Abstract
Recombinant adeno-associated virus (rAAV)-based gene therapy has entered a phase of clinical translation and commercialization. Despite this progress, vector integrity following production is often overlooked. Compromised vectors may negatively impact therapeutic efficacy and safety. Using single molecule, real-time (SMRT) sequencing, we can comprehensively profile packaged genomes as a single intact molecule and directly assess vector integrity without extensive preparation. We have exploited this methodology to profile all heterogeneic populations of self-complementary AAV genomes via bioinformatics pipelines and have coined this approach AAV-genome population sequencing (AAV-GPseq). The approach can reveal the relative distribution of truncated genomes versus full-length genomes in vector preparations. Preparations that seemingly show high genome homogeneity by gel electrophoresis are revealed to consist of less than 50% full-length species. With AAV-GPseq, we can also detect many reverse-packaged genomes that encompass sequences originating from plasmid backbone, as well as sequences from packaging and helper plasmids. Finally, we detect host-cell genomic sequences that are chimeric with inverted terminal repeat (ITR)-containing vector sequences. We show that vector populations can contain between 1.3% and 2.3% of this type of undesirable genome. These discoveries redefine quality control standards for viral vector preparations and highlight the degree of foreign products in rAAV-based therapeutic vectors.
Keywords: AAV-GPseq, recombinant adeno-associated virus, single molecule real-time sequencing, rAAV-ITR, gene therapy vector QC
Introduction
Recombinant adeno-associated viruses (rAAVs) have recently become an attractive delivery vehicle for the expression of therapeutic gene products. The specific need for clinical grade vectors for human application demands rigorous quality control (QC) tests to assess vector purity and integrity. Unfortunately, current standard QC protocols are primarily limited to the titration and quantification of vector by qPCR analysis, verification of genome size by native or alkaline (denaturing) agarose-gel electrophoresis, and characterization of viral purity by silver-stained polyacrylamide gel electrophoresis.1 These methods do not characterize the prevalence, compositions, or structures of fragmented genomes. Heterogeneous populations composed of smaller than unit-length genomes were originally observed in wild-type AAV (wtAAV) as a consequence of abortive replication that generate a pool of defective interfering (DI) particles.2, 3 A handful of studies have used high-throughput sequencing approaches to profile packaged single-stranded (ss)AAV genomes to assess the extent of “error-prone” genome encapsidation during rAAV production.4, 5 However, these methods fall short in their ability to interrogate entire genomes from 5′ inverted terminal repeat (ITR) to 3′ ITR as a single intact molecule. There is also a lack of effective and standardized methodologies for detailing the nature and abundance of erroneously packaged sequences originating from the host-cell genome of packaging cell lines or viral fragments originating from Ad-helper and rep/cap constructs, despite more than 10 years of documentation.6 Furthermore, the mechanisms underlying many of these events are not fully understood.
The need to profile heterogeneic rAAV genome populations has taken on particular significance, since we have recently shown that inclusion of sequences that contain secondary structures in the form of short-hairpin DNAs promote the generation of truncated packaged genomes.7 This is especially critical since we demonstrated that rAAVs designed to deliver short-hairpin RNAs (shRNAs), which have inherent secondary structure, exhibit a high degree of genome truncations. A consequence of replication stalling followed by strand-switching events,8 truncated genomes predominantly exist as self-complementary strands with a hairpin loop terminating at one end.7 At the other end, truncated genomes harbor two ITR free ends, similar to self-complementary (sc)AAV genomes.9, 10 We deduced that when self-complementary sequences are ligated to a single-stranded DNA adaptor loop at the free end, they become circular single-stranded molecules that are ideal for single molecule, real-time (SMRT) sequencing.11 This ability allowed us, for the first time, to profile the heterogeneous outcomes of rAAVs carrying shRNA cassettes on a single-vector scale.
One of the major advantages of SMRT sequencing over short-read sequencing platforms is that relatively long DNA fragments (≥500 bp) do not need to be reconstructed from fragments in silico to determine the composition of the template molecules, allowing for the interrogation of full-length and truncated vector genomes together. The approach ensures that only single-affixed polymerases in each zero-mode waveguide (ZMW) at the bottom of the SMRT cell are evaluated, thus achieving single-vector resolution. In addition, SMRT sequencing benefits from the use of a phi29 polymerase derivative, which exhibits strand-displacement activity, making it the most favorable platform for efficient processivity through the notoriously difficult to sequence ITR structure.
Here, we fully explore the utility of direct SMRT sequencing of vector genome populations, aptly named AAV-genome population sequencing (AAV-GPseq), to profile rAAVs prepared by the HEK293 cell-triple transfection method.1 Self-complementary genomes were specifically profiled to demonstrate the diverse applications of AAV-GPseq. We show that the introduction of an enzyme-digested Lambda-phage DNA (λDNA) spike-in can normalize read counts by length to overcome SMRT sequencing molecular loading bias and to accurately assess the relative abundance of truncated genome populations. Using AAV-GPseq, we also detect encapsidated, DNaseI-resistant bacterial sequences originating from reverse packaging events, as well as detection of adenoviral helper and Rep/Cap-construct sequences packaged into virions. This approach was also able to identify sequences originating from the host-cell genome. Importantly, we show that many of these undesired sequences are chimeric with vector-ITR sequences. Finally, the molecular characterization and quantitation of error-prone rAAV genome replication and packaging events is now possible with AAV-GPseq and can be easily adapted for research-grade and clinical vector manufacturing QC pipelines.
