Abstract
Adeno-associated virus (AAV) has provided the gene therapy field with the most powerful in vivo gene delivery vector to realize safe, efficacious, and sustainable therapeutic gene expression. Because many clinically relevant properties of AAV-based vectors are governed by the capsid, much research effort has been devoted to the development of AAV capsids for desired features. Here, we combine AAV capsid discovery from nature and rational engineering to report an AAV9 capsid variant, designated as AAV9.HR, which retains AAV9’s capability to traverse the blood-brain barrier and transduce neurons. This variant shows reduced transduction in peripheral tissues when delivered through intravascular (IV) injection into neonatal mice. Therefore, when IV AAV delivery is used to treat CNS diseases, AAV9.HR has the advantage of mitigating potential off-target effects in peripheral tissues compared to AAV9. We also demonstrate that AAV9.HR is suitable for peripheral tissue-detargeted CNS-directed gene therapy in a mouse model of a fatal pediatric leukodystrophy. In light of recent success with profiling diversified natural AAV capsid repertoires and the understanding of AAV capsid sequence-structure-function relationship, such a combinatory approach to AAV capsid development is expected to further improve vector targeting and expand the vector toolbox for therapeutic gene delivery.
Keywords: AAV capsid, adeno-associated virus, CNS disease, gene therapy, tissue detargeting
Introduction
Adeno-associated virus (AAV) is a small, non-pathogenic virus found in many mammals, including humans. The simple architecture of AAV comprises an icosahedral capsid of ∼26 nm in diameter and a single-stranded DNA genome of ∼4.7 kb.1 Due largely to its superb in vivo tissue tropism for both dividing and non-dividing cells, recombinant AAV (rAAV) has been developed as a gene delivery vector, drawing considerable interest particularly in the gene therapy field. rAAV development has been greatly facilitated by the discovery of numerous AAV capsid serotypes and variants from both human and nonhuman primate (NHP) tissues, which collectively provide a toolbox for targeting a variety of tissue and cell types.2 These naturally existing capsids also serve as lead templates for rational capsid engineering guided by structure-function analysis, such as surface decoration with specific receptor ligands3 or alteration of specific residues involved in cellular interaction.4 In addition, directed evolution from random DNA shuffling or mutagenesis libraries also yielded capsids that have clinically relevant features.5
Among all the AAV serotypes and variants tested so far, AAV9 has gained popularity in gene therapy applications due to its high tropism in tissues, such as liver, heart, and skeletal muscle following intravascular (IV) delivery.6 In addition, its capability of traversing the blood-brain barrier (BBB) and transducing cells residing in the CNS opens new avenues for CNS gene therapy.7, 8 For example, IV delivery of rAAV9 expressing the human ASPA gene proved to be efficacious in rescuing most disease phenotypes in a mouse model of ASPA deficiency (Canavan disease), a white matter neurodegenerative disorder.9, 10 Recently, a clinical trial for spinal muscular atrophy by IV-delivered rAAV9 showed early promising results.11 In parallel with pursuing stronger transduction in target tissues following IV delivery of rAAV, de-targeting specific tissues is often a desired feature to diminish potential off-target toxicity.12 Several approaches have been taken in this regard. First, a tissue-specific promoter can be utilized to restrict gene expression.13 Second, microRNA (miRNA) binding sites that are recognized by endogenous tissue-specific miRNAs can be engineered in the transgene 3′ UTRs, which will silence transgene expression in that tissue.14 Third, a vector library obtained by capsid engineering can be screened for a desired transduction profile.15 The most significant limitation to systemic rAAV gene therapy in numerous clinical trials has been the effector T cell response to the input rAAV capsid proteins.16 Thus, only those de-targeting strategies that decrease exposure of the liver to capsids, rather than those that limit transgene expression alone, are likely to result in substantial improvements in the efficacy and safety profile.
Research on the AAV capsid structural biology has provided valuable knowledge in understanding its characteristics and guiding vector development for gene therapy.17 The AAV capsid consists of 60 copies of viral protein (VP) subunits, with VP1, VP2, and VP3 at 1:1:10 ratio. The capsid structure of a handful of wild-type (WT) AAV serotypes has been determined by cryo-electron microscopy (cryo-EM) followed by three-dimensional image reconstruction and/or X-ray crystallography.17 VP3 monomers from all examined serotypes share a conserved β-barrel core consisting of eight antiparallel β sheets. The large loops connecting the β strands contain nine variable regions (VR-I to -IX) that are accessible from the solvent side for interactions with cellular receptors and antibodies. The VRs are highly diverged between serotypes and likely determine capsid characteristics, such as tropism and immunogenicity. One noticeable feature of all known AAV capsid structures is the protrusions surrounding the icosahedral three-fold axes of symmetry, where most VRs are located. Therefore, the three-fold protrusions play an important role in shaping the unique properties of each variant capsid.
For AAV9, the primary receptor is cell surface glycans with terminal β-galactose.18, 19 The galactose binding domain on AAV9 capsid has been mapped to a pocket at the base of the three-fold protrusions formed by N470, D271, N272, Y446, and W503.20 The neighboring 512NGR514 tripeptide motif has been implicated in cell entry in vivo through interaction with integrin.21 In addition, the 37/67-kilodalton laminin receptor (LamR) expression level plays a role in AAV9-based vector transduction in vitro, and residues 489–545 and 591–621 of AAV9 VP are equivalent to AAV8 VP residues that interact with LamR in a yeast two-hybrid binding assay.22 A distinctive feature of AAV9 compared with other serotypes, such as AAV1, is its delayed blood clearance, which is attributable to residues 456–568 and is suggested to contribute to AAV9’s robust cardiac transduction in mice.23 When mapped to the AAV9 capsid structure,24 most residues involved in receptor binding and blood clearance are located at or near the three-fold protrusions, highlighting their important role in mediating cellular interactions and determining capsid properties.
Here, taking an approach combining AAV capsid discovery from nature and rational design, we report the AAV9.HR capsid that differs from AAV9 by two amino acid residues that are located at the three-fold protrusions. Following IV delivery, this capsid is able to transduce neurons in broad brain regions with a peripheral tissue de-targeting property and is suitable for CNS gene therapy.
Results
Discovery of AAV Capsid Variants
In an attempt to discover naturally existing AAV capsid sequences from NHP, we extracted total DNA and RNA from chimpanzee tissues, including brain, liver, lung, and spleen. We found several previously reported AAV serotype capsid sequences, including AAV3B, 4, 5, and 9. In addition, we discovered 121 variants with capsid sequences similar to AAV1, 3B, 4, 5, 6, and 9 (Figure S1). Forty-seven of these sequences were variants of AAV9 differing from the WT AAV9 capsid sequence by no more than nine amino acid residues (Table S1). The AAV9 capsid shows broad and strong tissue tropism in vivo and is able to traverse the BBB following systemic delivery. Therefore, we decided to focus on the characterization of one of these AAV9 variants in this study, hypothesizing that a minimal number of capsid changes may alter the tropism properties of AAV9, thus allowing for residue function analysis and generating useful vectors for in vivo gene delivery.
The CLv-D8 Capsid Variant Transduced Liver Much Less Efficiently Than AAV9
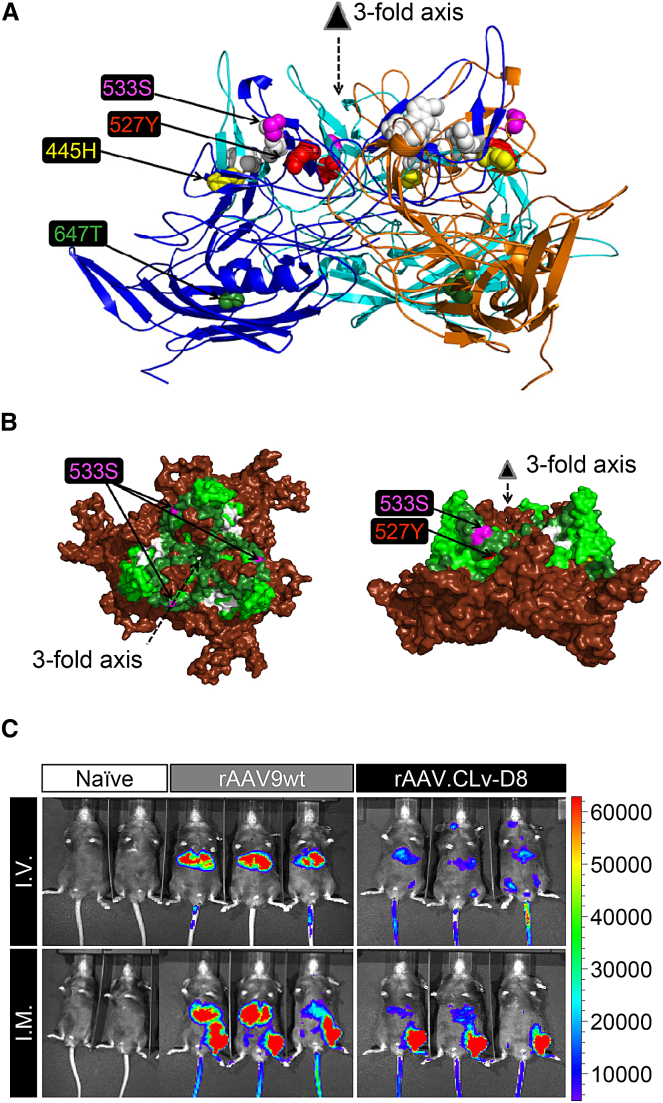
Among the AAV9 variants, we noticed the conservation of four residue changes among multiple isolates, namely Y445H, H527Y, R533S, and I647T (AAV9 residue type in front; AAV9 VP1 numbering; Table S1). We modeled these four residue changes onto the AAV9 capsid crystal structure.24 As shown in Figure 1A, three of the four residues cluster near the three-fold axis. The 445H resides at the interface of two three-fold-related VP monomers, and 647T is located in the conserved βH strand and buried within the eight-stranded β-barrel motif. These two residues are not visible from the capsid surface view (Figure 1B). The 527Y and 533S residues are located at the protrusions surrounding the three-fold axis (Figure 1B), which have been shown to have an impact on receptor binding, including galactose and LamR, and blood clearance of rAAV vectors.20, 22, 23 Therefore, we decided to further examine the CLv-D8 (clone D8 from chimpanzee liver) variant that harbors all four residue changes along with the 527Y and 533S changes.
Figure 1.
Capsid Structure Characteristics and In Vivo Transduction Profile of rAAV.CLv-D8
(A) Side view of a trimer comprising three copies of AAV9 VP protein.24 The four residues in CLv-D8 that differ from AAV9 are modeled in spheres and labeled with color. White spheres are residues involved in galactose binding.20 (B) Surface model of the trimer shown in (A) presented in top view (left) or side view (right) is shown. The 533S and 527Y residues found in CLv-D8 are labeled when visible. Residues in dark green are involved in LamR binding.22 Residues in lime green have impact on blood clearance kinetics.23 Residues in white are involved in galactose binding.20 (C) In vivo imaging of luminescence at four weeks after intravascular (IV) or intramuscular (IM) injection of 1.0E+11 genome copies (GCs) of luciferase-expressing rAAV packaged into the wild-type AAV9 capsid (rAAV9wt; n = 3) or the CLv-D8 capsid (rAAV.CLv-D8; n = 3) is shown. In the IM delivery, rAAVs were injected into the left tibialis anterior muscle. Naive mice that did not receive any rAAV serve as background controls (n = 2). Luminescence intensity is presented on a color-coded scale shown on the right in the unit of photons/s/cm2/sr.
To characterize the in vivo tissue transduction profile of AAV.CLv-D8 compared with AAV9 (AAV9wt hereafter), we packaged a single-stranded recombinant AAV genome expressing firefly luciferase (Fluc) into both capsids and compared in vivo transduction following IV injection of 1 × 1011 genome copies (GCs) into adult mice. Whereas rAAV9wt-Fluc yielded strong Fluc expression in the liver as previously reported,6 rAAV.CLv-D8-Fluc-treated mice displayed reduced bioluminescence from the liver (Figure 1C). Delivery of the same amount of the vector genomes by intramuscular (IM) injection at the left tibialis anterior muscle (LTA) also showed reduced liver transduction with the rAAV.CLv-D8-Fluc vector, whereas the signal from LTA was comparable between the two vectors (Figure 1C).
The AAV9.HR Capsid Mutant Recapitulates the Liver De-targeting Property of CLv-D8
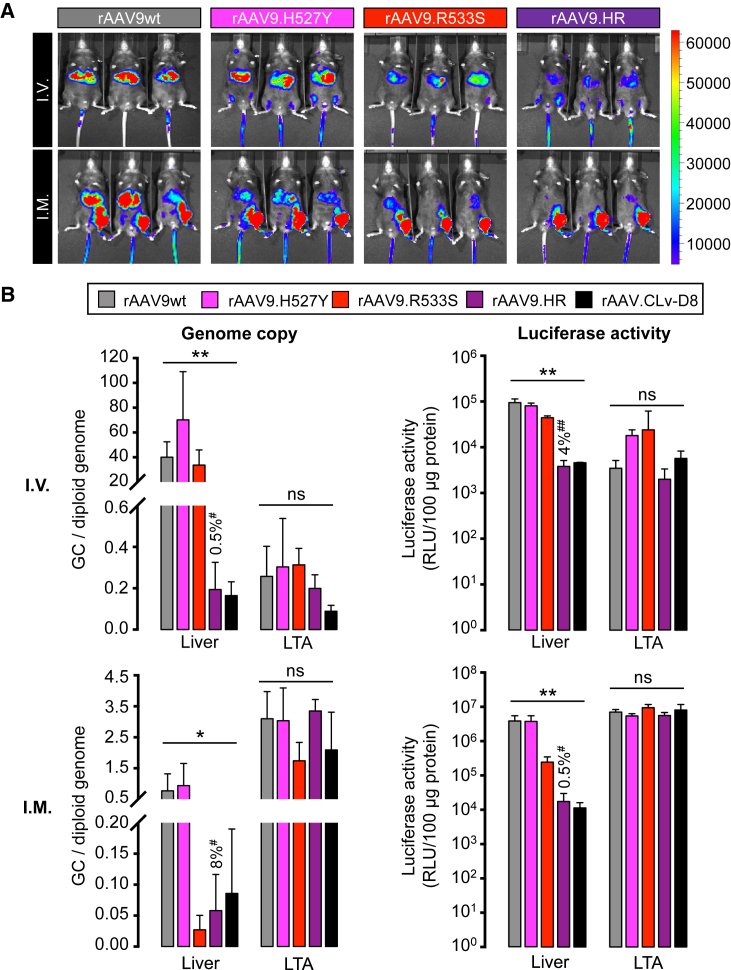
To examine the contributions of the surface-exposed residues 527Y and 533S to the reduced liver transduction profile of AAV.CLv-D8 relative to AAV9wt, we made AAV9 capsid “single mutants,” carrying either residue changes, and “double mutants,” carrying both residue changes. By in vivo imaging, the single-mutant vector rAAV9.R533S-Fluc showed reduced liver transduction compared with rAAV9wt-Fluc following IV or IM injection of the same dose (Figure 2A). Notably, the double-mutant vector rAAV9.HR-Fluc, containing the H527Y and R533S mutations, showed similarly reduced liver transduction as the rAAV.CLv-D8-Fluc vector (Figure 2A). Single changes at the 445 and 647 positions, as well as double-mutant combinations containing them, did not show reduced liver expression and were not studied further (data not shown).
Figure 2.
rAAV9.HR Showed Reduced Liver Tropism Relative to rAAV9wt following IV or IM Delivery in Mice
(A) In vivo imaging of luminescence at four weeks after IV or IM injection of 1.0E+11 GC of luciferase-expressing rAAV packaged into WT AAV9 or each of three AAV9 capsid mutants, namely H527Y (rAAV9.H527Y; n = 3), R533S (rAAV9.R533S; n = 3), or H527Y/R533S (rAAV9.HR; n = 3). Luminescence intensity is presented by a color-coded scale shown on the right in the unit of photons/s/cm2/sr. The rAAV9wt-treated mice shown in Figure 1 are included for comparison. (B) Bar graphs show molecular characterization of genome abundance and transduction of different rAAV vectors in the liver and left tibialis anterior (LTA) muscle at four weeks after IV (top panels) or IM (bottom panels) delivery (n = 3). rAAV genome copy data (left panels) are presented in genome copies per diploid genome (GC/diploid genome). Luciferase activity data are presented in relative light units per 100 μg of total protein (RLU/100 μg protein). Values shown above the rAAV9.HR bars denote the percentage relative to the rAAV9wt group (defined as 100%). All values are presented as mean and SD. *p < 0.05 and **p < 0.01 by one-way ANOVA; ns, not significant by one-way ANOVA; #p < 0.05 and ##p < 0.01 by Tukey’s multiple comparisons test against the rAAV9wt group.
After in vivo imaging, we sacrificed the mice and harvested liver and LTA muscle to quantitatively measure rAAV genome abundance by qPCR and transduction efficiency by luciferase activity in tissue lysates (Figure 2B). Both assays corroborated the results of in vivo imaging. Specifically, following IV injection of the rAAV9.HR vector, the rAAV genome copy (GC) number in liver was only 0.5% of rAAV9wt vector levels, suggesting that the AAV9.HR capsid mutant had significantly reduced tropism in this organ compared to WT. Correspondingly, the luciferase activity in the liver was reduced to 4.0% of WT. The decreases were comparable to observations with the rAAV.CLv-D8 vector (Figure 2B). In contrast, there was no significant difference among the vectors tested in LTA muscle transduction (Figure 2B, top panels). Following IM injection, similar liver de-targeting profile was observed with the rAAV9.HR vector, 8% of GC number and 0.5% of luciferase activity relative to WT (Figure 2B, bottom panels). The genome copies and transduction level in LTA muscle (the injected muscle) again did not show significant difference among the vectors (Figure 2B, bottom panels). For the single-mutant vectors, only rAAV9.R533S showed significant liver de-targeting following IM injection but did not fully recapitulate the characteristics of the rAAV.CLv-D8 vector following IV injection as the rAAV9.HR vector did (Figures 1C and 2). Therefore, we conclude that both the 527Y and 533S residues in the CLv-D8 capsid variant synergistically contribute to its liver de-targeting property in mice.
Several reports suggested that the blood clearance kinetics of rAAV have an impact on peripheral tissue tropism following IV delivery.23, 25 Moreover, residues 456–568 of AAV9 VP have been shown to determine its delayed blood clearance kinetics.23 Both residues 527Y and 533S are located within this region. However, we did not observe any significant difference in blood clearance among rAAV9wt, rAAV9.H527Y, rAAV9.R533S, rAAV9.HR, and rAAV.CLv-D8 (Figure S2). Although rAAV9.HR and rAAV.CLv-D8 performed similarly in the gene delivery, transduction, and blood clearance assays, the titers of rAAV.CLv-D8 preparations were consistently lower than rAAV9.HR preparations when the same genome was packaged (Figure S3), indicating a potential advantage of the engineered AAV9.HR capsid over the naturally existing CLv-D8 isolate. Therefore, the transduction pattern of AAV9.HR was further characterized.
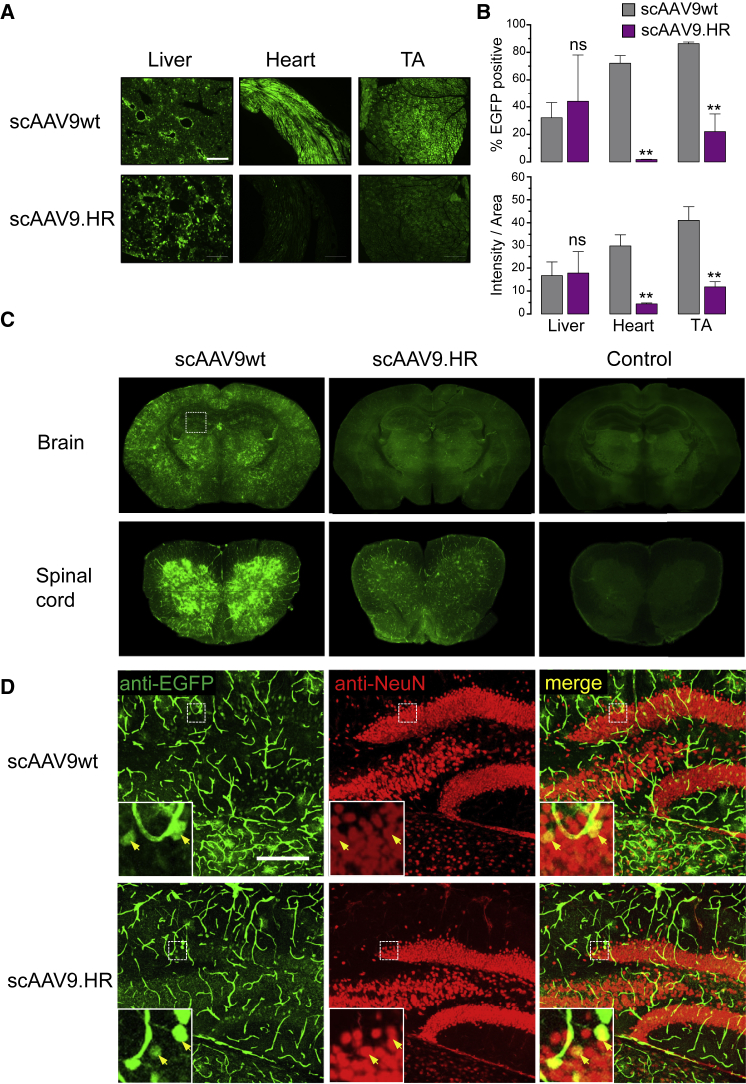
scAAV9.HR-EGFP Vector Displayed Reduced Transduction of Both Peripheral and CNS Tissues by High-Dose IV Injection into Adult Mice
The ability to cross the BBB and to transduce the cells within CNS following high-dose IV delivery is an attractive feature of rAAV9 that holds great promise for CNS gene therapy. However, this CNS-targeted treatment regimen will highly transduce major peripheral organs, such as the liver, which may cause detrimental off-target effects. To test whether rAAV9.HR can also reach the CNS while de-targeted from peripheral organs, we treated young adult mice with high-dose (4.0E+12 GC) self-complementary (sc)AAV9wt or scAAV9.HR carrying the same EGFP-expressing genome by IV delivery and compared EGFP expression in tissue sections. As shown in Figures 3A and 3B, EGFP fluorescence in heart and tibialis anterior (TA) was greatly reduced with scAAV9.HR compared to scAAV9wt, whereas there was no statistically significant difference in the liver. In the adult CNS, scAAV9wt-EGFP yielded widespread transduction in the brain and in the gray matter of the spinal cord as previously described,7, 8 whereas the scAAV9.HR-EGFP vector led to much weaker transduction (Figure 3C). By performing immunofluorescence co-staining, we observed that transduction occurred in the hippocampal neurons (Figure 3D) in addition to vasculature (Figure S4), confirming the ability of high-dose rAAV9 to traverse adult BBB. Neuronal transduction was noticeably reduced with the scAAV9.HR-EGFP vector at the same dose (Figure 3D).
Figure 3.
Comparison of scAAV9wt and scAAV9.HR Transduction Profiles following IV Delivery of High Dose (4.0E+12 GC) into Young Adult Mice (Six Weeks Old)
(A) Representative fluorescence images showing native EGFP fluorescence in tissue sections of liver, heart, and TA muscle at three weeks after treatment with self-complementary (sc) EGFP-expressing vectors packaged into AAV9 capsid (scAAV9wt) or AAV9.HR capsid (scAAV9.HR). The scale bar represents 500 μm. (B) Quantification of EGFP signal in tissue sections is shown. EGFP signal is quantified by percentage of EGFP-positive pixels (% EGFP positive, top panel) or the average integrated intensity calculated as the ratio of total intensity over area (intensity/area, bottom panel). n = 3 for each group. Data are presented as mean and SD. **p < 0.01 and ns, not significant by Student’s t test. (C) Representative immunofluorescence images show anti-EGFP signal in brain and spinal cord (lumbar region) sections. Untreated samples processed in the same manner as the treated samples serve as fluorescence background control. (D) Representative confocal immunofluorescence images show anti-EGFP signal (green), neurons (red, labeled as NeuN-positive), and merged signal (yellow) in the hippocampus (squared region in C). The scale bar represents 200 μm.
Overall, when compared with the parental AAV9 capsid, the AAV9.HR double mutant capsid shows reduced in vivo transduction in a subset of peripheral tissues and CNS following high-dose IV delivery of sc vectors into young adult mice.
