cl, 4mM MgSO4, 1mM EDTA, 0.25mM ADP, 0.15M NaCl, 5mM ε-amino caproic acid and 5mM benzamidine HCl, 1 mM PMSF, pH 8.0. The specific activity (u/mg) and percent oligomycin sensitive ATPase using wild type enzyme was 10:42%, 59:78%, and 55:85% for reconstitution using E. coli polar lipids, DMPC an POPC/CL, respectively.
Cryo-electron microscopy data acquisition
Purified yeast F1Fo reconstituted in nanodiscs at the concentration of 1 mg/ml (2.5 µl) was applied to a glow-discharged Quantifoil holey carbon grid (1.2/1.3, 400 mesh), and blotted for 3 s with ~91% humidity before plunge-freezing in liquid ethane using a Cryoplunge 3 System (CP3, Gatan). For cryo-EM of F1Fo with oligomycin, 60 mM stock solution of oligomycin in dimethyl sulfoxide was added to a final concentration of 30 µM, and incubated on ice for 30 min. The mixture (2.5 µl) was applied to a grid, blotted and plunge-frozen. Cryo-EM data were recorded on a 300 kV Polara electron microscope (FEI) at Harvard Medical School. All cryo-EM movies were manually recorded with a K2 Summit direct electron detector (Gatan) in super-resolution counting mode using UCSFImage4 (40). Details of the EM data collection parameters are listed in Table 1.
EM image processing
EM data were processed as previously described (41). Dose-fractionated super-resolution movies collected using the K2 Summit direct electron detector were binned over 2 × 2 pixels, and subjected to motion correction using the program MotionCor2 (42). A sum of all frames of each image stack was calculated following a dose-weighting scheme, and used for all image-processing steps except for defocus determination. CTFFIND4 (43) was used to calculate defocus values of the summed images from all movie frames without dose weighting. Particle picking was performed using a semi-automated procedure with SAMUEL and SamViewer (44). Two- and three-dimensional (2D and 3D) classification and 3D refinement were carried out using “relion_refine_mpi” in RELION (45). Masked 3D classification focusing on Fo with residual signal subtraction was performed following a previously described procedure (46). All refinements followed the gold-standard procedure, in which two half data sets were refined independently. The overall resolutions were estimated based on the gold-standard criterion of Fourier shell correlation (FSC) = 0.143. Local resolution variations were estimated from two half data maps using ResMap. The amplitude information of the final maps were corrected by applying a negative B-factor using the program bfactor.exe.
Model refinement
The initial model was derived from the crystal structure of F1 ATPase (PDB, 2WPD) and the c10-ring from our previous crystal structure (PDB, 3U2F). Models for subunits OSCP, b, F6 and subunit d were initiated as homology models based on bovine structures (PDB, 2CLY and 5FIK). Models for subunits a, A6L/subunit 8, f, j and N-terminal part of subunit b were derived from the cryo-EM structure (PDB, 6B2Z). Initial models were rigid-body fitted to our cryoEM maps, extensively rebuilt in Coot (47) and refined in Refmac (48) using the script refine_local, and subsequently, using real space refinement in Phenix (49).
Final models were validated with statistics from Rama-chandran plots, MolProbity scores, and EMRinger scores (Table 1). MolProbity and EMRinger scores were calculated as described (50, 51).
Molecular dynamics simulations
All simulations were performed with NAMD2 (52), using CHARMM36 force-field for proteins and lipids (53, 54), periodic boundary conditions and constant temperature (298 K) and pressure (1 atm). Long-range electrostatic interactions were calculated using PME, with a real-space cut-off of 12 Å. Van der Waals interactions were computed with a Lennard-Jones potential, cut-off at 12 Å with a smooth switching function taking effect at 10 Å. Simulations were carried out for two systems: a complex of the yeast c10-ring with 4 oligomycin molecules bound (PDB ID: 4F4S), embedded in a hydrated, pre-equilibrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatydylcholine (POPC) lipid bilayer; and a system of the same components, with one oligomycin molecule dissociated from the c10-ring and free in the lipid bilayer. Force-field parameters for oligomycin were derived using the GAAMP server (55). The c10-ring complex was embedded in the POPC membrane using GRIFFIN (56), as previously described (57). Counter-ions were added to neutralize the charge of the simulation system. The simulation systems were equilibrated following a staged protocol whereby positional and conformational restraints acting of the protein-inhibitor complex are gradually weakened over 200 ns. The equilibrated systems were then used as the starting point for the analysis of the oligomycin binding free-energy.
