Abstract
The preservation of genome stability is fundamental for every cell. Genomic integrity is constantly challenged. Among those challenges are also non-canonical nucleic acid structures. In recent years, scientists became aware of the impact of G-quadruplex (G4) structures on genome stability. It has been shown that folded G4-DNA structures cause changes in the cell, such as transcriptional up/down-regulation, replication stalling, or enhanced genome instability. Multiple helicases have been identified to regulate G4 structures and by this preserve genome stability. Interestingly, although these helicases are mostly ubiquitous expressed, they show specificity for G4 regulation in certain cellular processes (e.g., DNA replication). To this date, it is not clear how this process and target specificity of helicases are achieved. Recently, Mms1, an ubiquitin ligase complex protein, was identified as a novel G4-DNA-binding protein that supports genome stability by aiding Pif1 helicase binding to these regions. In this perspective review, we discuss the question if G4-DNA interacting proteins are fundamental for helicase function and specificity at G4-DNA structures.
Keywords: DNA secondary structures, Pif1 helicase, Genome stability, S. cerevisiae
Introduction
Secondary DNA structures, such as G-quadruplex (G4) structures, are hypothesized to hamper DNA and RNA-related processes due to their high thermodynamic stability and by this challenging genome integrity (reviewed in Tarsounas and Tijsterman 2013). G4 structures are guanine-rich, four-stranded structures that can form within nucleic acids, if a defined nucleotide sequence, called G4 motif is present (Gellert et al. 1962).
Multiple analyses demonstrate that, once formed, G4 structures positively and negatively influence biological processes (reviewed in Bochman et al. 2012; Maizels and Gray 2013), such as DNA replication (Foulk et al. 2015; Valton et al. 2014), transcription (Law et al. 2010; Siddiqui-Jain et al. 2002), and translation (Morris et al. 2010). Currently, they are discussed as a molecular fine-tuning mechanism of the cell that influences specific processes (David et al. 2016; Nguyen et al. 2014; Tang et al. 2016). However, due to their stability, the formation and unfolding is predicted to be slow (Hazel et al. 2004; Lane et al. 2008). If G4 structures are a regulatory tool in the cell, fast and efficient regulation mechanisms are required. To this date, multiple helicases have been identified that can unwind G4 structure in vitro (Sauer and Paeschke 2017). Interestingly, in vivo experiments support the idea that most G4 structure-regulating helicases are specific for a given cellular process. For example, FANCJ, the helicase linked to the human disorder Fanconi anemia, as well as Pif1 helicases are shown to regulate G4 formation during DNA replication (Castillo Bosch et al. 2014; Paeschke et al. 2011, 2013; Piazza et al. 2017; Ribeyre et al. 2009; Wu and Spies 2016). In the absence of Pif1 in yeast, or FANCJ in humans, deletion or mutations occur at G4 motifs and genome instability accumulates. Therefore, it is not surprising that mutations in many G4-regulating helicases are linked to human genetic disorders (reviewed in Maizels 2006; Mendoza et al. 2016).
In addition to helicases, other machineries/proteins have been shown to support the disruption of formed G4 structures such as translesion synthesis proteins, homologous recombination (HR), or telomerase holoenzyme (Edwards et al. 2014; Paeschke et al. 2008; van Kregten and Tijsterman 2014). How these proteins act specifically at G4 structures or how they communicate with helicases is not clear, yet. Although the regulation of G4 structures is very efficient and G4 structures have an impact on all stages of the cell cycle, it seems that the proteins/helicases that regulate G4 disruption are highly process specific. For example, Pif1 helicase has been shown to act specifically at formed G4 structures during DNA replication, whereas the RecQ helicases are proposed to act at G4 structures during telomere maintenance (reviewed in Sauer and Paeschke in press). To this date, it is not completely understood how this specificity is achieved; one hypothesis is that G4 structure-interacting proteins support helicase binding and thereby increase helicase specificity. In recent years, many scientists performed global approaches to identify novel G4 interacting proteins (reviewed in Mishra et al. 2016).
Mms1 is a novel factor supporting Pif1 helicase binding to G4s
Using a pull-down-based approach, we identified in yeast, among known G4 interacting proteins, Mms1 as a novel G4-interacting protein (Wanzek et al. 2017). We demonstrated that Mms1 acts together with Rtt101 and Mms22 and forms an ubiquitin ligase complex, Rtt101Mms1/Mms22 (Zaidi et al. 2008). All three components are required for replication fork progression after treating cells with the alkylating agent methyl methanesulfonate (MMS), which causes replicative stress (Luke et al. 2006; Vaisica et al. 2011). In line with our observations, the previous studies have shown that Mms1 and Mms22 are required for HR at stalled forks but not at HO-induced double-strand-break (DSB) sites (Duro et al. 2008; Zaidi et al. 2008).
The replication machinery slows at G4 structures (Anand et al. 2012; Valton and Prioleau 2016), especially if G4 structures are not unwound by a DNA helicase, e.g., Pif1 (Paeschke et al. 2011). We speculated that in S cerevisiae, the ubiquitin ligase Rtt101Mms1/Mms22 is also involved in replication fork progression at G4 motifs. Chromatin immunoprecipitation (ChIP) experiments coupled with deep-sequencing analyses supported the hypothesis. Analyses revealed that the guanine-rich Mms1-binding motif can form “relaxed” G4 structures in vitro. These “relaxed” G4 motifs form less stable G4 structures, because they only harbor two guanines per G-tract (GGN1–8GGN1–8GGN1–8GG) as compared to three or more in canonical G4 structures (e.g.: GGGN1–7GGGN1–7GGGN1–7GGG) (Gellert et al. 1962). Interestingly, “relaxed” G4 motifs that are bound by Mms1 are located on the lagging strand template for DNA replication (Wanzek et al. 2017). In addition, detailed analyses of Mms1 target regions showed that they are also targeted by Pif1 (Wanzek et al. 2017).
Previously, it was shown that Pif1, a 5′–3′ DNA helicase, is a robust unwinder of G4 structures in vitro (Ribeyre et al. 2009). Using two different methods in yeast, it was demonstrated that in Pif1 mutant cells (pif1-m2), replication slows at G4 motifs (Paeschke et al. 2011). Furthermore, different genetic analyses revealed that Pif1 preserves genome stability at G4 motifs, and accordingly, deletions and/or mutations occur at these motifs in the absence of Pif1 (Lopes et al. 2011; Paeschke et al. 2013). This led to the model that Pif1 unwinds G4 structures in vivo and by this supports replication fork progression and genome stability. Although these studies nicely support each other, they also harbor controversial aspects. Under one experimental setting, Pif1 acts equally well on G4 motifs on both strands of the replication fork machinery. In contrast, studies that used a repetitive G4 motif from a human minisatellite (CEB1) in yeast showed that Pif1 acts only at G4 motifs located on the leading strand template for DNA replication (Lopes et al. 2011; Paeschke et al. 2013).
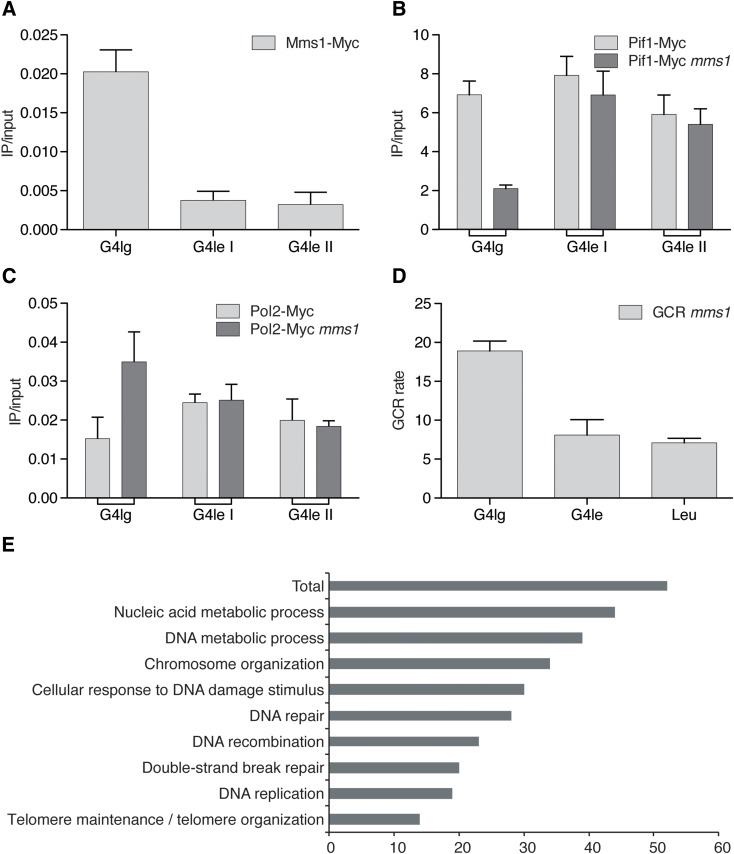
The newest published results shed some light into this discussion. They revealed that Mms1 supports Pif1 binding to specific G4 structures located on the lagging strand (Wanzek et al. 2017). Without Mms1, Pif1 binding is reduced, replication slows at G4 motifs, and G4-dependent genome instability is observed. The effects are similar to effects reported in Pif1 mutant cells (Paeschke et al. 2011, 2013; Piazza et al. 2012). Mms1 does not bind to G4 motifs on the leading strand (Fig. 1a). Expectedly, in the absence of Mms1, Pif1 binding to G4s on the leading strand was unaffected (Fig. 1b). In addition, no replication pausing and consequently no increased genome instability by GCR were observed, if the G4 motifs was located on the leading strand (Fig. 1c, d). Interaction network analyses showed a significant overlap between Pif1 and Mms1 (50 proteins interacted genetically and 2 proteins physically with both proteins, Fig. 1e). These results provide information in the discussion, whether G4 motifs on the leading, on the lagging strand, or both cause genome instability. It is probably depending on the substrate, the genetic surrounding, and consequently the associated proteins which determine the fate of the G4 motif and Pif1. In practice, this means that repeats (e.g., CEB1) are targeted by a different set of proteins than other G4 motifs. Furthermore, the current data indicate that specific G4 motifs on the lagging strand are targeted by Mms1 and consequently Pif1, but provide no information about other G4 motifs on both leading and lagging strand. In addition, because Mms1 acts here without the Ubiquitin complex, which is an unusual setting for Mms1, it is likely that other proteins are involved in this G4 regulation process. These data and consequent speculation exhibit that disruption of G4 structures is not as trivial as originally anticipated. Presumably a whole complex of proteins acts together to control G4 unwinding at different genomic settings.
Fig. 1.
Mms1 does not support Pif1 function at all targets. a Mms1 does not bind to G4 motifs on the leading strand. ChIP experiments, using endogenous Myc-tagged Mms1 at specific loci, as well as genome wide analyses (Wanzek et al. 2017) revealed that Mms1 does not bind to G4 motifs on the leading strand (G4lg). Here and in all subsequent ChIP experiments, plotted IP/input values are mean values ± standard deviation (SD) of three biological replicates. b Pif1 binding is not affected at G4 motifs on the leading strand. ChIP experiments using endogenously Myc-tagged Pif1 were performed in WT (light grey) and mms1Δ (dark grey) cells. Pif1 binding was reduced at a specific G4 motif on the lagging strand (G4lg) in mms1Δ cells (Wanzek et al. 2017), whereas binding was unaffected at G4 motifs on the leading strand (G4le). c Replication fork progression is not dependent on Mms1 at all G4 motifs. DNA Pol2 binding to G4 motifs was analyzed by ChIP using endogenous Myc-tagged Pol2 and qPCR in WT (light grey) and mms1Δ (dark grey) cells. As previously (Wanzek et al. 2017), DNA Pol2 binding was enhanced in the absence of Mms1 at G4 motifs on the lagging strand (G4lg), but no difference in DNA Pol2 binding was observed at other G4 motifs, located on the leading strand template (G4le). d G4 motifs on the leading strand did not increase the GCR rate in mms1Δ cells. The GCR rate was determined in wild-type and mms1Δ cells. Plotted data are normalized to WT. All inserts were integrated using the LEU2 marker. Inserts are: G4 motif on the lagging strand template (G4lg) (Wanzek et al. 2017), G4 motif on the leading strand template (G4le), as well as the LEU2 marker alone. e Analysis of the 52 joined genetic and physical interaction of Pif1 and Mms1 by processes
G4 structures are discussed to play a positive role in different biological processes such as transcriptional regulation, where they supposed to serve as a loading scaffold for proteins (Bochman et al. 2012). Multiple analyses have now shown that G4 motifs fold into G4 structures in vivo (Biffi et al. 2013, 2014; Paeschke et al. 2005; Schaffitzel et al. 2001) and that G4 motifs are conserved throughout evolution (Capra et al. 2010; Nakken et al. 2009). This and other data suggest that G4 structures have a biological function and fold on purpose and not by chance. Even though it is not fully understood yet, how G4 structures form efficiently and specifically, recent data imply that Mms1 plays a role during G4 structure formation, because it binds to G4 motifs throughout the whole cell cycle (Wanzek et al. 2017) where it has the potential to recruit other G4 modulators.
According to the Saccharomyces Database (SGD) and further publications, there are with Est1, Est2, Rap1, Mre11, Rad50, Xrs2, Pif1, Rrm3, Sgs1, Yku70, Rif1, Hop1, Kem1, Stm1, and Sub1; at least 15 proteins known to bind to DNA G4 structures in S. cerevisiae (Cherry et al. 2012; Fisher et al. 2004; Kanoh et al. 2015; Liu and Gilbert 1994; Lopez et al. 2017; Muniyappa et al. 2000; Sun et al. 1999; Van Dyke et al. 2004). Comparing genetic and physical interaction maps of those proteins shows that 5 proteins are genetically or physically linked to Mms1 in S. cerevisiae. A genetic/physical interaction does not proof a link, but a potential idea on how to address G4 regulation and unfolding in the near future. The identification of proteins that aid helicase binding and function will be interesting by itself but will also be beneficial for medical aspects. G4 structures are discussed as novel therapeutically targets to regulate biological processes (Balasubramanian and Neidle 2009; Bates et al. 2017; Cogoi and Xodo 2016). The current research focuses on chemical components (ligands) that induce or stabilize G4 structures in vivo (Salgado et al. 2015). The aim is to influence/block specific processes by inducing/stabilizing G4 structures. Due to the high number of G4 motifs in the cell over 500/300,000 in S. cerevisiae/human (Capra et al. 2010; Chambers et al. 2015; Huppert and Balasubramanian 2005), the current challenge is the specificity of these ligands to certain and not all G4s. Targeting the functional helicases at the right spatiotemporal moment will be an additional difficulty due to their multifunctional nature. However, a promising future strategy to gain specificity could be to target the aiding proteins instead of a multifunctional helicase.
Acknowledgements
We thank Stefan Juranek for the fruitful discussions and careful reading of the manuscript. We also like to thank Katharina Wanzek who started the Mms1 project. We apologize to all colleagues whose work could not be cited due to space limitations. Research in the Paeschke laboratory is supported by the Emmy-Noether Program of the Deutsche Forschungsgemeinschaft (DFG) as well as a Starting grant from the European Research Council (ERC Stg Grant: 638988-G4DSB).
References
- Anand RP, Shah KA, Niu H, Sung P, Mirkin SM, Freudenreich CH. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012;40:1091–1105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr Opin Chem Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PJ, Reyes-Reyes EM, Malik MT, Murphy EM, O’, Toole MG, Trent JO. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: uses and mechanisms. Biochim Biophys Acta. 2017;1861:1414–1428. doi: 10.1016/j.bbagen.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nature chemistry. 2014;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo Bosch P, Segura-Bayona S, Koole W, van Heteren JT, Dewar JM, Tijsterman M, Knipscheer P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014;33:2521–2533. doi: 10.15252/embj.201488663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700-705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S, Xodo LE. G4 DNA in ras genes and its potential in cancer therapy. Biochim Biophys Acta. 2016;1859:663–674. doi: 10.1016/j.bbagrm.2016.02.002. [DOI] [PubMed] [Google Scholar]
- David AP, Margarit E, Domizi P, Banchio C, Armas P, Calcaterra NB. G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 2016;44:4163–4173. doi: 10.1093/nar/gkw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duro E, Vaisica JA, Brown GW, Rouse J. Budding yeast Mms22 and Mms1 regulate homologous recombination induced by replisome blockage. DNA Repair (Amst) 2008;7:811–818. doi: 10.1016/j.dnarep.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Edwards DN, Machwe A, Wang Z, Orren DK. Intramolecular telomeric G-quadruplexes dramatically inhibit DNA synthesis by replicative and translesion polymerases, revealing their potential to lead to genetic change. PloS One. 2014;9:e80664. doi: 10.1371/journal.pone.0080664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher T, Taggart A, Zakian V. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- Foulk MS, Urban JM, Casella C, Gerbi SA. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015;25:725–735. doi: 10.1101/gr.183848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, Davies DR (1962) Helix formation by guanylic acid. Proc Natl Acad Sci USA 48:2013–2018 [DOI] [PMC free article] [PubMed]
- Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh Y, Matsumoto S, Fukatsu R, Kakusho N, Kono N, Renard-Guillet C, Masuda K, Iida K, Nagasawa K, Shirahige K, Masai H. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat Struct Mol Biol. 2015;22:889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I, Higgs DR, Gibbons RJ. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378 . doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Liu Z, Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CR, Singh S, Hambarde S, Griffin WC, Gao J, Chib S, Yu Y, Ira G, Raney KD, Kim N. Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 2017;45:5850–5862. doi: 10.1093/nar/gkx201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol CB. 2006;16:786–792. doi: 10.1016/j.cub.2006.02.071. [DOI] [PubMed] [Google Scholar]
- Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- Maizels N, Gray LT. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza O, Bourdoncle A, Boule JB, Brosh RM, Jr, Mergny JL. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tawani A, Mishra A, Kumar A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- Muniyappa K, Anuradha S, Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol Cell Biol. 2000;20:1361–1369. doi: 10.1128/MCB.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakken S, Rognes T, Hovig E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009;37:5749–5756. doi: 10.1093/nar/gkp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen GH, Tang W, Robles AI, Beyer RP, Gray LT, Welsh JA, Schetter AJ, