Abstract
Endometriosis is a disease in which endometriotic tissue occurs outside the uterus. Its pathogenesis is still unknown. The most widespread hypothesis claims that ectopic endometrium appears as a result of retrograde menstruation and its insufficient elimination by immunocytes. Some reports have shown expression of non-classical HLA-G molecules on ectopic endometrium. HLA-G is recognized by KIR2DL4, LILRB1 and LILRB2 receptors on natural killer (NK) and other cells. These receptors are polymorphic, which may affect their activity. In this study we investigated whether HLA-G, KIR2DL4, LILRB1 and LILRB2 polymorphisms may influence susceptibility to endometriosis and disease progression. We used polymerase chain reaction (PCR), PCR-restriction fragment length polymorphism (PCR-RFLP) and allelic discrimination methods with TaqMan SNP Genotyping Assays for typing of 276 patients with endometriosis and 314 healthy fertile women. The HLA-G rs1632947:GG genotype was associated with protection against the disease and its severe stages; HLA-G rs1233334:CT protected against progression; LILRB1 rs41308748:AA and LILRB2 rs383369:AG predisposed to the disease and its progression. No effect of KIR2DL4 polymorphism was observed. These results support the role of polymorphisms of HLA-G and its receptors LILRB1 and LILRB2 in susceptibility to endometriosis and its progression.
Electronic supplementary material
The online version of this article (10.1007/s00438-017-1404-3) contains supplementary material, which is available to authorized users.
Keywords: Endometriosis, KIR2DL4, LILRB, HLA-G
Introduction
Endometriosis is an estrogen-dependent gynecological disease, affecting about 10% of women in reproductive age. It is associated with the occurrence of endometrium outside the uterus. Endometriotic lesions can be found mainly in the ovaries and pelvic peritoneum, but also in the rectovaginal septum, and at more distant locations such as the lung, liver, and pancreas, and even in scars after operative surgery (Ahn et al. 2015; Serdar and Bulun 2009; Gupta et al. 2016; Parkin and Fazleabas 2016; Vercellini et al. 2014). In addition, endometriotic lesions may undergo malignant transformation (Worley et al. 2013). The etiopathology of endometriosis is still poorly understood. One hypothesis of endometriosis development is Sampson’s theory of retrograde menstruation (Sampson 1927; Dastur et al. 2010). According to this theory, retrograde menstruation may result in implantation, survival and growth of endometrial cell foci in the peritoneal cavity. The mechanism(s) of this phenomenon is unknown; it is plausible, however, that it may be due to insufficient elimination of endometrial cells by the local immune system. Indeed, women with endometriosis were found to have reduced activity of natural killer (NK) cells (Oosterlynck et al. 1992; Maeda et al. 2012; Eidukaite and Tamosiunas 2008; Tariverdian et al. 2009). These cells, granular cytotoxic lymphocytes, have been found not only in the peripheral blood, but also in the peritoneal fluid (Eidukaite and Tamosiunas 2008; Králíčková and Vetvicka 2015; Kawashima et al. 2009). A defect of the NK activity in the recognition and lysis of implanted endometrial cells may be thus one of the crucial mechanisms in the initiation and progression of endometriosis. NK cell activity is regulated by different receptors—with activating or inhibitory action—such as killer immunoglobulin-like receptors (KIRs) and leukocyte immunoglobulin-like receptors (LILRs) (Maeda et al. 2012; Králíčková and Vetvicka 2015; Borges et al. 1997; van der Touw et al. 2017). KIR and LILR recognize class I human leukocyte antigens (HLA), among them HLA-G. HLA-G is expressed by placental trophoblasts, and it is known as a crucial factor in maintaining pregnancy. However, it may also be expressed on ectopic endometrial tissue in the peritoneal cavity and be recognized by immune cells via its receptors: KIR2DL4 (of both inhibitory and activating potential), and inhibitory LILRB1 and LILRB2 (Maeda et al. 2012; Kawashima et al. 2009; Wang et al. 2008; Hudson and Allen 2016; Kang et al. 2016). Moreover, HLA-G up-regulates LILRB1, LILRB2 and KIR2DL4 expression in antigen-presenting cells, NK cells, and T cells (LeMaoult et al. 2005).
Previous GWA studies of endometriosis have implicated WNT (wingless-type MMTV integration site) signaling and oestrogen responsive genes, genes involved in the actin cytoskeleton and cellular adhesion (Rahmioglu et al. 2014; Nyholt et al. 2012), the CDKN2BAS locus encoding the cyclin-dependent kinase inhibitor 2B antisense RNA (Uno et al. 2010), and four single nucleotide polymorphisms (SNPs) located in and around interleukin 1α (Adachi et al. 2010). Most of the identified GWAS variants were non-coding. The most recently published studies by Sapkota et al. (2017a, b) have evaluated the potential role of coding variants in endometriosis risk by large exome-array analysis. However, their results did not identify any coding variants with MAF > 0.01, with moderate or large effect sizes in endometriosis pathogenesis. They provide genome-wide significant evidence for association with a splice variant (rs13394619) in the GREB1 (Growth Regulation By Estrogen In Breast Cancer 1) locus in women with European ancestry. Moreover, the 19 SNPs identified in endometriosis explain up to 5.19% of variance in endometriosis, suggesting that many more variants remain to be detected. On the other hand, we focused rather on genes important for innate immune response. In our previous paper we found an association of NK cell receptor KIR2DS5 gene and its potential ligand HLA-C C2 with endometriosis (Nowak et al. 2015a). Here, we analyzed other genes which may be involved in immune control of extra-uteral endometrial tissue. We examined the SNPs which may be associated with gene expression or splicing and therefore they could have potential influence on the receptor-ligand interaction between immune cells and ectopic endometrium.
Therefore, the aim of this retrospective study was to evaluate the association of the SNPs in genes coding for KIR2DL4, LILRB1 and LILRB2 receptors and their ligand HLA-G with susceptibility to and severity of endometriosis as potential non-invasive markers for the diagnosis of this disease.
Materials and methods
Study groups
The present study included 590 women from the Polish population who were enrolled during the period from 2005 to 2016. The study was approved by the Local Bioethics Committees at the Medical University of Wroclaw, Polish Mothers’ Memorial Hospital–Research Institute in Łódź, and the Medical University of Warsaw, Poland. Informed consent was obtained from all individual participants included in the study.
Endometriosis was diagnosed in 276 women. The patients were recruited at several Polish clinics: the First and Second Department of Obstetrics and Gynecology, Medical University of Warsaw; the Department of Surgical, Endoscopic and Oncologic Gynecology and the Department of Gynecology and Gynecologic Oncology in Polish Mothers’ Memorial Hospital–Research Institute in Łódź; and Gameta Hospital in Rzgów. The mean age of affected women was 33.02 ± 7.03 years. The diagnosis was based on laparoscopic surgery and confirmed by histopathological examination.
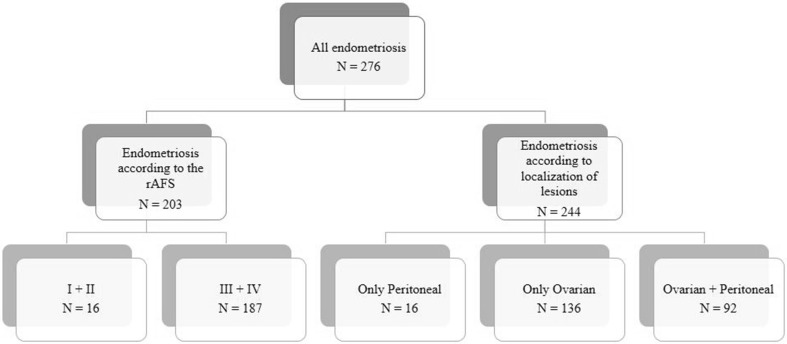
The patients were classified and analyzed according to the stage of the disease (American Fertility Society 1985) or according to the localization of the endometriotic lesions (Fig. 1). For 22 patients with endometriosis, detailed information on rAFS stage and lesion localization were not available.