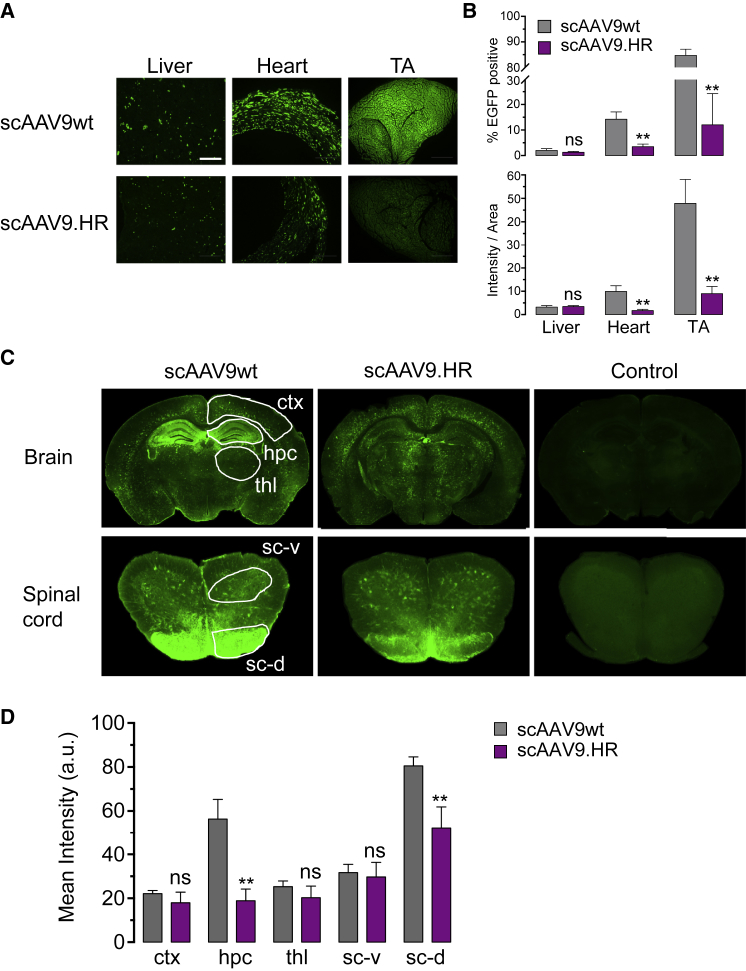
IV Delivery of scAAV9.HR-EGFP to Neonatal Mice Resulted in Widespread Neuronal Transduction in the CNS with Peripheral Tissue De-targeting Compared with scAAV9wt-EGFP
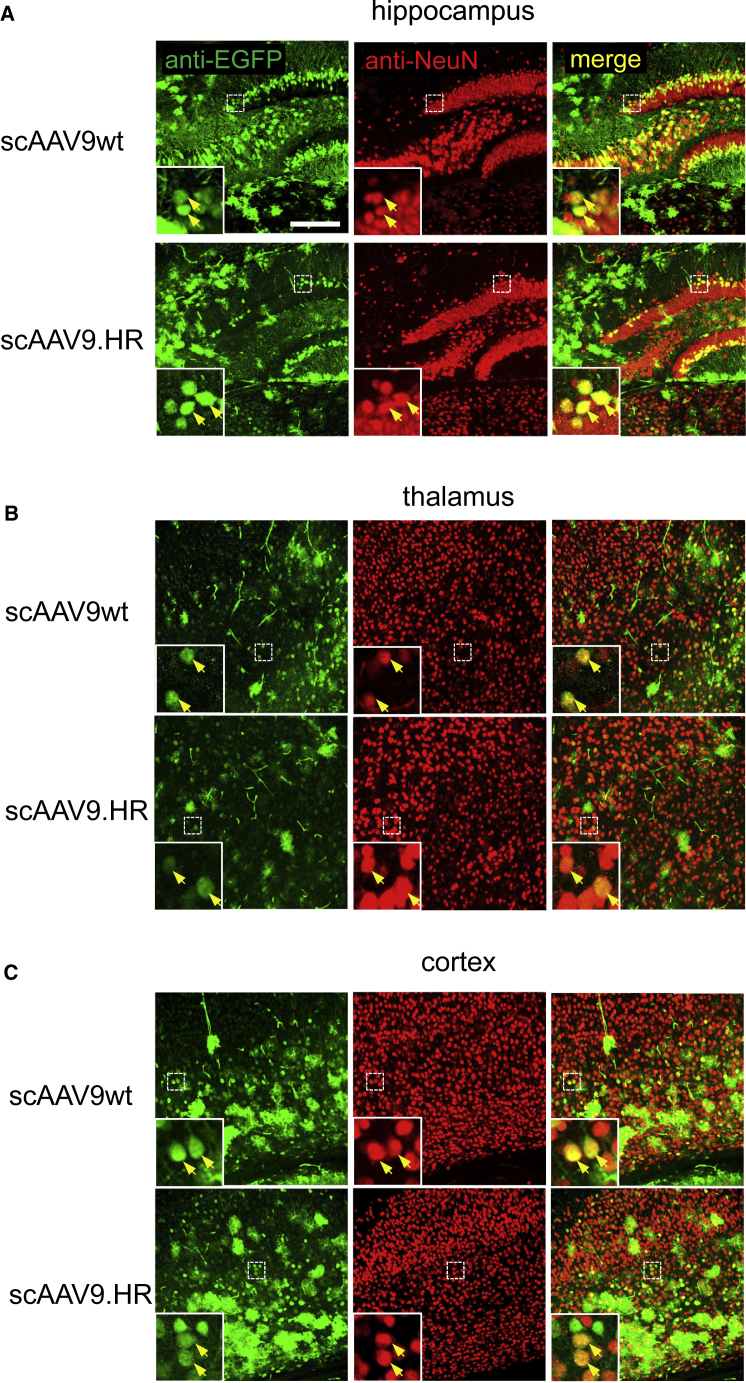
rAAV9 vectors transduced neurons more efficiently following IV delivery into neonatal mice than into adult mice.7 Therefore, we next injected 4.0E+11 GC of scAAV9wt-EGFP or scAAV9.HR-EGFP into postnatal day 1 (P1) pups and compared transduction profile of the two vectors in both peripheral tissues and CNS. Consistent with the results in young adult mice, we observed comparable EGFP expression in the liver but significantly reduced EGFP expression in the heart and TA muscle with scAAV9.HR (Figures 4A and 4B). One should note that the sparse EGFP-expressing cells in the liver was likely attributable to the dilution of rAAV genomes along with rapid hepatocyte division after birth.26 In both brain and spinal cord, scAAV9.HR-EGFP showed generally comparable transduction with scAAV9wt-EGFP regarding spread and efficiency, except for the hippocampus in the brain and the dorsal portion of the spinal cord, where transduction by scAAV9.HR-EGFP was weaker (Figures 4C and 4D). Immunofluorescence labeling and confocal microscopy confirmed that both vectors resulted in neuronal transduction in various brain regions, including hippocampus, thalamus, and cortex (Figure 5). In addition, some cells transduced by either scAAV9wt or scAAV9.HR appear to be astrocytes based on morphology, consistent with previous reports that AAV9 has high tropism for astrocytes.7, 8
Figure 4.
Comparison of scAAV9wt and scAAV9.HR Transduction Profiles following IV Delivery of High Dose (4.0E+11 GC) into Newborn Mice (P1)
(A) Representative fluorescence images showing native EGFP fluorescence in tissue sections of liver, heart, and TA muscle at three weeks after treatment with EGFP-expressing vectors packaged into AAV9 capsid (scAAV9wt) or AAV9.HR capsid (scAAV9.HR). The scale bar represents 500 μm. (B) Quantification of EGFP signal in tissue sections is shown. EGFP signal is quantified by the percentage of EGFP-positive pixels (% EGFP positive, top panel) or the average integrated intensity calculated as the ratio of total intensity over area (intensity/area, bottom panel). n = 4 for each group. **p < 0.01 and ns, not significant by Student’s t test. (C) Representative immunofluorescence images show EGFP signal in the brain and spinal cord (lumbar region) sections. Untreated samples processed in the same manner as the treated samples serve as fluorescence background control. Cerebral cortex (ctx), hippocampus (hpc), thalamus (thl), ventral horn of spinal cord (sc-v), and dorsal horn of spinal cord (sc-d) are highlighted and subjected to signal quantification. (D) Quantification of the immunofluorescence signal of the CNS regions shown in (C) is shown (n = 4). **p < 0.01 and ns, not significant by Student’s t test. Data are presented as mean and SD.
Figure 5.
scAAV9.HR and scAAV9wt Transduced Neurons in Various Brain Regions following IV Delivery of 4.0E+11 GC into Newborn Mice (P1)
Representative confocal immunofluorescence images showing anti-EGFP signal (green), neurons (red, labeled as NeuN-positive), and merged signal (yellow) in the hippocampus (A), thalamus (B), and cortex (C). The scale bar represents 200 μm.