Results
AAV-GPseq Can Interrogate Full-Vector scAAV Genome Sequences from ITR-to-ITR with Single-Vector Genome Resolution
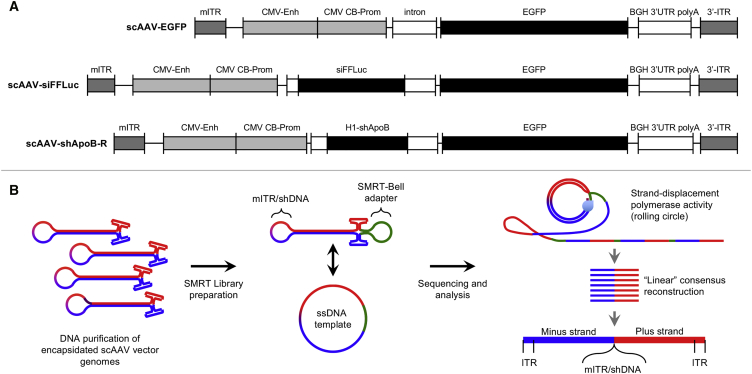
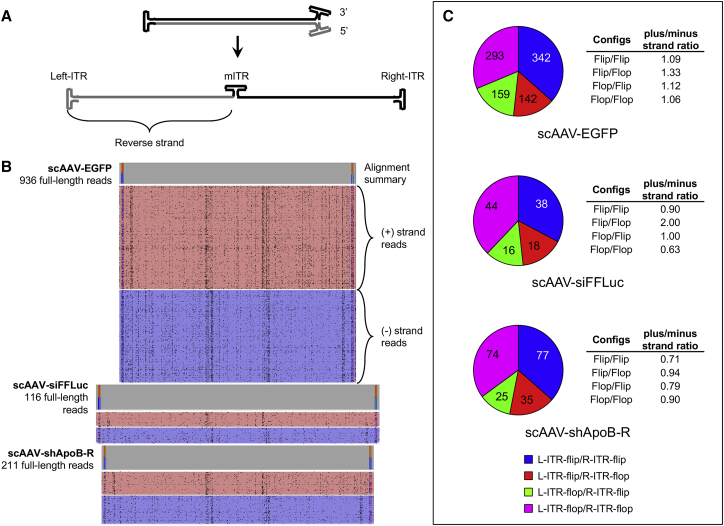
To test whether SMRT sequencing can be performed on individual vector molecules as an unbroken strand from ITR-to-ITR, we profiled three scAAV genomes (Figure 1A). The first is a conventional scAAV vector harboring the EGFP transgene driven by the chicken-β-actin/CMV promoter (scAAV-EGFP). The second and third are similar to scAAV-EGFP but contain shRNA cassettes designed to knock down the expression of either the firefly luciferase (FFLuc) gene or the Apolipoprotein B (ApoB) gene (scAAV-siFFLuc and scAAV-shApoB-R, respectively). To interrogate scAAV vector genome populations, virions were proteolyzed to release genomes. Following DNA nick and end repair, vectors were directly ligated to SMRTbell adaptor at the open end of the molecule, generating a circular single-strand DNA template library ideal for SMRT sequencing. Libraries were loaded onto SMRT cells by diffusion and subjected to standard Pacbio real-time sequencing (Figure 1B; see Materials and Methods). The resulting high-quality linear-consensus sequences that passed CCS2-defined quality scoring (Table 1) were aligned to the appropriate custom reference sequences reflecting a single-stranded linearized molecule stretching from the 5′ ITR to the 3′ ITR, with the mutant ITR (mITR) at the center of the sequence (Figure 2A). Upon visualizing only fully aligned reads, we immediately noticed that the abundance of full-length reads was much lower for vectors harboring shRNA cassettes (scAAV-siFFLuc and scAAV-shApoB-R) (Figure 2B). This outcome is in agreement with our previous finding that inclusion of short hairpin DNA (shDNA) sequences result in the generation of shorter than unit-length molecules and a reduction in full-length molecules as a consequence.7 We also noticed that sequences align in the forward or reverse orientations at near 1:1 ratios (Figure 2B, red and blue aligned reads, respectively). This observation coincides with previous findings that plus (+) stranded and minus (−) stranded genomes are packaged into capsids at equal ratios.12 Even more striking is the ability to detect the distribution of ITR flip and flop orientations.13 Several studies have shown that ITR orientations are established during genome replication and that ITR flip/flop configurations are established independently of each other.14, 15 Replication models for wtAAV suggest that over several rounds of replication, the four possible configurations: flip/flip, flip/flop, flop/flip, and flop/flop reach a 1:1:1:1 steady-state ratio.16 For the first time, AAV-GPseq enables us to directly identify and quantitate the distribution of packaged plus/minus strands and flip/flop configurations in rAAV preparations. Interestingly, we observed that ratios for flip/flop distribution for all three scAAV vectors are closer to 2.3:1:1:2.3 (Figure 2C). Based on a simplified rolling hairpin replication model as described by Cotmore and Tattersall,17 we have predicted the replication outcomes for 15 generations starting from a single plasmid by computational modeling (Figures S1 and S2). We speculate that since ITR resolution and replication cannot initiate at the mITR, the distribution is shifted toward flip/flip and flop/flop configurations. Interestingly, this model predicts that the steady-state levels of flip/flop configurations are 2:1:1:2, only slightly different from our observed distribution (Figure S2G). It is plausible that only a few replication rounds occur after plasmid rescue, resulting in this difference. The model also predicts that strandness (plus/minus) ratios for each flip/flop configuration do not reach 1:1 until the 10th generation (Figure S2H). The observation that the plus/minus ratios deviate from 1.00 for the majority of flip/flop configurations in our test vectors certainly support this notion. However, the sample size here may be too low to reach any definitive conclusions.
Figure 1.
Single-Particle Resolution Profiling of scAAV Genomes by AAV-Gpseq
(A) Diagrams of scAAV genomes: scAAV-EGFP, scAAV-siFFLuc, and scAAV-shApoB-R. (B) Purified scAAV genomes were subjected to SMRT library preparation by single-adaptering to SMRTbell adapters (green). The loops opposite the ITRs represent either the mITRs or shDNAs that form the terminal loop. Plus (+) and minus (−) strands of the molecule are depicted in red and blue, respectively. Single molecule, real-time sequencing is achieved by strand-displacement polymerase activity, generating linear consensus reads. Each read therefore reflects the intact scAAV molecule from 5′ ITR to 3′ ITR.
Table 1.
Percentage of Chimeras in Heterogeneous Genome Populations Post-normalization of Reads
| Construct | Hg38 (Human Genome) | pAdDeltaF6 (Helper Plasmid) | pAAV2-9 (Packaging Plasmid) |
|---|---|---|---|
| scAAV-EGFP | 1.32% | 1.98% | 1.52% |
| scAAV-siFFLuc | 1.77% | 0.23% | 0.65% |
| scAAV-shApoB-R | 2.31% | 2.73% | 2.12% |
Figure 2.
Reads from SMRT Sequencing Yield Full-Length Vector Genomes from 5′ ITR to 3′ ITR
(A) Diagram of a generic scAAV genome depicted as an intramolecular double-stranded molecule (top) and a single-stranded linear molecule (bottom). (B) Reads of the three test vectors were aligned to a custom reference sequence as a linear molecule where both “forward” and “reverse” strands are connected by the mITR sequence. Reads are grouped by strandness (forward, red; reverse, blue) and reflect plus (+) and minus (−) strand packaging. Alignment summaries are depicted above the read alignments. Alignment positions that differ from consensus by a frequency of more than 0.05 are highlighted. Here, only the ITRs exhibit significant differences and reflect flip and flop orientations. The highest frequency of sequencing errors inherent to SMRT sequencing are single-base deletions at poly(G) or poly(C) nucleotides and are marked in IGV as black dashes and show up as speckles in this collapsed display. (C) Pie charts displaying the distribution of flip and flop vector combinations. Read counts and the plus-to-minus strand ratios are displayed for each flip/flop configuration.
AAV-GPseq Can Assess the Relative Abundances of Heterogeneous Populations of Vector Genomes
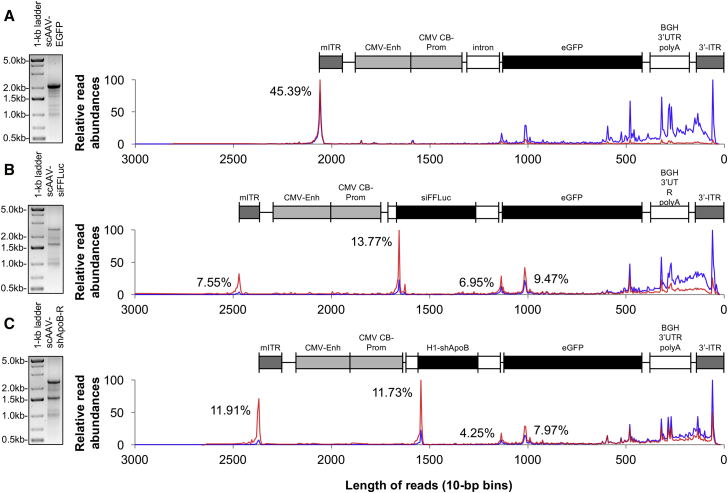
Initial analyses of scAAV-EGFP, scAAV-siFFLuc, and scAAV-shApoB-R vectors by sequence length showed a weak correlation to what we observed of heterogeneous genomes as detected by ethidium bromide (EtBr)-stained agarose-gels (Figure 3). Strikingly, the majority of reads were overrepresented by species with lengths that were less than 500 bp. The discrepancy between agarose-gel analyses and read distribution by SMRT sequencing was somewhat anticipated based on our previous work that showed SMRT cell loading led to size-representation bias.7 To overcome any possible discrepancy, we reasoned that DNA fragments of known lengths can be used as spike-ins to normalize for abundance differences to obtain a much more accurate assessment of heterogeneic population representation. For each vector genome preparation, we supplemented each sample with 10% (by mass) BstEII digested λDNA. Read-length profiles of diffusion loaded λDNA reveals a heavy bias toward the representation of smaller molecules (Figure S3), with the frequency of detection decaying exponentially as fragment lengths increase. By fitting these values to a polynomial-spline as a normalization function, we transformed observed read lengths by their expected abundances to yield adjusted abundance values (Figures S3B–S3D). Following abundance transformation, traces for all three of the vector genome populations now correlate more with their respective agarose-gel results (Figure 3, right panels). More importantly, we are now able to calculate the relative abundances of full-length molecules versus truncated species. By stacking diagrams of the appropriate vector genome over their respective abundance trace, we may also predict the hotspots for intramolecular strand-switching events,7 which give rise to these truncated species (Figure 3, right panels). Surprisingly, even though agarose-gel analysis indicated that the scAAV-EGFP full-length species (2.1-kb band) is the predominant packaged vector (Figure 3A), analysis by AAV-GPseq suggests that the 2.1-kb molecular form only makes up 45.39% of all vector-mapped reads. Similarly, scAAV-siFFLuc and scAAV-shApoB-R vectors also resulted in an extremely low percentage of full-length species (7.55% and 11.91%, respectively) (Figures 3B and 3C). This unexpectedly low abundance of full-length forms compared to agarose-gel assessments is attributed to the strikingly high abundance of reads that were below 500 bp in length and were not visible by EtBr staining.
Figure 3.
Abundance Assessment of Heterogeneous AAV Genome Populations
(A–C) Relative counts of SMRT sequencing reads of (A) scAAV-EGFP, (B) scAAV-siFFLuc, and (C) scAAV-shApoB-R vectors distributed by length/2 (both forward and reverse strands are sequenced for single-adaptered scAAV genomes). Each vector preparation is accompanied with EtBr-stained agarose gels to demonstrate the distribution of visually detectable heterogeneous vector genomes. Traces indicate the distribution of truncated genomes before (blue) and after (red) normalization to BstEII-digested λDNA spike-ins. Traces are scaled to the highest peak set to 100 for ease of comparison. Each trace is aligned to their respective to-scale vector genome diagram from mITR to wtITR (3′ ITR). Normalized read abundances for major peaks are also displayed as a percentage of all reads.