Fig. 1.
Flowchart of the study population
The control group consisted of 314 fertile women. Among them 219 had at least two healthy-born children with the same partner without a history of spontaneous miscarriage and immunological or endocrinological diseases. Ninety-five women had at least one child. The mean age of fertile patients was 32.29 ± 5.81 years. The control group was recruited in the First Chair and Clinic of Obstetrics and Gynecology and the Department of Medical Genetics, University of Warsaw.
DNA preparation and genotyping
Genomic DNA was isolated from 5 mL of the peripheral blood samples collected during the patient’s admittance to the hospital using the Invisorb Spin Blood Midi Kit (Invitek, Berlin, Germany) according to the producer’s instructions.
HLA-G genotyping was conducted in three sequence positions. To detect the 14 base pair insertion/deletion (rs371194629:c.*65_*66insATTTGTTCATGCCT) in the 3′ untranslated region (UTR) we used the polymerase chain reaction with sequence-specific primers (PCR-SSP) method. The rs1632947:A>G polymorphism was distinguished by real-time PCR. Details of the genotyping of these two polymorphisms have been previously described by Wiśniewski et al. (2010, 2015). The genotyping of the triallelic rs1233334:G>C/T was performed on a 7300 Real-Time PCR System (Applied Biosystems) using Applied Biosystems (Foster City, CA) ready-made Assay-on-Demand including two primers—forward 5′-ACTGTCTGGGAAAGTGAAACTTAAGAG-3′ and reverse 5′-AATGTGACTTTGGCCTGTTGGTATA-3′—and two fluorescently labeled probes: 5′-VIC-CTTTGTGAGTCGTGTTGTA-NFQ-3′ and 5′-FAM-CTTTGTGAGTCCTGTTGTA-NFQ-3′. The 10-μl reaction mixture contained ~ 20 ng of genomic DNA, 1 × TaqMan Universal PCR Master Mix, No AmpErase Uracil N-Glycosylase (UNG) (Applied Biosystems), primers and probes. PCR conditions were as follows: 95 °C for 10 min and (95 °C for 15 s, 60 °C for 1 min) × 40. This genotyping was confirmed by direct sequencing. Fig S1 shows the distribution of representative results in the scatter plot from the real-time PCR of HLA-G rs1233334:G>C/T SNP genotyping.
There are two variants of KIR2DL4 with 9 or 10 consecutive adenines in the gene sequence. The deletion of one adenine in exon 7 contributes to the frame shift; therefore the 9A allele encodes the soluble form of the receptor with a missing transmembrane domain or truncated cytoplasmic tail. The 10A allele determines the membrane-bound receptor (Nowak et al. 2015b; Goodridge et al. 2007, 2009). The 10A/9A insertion/deletion in the 9620 position (rs11410751) of the KIR2DL4 gene has been previously found in complete linkage disequilibrium with the rs649216:T>C of the gene (r 2 = 1) in our population (Nowak et al. 2015b). The T allele of the rs649216 corresponded to the 9A allele of the rs11410751, while rs649216:C corresponded to the variant with the 10A allele. Therefore, we decided to use the PCR method and restriction fragment length polymorphism (RFLP) with EarI digestion for testing of the rs649216:T>C KIR2DL4 polymorphism, instead of the high resolution melting (HRM) method, which we found more expensive and troublesome than PCR-RFLP. Detailed protocols about these methods were published previously (Nowak et al. 2015b).
Genotyping of the rs41308748:G>A polymorphism in the LILRB1 gene as well as the rs383369:G>A polymorphism and the rs7247538:C>T polymorphism in the LILRB2 gene was carried out using PCR-RFLP. The restriction enzymes used in this study were as follows: AciI, TaiI and Hpy166II, respectively. The rs1061680:T>C in LILRB1 was genotyped using the allelic discrimination method with TaqMan SNP Genotyping Assay (C_9491145_10) on a 7300 Real-Time PCR System (Applied Biosystems). Primer sequences, annealing temperatures, restriction enzymes and reaction conditions for LILRB1 and LILRB2 genotyping are listed in Table S1. Reference samples for all tested SNPs were sequenced by an external company (Genomed, Poland). Detailed information of all tested polymorphisms and their potential functions is summarized in Table 1.
Table 1.
Summary of the tested SNPs
| Chr | Gene | dbSNP ID | Position (bp) | Accession number | Reference sequence | Functional region | Potential effect |
|---|---|---|---|---|---|---|---|
| 6 | HLA-G | rs1632947 | 29,826,881 | NC_000006.12 | XM_017010817.1:c-964G>A | Promoter | Gene expression (Castelli et al. 2014) |
| 6 | HLA-G | rs1233334 | 29,827,120 | NC_000006.12 |
XM_005249055.1:c.-725G>C XM_005249055.1:c.-725G>T |
Promoter | Gene expression (Hviid et al. 2004, 2006) |
| 6 | HLA-G | rs371194629 | 29,830,804–29,830,805 | NC_000006.12 | NM_002127.5:c.*65_*66insATTTGTTCATGCCT | 3′UTR of exon 8 | mRNA stability; splicing; microRNA targeting (Castelli et al. 2014); the14 bp insertion allele is associated with lower concentration of soluble HLA-G (Chen et al. 2008) |
| 19 | LILRB1 | rs41308748 | 54,636,725 | NC_000019.10 | NM_006669.6:c.1807-7G>A | Intronic | Splicing |
| 19 | LILRB1 | rs1061680 | 54,632,001 | NC_000019.10 | NM_006669.6:c.425T>C | Non-synonymous, extracellular D2 domain, NP_006660.4:p. 142 Ile142Thr |
Receptor-ligand interaction (Davidson et al. 2010; Kuroki et al. 2005) |
| 19 | LILRB2 | rs7247538 | 54,278,869 | NC_000019.10 | NM_001080978.3:c.898C>T | Non-synonymous,NP_001074447.2:p. 300 His300Tyr |
Splicing |
| 19 | LILRB2 | rs383369 | 54,280,275 | NC_000019.10 | NM_001080978.3:c.59G>A | Signal peptide NP_001074447.2:p. 20 Arg20His | Gene expression (Hirayasu et al. 2008) |
| 19 | KIR2DL4 | rs649216 | 54,813,180 | NC_000019.10 | NM_001080772.1:c.762T>C |
NP_001074241.1:p. 254 Phe254 |
In complete LD with rs11410751(NC_000019.10:g.54813228_54813229insA), which determines the encoding of soluble or membrane-bound KIR2DL4 receptor (Nowak et al. 2015b) |
Chr Chromosome; Genomic position is shown relative to GRCh38.p7; SNP IDs are according to dbSNP (rs, http://www.ncbi.nlm.nih.gov/SNP); c.*65_*66insATTTGTTCATGCCT was earlier described as 14 bp ins/del in 3′UTR of the HLA-G gene (Wiśniewski et al. 2010, 2015); NM_006669.6:c.1807-7G>A was earlier described as 5651 G>A (rs41308748) (Wiśniewski et al. 2015; Nowak et al. 2016); NM_006669.6:c.425T>C was earlier described as 927 T>C (rs1061680) (Davidson et al. 2010), and were relative to the translation start site
Statistical analysis
SNP frequencies were estimated by direct counting. The statistical significance of differences in genotype and allele frequencies between the control group and patients was estimated using the two-sided Fisher’s exact test and by the Chi-square test with the appropriate degrees of freedom, χ 2 df (df=(m − 1) × (n − 1), where m = number of rows, n = number of columns). A p value of less than 0.05 was required to reject the null hypothesis, which assumes that there is no difference in the distribution of genotypes and alleles between the control group and patients. If P < 0.05, it was corrected (P corr.) by the number of comparisons using Bonferroni correction. For 2 × 2 tables the odds ratio (OR) and 95% confidence interval for it were also calculated. Statistical analysis was performed using the software package GraphPad InStat version 3.06 (San Diego, CA, USA). Hardy–Weinberg equilibrium was checked using the Chi-square test with one degree of freedom for each SNP.