The inhibitor binding free-energy was calculated as ΔGb = −kBT ln N + ΔGint + ΔGr + ΔGt, where N is the number of available binding sites on the c10-ring, ΔGint is free-energy difference between the associated and dissociated complex, and ΔGr and ΔGt denote the free-energy penalties due to the loss of rotational and translational entropy of the ligand upon binding, respectively (kB is the Boltzmann constant and T the temperature). ΔGint was, in part, derived from calculations of the free-energy change associated with decoupling oligomycin from the rest of the molecular system, in both the bound and unbound states. Analogous calculations were carried out whereby oligomycin is recoupled to the system. These calculations used the Free-Energy Perturbation method, whereby the decoupling and re-coupling process is induced through a parameter λ that scales the non-bonded interactions between the ligand and its environment. The parameter λ varied gradually and step-wise between 1 and 0, and vice versa, in 54 consecutive simulations. For each value of λ, in either direction, an incremental free-energy change was estimated from a simulation of 1 ns, after a 200-ps equilibration stage, in the case of bound oligomycin; for the dissociated ligand, the sampling and equilibration times per intermediate were 1.5ns and 300ps, respectively. The resulting values for this decoupling free-energy are 39.8 kcal/mol and 32.8 kcal/mol for the bound and unbound ligand, respectively. Intramolecular interactions thus result in a free-energy contribution that favors binding, i.e., ΔΔGFEP = −7.0 kcal/mol.
For computational efficiency, during the abovementioned decoupling/recoupling simulations, the internal conformation of the oligomycin molecule considered was preserved using a harmonic RMSD restraint, defined relative to that in the equilibrated systems, with a force constant k = 9.6 kcal/mol Å2. Hence, ΔGint = ΔΔGFEP + ΔΔGRMSD, where the latter term is the difference in the free-energy cost of imposing the conformational restraint on the bound versus the unbound ligand. This contribution was computed separately, through an additional set of simulations whereby k is scaled down gradually, using a parameter α that varies step-wise from 1 to 0, and vice versa. Specifically, 50 simulations of 1 ns each were carried out in each direction for the bound and unbound states. From each of these simulation series, the value of ΔGRMSD can be derived using the expressions:
where 〈…〉 denotes a time-average. The calculated values of ΔGRMSD are 0.5 kcal/mol and 0.9 kcal/mol for the bound and unbound ligand, respectively; that is, ΔΔGRMSD = 0.4 kcal/mol, i.e., binding is only marginally disfavored due to loss of conformational dynamics of the inhibitor. The resulting value of ΔGint is thus −6.6. kcal/mol.
When dissociated from the c10-ring, oligomycin adopts primarily an upright orientation, i.e., its rotational freedom relative to the membrane plane is restricted; however, the molecule rotates freely around the membrane perpendicular. The free-energy cost associated with the loss of rotational entropy upon binding can be therefore estimated as ΔGr = −kBTln[Δθ/2π], where Δθ is the rotational fluctuation of the bound ligand perpendicularly to the membrane; from our simulations we estimate that Δθ ~0.44 rad (or 25 degrees), and thus ΔGr = 1.6 kcal/mol.
Similarly, to estimate the free-energy cost associated with the loss of translational entropy upon binding, we used the expression ΔGt = −kBTln[D°ΔA], where ΔA represents the translational freedom of the bound ligand on the membrane plane when bound to the protein, while D° is the ligand area-density in the surrounding medium, akin to the standard concentration C° used in the context of three-dimensional bimolecular association. Although there is no analogous standard for D°, the mole-fraction of the inhibitor in the lipid membrane may be used to define a reference state. For example, an assumed mole-fraction of 0.01, i.e., 1 inhibitor per 100 lipids, translates into D° = 7,000 Å2, for an area-per-lipid of ~702. From our simulations we estimate that ΔA = 4 Å2, and therefore an oligomycin mole-fraction of 0.01 implies ΔGt = 4.4 kcal/mol.
Primary sequence analysis
Primary sequence alignment was done using COBALT (58).
Figure images
Many of the images presented in the figures were made with the assistance of Pymol (59).
Supplementary Material
Movie 1. The architecture of the monomeric form of the yeast ATP synthase.
Movie 2. Rotation of ATP synthase around the x and y-axes.
Movie 3. Views of first, the proton entry pathway and then the proton exit pathway.
Acknowledgments
Many thanks to John Rubinstein for providing a copy of his manuscript and the model for yeast Fo before publication. Funding: The work was supported by a grant from NIH R01GM66223 to DMM. WZ and JFG are funded by the Division of Intramural Research of the National Heart, Lung and Blood Institute.