AAV-GPseq Reveals Vector Populations Carrying Plasmid Backbone Sequences
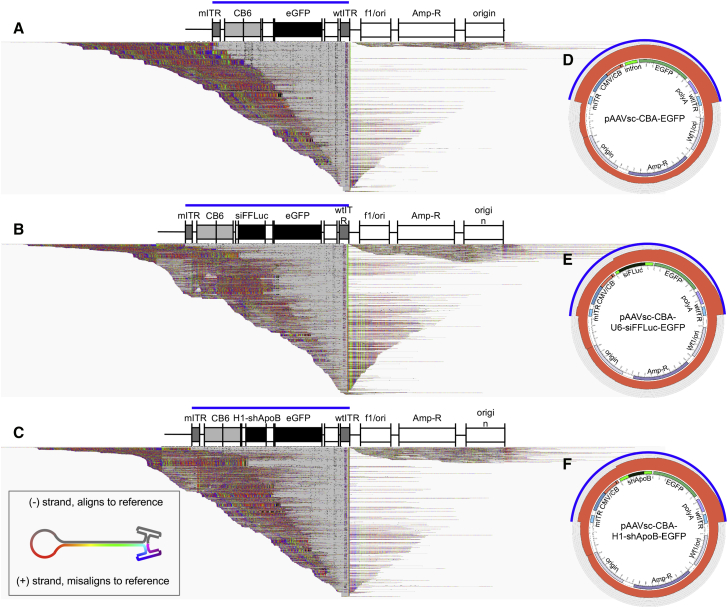
We next assessed the coverage of the reads across the vector plasmid to evaluate the ability of SMRT sequencing to detect packaging of genomes encompassing regions beyond the ITRs (i.e., plasmid backbone sequences).18 Since each scAAV molecule is actually a linear self-complementary sequence with two ITRs at the open ends of the vector genome, only one-half of the molecule will properly align to a vector reference, while the other complementary strand should not. To demonstrate this effect, alignments were displayed on the Integrative Genome Viewer (IGV) browser to include the segments of the reads that do not align to the reference, also known as “soft-clipped” bases (Figure 4A–4C, colored portions of alignments).19 In addition, visualizing alignments on a circular plasmid reference confirmed that a minority population of reads indeed encompass sequences ranging beyond the mITR and wild-type ITR (wtITR) regions for all three test vector genomes (Figures 4D–4F). We attributed the origins of these species to reversed-packaged genomes6 or from larger-than-unit-length molecules that package sequences beyond the mITR.20 We also confirmed that the shorter-than-full-length sequences from scAAV-siFFLuc and scAAV-shApoB-R vector preparations shown in Figures 3B and 3C indeed map from the wtITR region to the shDNA sequences (Figures 4B and 4C). Finally, we observed that the majority of truncated genomes with sizes under 500 bp in length also span the wtITR region (i.e., gray segments of the linear alignments all overlap with the wtITR sequence) (Figures 4A–4C). This latter finding initially suggested that vectors containing only AAV ITRs could be packaged into capsids. However, we questioned whether this interpretation was accurate. Fragment analysis of purified vector genomes by capillary electrophoresis demonstrates that the small molecular weight species, which are under 500 bp, are at background levels of detection or non-existent for all vectors tested (Figure S4). Although SMRT sequencing relies on the strand-displacing polymerase derived from phi29, which should be processive through sequences with high secondary structures, we aimed to assess whether replication error during SMRT sequencing can account for the high abundance of short reads identified by AAV-GPseq.
Figure 4.
Alignments of Heterogeneous Vector Populations to the pCis-Plasmid Reference
(A–C) IGV displays illustrating the linear alignments of (A) scAAV-EGFP, (B) scAAV-siFFLuc, and (C) scAAV-shApoB-R vectors across their respective pCis-plasmid references. Alignments are displayed with soft-clipped bases toggled on to demonstrate that each self-complementary molecule should align half of its genome to the reference (gray), while the remaining half should not (colored segments). Alignments are displayed with a 40-read maximum read count per 50-bp window size downsampling. (D–F) Circos plots of scAAV-EGFP (D), scAAV-siFFLuc (E), and scAAV-shApoB-R (F) vectors demonstrate the coverage of reads across a circular plasmid. The relative thickness of the track radius (red) directly reflects coverage of reads across the plasmid reference (log-scale). Regions expected to be packaged into AAV capsids are denoted by blue bars (linear alignments) and blue arch (circus plots). Alignments beyond these regions are referred to as non-vector sequences.
We digested our three vector plasmid constructs with PacI, which cuts directly outside of the mITR and wtITR sequences, and subjected gel-purified, ITR-bearing restriction fragments to the AAV-GPseq pipeline. Strikingly, many of the reads recovered from this analysis were less than 500 bp in length and specifically mapped to either the mITR or the wtITR regions (Figure S5). This unexpected result indicated that there is some inherent error associated with SMRT sequencing when it encounters AAV-ITR sequences. At this time, it is not clear whether these reads are produced from intramolecular strand-switching events during sequencing or whether they originate from fragmented material during library preparation steps. Regardless, these data suggest that AAV-GPseq cannot accurately interrogate encapsidated vector genomes that are smaller than 500 bp, due to the high frequency of truncated reads at ITR sequences generated by the technique itself. These data demonstrate that the operational molecular range for AAV-GPseq to profile heterogeneous scAAV genome populations is between 2.4 kb and 0.5 kb, where 2.4 kb is the maximum packaging size for self-complementary vectors.
AAV-GPseq Detects Packaging of Non-vector Genome Sequences
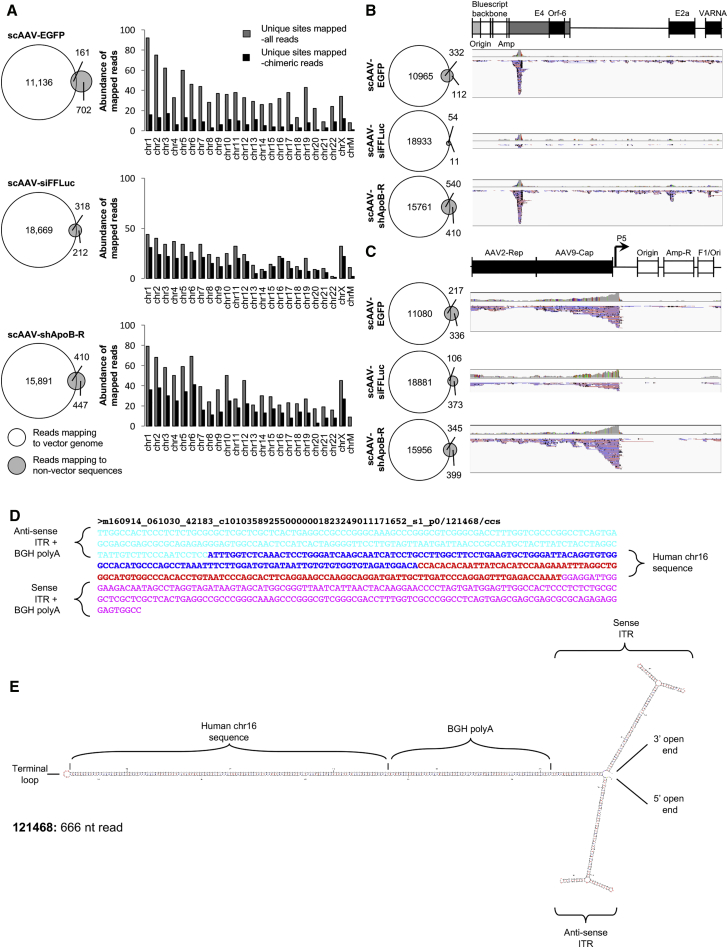
The packaging of non-vector genomes has long been shown to occur in rAAV preparations.6 We therefore asked whether AAV-GPseq could also be tailored to identify and quantitate the abundance of particles packaged with non-vector genome sequences. We first addressed whether any reads were associated with host-cell genomic sequence. Since the triple-transfection procedures for rAAV packaging were carried out in HEK239 cells, reads were mapped to the human genome (hg38 build) to assess host-cell genomic sequence encapsidation (Figure 5). Indeed, a relatively high percentage of reads mapped to the hg38 genome (scAAV-EGFP, 7.19%; scAAV-siFFLuc, 2.76%; and scAAV-shApoB-R, 5.12%). Evaluation of read distribution across the human genome did not reveal any clear trends for specific chromosomes. The only general trend observed for all three vector-preparations was that the largest chromosome (chr1) exhibited the highest frequency of mapped reads, while shorter chromosomes tended to have less mapped reads, suggesting at a more randomized distribution of host-cell genomic packaging across the genome (Figure 5A).