rAAV9.HR-Mediated ASPA Gene Replacement Therapy Rescued Canavan Disease Phenotypes in a Mouse Model of the Disease
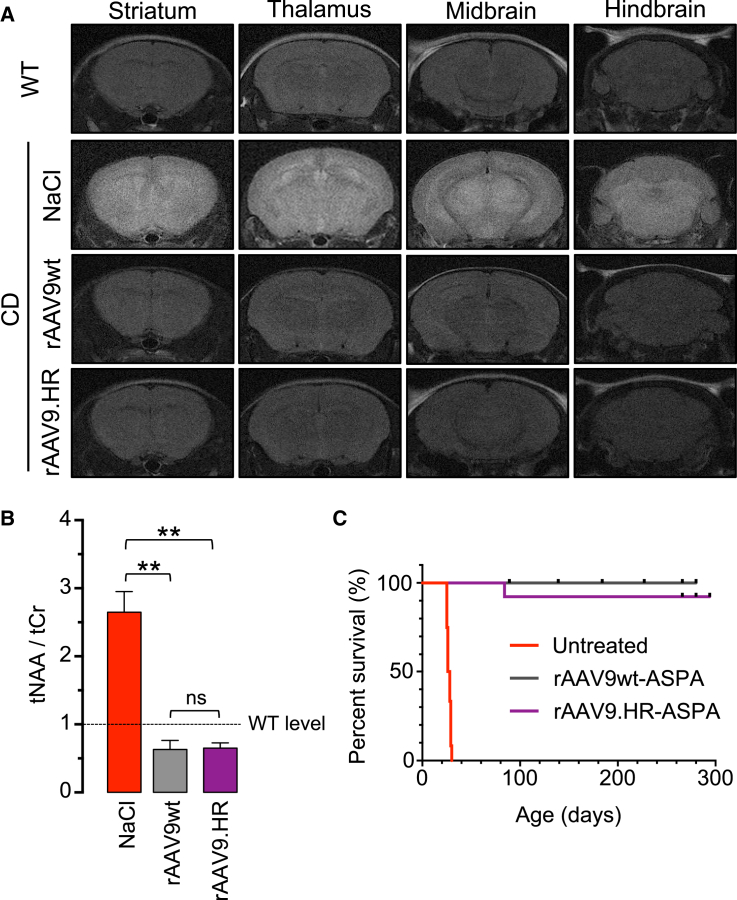
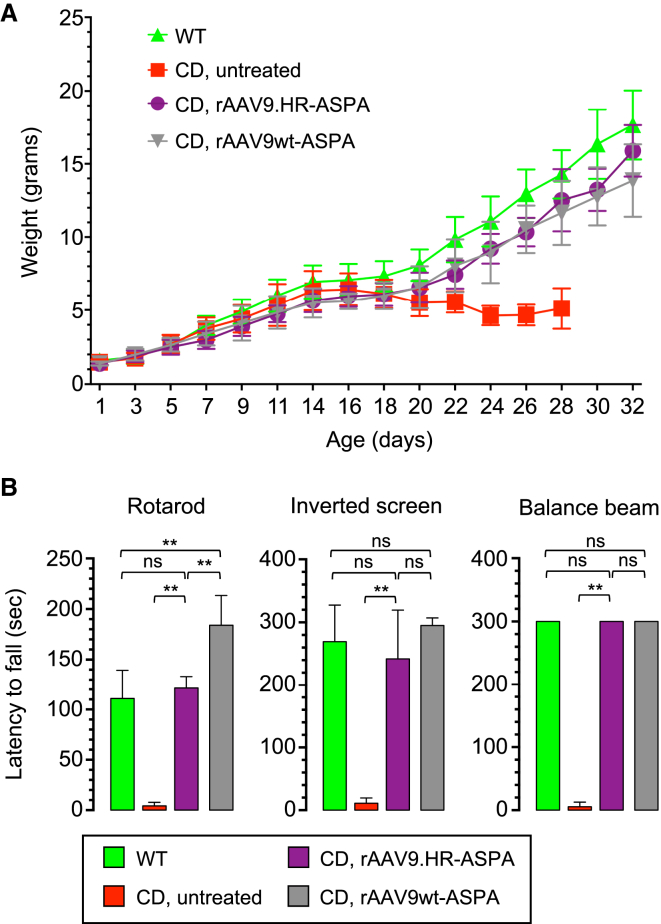
Encouraged by the finding that, following IV delivery into neonatal mice, the AAV9.HR capsid mutant is able to achieve comparable transduction to AAV9wt in most CNS compartments examined but reduced transduction in the heart and TA muscle, we next asked whether a therapeutic rAAV9.HR vector for treating a CNS disease is efficacious. To this end, we constructed a single-stranded rAAV genome expressing an optimized human ASPA gene (see Materials and Methods) and packaged into the AAV9wt or AAV9.HR capsid for gene replacement therapy in a mouse model of Canavan disease (CD). We injected 4.0E+11 GC of each vector into P1 CD mice and evaluated their therapeutic efficacy by a series of established measurements.9, 27 First, the saline-treated control mice showed signs of brain edema as revealed by T2-weighted MRI; both vectors completely corrected this phenotype (Figure 6A). Second, both vectors completely cleared brain N-acetylaspartic acid (NAA) accumulation due to ASPA deficiency seen in the saline-treated control CD mice as determined by magnetic resonance spectroscopy (MRS) (Figure 6B). Third, both vectors rescued the early lethality seen in the control CD mice and extended lifespan up to nine months, which was the duration of this study (Figure 6C). Fourth, treatment with rAAV9.HR-ASPA corrected the body weight loss observed in control CD mice during their short lifespan (Figure 7A) and over an extended period of time (Figure S5A). Fifth, treatment with rAAV9.HR-ASPA also restored the motor function to WT level, as revealed by three motor function tests, namely the rotarod test, the inverted screen test, and the balance beam test (Figure 7B). The CD mice receiving rAAV9wt-ASPA outperformed the WT mice in the rotarod test, resulting in a super-mouse phenotype as we previously reported.10 Importantly, the motor function improvement proved to be sustainable up to 180 days, the last time point when such tests were performed (Figure S5B). These results demonstrate that the AAV9.HR capsid mutant is suitable for vector development to target CNS diseases.
Figure 6.
IV Delivery of rAAV9.HR-ASPA at P1 to CD Mice Corrected Brain Pathology and Extended Survival as Efficiently as rAAV9wt-ASPA
(A) Coronal series of T2-weighted MRI brain images obtained from an untreated wild-type mouse (WT, P42) and Canavan disease (CD) mice treated with 0.9% NaCl (NaCl, P27), rAAV9wt-ASPA (rAAV9wt, P42), or rAAV9.HR-ASPA (rAAV9.HR, P42). Note that treatment with either rAAV9wt or rAAV9.HR in CD mice completely normalized the hyperintensity seen in the NaCl-treated CD mouse. (B) Bar graph shows NAA levels normalized to creatine levels (tNAA/tCr; determined by MRS) in the brains of CD mice treated as described in (A) (n = 3). **p < 0.01 and ns, not significant by Tukey’s multiple comparisons test following one-way ANOVA. Dashed line indicates WT level. Data are presented as mean and SD. (C) Kaplan-Meier survival curves show the percentage of survived mice over time course, including CD mice that were left untreated (red, n = 12), treated with rAAV9wt-ASPA (gray, n = 12), or treated with rAAV9.HR-ASPA (purple, n = 13).
Figure 7.
IV Delivery of rAAV9.HR-ASPA and rAAV9wt-ASPA at P1 to CD Mice Rescued Body Weight Loss and Behavioral Deficits
(A) Body weight curves plotted against age of untreated WT mice (green), untreated CD mice (red), and CD mice treated with rAAV9.HR-ASPA (purple) or rAAV9wt-ASPA (gray). n = 10 mice per group. (B) Motor function performance of untreated WT mice (green), untreated CD mice (red), and CD mice treated with rAAV9.HR-ASPA (purple) or rAAV9wt-ASPA (gray) in three behavioral tests, including rotarod test, inverted screen test, and balance beam test is shown. All tests were performed with P27 mice (n = 6 per group per test). One-way ANOVA was performed (p < 0.01). **p < 0.01 and ns, not significant by Tukey’s multiple comparisons test. Data are presented as mean and SD.
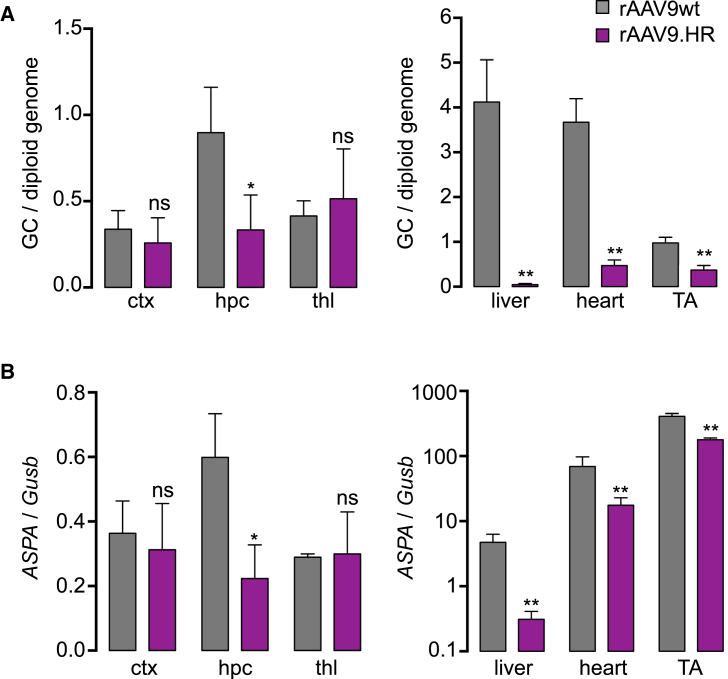
In a subset of treated CD mice, we quantified gene delivery efficacy of both ASPA vectors in various tissues. Consistent with the data of P1 injection with EGFP vectors (Figures 4C and 4D), the rAAV9.HR-ASPA and rAAV9wt-ASPA resulted in comparable genome delivery and expression in the cerebral cortex and thalamus, whereas rAAV9wt-ASPA was superior in hippocampus by both measurements (Figure 8). We also found de-targeting of rAAV9.HR-ASPA in all peripheral tissues examined, including liver, heart, and TA muscle (Figure 8). This is in contrast to the performance of the scAAV9.HR-EGFP vector that showed de-targeting in the heart and TA muscle, but not liver (Figures 4A and 4B).
Figure 8.