Results
HLA-G polymorphisms are associated with endometriosis
We found lower representation of rs1632947:GG genotype in patients with endometriosis than in controls (P = 0.04, P corr . = 0.12, OR = 0.61, 95% CI = 0.39–0.96; Table 2). Limitation of our analysis to patients with known localization of lesions gave similar results but remaining even after correction (P = 0.009, P corr. = 0.027, OR = 0.53, 95% CI = 0.33–0.85; Table S2).
Table 2.
HLA-G genotype and minor allele frequencies in women from Control and Endometriosis groups
| Genotype | Control (%) | Patients (%) | Patients vs control | ||||
|---|---|---|---|---|---|---|---|
| P | OR | 95% CI | Test for independence | ||||
| p | χ2 | ||||||
| rs371194629:ins/del | N = 314 | N = 276 | |||||
| Del/del* | 113 (35.99) | 97 (35.14) | 1 | 0.56 | 1.15 | ||
| Ins/del | 149 (47.45) | 124 (44.93) | 0.93 | 0.97 | (0.68–1.39) | ||
| Ins/ins | 52 (16.56) | 55 (19.93) | 0.41 | 1.23 | (0.77–1.96) | ||
| Minor allele ins | 253 (40.29) | 234 (42.39) | |||||
| H-W | 0.81 | 0.18 | |||||
| rs1632947:G>A | N = 314 | N = 276 | 0.08 | 4.97 | |||
| AA* | 63 (20.06) | 76 (27.54) | 1 | ||||
| AG | 157 (50.00) | 131 (47.46) | 0.08 | 0.69 | (0.46–1.04) | ||
| GG | 94 (29.94) | 69 (25.00) | 0.04 a | 0.61 | (0.39–0.96) | ||
| Minor allele A | 283 (45.06) | 283 (51.27) | |||||
| H-W | 0.86 | 0.41 | |||||
| rs1233334:G>C/T | N = 314 | N = 276 | 0.65 | 2.48 | |||
| CC* | 215 (68.47) | 188 (68.12) | 1 | ||||
| CG | 79 (25.16) | 70 (25.36) | 1.00 | 1.01 | (0.70–1.47) | ||
| GG | 8 (2.55) | 6 (2.17) | 1.00 | 0.86 | (0.29–2.52) | ||
| GT | 1 (0.32) | 4 (1.45) | 0.19 | 4.57 | (0.51–41.31) | ||
| CT | 11 (3.50) | 8 (2.90) | 0.82 | 0.83 | (0.33–2.11) | ||
| TT | 0 (0.00) | 0 (0.00) | – | – | – | ||
| Minor allele T | 12 (1.91) | 12 (2.17) | |||||
| H-W | 0.51 | 0.24 | |||||
H–W Hardy–Weinberg equilibrium, P probability, OR odds ratio, 95% CI 95% confidence interval from two-sided Fisher’s exact test, χ 2 df = 2 p Chi-square test for independence with two degrees of freedom for polymorphisms 14 bp ins/del (rs371194629:insATTTGTTCATGCCT/del) in 3′UTR and rs1632947:G>A, χ 2 df = 4 p Chi-square test for independence with four degrees of freedom for the polymorphism rs1233334:G>C/T
*Reference
a P corr. = 0.12
Comparison of patients with minimal and mild (I + II) with moderate and severe (III + IV) endometriosis revealed a protective effect of rs1632947:GG genotype (P = 0.04, OR = 0.2, 95% CI = 0.04–0.97), and, in addition, of rs1233334:CT genotype (P = 0.04, OR = 0.09, 95% CI = 0.01–0.62; Table 3). These associations lost significance after correction (P corr. = 0.12 for both comparisons).
Table 3.
HLA-G genotype frequencies in women depending on the severity of endometriosis
| Genotype | E I + II (%) | E III + IV (%) | E III + IV vs E I + II | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | OR | 95% CI | Test for independence | Test for trend | |||||
| p | χ2 | p | χ2 | ||||||
| rs371194629:ins/del | N = 16 | N = 187 | |||||||
| Del/del* | 8 (50.00) | 62 (33.16) | 1 | 0.24 | 2.89 | 0.52 | 0.41 | ||
| Ins/del | 4 (25.00) | 87 (46.52) | 0.13 | 2.81 | (0.81–9.74) | ||||
| Ins/ins | 4 (25.00) | 38 (20.32) | 1.00 | 1.23 | (0.35–4.35) | ||||
| Minor allele ins | 12 (37.50) | 163 (43.58) | |||||||
| rs1632947:G>A | N = 16 | N = 187 | |||||||
| AA* | 2 (12.50) | 56 (29.95) | 1 | 0.05 | 5.86 | 0.022 | 5.26 | ||
| AG | 6 (37.50) | 87 (46.52) | 0.71 | 0.52 | (0.10–2.66) | ||||
| GG | 8 (50.00) | 44 (23.53) | 0.04 a | 0.20 | (0.04–0.97) | ||||
| Minor allele A | 10 (31.25) | 199 (53.21) | |||||||
| rs1233334:G>C/T | N = 16 | N = 187 | |||||||
| CC* | 8 (50.00) | 132 (70.59) | 1 | 0.05 | 9.27 | 0.013 | 6.15 | ||
| CG | 5 (31.25) | 44 (23.53) | 0.33 | 0.53 | (0.17–1.72) | ||||
| GG | 1 (6.25) | 5 (2.67) | 0.32 | 0.30 | (0.03–2.91) | ||||
| GT | 0 (0.00) | 3 (1.60) | 1.00 | 0.45 | (0.02–9.43) | ||||
| CT | 2 (12.50) | 3 (1.60) | 0.04 b | 0.09 | (0.01–0.62) | ||||
| TT | 0 (0.00) | 0 (0.00) | |||||||
| Minor allele T | 2 (6.25) | 6 (1.60) | |||||||
E I + II endometriosis I + II, E III + IV endometriosis III + IV, P probability, OR odds ratio, 95% CI 95% confidence interval from two-sided Fisher’s exact test, χ 2 df = 2 p Chi-square test for independence with two degrees of freedom for the 14 bp ins/del (rs371194629:insATTTGTTCATGCCT/del) in 3′UTR and rs1632947:G>A polymorphisms, χ 2 df = 4 p Chi-square test for independence with four degrees of freedom for the polymorphism rs1233334:G>C/T, χ 2 df = 1 p Chi-square test for trend with one degree of freedom for all tested polymorphisms; *Reference
a P corr. = 0.12
b P corr. = 0.12
Analysis of peritoneal vs ovarian localization of lesions showed protective effects of rs1632947:GG genotype against ovarian endometriosis (P = 0.028, P corr. = 0.08, OR = 0.16, 95% CI = 0.03–0.84), whereas rs1233334:CT genotype gave a significant result only for peritoneal vs ovarian plus peritoneal endometriosis (P = 0.01, P corr. = 0.03, OR = 0.02, 95% CI = 0.001–0.53). Analysis of all rs1233334 genotypes revealed even higher significance (p = 0.006, χ2 = 14.35; Table 4). On the other hand, no association with any form of endometriosis was found for the 14 base pair insertion/deletion polymorphism (rs371194629) in the HLA-G gene (Tables 2, 3, 4 and Table S2).
Table 4.