Figure 5.
Detection of Encapsidated Non-vector Genomes
(A) Alignment of SMRT sequence reads for each test vector preparation to the human reference genome (hg38). Venn diagrams display the number of reads mapping to the vector genome (white circles) and to the human genome (gray circles). Histograms display the abundance of uniquely mapped sites on each chromosome (gray bars) and the abundance of unique sites that are mapped by reads that also contain vector genome sequences (chimeras, black bars). (B) Alignment data of reads mapping to the Ad-helper plasmid and (C) to the AAV-Rep/Cap plasmid (note: the conventional AAV2/9 construct contains the p5 TATA-less promoter placed downstream of the Rep/Cap coding sequence). Venn diagrams again display reads mapping to the vector genome (white) and to either the Ad-helper or AAV-rep/cap plasmids (gray). Right, IGV displays showing individual read alignments to their respective references diagrammed above as a linear strand. Reads mapping in the forward and reverse orientations are indicated in red and blue, respectively. Reads mapping uniquely to vector backbones were masked, since common sequences between the pCis, Ad-helper (pAdDeltaF6), and Rep-Cap (pAAV2/9) plasmids cannot be differentiated from each other. (D) Selected hg38-vector chimeric read (121468) showing recombination between the BGH poly(A) of the vector genome and host-cell sequence mapping to chr16. (E) mFold display of the selected read demonstrating molecular self-complementation.
We next explored the abundance of reads associated with the Ad-helper plasmid or the AAV2/9 packaging plasmid. Since both of these plasmids are derived from a Bluescript backbone, and share sequence similarities to the pCis plasmid, there is no way to discern whether sequences containing these aspects originate from the pCis plasmid sequences in cis, or from the Ad-helper or AAV2/9 packaging plasmid constructs in trans. We therefore masked these common sequences from this analysis. Nonetheless, many reads indeed mapped to the other plasmid constructs with varying degrees between vector preparations (Figures 5B and 5C).
Detection and Characterization of Chimeric Reads
We were initially cautious to conclude that all sequences detected by AAV-GPseq were truly encapsidated. Despite extensive benzonase nuclease treatment during rAAV purification process followed by DNaseI treatment before extraction of viral DNAs, we still could not rule out contaminating DNAs as sources of packaged non-vector sequences. However, it is important to note that vector genome packaging relies on the recognition of the Rep binding element (RBE) within AAV-ITRs. Furthermore, the passive packaging of random sequences into AAV has yet to be formally proven. There are therefore two possible explanations for encapsidation of non-vector sequences: (1) contaminating sequences detected by AAV-GPseq have RBE-like motifs, or (2) vector genomes have recombined with host genomic sequences to yield chimeric vector genomes. Upon investigating these two possibilities, we discovered that a large portion of sequences mapping to hg38 also mapped to the vector genome. This finding revealed a class of chimeric genomes packaged into rAAV particles. Importantly, these chimeric sequences all contain ITR sequence (Figure S6), supporting the hypothesis that host-genomic sequences can be packaged into capsids via recognition of RBE sequences gained by recombining with ITR sequences during production. To accurately assess the abundance of these chimeric reads among the vector genomes, we normalized these reads to the λDNA spike-in as described above. After normalization, the percentages of chimeras were calculated as scAAV-EGFP, 1.32%; scAAV-siFFLuc, 1.77%; and scAAV-shApoB-R, 2.31% (Table 1). Furthermore, many vector genomes mapping to the Ad-helper and the packaging plasmid constructs also appeared to be chimeric genomes (Figures 5B and 5C; Table 1). Surprisingly, we observed that chimeric sequences did not map randomly to construct regions. Instead, they are enriched at transgene promoter sequences (Figures 5B, E4 promoter region, and Figure 5C, p5 promoter region). These read enrichments suggest that chimeric reads are a result of vector genomes recombining to sequences that favor gene promoter regions.
As stated above, chimeric genomes described here are of biological importance, since they contain intact ITR sequences and present a means to be packaged into AAV capsids and transduced into cells in vivo. Furthermore, with intact ITRs, these non-vector sequences can be reconfigured to form stabilized circular molecules, which can persist in non-dividing cells. We therefore aimed to leverage the advantage of AAV-GPseq to assess intact vector genome sequences to characterize the composition of individual chimeric molecules. When mapped to the human genome, we immediately noticed that many reads aligned twice to the same regions (Figures S8A and S8B). For example, of the 13 chimeric reads that map to chromosome 16, six chimeras map twice and one chimera maps four times to the same genomic position. Analyses of reads attributed to chimeric species, as well as those that exclusively map to the human genome, indicate that foreign DNA can be packaged as self-complementary sequences that are similar to the configurations of scAAVs (Figures 5D, 5E, S8D–S8I, and S9). Furthermore, these chimeras display a diversity of forms. For example, Figure S8H depicts a vector genome that is a product of six recombination events, incorporating two separate human genomic sequences and four different regions of packaging plasmid sequences. Although this is an unprecedented display of recombination for packaged vectors, only 7.48% of chimeric reads exhibit multiple recombination events between different genomic sources (Figure S8J).