Quantification of rAAV Genome Copies and Transgene Expression following IV Delivery to CD Mice
(A) Genome copy numbers in ctx, hpc, thl, liver, heart, and tibialis anterior muscle (TA) in CD mice that were treated with either rAAVwt-ASPA or rAAV.HR-ASPA at 4.0E+11 GC at P1. Mice were euthanized four weeks after treatment. Data are presented as genome copies per diploid genome (GC/diploid genome). **p < 0.01 and ns, not significant by Student’s t test. (B) ASPA gene expression (cDNA) normalized to Gusb expression in the same tissues of the same mice described in (A) is shown. **p < 0.01 and ns, not significant by Student’s t test. All data are presented as mean and SD.
Discussion
The AAV9.HR capsid described in this study will be useful in various gene therapy applications requiring de-targeting in specific tissues. First, following IM delivery, rAAV9.HR has tropism and transduction in the injected muscle as high as rAAV9, but liver tropism and transduction is diminished (Figure 2). This makes AAV9.HR an attractive capsid for muscle-directed gene delivery when unwanted liver transduction may cause toxicity. In addition, muscle-specific promoter and a miRNA de-targeting strategy can be utilized along with the AAV9.HR capsid to further restrict gene expression in the muscle. Second, following IV delivery into neonates, AAV9.HR vectors packaging either a sc genome (Figures 4C and 4D) or a single-stranded (ss) genome (Figure 8) yielded widespread brain transduction as efficiently as AAV9wt except for hippocampus, but transduction in peripheral tissues is greatly reduced. This feature of AAV9.HR will reduce the risk of off-target toxicity in peripheral tissues when the vector is applied for CNS gene therapy. We observed peripheral tissue de-targeting in the heart and skeletal muscle with the scAAV9.HR-EGFP vector; in contrast, de-targeting occurred in the liver, heart, and skeletal muscle with the ss rAAV.HR-ASPA vector. We postulate that the difference in liver de-targeting between the two vectors may be due to the genome configuration. Consistent with this notion, the ss rAAV9.HR-Fluc vector also showed liver de-targeting compared with rAAV9wt-Fluc following IV delivery (Figure 2).
With the exception of the change at amino acid position 647, the CLv-D8 amino acid differences to WT AAV9 are located on the protrusions that surround the icosahedral 3-fold axis. This capsid region has been implicated as a determinant of receptor binding and cellular transduction for numerous AAVs, including AAV9.18, 19, 20, 22, 23 The surface location of H527Y and R533S and their effect on tissue tropism highlights their potential role in similar functions on the AAV capsid. The mechanism underlying the tissue de-targeting property conferred by the H527Y and R533S mutations following a systemic route remains to be studied. Nevertheless, the notion that AAV9.HR is a viable capsid for CNS gene therapy is fortified by the successful gene replacement therapy in a mouse model of Canavan disease, a fatal neurological disease affecting broad brain regions (Figures 6 and 7).
Following IV delivery, the two-residue change in AAV9.HR compared with AAV9, i.e., H527Y+R533S, severely impairs brain transduction in young adult mice, but not so much in neonatal mice (Figures 3 and 4). We also notice that IV rAAV9wt and rAAV9.HR vectors transduce massive structures resembling blood vessels in the young adult brain but much less so in the neonatal brain (Figures 3 and 4). Despite productive effort in characterizing AAV9’s receptor binding domain,20 its capsid feature contributing to crossing the BBB from vasculature remains unknown, although transcytosis across the endothelial cells of brain blood vessel is a plausible candidate mechanism.28, 29 We speculate that the AAV9wt and/or AAV9.HR may employ different routes to cross the BBB in adult brain and in neonatal brain, which may also reflect potential age-dependent features of the BBB.
In summary, we demonstrate the feasibility of a combinatory approach to rAAV capsid development, employing WT AAV capsid discovery from nature and rational engineering guided by sequence and structural analysis. A handful of naturally occurring serotype capsids found in human and NHP lay the foundation of rAAV-mediated gene therapy,2 some of which are becoming the vectors of choice for different in vivo gene transfer applications nowadays. In the future, designing AAV capsids with clinically desired properties will be a continuing stream of effort. There exists a large repertoire of AAV capsid in nature, which can be captured in a high-throughput manner.30 These capsid sequences survive in vivo natural evolution31 and likely possess important features relevant to therapeutic vector development, such as being able to encapsidate DNA and tissue tropism. Therefore, these capsids can serve as lead candidates for further engineering. A better understanding of the capsid sequence, structure, and function allows for selecting a manageable subset of natural variants for biological characterization based on computational predictions, which will accelerate the development of AAV vectors against a myriad of clinical targets.
Materials and Methods
Animal Use
Snap-frozen post mortem tissues from a chimpanzee were obtained from the University of Louisiana at Lafayette New Iberia Research Center tissue bank, including brain, liver, lung, and spleen. This animal died from unrelated health issues and underwent necropsy. All mice were bred in house and maintained on a 12-hr light cycle. Only male C57BL/6 WT mice were used due to differences in transduction between genders.32 For IV delivery, 6-week-old mice were injected with rAAV through tail vein, whereas P1 pups were injected through facial vein. For IM delivery, 6-week-old mice were injected at the left tibialis anterior muscle. All injections were performed using 28.5-gauge needles. The CD mice were described previously.33 Briefly, the Aspa gene is disrupted by a targeted insertion of neo gene in exon 4. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and the Institutional Biosafety Committee (IBC) at the University of Massachusetts Medical School.
Isolating AAV Capsid Variants from Chimpanzee Tissues
The chimpanzee tissues were manually ground into powder in liquid nitrogen using mortar and pestle. Total DNA was extracted from each tissue using the QIAamp DNA Mini Kit (QIAGEN; cat no. 51306). Total RNA was extracted using the TRIzol Reagent (Thermo Fisher Scientific; cat no. 15596018), followed by reverse transcription into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific; cat no. 4368814). Both DNA and cDNA samples were used as templates to perform AAV cap gene PCR using the Platinum PCR SuperMix High Fidelity kit (Thermo Fisher Scientific; cat no. 12532016). The primers bind to conserved sequences found in AAV genome. The forward primer is as follows: 5′-GCCGACAGGTACCAAAACAAATGT-3′. The reverse primer is as follows: 5′-GAAACGAATTAACCGGTTTATTGATTAA-3′. This PCR yielded products of approximately 2.5 kb pairs (kb) covering the entire cap gene. After being resolved on agarose gel, PCR product of the expected size was excised and purified using the QIAquick Gel Extraction Kit (QIAGEN; cat no. 28706) and subjected to TOPO cloning using the pCR4-TOPO Vector (Thermo Fisher Scientific; cat no. K457502). Plasmid in single colonies of bacteria cells was expanded and sequenced by Sanger method. The resulting DNA sequence was “translated” into amino acid sequences, which were aligned with the capsid protein sequences of AAV serotypes 1, 3B, 4, 5, 6, and 9. One of the resulting variants is named as CLv-D8 (chimpanzee liver, clone D8), which is the focus of this study.
Plasmid Constructs and rAAV Vector Production
The firefly luciferase (Fluc)-expressing construct contains a cytomegalovirus enhancer fused with chicken β-actin promoter (CMV/CB) driving the Fluc gene. The EGFP-expressing construct contains CMV/CB driving the EGFP gene. Plasmids expressing mutated CLv-D8 capsid were generated by mutating the isolated CLv-D8 cap gene using the QuikChange II Site-Directed Mutagenesis Kit (Agilent; cat no. 200523). The construct expressing human ASPA contains CMV/CB driving a codon-optimized human ASPA cDNA. The Kozak sequence (GCCACC) is added upstream of the start codon sequence to enhance gene expression. The Fluc- and ASPA-expressing constructs were packaged as single-stranded vectors. The EGFP-expressing construct was packaged as a sc vector. rAAV vectors were packaged using a standard triple-transfection protocol as previously described34 and purified by CsCl sedimentation. Vector titer was determined by qPCR for genome titer and gel electrophoresis followed by silver staining for capsid titer.
In Vivo Imaging
D-luciferin (Gold Biotechnology; cat no. LUCK-1G) was dissolved in sterile PBS, and aliquots were stored under −80°C until use. Mice were anesthetized by isoflurane throughout the procedure. 250 μL D-luciferin solution was injected into each animal by intraperitoneal injection. Ten minutes later, luminescence was measured using the Xenogen IVIS 100 in vivo imaging system in the unit of photons/s/cm2/sr (PerkinElmer, Waltham, MA).