HLA-G genotype frequencies in women from endometriosis groups depending on the localization of lesions
| Genotype | Endometriosis peritoneal only | Endometriosis ovarian only | Endometriosis ovarian + peritoneal | Endometriosis peritoneal only vs endometriosis ovarian only | Endometriosis peritoneal only vs endometriosis ovarian + peritoneal | Endometriosis ovarian only vs endometriosis ovarian + peritoneal | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | OR | 95% CI | Test for independence | P | OR | 95% CI | Test for independence | P | OR | 95% CI | Test for independence | |||||||
| p | χ2 | p | χ2 | p | χ2 | |||||||||||||
| rs371194629:ins/del | N = 16 | N = 136 | N = 92 | |||||||||||||||
| Del/del* | 8 (50.00) | 43 (31.62) | 34 (36.95) | 1 | 0.23 | 2.97 | 1 | 0.38 | 1.94 | 1 | 0.70 | 0.72 | ||||||
| Ins/del | 4 (25.00) | 30 (22.06) | 18 (19.57) | 0.76 | 1.40 | (0.38–5.06) | 1.00 | 1.06 | (0.28-4.00) | 0.57 | 0.76 | (0.36–1.59) | ||||||
| Ins/ins | 4 (25.00) | 63 (46.32) | 40 (43.48) | 0.12 | 2.93 | (0.83–10.35) | 0.22 | 2.35 | (0.65–8.50) | 0.54 | 0.80 | (0.44–1.46) | ||||||
| Minor allele ins | 12 (37.50) | 156 (57.35) | 98 (53.26) | |||||||||||||||
| rs1632947:G>A | N = 16 | N = 136 | N = 92 | |||||||||||||||
| AA* | 2 (12.50) | 44 (32.35) | 26 (28.26) | 1 | 0.042 | 6.33 | 1 | 0.28 | 2.57 | 1 | 0.29 | 2.49 | ||||||
| AG | 7 (43.75) | 67 (49.26) | 41 (44.57) | 0.48 | 0.44 | (0.90–2.19) | 0.47 | 0.45 | (0.09–2.34) | 1.00 | 1.04 | (0.56–1.93) | ||||||
| GG | 7 (43.75) | 25 (18.38) | 25 (27.17) | 0.028 a | 0.16 | (0.03–0.84) | 0.15 | 0.27 | (0.05–1.45) | 0.19 | 1.69 | (0.81–3.54) | ||||||
| Minor allele A | 11 (34.38) | 155 (56.99) | 129 (47.43) | |||||||||||||||
| rs1233334:G>C/T | N = 16 | N = 136 | N = 92 | |||||||||||||||
| CC* | 7 (43.75) | 101 (74.26) | 64 (69.57) | 1 | 0.07 | 8.80 | 1 | 0.006 | 14.35 | 1 | 0.29 | 5.02 | ||||||
| CG | 6 (37.50) | 28 (20.59) | 23 (25.00) | 0.08 | 0.32 | (0.10–1.04) | 0.19 | 0.42 | (0.13–1.38) | 0.51 | 1.30 | (0.69–2.45) | ||||||
| GG | 1 (6.25) | 2 (1.47) | 3 (3.26) | 0.20 | 0.14 | (0.01–1.72) | 0.37 | 0.33 | (0.03–3.60) | 0.38 | 2.37 | (0.38–14.56) | ||||||
| GT | 0 (0.00) | 1 (0.74) | 2 (2.17) | 1.00 | 0.22 | (0.01–5.93) | 1.00 | 0.58 | (0.03–13.30) | 0.56 | 3.16 | (0.28–35.54) | ||||||
| CT | 2 (12.50) | 4 (2.94) | 0 (0.00) | 0.07 | 0.14 | (0.02–0.89) | 0.01 b | 0.02 | (0.001–0.53) | 0.30 | 0.17 | (0.01–3.30) | ||||||
| TT | 0 (0.00) | 0 (0.00) | 0 (0.00) | – | – | – | – | – | – | – | – | – | ||||||
| Minor allele T | 2 (6.25) | 5 (1.84) | 2 (1.09) | |||||||||||||||
P probability, OR odds ratio, 95% CI 95% confidence interval from two-sided Fisher’s exact test, χ 2 df = 2 p Chi-square test for independence with two degrees of freedom for the 14 bp ins/del (rs371194629:ins ATTTGTTCATGCCT/del) in 3′UTR and rs1632947:G>A polymorphisms, χ 2 df = 4 p Chi-square test for independence with four degrees of freedom for the polymorphism of the rs1233334:G>C/T
*Reference
a P corr. = 0.08
b P corr. = 0.03
LILRB1 and LILRB2 but not KIR2DL4 polymorphisms are associated with endometriosis
LILRB1 rs41308748:G>A polymorphism was distributed differently between patients and controls (P = 0.0048, P corr. = 0.024, OR = 4.62, 95% CI = 1.52–14.02 for AA genotype, and p = 0.0035, χ 2 = 11.33 for all genotypes; Table 5). Similar results were found by analysis according to the stage of the disease (P = 0.007, P corr. = 0.035, OR = 4.8, 95% CI = 1.52–15.15 for AA, and p = 0.007, χ 2 = 9.93 for all genotypes), and localization of lesions (P = 0.011, P corr. = 0.055, OR = 4.24, 95% CI = 1.36–13.21 for AA and p = 0.01, χ 2 = 9.12 for all genotypes; Table S3). The frequency of other examined SNPs did not differ between analyzed groups (Table S3).
Table 5.
LILRB1, LILRB2 and KIR2DL4 genotype and minor allele frequencies in women from Control and Endometriosis groups
| Genotype | Control (%) | Patients (%) | Patients vs Control | ||||
|---|---|---|---|---|---|---|---|
| P | OR | 95% CI | Test for independence | ||||
| p | χ2 | ||||||
| LILRB1 rs41308748:G>A | N = 314 | N = 272 | |||||
| GG* | 261 (83.12) | 226 (83.09) | 1 | 0.0035 | 11.33 | ||
| GA | 49 (15.61) | 30 (11.03) | 0.18 | 0.71 | (0.43–1.15) | ||
| AA | 4 (1.27) | 16 (5.88) | 0.0048 a | 4.62 | (1.52–14.02) | ||
| Minor allele A | 57 (9.08) | 62 (11.40) | |||||
| H-W | 0.33 | 0.00 | |||||
| LILRB1 rs1061680:T>C | N = 314 | N = 272 | |||||
| TT* | 191 (60.82) | 176 (64.71) | 1 | 0.14 | 3.88 | ||
| TC | 112 (35.67) | 80 (29.41) | 0.18 | 0.76 | (0.54–1.10) | ||
| CC | 11 (3.51) | 16 (5.88) | 0.32 | 1.58 | (0.71–3.50) | ||
| Minor allele C | 134 (21.34) | 112 (20.59) | |||||
| H-W | 0.27 | 0.097 | |||||
| LILRB2 rs383369:G>A | N = 314 | N = 272 | |||||
| AA* | 226 (71.97) | 186 (68.38) | 1 | 0.23 | 2.91 | ||
| AG | 82 (26.12) | 84 (30.88) | 0.27 | 1.25 | (0.87–1.79) | ||
| GG | 6 (1.91) | 2 (0.74) | 0.31 | 0.41 | (0.08–2.03) | ||
| Minor allele G | 94 (14.97) | 88 (16.18) | |||||
| H-W | 0.65 | 0.022 | |||||
| LILRB2 rs7247538:C>T | N = 314 | N = 272 | |||||
| TT* | 107 (34.08) | 94 (34.56) | 1 | 0.58 | 1.10 | ||
| CT | 146 (46.50) | 134 (49.26) | 0.85 | 1.05 | (0.73–1.50) | ||
| CC | 61 (19.42) | 44 (16.18) | 0.47 | 0.82 | (0.51–1.32) | ||
| Minor allele C | 268 (42.68) | 222 (40.81) | |||||
| H-W | 0.38 | 0.74 | |||||
| KIR2DL4 rs649216:T>C | N = 314 | N = 276 | |||||
| TT* | 103 (32.80) | 93 (33.70) | 1 | 0.26 | 2.70 | ||
| CT | 150 (47.77) | 116 (42.03) | 0.45 | 0.86 | (0.59–1.24) | ||
| CC | 61 (19.43) | 67 (24.28) | 0.42 | 1.22 | (0.78–1.90) | ||
| Minor allele C | 272 (43.31) | 250 (45.29) | |||||
| H-W | 0.63 | 0.012 | |||||
H–W Hardy–Weinberg equilibrium, P probability, OR odds ratio, 95% CI 95% confidence interval from two-sided Fisher’s exact test, χ 2 df = 2 p Chi-square test for independence with two degrees of freedom for all tested polymorphisms; For four samples from the endometriosis group we could not perform LILRB1 and LILRB2 genotyping because of a lack of DNA
*Reference
a P corr. = 0.024
LILRB2 rs383369:AG genotype was almost five times more frequent in severe stages (III + IV) of endometriosis than in milder (I + II) stages (P = 0.043, P corr. = 0.215, OR = 7.02, 95% CI = 0.90–54.43, Table 6). A similar, albeit no significant difference was seen in comparison of peritoneal only with peritoneal + ovarian endometriosis (P = 0.09, OR = 3.8, 95% CI = 0.81–17.77; Table S4).