Footnotes
Author contributions: A.P.S. designed and performed experiments and helped in the writing, M. Luo performed EM data collection and image processing and helped in the writing, W. Z. performed the computational work and helped in the writing, J.S. helped in the data processing and refinement and helped in the writing, D. B. helped in the design and performed biochemical experiments, M.G.C. helped in the cryo-EM experiments, J. F-G designed and supervised the computational work and helped in the writing, M. Liao supervised the EM studies and helped in the writing. D.M.M. devised and supervised experiments, analyzed data, and helped in writing the manuscript.
Competing interests: None.
Data and materials availability: Four three-dimensional cryo-EM density maps of the yeast mitochondrial ATP synthase in nanodiscs have been deposited in the Electron Microscopy Data Bank under accession numbers: EMD-7548 (F1Fo); EMD-7549 (Fo); EMD-7546 (F1Fo with oligomycin) and EMD-7547 (Fo with oligomycin). The corresponding atomic coordinates for the atomic models have been deposited in the Protein Data Bank under accession numbers 6CP6 (F1Fo); 6CP7 (Fo); 6CP3 (F1Fo with oligomycin) and 6CP5 (Fo with oligomycin).
References
- 1.Xu T, Pagadala V, Mueller DM. Understanding structure, function, and mutations in the mitochondrial ATP synthase. Microb. Cell. 2015;2:105–125. doi: 10.15698/mic2015.04.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: Identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daum B, Nicastro D, Austin J, II, McIntosh JR, Kühlbrandt W. Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell. 2010;22:1299–1312. doi: 10.1105/tpc.109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker LA, Watt IN, Runswick MJ, Walker JE, Rubinstein JL. Arrangement of subunits in intact mammalian mitochondrial ATP synthase determined by cryo-EM. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11675–11680. doi: 10.1073/pnas.1204935109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 7.Kabaleeswaran V, Puri N, Walker JE, Leslie AG, Mueller DM. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 2006;25:5433–5442. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dautant A, Velours J, Giraud MF. Crystal structure of the Mg•ADP-inhibited state of the yeast F1c10-ATP synthase. J. Biol. Chem. 2010;285:29502–29510. doi: 10.1074/jbc.M110.124529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 10.Dickson VK, Silvester JA, Fearnley IM, Leslie AG, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H, Bueler SA, Rubinstein JL. Atomic model for the dimeric Fo region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinothkumar KR, Montgomery MG, Liu S, Walker JE. Structure of the mitochondrial ATP synthase from Pichia angusta determined by electron cryo-microscopy. Proc. Natl. Acad. Sci. U.S.A. 2016;113:12709–12714. doi: 10.1073/pnas.1615902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vik SB, Long JC, Wada T, Zhang D. A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim. Biophys. Acta. 2000;1458:457–466. doi: 10.1016/S0005-2728(00)00094-3. [DOI] [PubMed] [Google Scholar]
- 14.Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 15.Junge W, Lill H, Engelbrecht S. ATP synthase: An electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 1997;22:420–423. doi: 10.1016/S0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 16.Symersky J, Pagadala V, Osowski D, Krah A, Meier T, Faraldo-Gómez JD, Mueller DM. Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation. Nat. Struct. Mol. Biol. 2012;19:485–491. doi: 10.1038/nsmb.2284. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogoryelov D, Yildiz O, Faraldo-Gómez JD, Meier T. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 2009;16:1068–1073. doi: 10.1038/nsmb.1678. [DOI] [PubMed] [Google Scholar]
- 18.Mitome N, Ono S, Sato H, Suzuki T, Sone N, Yoshida M. Essential arginine residue of the Fo-I subunit in FoF1-ATP synthase has a role to prevent the proton shortcut without c-ring rotation in the Fo proton channel. Biochem. J. 2010;430:171–177. doi: 10.1042/BJ20100621. [DOI] [PubMed] [Google Scholar]
- 19.Klusch N, Murphy BJ, Mills DJ, Yildiz Ö, Kühlbrandt W. Structural basis of proton translocation and force generation in mitochondrial ATP synthase. eLife. 2017;6:e33274. doi: 10.7554/eLife.33274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allegretti M, Klusch N, Mills DJ, Vonck J, Kühlbrandt W, Davies KM. Horizontal membrane-intrinsic α-helices in the stator a-subunit of an F-type ATP synthase. Nature. 2015;521:237–240. doi: 10.1038/nature14185. [DOI] [PubMed] [Google Scholar]