Chimeric Host-Cell Genomic Reads Enrich at Promoter Sequences
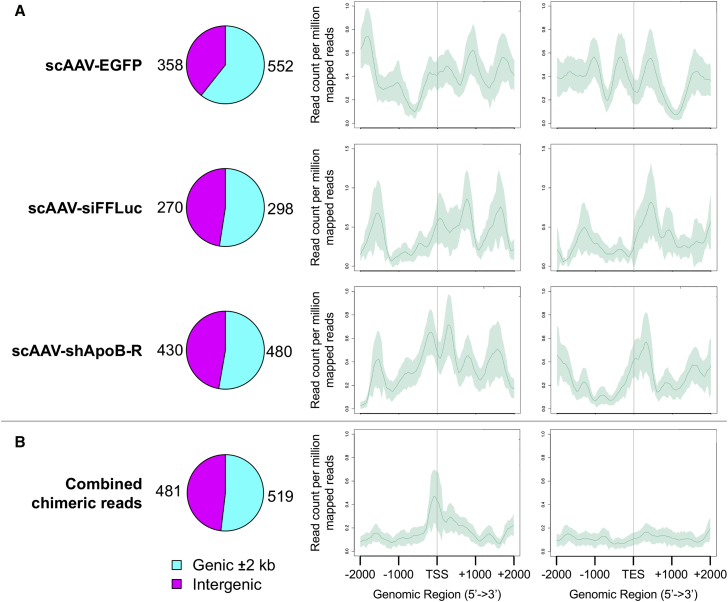
The relatively high percentage of ITR-bearing vectors that are chimeric with host-genomic sequences signifies a potential cause for concern, since vectors encapsidating genomic sequences of the host-packaging cell could lead to unanticipated issues. To further investigate the host-genomic sequences that are being packaged, we assessed whether these chimeric reads map to gene regions or non-genic (intergenic) regions (Figure 6). We found that for all three rAAV preparations, more than 50% of host-cell chimeric reads map to gene bodies ±2 kb. In the case of scAAV-CB6-EGFP construct, 60.6% of the chimeric reads map to or within the proximity of genes.
Figure 6.
Chimeric Reads Are Associated with Promoter Sequences
(A) Distribution of AAV-GPseq reads that map to hg38. Pie charts displaying reads distributed by their association with gene bodies ±2 kb (cyan) or intergenic regions of the genome (magenta). Aggregation plots show the distribution of reads that map within ±2 kb of transcriptional start sites (TSS) or transcriptional end sites (TES) of all annotated genes (Refseq annotations). (B) Due to the low abundance of chimeric AAV-GPseq reads, data for all three vectors were combined and subjected to read aggregation analysis at the TSS and TES of genes. Traces represent the mean read counts per million mapped reads. Shaded areas represent ±SD.
In Figure 5, we demonstrated that chimeric sequences that capture packaging vectors tended to map to promoter regions of the adenoviral helper and Rep genes. We therefore speculated whether this feature was also true for chimeras containing host-cell genomic sequences. All reads that mapped to hg38 were first aggregated and plotted in a 4-kb window (±2 kb) surrounding transcriptional start sites (TSSs) or transcriptional end sites (TESs) (Figure 6A). Despite the low representation of reads mapping to hg38, we noticed that in all three cases, reads mapping to the TSS or the TES exhibited periodic aggregation patterns across the defined genomic range. This pattern is similar in nature to the periodic positioning of nucleosomes detected at promoters by ChIP-seq analysis or by micrococcal nuclease (MNase) hypersensitivity.21 Although, the difference here is that the periodic spacing is far greater, with ∼500 bp per interval. When chimeric molecules containing ITR sequences were specifically assessed, we found that the combined chimeric reads from all vector preparations show a significant peak of reads aggregating at the TSS, while the TES lacked any significant peaks (Figure 6B). This data suggests that host-cell genome vector chimeras also tend to be associated with promoter regions.
Discussion
Clinical rAAV efficacy and safety have become crucial focal points for vector design considerations. To date, the ability to assess vector genome integrity of encapsidated DNA for clinical and basic research has mainly relied on agarose-gel electrophoresis, Southern blot analysis, and PCR techniques. These methods fall short since they cannot decipher the composition of individual vector genomes, making in-depth profiling of heterogeneous populations difficult. This type of precise characterization is critical, since it has long been known that wtAAVs package DI particles.2, 3 It has been hypothesized that these DI particles increase viral fitness by eliciting immune-response with inert virions in the host to favor survival of the host species and hence perpetuation of the virus.22 This attribute may have translated into undesirable rAAV vector populations consisting of truncated and/or chimeric genomes. The rAAV-gene therapy field is in need of new techniques that not only detect the encapsidation of undesirable genomes but can also offer clues to improve vector homogeneity. Although not all designs that lead to truncated genomes necessarily compromise transgene expression,7 the increasing interest of rAAV vectors for clinical applications still necessitates a gold standard for assessing the uniformity of gene therapy vector products.
Reliable use of qPCR analysis to profile encapsidated scAAV genomes have shown that they can exhibit as much as 26% of virions containing backbone plasmid sequences.20 However, these approaches only address limited aspects of rAAV heterogeneity. Until recently, methods to easily quantitate the frequency of erroneously packaged genomes were not practical for implementation into QC pipelines. Platforms such as Helicos Biosciences single-molecule sequencing (SMS) and Illumina-based deep sequencing were developed to determine the prevalence of less-than-full-length molecules and the extent of reverse packaging for ssAAVs, respectively.4, 5 Unfortunately, these high-throughput methods also fall short of capturing fully intact vector sequences. The capacity of AAV-GPseq to be processive through ITR structures is a major advantage over previous platforms and has allowed for the first time the means to profile vector heterogeneity with full-vector genome resolution.
With AAV-GPseq, we have shown that certain reads that map to non-vector sequences are chimeric to vector genomes with intact ITR sequences. This is a striking finding, since chimeric sequences have the means to actively package into capsids by binding Rep, confirming that some particles containing non-vector sequences are not a consequence of passive packaging of fragmented DNAs or packaging of DNAs with RBE-like sequences. This new finding may be a cause for concern since packaging of host-genome, rep/cap, or Ad-helper sequences may result in toxicity for transduced cells. This fear is lessened by our finding that the majority of non-vector sequences are on average 500 bp in size or smaller, and reads encompassing entire genes (host-cell, Ad-helper, AAV-rep/cap, or bacterial genes) were not detected. However, we know that sequence coverage is biased toward smaller molecules. Read abundances after λDNA normalization suggests that longer chimeric reads may be underrepresented (Figure S7A). Thus, full representation of particles that are chimeric and package longer fragments of DNAs is a limitation for AAV-GPseq.
Our study revealed that chimeric sequences tend to map to promoter sequences. We have hypothesized that short-hairpin structures in vectors may promote replication stalling.7 In turn, intramolecular-strand switching may occur as a consequence. Coincidently, similar stalling events at replication fork barriers (RFBs) within promoter sequences are both features of prokaryotic and eukaryotic genomes23 and are known hotspots for recombination. It is plausible that replication stalling at host-cell promoter regions and rAAV genomes may promote recombination by intermolecular strand switching, leading to the production of chimeric vector sequences. Unfortunately, we did not observe any clear motifs that may drive the formation of chimeric genomes, nor were there commonalities that defined the packaging of foreign DNAs that lack ITR elements. Further exploration into these phenomena is crucial for understanding AAV biology as well as the safety of rAAV for clinical use.
We also note it is more than possible that additional genomic species are not detected by AAV-GPseq, since the SMRT sequencing methodology may limit full representation of rAAV genomes that are encapsidated into rAAV particles. Notably, we have yet to overcome the inability to quantitate the packaging of vector genomes under 500 bp in size, since we discovered that subjecting linearized cis-plasmid DNA to SMRT sequencing resulted in the overrepresentation of shortened reads that overlapped ITRs (Figure S5). Incidentally, previous profiling of ssAAV genomes by SMS suggested that capsids can be packaged with DI particles that contain only ITR sequences.4 Initial interpretation of our own data seemed to lend support for these genome species. However, we concluded that many of these smaller read fragments might be artifacts of the sequencing strategy. In reflection, the high thermostability of ITRs may also have impacted SMS analyses of ssAAVs, since coverage of 5′ ends of genomes requires in vitro extension of viral genomes with DNA polymerase.4
Other possible considerations to take note of when accounting for non-represented genome species are the populations of genomes that fail to properly ligate to a SMRTbell adaptor. Whether the inherent structure of scAAV genomes can impact any of these crucial aspects of SMRT library preparation and sequencing, requires careful exploration. Lastly, the current format for AAV-GPseq unfortunately cannot be applied directly to ssAAV genomes, since the ssAAV genome on its own cannot serve as a self-complementary double-stranded template for SMRT sequencing. However, it should be noted that the current scAAV platforms exhibit several advantages—among the most significant are their higher stabilities upon transduction of in vivo tissues, and their ability to bypass the rate-limiting step of single-strand to double-strand conversion.10 Owing to these benefits, strategies using scAAVs are currently undergoing promising clinical trials, which range from gene replacement therapies for hemophilia B and spinal muscular dystrophy (SMA),24, 25 to the more than 20 siRNA approaches for targeting disease-related genes.26 Therefore, AAV-GPseq’s ability to specifically profile scAAV genomes provides a much-needed means for quality assessment for these potentially powerful therapies. Further development of the AAV-GPseq workflow to include methods for direct adaptering of single-stranded vector genomes is underway and will ensure that all clinical rAAVs are safe and efficacious for treating human diseases.
Materials and Methods
Vector Constructs
The pscAAV-CB-EGFP, pAAVsc-CB6-PI-siFFLuc-inverted-EGFP, and pH1-shApob-R constructs used in this study are described elsewhere.7 All vectors were generated, purified, and titrated as described previously.1 Purified viral vectors were digested with DNaseI, and viral DNAs were extracted following procedures for extraction of recombinant adenovirus genomic DNA.1 Vector DNAs were subjected to standard agarose electrophoresis, 2% agarose (Fisher Scientific, Waltham, MA) in 0.5× Tris-Borate-EDTA (TBE) (Fisher) and EtBr staining (Fisher). Fragment analysis of purified vector genomes by capillary electrophoresis was performed by The Deep Sequencing Core Facility at University of Massachusetts Medical School (Worcester, MA).
SMRT Sequencing and Data Analysis
Viral DNA library preparation and sequencing were performed as described previously with slight modifications.7 DNA from purified rAAV preparations was spiked with 10% λDNA digested by BstEII (NEB, Ipswich) for normalization. DNAs were subjected to DNA nick and end repair, followed by direct ligation to SMRTbell adapters at a 1:1 adaptor-to-vector molecular ratio, 1.8× AMPurePB bead purification, and sequenced on a Pacific Biosciences RSII Instrument running the SMRT Analysis v2.3 software packages at the Deep Sequencing Core Facility at University of Massachusetts Medical School. Of note, the standard PacBio SMRTbell library construction efficiency for linear double-stranded DNA fragments is ∼30%–36%, depending on the size of insert. For the libraries constructed on scAAV genomes, the overall ligation efficiency was ∼14%–17%, approximately 49.0%–56.6% of standard libraries. To ensure maximum output of reads to define high-quality consensus reads, 6-hr movies were performed. Since our self-complementary genomes have SMRTbell adapters at only one end of the molecule versus conventional SMRTbell libraries with two, each molecule upon strand-displacement sequencing will only generate forward reads separated by the SMRTbell adaptor sequence instead of alternating forward and reverse reads separated by the adapters. Therefore, to read these specific libraries, the circular consensus algorithm in SMRT Analysis 2.3 is not acceptable. Instead, we employed the CCS2 algorithm that performs a single-molecule consensus of reads regardless of strandedness (i.e., it does not force a plus or minus strand to each read pair and will align each read independent of strand orientation) (N.L. Hepler, et al., 2016, Adv. Genome Biol. Technol., conference). The following parameters were used: --minSnr=3.75 --minPasses=2 --minZScore=-10. The modified bam output file was converted to fastq format for downstream analysis using bam2fastq, a component of SMRT Link v3.0. Reads were de-multiplexed and aligned to custom reference sequences as described in Results using BWA-MEM on the Galaxy web-based platform for genome data analysis.27, 28, 29 Data was visualized using Integrative Genomes Viewer (IGV) version 2.3.61.19 Alignments to the human genome (hg38), are displayed as tracks on the University of California Santa Cruz (UCSC) Genome Browser.30, 31 It should be noted that since scAAV-siFFLuc and scAAV-shAboB-R vectors contain human sequences (U6 and H1 promoters, respectively), sequences mapping to these regions were removed from the analysis. Circos plots were also employed to visualize aligned reads.32 Venn diagrams were drawn using eulerAPE.33 Secondary structures of selected reads were visualized by mfold.34 Constraints to force base-pairing of ITR regions were used. Other parameters were set to default. Aggregation plots were generated using ngs.plot (version 2.41).35
Read Count Normalization
To distinguish reads associated with the λDNA spike-in versus the vector genome DNA pool, reads were simply mapped to either the λ-phage genome or the respective vector genome sequence. To determine the relative abundances of genomes in libraries, reads that aligned to the Lambda phage reference were tabulated by size. A parabolic-spline of the λDNA was defined by the count distribution of the read lengths using the R package, smooth.spline(). The raw read abundances of vector genomes of different sizes were fitted to λDNA defined parabolic-spline.
Data Reporting
The datasets generated during and/or analyzed during the current study are available in the NCBI Sequence Read Archive (SRA) under the SubmissionID: SUB2583306, BioProject: PRJNA383145.
Author Contributions
P.W.L.T. designed, conducted, and interpreted the bioinformatics analysis. J.X. and G.G. conceived and directed the project, supervised the design of the rAAV vectors, and interpreted the data. K.F. conducted the generational rolling-hairpin replication modeling. M.S., C.H., M.W., D.W., and M.L.Z. helped to develop the SMRT sequencing strategy and interpreted the primary quality assessments. Q.S. generated the vectors. P.W.L.T., J.X., and G.G. wrote the manuscript with significant contributions from M.S., C.H., M.W., and M.L.Z.
Conflicts of Interest
G.G. is a co-founder of Voyager Therapeutics and holds equity in the company. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics and other biopharmaceutical companies. M.S., C.H., and M.W. are full-time employees of Pacific Biosciences, a company commercializing SMRT sequencing technologies. All other authors have no disclosures.
Acknowledgments
This work was supported by Public Health Service grants 1R01NS076991-05, R01 HL097088, 1P01AI100263-05, and 4P01HL131471-01 from the NIH and an internal grant from University of Massachusetts Medical School to G.G. We thank Dr. Ellen Kittler and the UMass Deep Sequencing Core for their advice and execution of SMRT sequencing pipelines and Dr. Robert Kotin for critical advice.
Footnotes
Supplemental Information includes nine figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.02.002.
Supplemental Information
References
- 1.Gao G., Sena-Esteves M. Introducing genes into mammalian cells: viral vectors. In: Green M.R., Sambrook J., editors. Molecular Cloning: A Laboratory Manual. Volume 2. Cold Spring Harbor Laboratory Press; 2012. pp. 1209–1313. [Google Scholar]
- 2.Hauswirth W.W., Berns K.I. Adeno-associated virus DNA replication: nonunit-length molecules. Virology. 1979;93:57–68. doi: 10.1016/0042-6822(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 3.Laughlin C.A., Myers M.W., Risin D.L., Carter B.J. Defective-interfering particles of the human parvovirus adeno-associated virus. Virology. 1979;94:162–174. doi: 10.1016/0042-6822(79)90446-x. [DOI] [PubMed] [Google Scholar]
- 4.Kapranov P., Chen L., Dederich D., Dong B., He J., Steinmann K.E., Moore A.R., Thompson J.F., Milos P.M., Xiao W. Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum. Gene Ther. 2012;23:46–55. doi: 10.1089/hum.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lecomte E., Tournaire B., Cogné B., Dupont J.B., Lindenbaum P., Martin-Fontaine M., Broucque F., Robin C., Hebben M., Merten O.W. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol. Ther. Nucleic Acids. 2015;4:e260. doi: 10.1038/mtna.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright J.F. Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther. 2008;15:840–848. doi: 10.1038/gt.2008.65. [DOI] [PubMed] [Google Scholar]
- 7.Xie J., Mao Q., Tai P.W.L., He R., Ai J., Su Q., Zhu Y., Ma H., Li J., Gong S. Short DNA hairpins compromise recombinant adeno-associated virus genome homogeneity. Mol. Ther. 2017;25:1363–1374. doi: 10.1016/j.ymthe.2017.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward P., Berns K.I. In vitro replication of adeno-associated virus DNA: enhancement by extracts from adenovirus-infected HeLa cells. J. Virol. 1996;70:4495–4501. doi: 10.1128/jvi.70.7.4495-4501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty D.M., Fu H., Monahan P.E., Toulson C.E., Naik P., Samulski R.J. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Ma H.I., Li J., Sun L., Zhang J., Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 11.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 12.Berns K.I., Adler S. Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J. Virol. 1972;9:394–396. doi: 10.1128/jvi.9.2.394-396.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spear I.S., Fife K.H., Hauswirth W.W., Jones C.J., Berns K.I. Evidence for two nucleotide sequence orientations within the terminal repetition of adeno-associated virus DNA. J. Virol. 1977;24:627–634. doi: 10.1128/jvi.24.2.627-634.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K.C., Tyson J.J., Lederman M., Stout E.R., Bates R.C. A kinetic hairpin transfer model for parvoviral DNA replication. J. Mol. Biol. 1989;208:283–296. doi: 10.1016/0022-2836(89)90389-6. [DOI] [PubMed] [Google Scholar]
- 15.Lusby E., Bohenzky R., Berns K.I. Inverted terminal repetition in adeno-associated virus DNA: independence of the orientation at either end of the genome. J. Virol. 1981;37:1083–1086. doi: 10.1128/jvi.37.3.1083-1086.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyson J.J., Chen K.C., Lederman M., Bates R.C. Analysis of the kinetic hairpin transfer model for parvoviral DNA replication. J. Theor. Biol. 1990;144:155–169. doi: 10.1016/s0022-5193(05)80316-9. [DOI] [PubMed] [Google Scholar]
- 17.Cotmore S.F., Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 18.Chadeuf G., Ciron C., Moullier P., Salvetti A. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol. Ther. 2005;12:744–753. doi: 10.1016/j.ymthe.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnödt M., Schmeer M., Kracher B., Krüsemann C., Espinosa L.E., Grünert A., Fuchsluger T., Rischmüller A., Schleef M., Büning H. DNA minicircle technology improves purity of adeno-associated viral vector preparations. Mol. Ther. Nucleic Acids. 2016;5:e355. doi: 10.1038/mtna.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell O., Tiwari V.K., Thomä N.H., Schübeler D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 22.Dimmock N.J., Easton A.J. Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals? J. Virol. 2014;88:5217–5227. doi: 10.1128/JVI.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labib K., Hodgson B. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 2007;8:346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj D., Davidoff A.M., Nathwani A.C. Self-complementary adeno-associated viral vectors for gene therapy of hemophilia B: progress and challenges. Expert Rev. Hematol. 2011;4:539–549. doi: 10.1586/ehm.11.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scoto M., Finkel R.S., Mercuri E., Muntoni F. Therapeutic approaches for spinal muscular atrophy (SMA) Gene Ther. 2017;24:514–519. doi: 10.1038/gt.2017.45. [DOI] [PubMed] [Google Scholar]
- 26.Borel F., Kay M.A., Mueller C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 2014;22:692–701. doi: 10.1038/mt.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010;Chapter 19 doi: 10.1002/0471142727.mb1910s89. Unit 19.10.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goecks J., Nekrutenko A., Taylor J., Galaxy Team Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Micallef L., Rodgers P. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS ONE. 2014;9:e101717. doi: 10.1371/journal.pone.0101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L., Shao N., Liu X., Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.