Table 6.
Comparison of the LILRB and KIR2DL4 polymorphisms in women depending on the severity of endometriosis
| Genotype | E I + II (%) | E III + IV (%) | E III + IV vs E I + II | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | OR | 95% CI | Test for independence | Test for trend | |||||
| p | χ2 | p | χ2 | ||||||
| LILRB1 rs41308748:G>A | N = 16 | N = 183 | |||||||
| GG* | 13 (81.25) | 150 (81.97) | 1 | 0.43 | 1.70 | 0.68 | 0.17 | ||
| GA | 3 (18.75) | 21 (11.48) | 0.44 | 0.61 | (0.16–2.31) | ||||
| AA | 0 (0.00) | 12 (6.55) | 0.60 | 2.24 | (0.13–40.02) | ||||
| Minor allele A | 3 (9.38) | 45 (12.30) | |||||||
| LILRB1 rs1061680:T>C | N = 16 | N = 183 | |||||||
| TT* | 12 (75.00) | 122 (66.67) | 1 | 0.63 | 0.93 | 0.38 | 0.76 | ||
| TC | 4 (25.00) | 53 (28.96) | 0.78 | 1.30 | (0.40–4.23) | ||||
| CC | 0 (0.00) | 8 (4.37) | 1.00 | 1.74 | (0.09–31.90) | ||||
| Minor allele C | 4 (12.5) | 69 (18.85) | |||||||
| LILRB2 rs383369:G>A | N = 16 | N = 183 | |||||||
| AA* | 15 (93.75) | 124 (67.76) | 1 | 0.06 | 5.49 | 0.024 | 5.10 | ||
| AG | 1 (6.25) | 58 (31.69) | 0.043 a | 7.02 | (0.90–54.43) | ||||
| GG | 0 (0.00) | 1 (0.55) | 1.00 | 0.37 | (0.01–9.58) | ||||
| Minor allele G | 1 (3.13) | 60 (16.39) | |||||||
| LILRB2 rs7247538:C>T | N = 16 | N = 183 | |||||||
| TT* | 6 (37.50) | 64 (34.97) | 1 | 0.51 | 1.34 | 0.69 | 0.16 | ||
| CT | 6 (37.50) | 91 (49.73) | 0.56 | 1.42 | (0.44–4.61) | ||||
| CC | 4 (25.00) | 28 (15.30) | 0.72 | 0.66 | (0.17–2.51) | ||||
| Minor allele C | 14 (43.75) | 147 (40.16) | |||||||
| KIR2DL4 rs649216:T>C | N = 16 | N = 187 | |||||||
| TT* | 6 (37.50) | 60 (32.09) | 1 | 0.23 | 2.95 | 0.62 | 0.25 | ||
| CT | 4 (25.00) | 85 (45.45) | 0.33 | 2.13 | (0.57–7.86) | ||||
| CC | 6 (37.50) | 42 (22.46) | 0.56 | 0.70 | (0.21–2.32) | ||||
| Minor allele C | 16 (50.00) | 169 (45.19) | |||||||
P probability, OR odds ratio, 95% CI 95% confidence interval from two-sided Fisher’s exact test, χ 2 df = 2 p Chi-square test for independence with two degrees of freedom for all tested polymorphisms, χ 2 df = 1 p Chi-square test for trend with one degree of freedom for all tested polymorphisms; *Reference; for four samples from the III + IV endometriosis group we could not perform LILRB1 and LILRB2 genotyping because of a lack of DNA
a P corr. = 0.215
Neither the other LILRB2 SNP (rs7247538:T>C) nor KIR2DL4 (rs649216:T>C) or LILRB1 (rs41308748:G>A and rs1061680:T>C) was distributed differently between mild and severe disease (Table 6). None of other polymorphisms was associated with localization of lesions (Table S4).
Discussion
In the present study we found that susceptibility to and the severity of endometriosis are associated with polymorphisms in the HLA-G, LILRB1 and LILRB2 genes. On the other hand, the disease was not associated with the KIR2DL4 polymorphism. The data on HLA-G expression in endometrial tissue from healthy individuals and patients with endometriosis are controversial. HLA-G has been detected on eutopic endometrial cells and peritoneal fluid cells in the menstrual phase of women with or without endometriosis (Kawashima et al. 2009); however, Barrier et al. (2006) found HLA-G protein and mRNA expression only in ectopic endometrial tissue but not in eutopic endometrium in women with or without endometriosis, independently of cycle stage. Notably, in an earlier study, Hornung et al. (2001) did not detect HLA-G in peritoneal fluid, ectopic and normal endometrial tissues and stromal cells from endometriosis patients or controls.
The HLA-G molecule exists as seven protein isoforms as a result of alternative splicing: four membrane-bound (HLA-G1, G2, G3, G4) and three soluble (HLA-G5, G6, G7) isoforms (Menier et al. 2010; Donadi et al. 2011; Castelli et al. 2014). Soluble HLA-G (sHLA-G) was found in the peritoneal fluid in similar concentrations in control subjects and in mild and severe endometriosis (Eidukaite and Tamosiunas 2008).
Several important regulatory motifs have been described in the promoter of the HLA-G gene, e.g. Enhancer-A (EnhA), the interferon-stimulated response element (ISRE) and the SXY module. All of them are mainly responsible for controlling gene expression by affecting transcription factor binding or promoter methylation (Donadi et al. 2011; Castelli et al. 2014; Persson et al. 2017; Verloes et al. 2017). These regions exhibit many polymorphic sites; among them, positions − 964, − 725 and − 716 (in the promoter) may affect expression of HLA-G (Donadi et al. 2011; Castelli et al. 2014; Persson et al. 2017; Verloes et al. 2017; Amodio et al. 2016; Ober et al. 2003). Indeed, we found here protective effects of rs1632947:GG (− 964GG) and rs1233334:CT (− 725CT) HLA-G genotypes on susceptibility to endometriosis and/or progression of the disease (Table 7). On the other hand, a 14 bp insertion/deletion in the 3′UTR (rs371194629) has an influence on both expression and alternative splicing of HLA-G (Verloes et al. 2017) and the level of sHLA-G (Chen et al. 2008). However, no association of this polymorphism with endometriosis was seen in our study. The reason why one polymorphism, rs1632947:GG genotype in the promoter region, increasing expression of HLA-G (Ober et al. 2006), seems to protect against endometriosis, whereas 14 bp deletion in 3′UTR (rs371194629), also increasing HLA-G expression (Verloes et al. 2017), had no effect, needs explanation by further experiments. No other reports on the role of HLA-G polymorphisms in endometriosis have been published so far. However,it is worth to mention that other class of MHC genes located near HLA-G (HLA-DQ and HLA-DRB1) have already been published in the context of endometriosis (Zong et al. 2002; Sundqvist et al. 2011; Sobalska-Kwapis et al. 2017).