Structural Modeling of the CLv-D8 Capsid Variant
The 3D model of CLv-D8 was generated in the Coot program35 by using the “simple mutation” function to introduce amino acid changes onto the WT AAV9 VP3 monomer (PDB accession number 3UX1). The four residues that differ between CLv-D8 and AAV9, amino acid positions 445, 527, 533, and 647, were mutated from tyrosine to histidine, histidine to tyrosine, arginine to serine, and isoleucine to threonine, respectively. The side chain orientations of these altered residues were adjusted to fit structurally allowed rotamers and also to avoid steric hindrance when necessary. The model was not energy minimized. The coordinate of the VP3 trimer was generated for the CLv-D8 model and WT AAV9 crystal structure using the online VIPERdb server by icosahedral matrix multiplication.36 The CLv-D8 model was compared to the WT AAV9 structure, and potential losses/gains of interactions due to the residue mutations were analyzed interactively in the Coot program.35 The images of the variant VP monomer and trimer ribbon representations were generated using PyMOL program.37
qPCR Assay to Determine rAAV GC Biodistribution
Mice were sacrificed by CO2 inhalation followed by cervical dislocation. Tissues of interest were immediately dissected, flash-frozen in liquid nitrogen, and stored under −80°C. Frozen tissues were ground in liquid nitrogen using mortar and pestle. Total DNA was extracted from an aliquot of tissue powder using the QIAamp DNA Mini Kit (QIAGEN; cat no. 51306), and DNA concentration was quantified by UV spectrometry using a Synergy HT microplate reader (BioTek, Winooski, VT). 100 ng of total DNA was used in a qPCR assay using a Taqman reagent targeting the rabbit β-globin polyadenylation signal (RBGpA) present in the rAAV genome. The rAAV GC number in tissue DNA was calculated by comparing against a standard curve generated using a known amount of plasmid DNA containing the RBGpA sequence and presented as GC per mouse diploid genome, assuming that 100 ng of mouse genomic DNA is equivalent to ∼3 × 104 mouse haploid genomes.
Luciferase Activity Assay in Tissue Lysate
An aliquot of tissue powder was resuspended in Passive Lysis Buffer (Promega; cat no. E1941) and underwent three times of freezing in a dry ice/ethanol bath and thawing in a 37°C water bath. This method is efficient in breaking cell membranes and extracting total protein.38 Cell debris was spun down, and clear cell lysate was transferred into a microcentrifuge tube and kept on ice. Protein concentration was determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; cat no. 23335). Luciferase activity was determined using the Luciferase Assay System (Promega; cat no. E1501). Luciferase activity was calculated as relative light units per 100 μg of total protein (RLU/100 μg protein).
Blood Clearance of rAAVs
Five microliters of blood was drawn from the tail vein at each time point as indicated and mixed with 995 μL of PBS. Two microliters of the diluted blood was directly used in a Taqman qPCR assay to quantify rAAV genome sequence, namely the RBGpA, by comparing against a standard curve. To generate the standard curve, the rAAV vectors of the same batch as used for injection were serially diluted with 0.5% naive mouse blood and included in the same qPCR assay.
EGFP Fluorescence Detection and Quantification in Peripheral Tissue Sections
Mice were sacrificed by perfusion with ice-cold PBS and 4% paraformaldehyde (PFA). Whole body was then immersed in 4% PFA. After overnight post-fixation, peripheral tissues of interest were cryo-preserved by being immersed in 10%, 20%, and 30% sucrose overnight for each gradient and embedded in O.C.T. Compound (Tissue-Tek; cat no. 4583). Tissues were cut into 8-μm sections using a cryostat (cross-section for heart and TA muscle; no specific orientation for liver) and mounted on a glass slide using the VECTASHIELD Mounting Medium with DAPI (Vector Laboratories; cat no. H-1200). EGFP fluorescence was imaged using an inverted fluorescence microscope (Leica Microsystems). EGFP signal in captured images was quantified using the ImageJ software (NIH).
Immunofluorescence in CNS Tissue Sections and Quantification of Anti-EGFP Signals
After overnight post-fixation as described above, brain and spinal cord were carefully dissected and immersed in 4% PFA overnight and then underwent cryo-preservation and embedding in O.C.T. in the same manner as for peripheral tissues as mentioned above. CNS tissues were cut into 40-μm coronal sections using a cryostat. Sections were co-stained using antibodies against EGFP (Thermo Fisher Scientific; cat no. A-11122; 1:1,000 diluted) and NeuN (Millipore; cat no. MAB377; 1:1,000 diluted) and proper fluorophore-labeled secondary antibodies derived from goat (Invitrogen). To stain for blood vessels, DyLight-labeled tomato lectin was used (Vector Laboratories; cat. no. DL-1178). Stained sections were mounted as described above. Immunofluorescence signal from entire section was captured using an upright fluorescence microscope with tilling function enabled (Leica Microsystems). Signal quantification was performed using the ImageJ software. Briefly, the regions of interest (ROIs), i.e., cortex, hippocampus, thalamus, spinal cord ventral, and dorsal horns, were selected, and the mean intensity within each ROI was automatically calculated by the software. Confocal microscopy (Leica Microsytems) was employed to capture both anti-EGFP and anti-NeuN signal in order to demonstrate co-localization.
MRI and MRS
Anesthesia was induced with isoflurane and maintained with 1.5% isoflurane using the Surgivet tabletop isoflurane vaporizer (Smiths Medical, Waukesha, WI, USA). Vital signs were monitored throughout the imaging session. Mice were imaged with a 4.7T/40-cm horizontal magnet (Oxford, UK) equipped with a Biospec Avance Bruker console (Bruker, Germany). A 1H radiofrequency mouse head coil (Bruker, Germany; inner diameter of 23 mm) was used for the experiments. T2-weighted image acquisition was performed using TurboRARE sequence with TR = 2,200 ms, TE = 36 ms, echo spacing = 12 ms, 8 averages, and rare factor = 8. Voxel PRESS was used for 1H magnetic resonance spectroscopy data acquisition (point resolved spectroscopy sequence; repetition time = 2,500 ms; echo time = 16 ms; number of averages = 512; voxel size = 3 × 3 × 3 mm). For proton spectra fitting, the LCModel (version 6.2-2B) was used. This model analyzed in vivo proton spectrum linearity combined with in vitro spectra from individual metabolite solutions39 and generated data as absolute fits (in institutional units) and SD%. SD was used to assess reliability of the fit. Inclusion criteria for the spectra were SD < 20% for NAA, creatine, and inositol.
Behavioral Tests
Overall motor performance was measured using accelerated rotarod (Roto-Rod Series 8; IITC Life Science, Woodland Hills, CA) for coordination function and endurance, balance beam for vestibular function and ataxia, and inverted screen for grip strength. The inverted screen and balance beam equipment was built by the University of Massachusetts Medical School Machine Shop.
Accelerated Rotarod
Mice were trained for two days prior to testing. On the day of testing, mice were acclimated for one minute on the beam. Each mouse was evaluated three times. The best value was counted. The acceleration and timing was set to 4–40 rpm over five minutes.
Balance Beam
A single test run lasted for up to five minutes. Briefly, mice were placed on the balance beam, and the time until drop-off was recorded. This was repeated three times, and the best value was counted.
Inverted Screen
A grid of 30 cm2 with 25 mm2 holes in horizontal position was used as initial setup. Mice were placed in the center and left unchallenged to acclimate for one minute. Next, the grid was brought to an angle of 125 degrees in order to force the mouse to hang upside down. The time until drop-off was recorded. The maximum duration of a single test run was five minutes.
ddPCR
DNA and RNA were extracted from various tissues using the AllPrep DNA/RNA Mini Kit (QIAGEN; cat. no. 80204). RNA was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific; cat no. 4368814). Taqman-based multiplexing droplet digital PCR (ddPCR) was performed using a QX200 ddPCR instrument (Bio-Rad). For both vector genome analysis and transgene expression analysis, a customized Taqman reagent targeting the codon-optimized human ASPA cDNA was used (Thermo Fisher Scientific; Assay ID APNKRZW). We used the Taqman reagents targeting mouse Tfrc genomic sequence (Invitrogen; cat. no. 4458367) and mouse Gusb cDNA sequence (Invitrogen; Assay ID Mm01197698_m1) as reference for vector genome analysis and transgene expression analysis, respectively.
Statistical Analysis
Two-tailed Student’s t test was used to compare between two groups. To compare among multiple groups, one-way ANOVA was used followed by Tukey’s multiple comparisons test. Statistical analysis was performed using the GraphPad Prism 7 software.
Author Contributions
Conceptualization, M.A.-M. and G.G.; Methodology, D.W., S.L., D.J.G., J.X., L.Z., and K.V.V.; Investigation, D.W., S.L., D.J.G., J.X., L.Z., J.L., K.T., K.V.V., L.R., Q.S., and R.H.; Resources, J.E.G., T.R.F., M.A.-M., and G.G.; Writing – Original Draft, D.W.; Writing – Review & Editing, K.V.V., J.E.G., T.R.F., M.A.-M., and G.G.; Visualization, D.W., S.L., D.J.G., and K.V.V.; Supervision, M.A.-M. and G.G.; Project Administration, D.W. and L.Z.; Funding Acquisition, M.A.-M. and G.G.
Conflicts of Interest
J.X. and G.G. hold the patent on the DNA sequences of the AAV capsid variants isolated from chimpanzee tissues. G.G. is a scientific co-founder of Voyager Therapeutics and holds equity in the company. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics and other biopharmaceuticals. M.A.-M. is a co-founder of StrideBio Therapeutics, a biopharmaceutical company with interest in developing AAV vectors for gene delivery applications. M.A.-M. is an SAB member for Voyager Therapeutics and AGTC and is a consultant to Intima Biosciences. M.A.-M. has sponsored research agreements with AGTC, Adverum Biotechnologies, and Voyager Therapeutics. These companies have interest in the development of AAV for gene delivery applications. M.A.-M. is an inventor of AAV patents licensed to various biopharmaceutical companies.
Acknowledgments
This work is primarily supported by an internal grant from the University of Massachusetts Medical School and partially supported by grants from the NIH (1P01AI100263-05, 1R01NS076991-05, R01HL097088, and 4P01HL131471-01 to G.G. and R01GM082946 to M.A.-M.).
Footnotes
Supplemental Information includes five figures and one table and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.03.004.
Contributor Information
Mavis Agbandje-McKenna, Email: mckenna@ufl.edu.
Guangping Gao, Email: guangping.gao@umassmed.edu.
Supplemental Information
References
- 1.Muzyczka N., Berns K. Parvoviridae: the viruses and their replication. In: Knipe D., editor. Volume 2. Lippincott, Williams and Wilkins; Philadelphia, PA: 2001. pp. 2327–2359. (Fields Virology). [Google Scholar]
- 2.Gao G., Zhong L., Danos O. Exploiting natural diversity of AAV for the design of vectors with novel properties. Methods Mol. Biol. 2011;807:93–118. doi: 10.1007/978-1-61779-370-7_4. [DOI] [PubMed] [Google Scholar]
- 3.Münch R.C., Muth A., Muik A., Friedel T., Schmatz J., Dreier B., Trkola A., Plückthun A., Büning H., Buchholz C.J. Off-target-free gene delivery by affinity-purified receptor-targeted viral vectors. Nat. Commun. 2015;6:6246. doi: 10.1038/ncomms7246. [DOI] [PubMed] [Google Scholar]
- 4.Asokan A., Conway J.C., Phillips J.L., Li C., Hegge J., Sinnott R., Yadav S., DiPrimio N., Nam H.J., Agbandje-McKenna M. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat. Biotechnol. 2010;28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 7.Foust K.D., Nurre E., Montgomery C.L., Hernandez A., Chan C.M., Kaspar B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duque S., Joussemet B., Riviere C., Marais T., Dubreil L., Douar A.M., Fyfe J., Moullier P., Colle M.A., Barkats M. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed S.S., Li H., Cao C., Sikoglu E.M., Denninger A.R., Su Q., Eaton S., Liso Navarro A.A., Xie J., Szucs S. A single intravenous rAAV injection as late as P20 achieves efficacious and sustained CNS Gene therapy in Canavan mice. Mol. Ther. 2013;21:2136–2147. doi: 10.1038/mt.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gessler D.J., Li D., Xu H., Su Q., Sanmiguel J., Tuncer S., Moore C., King J., Matalon R., Gao G. RedirectingN-acetylaspartate metabolism in the central nervous system normalizes myelination and rescues Canavan disease. JCI Insight. 2017;2:e90807. doi: 10.1172/jci.insight.90807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L., Kissel J.T., Prior T.W., Miranda C., Lowes L., Alfano L. Gene therapy for spinal muscular atrophy type 1 shows potential to improve survival and motor functional outcomes. Mol. Ther. 2016;24:S190. [Google Scholar]
- 12.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018 doi: 10.1089/hum.2018.015. Published online February 12, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salva M.Z., Himeda C.L., Tai P.W., Nishiuchi E., Gregorevic P., Allen J.M., Finn E.E., Nguyen Q.G., Blankinship M.J., Meuse L. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol. Ther. 2007;15:320–329. doi: 10.1038/sj.mt.6300027. [DOI] [PubMed] [Google Scholar]
- 14.Xie J., Xie Q., Zhang H., Ameres S.L., Hung J.H., Su Q., He R., Mu X., Seher Ahmed S., Park S. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol. Ther. 2011;19:526–535. doi: 10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulicherla N., Shen S., Yadav S., Debbink K., Govindasamy L., Agbandje-McKenna M., Asokan A. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol. Ther. 2011;19:1070–1078. doi: 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 17.Drouin L.M., Agbandje-McKenna M. Adeno-associated virus structural biology as a tool in vector development. Future Virol. 2013;8:1183–1199. doi: 10.2217/fvl.13.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell C.L., Vandenberghe L.H., Bell P., Limberis M.P., Gao G.P., Van Vliet K., Agbandje-McKenna M., Wilson J.M. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J. Clin. Invest. 2011;121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen S., Bryant K.D., Brown S.M., Randell S.H., Asokan A. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J. Biol. Chem. 2011;286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell C.L., Gurda B.L., Van Vliet K., Agbandje-McKenna M., Wilson J.M. Identification of the galactose binding domain of the adeno-associated virus serotype 9 capsid. J. Virol. 2012;86:7326–7333. doi: 10.1128/JVI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen S., Berry G.E., Castellanos Rivera R.M., Cheung R.Y., Troupes A.N., Brown S.M., Kafri T., Asokan A. Functional analysis of the putative integrin recognition motif on adeno-associated virus 9. J. Biol. Chem. 2015;290:1496–1504. doi: 10.1074/jbc.M114.608281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akache B., Grimm D., Pandey K., Yant S.R., Xu H., Kay M.A. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotchey N.M., Adachi K., Zahid M., Inagaki K., Charan R., Parker R.S., Nakai H. A potential role of distinctively delayed blood clearance of recombinant adeno-associated virus serotype 9 in robust cardiac transduction. Mol. Ther. 2011;19:1079–1089. doi: 10.1038/mt.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiMattia M.A., Nam H.J., Van Vliet K., Mitchell M., Bennett A., Gurda B.L., McKenna R., Olson N.H., Sinkovits R.S., Potter M. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012;86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen S., Bryant K.D., Sun J., Brown S.M., Troupes A., Pulicherla N., Asokan A. Glycan binding avidity determines the systemic fate of adeno-associated virus type 9. J. Virol. 2012;86:10408–10417. doi: 10.1128/JVI.01155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., Wang H., Bell P., McMenamin D., Wilson J.M. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum. Gene Ther. 2012;23:533–539. doi: 10.1089/hum.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed S.S., Schattgen S.A., Frakes A.E., Sikoglu E.M., Su Q., Li J., Hampton T.G., Denninger A.R., Kirschner D.A., Kaspar B. rAAV gene therapy in a Canavan’s disease mouse model reveals immune impairments and an extended pathology beyond the central nervous system. Mol. Ther. 2016;24:1030–1041. doi: 10.1038/mt.2016.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Pasquale G., Chiorini J.A. AAV transcytosis through barrier epithelia and endothelium. Mol. Ther. 2006;13:506–516. doi: 10.1016/j.ymthe.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Merkel S.F., Andrews A.M., Lutton E.M., Mu D., Hudry E., Hyman B.T., Maguire C.A., Ramirez S.H. Trafficking of adeno-associated virus vectors across a model of the blood-brain barrier; a comparative study of transcytosis and transduction using primary human brain endothelial cells. J. Neurochem. 2017;140:216–230. doi: 10.1111/jnc.13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu G., Luo L., Tai P.W.L., Qin W., Xiao Y., Wang C., Su Q., Ma H., He R., Wei Y. High-throughput sequencing of AAV proviral libraries from the human population reveals novel variants with unprecedented intra- and inter-tissue diversity. Mol. Ther. 2016;24:S4. [Google Scholar]
- 31.Gao G., Alvira M.R., Somanathan S., Lu Y., Vandenberghe L.H., Rux J.J., Calcedo R., Sanmiguel J., Abbas Z., Wilson J.M. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA. 2003;100:6081–6086. doi: 10.1073/pnas.0937739100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidoff A.M., Ng C.Y., Zhou J., Spence Y., Nathwani A.C. Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood. 2003;102:480–488. doi: 10.1182/blood-2002-09-2889. [DOI] [PubMed] [Google Scholar]
- 33.Matalon R., Rady P.L., Platt K.A., Skinner H.B., Quast M.J., Campbell G.A., Matalon K., Ceci J.D., Tyring S.K., Nehls M. Knock-out mouse for Canavan disease: a model for gene transfer to the central nervous system. J. Gene Med. 2000;2:165–175. doi: 10.1002/(SICI)1521-2254(200005/06)2:3<165::AID-JGM107>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 34.Gao G., Sena-Esteves M. Introducing genes into mammalian cells: viral vectors. In: Green M.R., Sambrook J.R., editors. Molecular Cloning. Fourth Edition. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2012. pp. 1209–1330. [Google Scholar]
- 35.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Carrillo-Tripp M., Shepherd C.M., Borelli I.A., Venkataraman S., Lander G., Natarajan P., Johnson J.E., Brooks C.L., 3rd, Reddy V.S. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLano W.L. DeLano Scientific; New York, NY: 2008. The PyMOL Molecular Graphics System. [Google Scholar]
- 38.Lu J., Jiang C. Detergents inhibit chloramphenicol acetyl transferase. Biotechniques. 1992;12:643–644. [PubMed] [Google Scholar]
- 39.Provencher S.W. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.