- 21.Leone V, Faraldo-Gómez JD. Structure and mechanism of the ATP synthase membrane motor inferred from quantitative integrative modeling. J. Gen. Physiol. 2016;148:441–457. doi: 10.1085/jgp.201611679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Assadi-Porter FM, Fillingame RH. Proton-translocating carboxyl of subunit c of F1Fo H+-ATP synthase: The unique environment suggested by the p Ka determined by 1H NMR. Biochemistry. 1995;34:16186–16193. doi: 10.1021/bi00049a034. [DOI] [PubMed] [Google Scholar]
- 23.Root MJ, MacKinnon R. Two identical noninteracting sites in an ion channel revealed by proton transfer. Science. 1994;265:1852–1856. doi: 10.1126/science.7522344. [DOI] [PubMed] [Google Scholar]
- 24.Morrill JA, MacKinnon R. Isolation of a single carboxyl-carboxylate proton binding site in the pore of a cyclic nucleotide-gated channel. J. Gen. Physiol. 1999;114:71–84. doi: 10.1085/jgp.114.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris TK, Turner GJ. Structural basis of perturbed p Ka values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 26.Koeppe RE, II, Stroud RM. Mechanism of hydrolysis by serine proteases: Direct determination of the pKa’s of aspartyl-102 and aspartyl-194 in bovine trypsin using difference infrared spectroscopy. Biochemistry. 1976;15:3450–3458. doi: 10.1021/bi00661a009. [DOI] [PubMed] [Google Scholar]
- 27.Valiyaveetil FI, Fillingame RH. On the role of Arg-210 and Glu-219 of subunit a in proton translocation by the Escherichia coli F0F1-ATP synthase. J. Biol. Chem. 1997;272:32635–32641. doi: 10.1074/jbc.272.51.32635. [DOI] [PubMed] [Google Scholar]
- 28.Valiyaveetil FI, Fillingame RH. Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 29.Symersky J, Osowski D, Walters DE, Mueller DM. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John UP, Nagley P. Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance. FEBS Lett. 1986;207:79–83. doi: 10.1016/0014-5793(86)80016-3. [DOI] [PubMed] [Google Scholar]
- 31.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: Linkages in substrate binding, spin state, uncoupling, and product formation. J. Biol. Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 32.Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 33.Pagadala V, Vistain L, Symersky J, Mueller DM. Characterization of the mitochondrial ATP synthase from yeast Saccharomyces cerevisae. J. Bioenerg. Biomembr. 2011;43:333–347. doi: 10.1007/s10863-011-9364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puri N, Lai-Zhang J, Meier S, Mueller DM. Expression of bovine F1-ATPase with functional complementation in yeastSaccharomyces cerevisae. J. Biol. Chem. 2005;280:22418–22424. doi: 10.1074/jbc.M411113200. [DOI] [PubMed] [Google Scholar]
- 36.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 38.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Chapter Eleven - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penefsky HS. A centrifuged-column procedure for the measurement of ligand binding by beef heart F1. Methods Enzymol. 1979;56:527–530. doi: 10.1016/0076-6879(79)56050-9. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Zheng S, Agard DA, Cheng Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J. Struct. Biol. 2015;192:174–178. doi: 10.1016/j.jsb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi W, Li Y, Yoon SH, Ernst RK, Walz T, Liao M. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature. 2017;549:233–237. doi: 10.1038/nature23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ru H, Chambers MG, Fu T-M, Tong AB, Liao M, Wu H. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife. 2015;4:e11182. doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barad BA, Echols N, Wang RY-R, Cheng Y, DiMaio F, Adams PD, Fraser JS. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, Mackerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang L, Roux B. Automated force field parameterization for non-polarizable and polarizable atomic models based on ab initio target data. J. Chem. Theory Comput. 2013;9:3543–3556. doi: 10.1021/ct4003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staritzbichler R, Anselmi C, Forrest LR, Faraldo-Gómez JD. GRIFFIN: A versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. J. Chem. Theory Comput. 2011;7:1167–1176. doi: 10.1021/ct100576m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou W, Leone V, Krah A, Faraldo-Gómez JD. Predicted structures of the proton-bound membrane-embedded rotor rings of the Saccharomyces cerevisiae and Escherichia coli ATP synthases. J. Phys. Chem. B. 2017;121:3297–3307. doi: 10.1021/acs.jpcb.6b08051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papadopoulos JS, Agarwala R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 59.The PyMOL Molecular Graphics System, version 2.0. Schrödinger, LLC; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. The architecture of the monomeric form of the yeast ATP synthase.
Movie 2. Rotation of ATP synthase around the x and y-axes.
Movie 3. Views of first, the proton entry pathway and then the proton exit pathway.