Table 7.
Summarized effect of HLA-G and LILRB polymorphisms on susceptibility to and severity of endometriosis
| Polymorphism | Associated genotype | Comparison | Table | Effect |
|---|---|---|---|---|
| HLA-G rs1632947:G>A | GG GG GG |
Patients vs control Patients according to the rAFS vs control Patients according to the localization of lesions vs control |
2 S2 S2 |
↓ ↓ ↓ |
| HLA-G rs1632947:G>A | GG | Severity III + IV vs I + II | 3 | ↓ |
| HLA-G rs1233334:G>C/T | CT | Severity III + IV vs I + II | 3 | ↓ |
| HLA-G rs1632947:G>A | GG | Ovarian only vs peritoneal only | 4 | ↓ |
| HLA-G rs1233334:G>C/T | CT | Ovarian + peritoneal vs peritoneal only | 4 | ↓ |
| CT | Ovarian only vs peritoneal only | 4 | ↓? | |
| LILRB1 rs41308748:G>A | AA AA AA |
Patients vs control Patients according to the rAFS vs control Patients according to the localization of lesions vs Control |
5 S3 S3 |
↑ ↑ ↑ |
| LILRB2 rs383369:G>A | AG AG |
Severity III + IV vs I + II Peritoneal only vs ovarian + peritoneal |
6 S4 |
↑ ↑? |
↓ protection, ↑ susceptibility
The putative role of HLA-G in the etiopathogenesis of endometriosis may be strengthened by our further observation that the disease is also associated with polymorphism in LILRB1 and LILRB2 genes coding for HLA-G receptors. NK cells express different levels of LILRB1 (Kirwan and Burshtyn 2005) and individuals vary in its positivity, ranging from 10 to 77% of NK cells, depending on gene polymorphism (Davidson et al. 2010).
The rs41308748:G>A polymorphism of the LILRB1 gene is an intronic SNP situated between the cytoplasmic tail and the 3′UTR sequence, which could have an influence on the splicing process. We found its association (AA genotype) with susceptibility to endometriosis (Table 7); therefore, studies on splicing variants in endometriosis would be desirable. We observed earlier a protective effect of the GA genotype in recurrent miscarriage, whereas the AA genotype had no effect (Nowak et al. 2016). The rs1061680:T>C is a non-synonymous SNP, located in the sequence encoding the extracellular D2 domain (Davidson et al. 2010). It is in strong linkage disequilibrium with another SNP (rs10423364:A>G) which is located in a potential transcription factor binding site (our in silico analysis) and may therefore affect gene expression. Thus, rs1061680:T>C may be a marker of rs10423364:A>G, and may also influence protein structure. However, in our present study we did not reveal its association with endometriosis.
The polymorphism rs7247538:C>T of LILRB2 changes histidine to tyrosine (p. His300Tyr) in the amino acid sequence of the protein. Our in silico analysis indicated that it may also have a possibly damaging influence on the splicing process. However, this polymorphism was not associated with endometriosis. The second tested SNP in the LILRB2 gene was the rs383369:G>A (p. Arg20His) and it has been located in the signal sequence region. The G allele of rs383369 has been associated with low expression levels of LILRB2 in Northeast Asians, where it has a high frequency; however, it is infrequent in Europeans (Hirayasu et al. 2008). In our population, almost all individuals possessed the alternative A allele, and GG homozygotes were virtually absent. Nevertheless, the AG heterozygotes had 7 times higher probability of having severe endometriosis than AA homozygotes (Table 6). It suggests, then, that lower LILRB2 expression may predispose to more severe stages of the disease.
KIR2DL4 has been considered to be also an HLA-G receptor (Rajagopalan and Long 2012, 2014). Its long cytoplasmic tail suggests an inhibitory function. However, it has only one immunoreceptor tyrosine inhibitory motif (ITIM) in the cytoplasmic tail and a positively charged arginine residue in its transmembrane region, allowing it to complex with the FcεRI-γ chain which transduces the activation signal upon ligand binding by KIR2DL4 (Kikuchi-Maki et al. 2005). However, the HLA-G/KIR2DL4 interaction has recently been questioned (Le Page et al. 2014). In addition, only one out of four individuals in our population possesses a functional receptor (Nowak et al. 2015b). The lack of functional KIR2DL4 may be compensated by the presence of LILRB1. Notably, LILRB1, despite its inhibitory potential, may also exert an activating effect through its immunoreceptor tyrosine-based switch motif (ITSM) (Li et al. 2009) and therefore substitute for KIR2DL4.
There are some limitations of our work. First, the group of subjects with minimal or mild endometriosis was small (16 individuals). This resulted from late diagnosis, as women often do not see their doctor until they suffer from infertility or the pain becomes unbearable. Second, protein expression of cell surface LILRB1, LILRB2 and KIR2DL4 as well as soluble or membrane HLA-G was not examined here. However, this will be a future direction of our research, with particular emphasis on expression of these molecules in endometriotic lesions in peritoneum vs ovary. Moreover, recently published GWAS analysis of potential protein-modifying genetic variants in 9000 endometriosis patients and 150,000 controls of European ancestry (Sapkota et al. 2017b) have not identified our proposed variants with endometriosis pathogenesis. However, variants which modify protein structure through amino acid substitutions or alter stop signals or splicing, particularly those with MAF < 0.05 have been implicated as important but not well covered in GWA studies. Moreover, only about 18% of endometriosis cases in Sapkota et al. (2017b) samples had moderate-to-severe disease while in our study these stages accounted to 92%, and therefore Sapkota et al. (2017b) analysis may not have adequate reference in severe cases. In addition, the cost of whole genome or exome sequencing methods limits large-scale studies and it still limits the selection of potential SNPs for testing.
In conclusion, our results suggest that HLA-G and its receptors LILRB1 and LILRB2, but not KIR2DL4, may play a role in elimination of ectopic endometrial cells and in development of the disease. Our data are novel, as this is the first report on this topic.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1 PCR conditions for LILRB1 (rs41308748:G>A) and LILRB2 (rs383369:G>A, rs7247538:C>T) genotyping (DOCX 13 KB)
Table S2 HLA-G genotype and minor allele frequencies in women from Control and Endometriosis groups (DOC 65 KB)
Table S3 LILRB1, LILRB2 and KIR2DL4 genotype and minor allele frequencies in women from Control and Endometriosis groups (DOC 75 KB)
Table S4 Comparison of the LILRB and KIR2DL4 polymorphisms in women depending on the localization of lesions (DOC 85 KB)
Fig S1 Scatter plot of representative results from the real-time PCR of HLA-G rs1233334:G>C/T SNP genotyping (TIF 244 KB)
Acknowledgements
The authors are grateful to patients and control volunteers for the donation of blood and their agreement to use their clinical data in this study.
Compliance with ethical standards
Funding
This study was funded by Ministry of Science and Higher Education (No. 2.P05A.069.28) and the National Science Centre (N N401 588340), Poland.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Aleksandra Bylińska, Karolina Wilczyńska equally contributed to this article.
Electronic supplementary material
The online version of this article (10.1007/s00438-017-1404-3) contains supplementary material, which is available to authorized users.
Contributor Information
Piotr Kuśnierczyk, Phone: +48-71-370-9976, Email: pkusnier@iitd.pan.wroc.pl.
Izabela Nowak, Phone: +48-71-370-9921, Email: izan@iitd.pan.wroc.pl.
References
- Adachi S, Tajima A, Quan J, et al. Meta-analysis of genome-wide association scans for genetic susceptibility to endometriosis in Japanese population. J Hum Genet. 2010;55:816–821. doi: 10.1038/jhg.2010.118. [DOI] [PubMed] [Google Scholar]
- Ahn SH, Monsanto SP, Miller C, Singh SS, Thomas R, Tayade C. Pathophysiology and immune dysfunction in endometriosis. Biomed Res Int. 2015;2015:795976. doi: 10.1155/2015/795976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Fertility Society Revised American Fertility Society classification of endometriosis. Fertil Steril. 1985;43:351–352. doi: 10.1016/S0015-0282(16)48430-X. [DOI] [PubMed] [Google Scholar]
- Amodio G, Canti V, Maggio L, Rosa S, Castiglioni MT, Rovere-Querini P, Gregori S. Association of genetic variants in the 3′UTR of HLA-G with recurrent pregnancy loss. Hum Immunol. 2016;77:886–891. doi: 10.1016/j.humimm.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrier BF, Kendall BS, Ryan CE, Sharpe-Timms KL. HLA-G is expressed by the glandular epithelium of peritoneal endometriosis but not in eutopic endometrium. Hum Reprod. 2006;21:864–869. doi: 10.1093/humrep/dei408. [DOI] [PubMed] [Google Scholar]
- Borges L, Hsu ML, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res. 2014;2014:734068. doi: 10.1155/2014/734068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Yan WH, Lin A, Xu HH, Zhang JG, Wang XX. The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens. 2008;72:335–341. doi: 10.1111/j.1399-0039.2008.01107.x. [DOI] [PubMed] [Google Scholar]
- Dastur AE, Tank PD, John A. Sampson and the origins of endometriosis. J Obstet Gynaecol India. 2010;60:299–300. doi: 10.1007/s13224-010-0046-8. [DOI] [Google Scholar]
- Davidson CL, Li NL, Burshtyn DN. LILRB1 polymorphism and surface phenotypes of natural killer cells. Hum Immunol. 2010;71:942–949. doi: 10.1016/j.humimm.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D, Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci. 2011;68:369–395. doi: 10.1007/s00018-010-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidukaite A, Tamosiunas V. Soluble HLA-G in the peritoneal fluid of women with endometriosis. Fertil Steril. 2008;89:465–467. doi: 10.1016/j.fertnstert.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, Hurley CK, Christiansen FT, Witt CS. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- Goodridge JP, Lathbury LJ, John E, Charles AK, Christiansen FT, Witt CS. The genotype of the NK cell receptor, KIR2DL4, influences INF gamma secretion by decidual natural killer cells. Mol Hum Reprod. 2009;15:489–497. doi: 10.1093/molehr/gap039. [DOI] [PubMed] [Google Scholar]
- Gupta D, Hull ML, Fraser I, Miller L, Bossuyt PM, Johnson N, Nisenblat V. Endometrial biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst Rev. 2016;4:CD012165. doi: 10.1002/14651858.CD012165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayasu K, Ohashi J, Tanaka H, Kashiwase K, Ogawa A, Takanashi M, Satake M, Jia GJ, Chimge NO, Sideltseva EW, Tokunaga K, Yabe T. Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in Northeast Asians. Am J Hum Genet. 2008;82:1075–1083. doi: 10.1016/j.ajhg.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung D, Fujii E, Lim KH, Vigne JL, McMaster MT, Taylor RN. Histocompatibility leukocyte antigen-G is not expressed by endometriosis or endometrial tissue. Fertil Steril. 2001;75:814–817. doi: 10.1016/S0015-0282(00)01791-X. [DOI] [PubMed] [Google Scholar]
- Hudson LE, Allen RL. Leukocyte Ig-like receptors—a model for MHC class I disease associations. Front Immunol. 2016;7:281. doi: 10.3389/fimmu.2016.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hviid TV, Larsen LG, Hoegh AM, Bzorek M. HLA-G expression in placenta in relation to HLA-G genotype and polymorphisms. Am J Reprod Immunol. 2004;52:212–217. doi: 10.1111/j.1600-0897.2004.00208.x. [DOI] [PubMed] [Google Scholar]
- Hviid TV, Rizzo R, Melchiorri L, Stignani M, Baricordi OR. Polymorphism in the 5′ upstream regulatory and 3′ untranslated regions of the HLA-G gene in relation to soluble HLA-G and IL-10 expression. Hum Immunol. 2006;67:53–62. doi: 10.1016/j.humimm.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Kang X, Kim J, Deng M, John S, Chen H, Wu G, Phan H, Zhang CC. Inhibitory leukocyte immunoglobulin-like receptors: immune checkpoint proteins and tumor sustaining factors. Cell Cycle. 2016;15:25–40. doi: 10.1080/15384101.2015.1121324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima M, Maeda N, Adachi Y, Takeuchi T, Yamamoto Y, Izumiya C, Hayashi K, Furihata M, Udaka K, Fukaya T. Human leukocyte antigen-G, a ligand for the natural killer receptor KIR2DL4, is expressed by eutopic endometrium only in the menstrual phase. Fertil Steril. 2009;91:343–349. doi: 10.1016/j.fertnstert.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol. 2005;174:3859–3863. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- Kirwan SE, Burshtyn DN. Killer cell Ig-like receptor-dependent signaling by Ig-like transcript 2 (ILT2/ CD85j/ LILRB1/ LIR-1) J Immunol. 2005;175:5006–5015. doi: 10.4049/jimmunol.175.8.5006. [DOI] [PubMed] [Google Scholar]
- Králíčková M, Vetvicka V. Immunological aspects of endometriosis: review. Ann Transl Med. 2015;3:153. doi: 10.3978/j.issn.2305-5839.2015.06.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki K, Tsuchiya N, Shiroishi M, et al. Extensive polymorphisms of LILRB1 (ILT2, LIR1) and their association with HLA-DRB1 shared epitope negative rheumatoid arthritis. Hum Mol Genet. 2005;14:2469–2480. doi: 10.1093/hmg/ddi247. [DOI] [PubMed] [Google Scholar]
- Le Page ME, Goodridge JP, John E, Christiansen FT, Witt CS. Killer Ig-like receptor 2DL4 does not mediate NK cell IFN-γ responses to soluble HLA-G preparations. J Immunol. 2014;192:732–740. doi: 10.4049/jimmunol.1301748. [DOI] [PubMed] [Google Scholar]
- LeMaoult J, Zafaranloo K, Le Danff C, Carosella ED. HLA-G up-regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J. 2005;19:662–664. doi: 10.1096/fj.04-1617fje. [DOI] [PubMed] [Google Scholar]
- Li C, Houser BL, Nicotra ML, Strominger JL. HLA-G homodimer-induced cytokine secretion through HLA-G receptors on human decidual macrophages and natural killer cells. Proc Natl Acad Sci. 2009;106:5767–5772. doi: 10.1073/pnas.0901173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Izumiya C, Taniguchi K, Matsushima S, Fukaya T. T role of NK cells and HLA-G in endometriosis. Front Biosci (Schol Ed) 2012;4:1568–1581. doi: 10.2741/s353. [DOI] [PubMed] [Google Scholar]
- Menier C, Rouas-Freiss N, Favier B, LeMaoult J, Moreau P, Carosella ED. Recent advances on the non-classical major histocompatibility complex class I HLA-G molecule. Tissue Antigens. 2010;75:201–206. doi: 10.1111/j.1399-0039.2009.01438.x. [DOI] [PubMed] [Google Scholar]
- Nowak I, Płoski R, Barcz E, Dziunycz P, Kamiński P, Kostrzewa G, Milewski Ł, Roszkowski PI, Senitzer D, Malejczyk J, Kuśnierczyk P. KIR2DS5 in the presence of HLA-C C2 protects against endometriosis. Immunogenetics. 2015;67:203–209. doi: 10.1007/s00251-015-0828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak I, Barcz E, Majorczyk E, Malinowski A, Wilczyński JR, Banasik M, Motak-Pochrzęst H, Kuśnierczyk P. Genetic polymorphism of KIR2DL4 in the Polish population. Tissue Antigens. 2015;85:450–457. doi: 10.1111/tan.12544. [DOI] [PubMed] [Google Scholar]
- Nowak I, Malinowski A, Barcz E, Wilczyński JR, Wagner M, Majorczyk E, Motak-Pochrzęst H, Banasik M, Kuśnierczyk P. Possible role of HLA-G, LILRB1 and KIR2DL4 gene polymorphisms in spontaneous miscarriage. Arch Immunol Ther Exp (Warsz) 2016;64:505–514. doi: 10.1007/s00005-016-0389-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR, Low SK, Anderson CA, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat Genet. 2012;44:1355–1359. doi: 10.1038/ng.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Aldrich CL, Chervoneva I, Billstrand C, Rahimov F, Gray HL, Hyslop T. Variation in the HLA-G promoter region influences miscarriage rates. Am J Hum Genet. 2003;72:1425–1435. doi: 10.1086/375501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Billstrand C, Kuldanek S, Tan Z. The miscarriage-associated HLA-G -725G allele influences transcription rates in JEG-3 cells. Hum Reprod. 2006;21:1743–1748. doi: 10.1093/humrep/del036. [DOI] [PubMed] [Google Scholar]
- Oosterlynck DJ, Cornillie FJ, Waer M, Vandeputte M, Koninckx PR. Women with endometriosis show a defect in natural killer activity resulting in a decreased cytotoxicity to autologous endometrium. Fertil Steril. 1991;56:45–51. doi: 10.1016/S0015-0282(16)54414-8. [DOI] [PubMed] [Google Scholar]
- Oosterlynck DJ, Meuleman C, Waer M, Vandeputte M, Koninckx PR. The natural killer activity of peritoneal fluid lymphocytes is decreased in women with endometriosis. Fertil Steril. 1992;58:290–295. doi: 10.1016/S0015-0282(16)55224-8. [DOI] [PubMed] [Google Scholar]
- Parkin KL, Fazleabas AT. Uterine leukocyte function and dysfunction: a hypothesis on the impact of endometriosis. Am J Reprod Immunol. 2016;75:411–417. doi: 10.1111/aji.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson G, Melsted WN, Nilsson LL, Hviid TVF. HLA class Ib in pregnancy and pregnancy-related disorders. Immunogenetics. 2017;69:581–595. doi: 10.1007/s00251-017-0988-4. [DOI] [PubMed] [Google Scholar]
- Rahmioglu N, Nyholt DR, Morris AP, Missmer SA, Montgomery GW, Zondervan KT. Genetic variants underlying risk of endometriosis: insights from meta-analysis of eight genome-wide association and replication datasets. Hum Reprod Update. 2014;20:702–716. doi: 10.1093/humupd/dmu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Long EO. KIR2DL4 (CD158d): an activation receptor for HLA-G. Front Immunol. 2012;3:258. doi: 10.3389/fimmu.2012.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Long EO. Comment on “Killer Ig-like receptor 2DL4 does not mediate NK Cell IFN-γ responses to soluble HLA-G preparations”. J Immunol. 2014;192:4003. doi: 10.4049/jimmunol.1400430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. Peritoneal endometriosis due to menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol. 1927;14:422–467. doi: 10.1016/S0002-9378(15)30003-X. [DOI] [Google Scholar]
- Sapkota Y, Steinthorsdottir V, Morris AP, et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat Commun. 2017;248:15539. doi: 10.1038/ncomms15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota Y, Vivo I, Steinthorsdottir V, et al. Analysis of potential protein-modifying variants in 9000 endometriosis patients and 150,000 controls of European ancestry. Sci Rep. 2017;7:11380. doi: 10.1038/s41598-017-10440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serdar E, Bulun MD. Mechanisms of disease: endometriosis. N Engl J Med. 2009;360:268–279. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- Sobalska-Kwapis M, Smolarz B, Słomka M, Szaflik T, Kępka E, Kulig B, Siewierska-Górska A, Polak G, Romanowicz H, Strapagiel D, Szyłło K. New variants near RHOJ and C2, HLA-DRA region and susceptibility to endometriosis in the Polish population-The genome-wide association study. Eur J Obstet Gynecol Reprod Biol. 2017;217:106–112. doi: 10.1016/j.ejogrb.2017.08.037. [DOI] [PubMed] [Google Scholar]
- Sundqvist J, Falconer H, Seddighzadeh M, Vodolazkaia A, Fassbender A, Kyama C, Bokor A, Stephansson O, Padyukov L, Gemzell-Danielsson K, D’Hooghe TM. Endometriosis and autoimmune disease: association of susceptibility to moderate/severe endometriosis with CCL21 and HLA-DRB1. Fertil Steril. 2011;95:437–440. doi: 10.1016/j.fertnstert.2010.07.1060. [DOI] [PubMed] [Google Scholar]
- Tariverdian N, Siedentopf F, Rücke M, Blois SM, Klapp BF, Kentenich H, Arck PC. Intraperitoneal immune cell status in infertile women with and without endometriosis. J Reprod Immunol. 2009;80:80–90. doi: 10.1016/j.jri.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K, Nakamura Y. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42:707–710. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- van der Touw W, Chen HM, Pan PY, Chen SH. LILRB receptor-mediated regulation of myeloid cell maturation and function. Cancer Immunol Immunother. 2017;66:1079–1087. doi: 10.1007/s00262-017-2023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercellini P, Viganò P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10:261–275. doi: 10.1038/nrendo.2013.255. [DOI] [PubMed] [Google Scholar]
- Verloes A, Spits C, Vercammen M, Geens M, LeMaoult J, Sermon K, Coucke W, Van de Velde H. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem Cell Res. 2017;19:118–127. doi: 10.1016/j.scr.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Wang F, Wen Z, Li H, Yang Z, Zhao X, Yao X. Human leukocyte antigen-G is expressed by the eutopic and ectopic endometrium of adenomyosis. Fertil Steril. 2008;90:1599–1604. doi: 10.1016/j.fertnstert.2007.06.073. [DOI] [PubMed] [Google Scholar]
- Wiśniewski A, Bilińska M, Klimczak A, Wagner M, Majorczyk E, Nowak I, Pokryszko-Dragan A, Kuśnierczyk P. Association of the HLA-G gene polymorphism with multiple sclerosis in a Polish population. Int J Immunogenet. 2010;37:307–311. doi: 10.1111/j.1744-313X.2010.00926.x. [DOI] [PubMed] [Google Scholar]
- Wiśniewski A, Kowal A, Wyrodek E, Nowak I, Majorczyk E, Wagner M, Pawlak-Adamska E, Jankowska R, Ślesak B, Frydecka I, Kuśnierczyk P. Genetic polymorphisms and expression of HLA-G and its receptors, KIR2DL4 and LILRB1, in non-small cell lung cancer. Tissue Antigens. 2015;85:466–475. doi: 10.1111/tan.12561. [DOI] [PubMed] [Google Scholar]
- Worley MJ, Welch WR, Berkowitz RS, Ng SW. Endometriosis-associated ovarian cancer: a review of pathogenesis. Int J Mol Sci. 2013;14:5367–5379. doi: 10.3390/ijms14035367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong L, Pan D, Chen W, He Y, Liu Z, Lin J, Xu A. Comparative study of HLA-DQA1 and HLA-DRB1 allele in patients with endometriosis and adenomyosis. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:49–51. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 PCR conditions for LILRB1 (rs41308748:G>A) and LILRB2 (rs383369:G>A, rs7247538:C>T) genotyping (DOCX 13 KB)
Table S2 HLA-G genotype and minor allele frequencies in women from Control and Endometriosis groups (DOC 65 KB)
Table S3 LILRB1, LILRB2 and KIR2DL4 genotype and minor allele frequencies in women from Control and Endometriosis groups (DOC 75 KB)
Table S4 Comparison of the LILRB and KIR2DL4 polymorphisms in women depending on the localization of lesions (DOC 85 KB)
Fig S1 Scatter plot of representative results from the real-time PCR of HLA-G rs1233334:G>C/T SNP genotyping (TIF 244 KB)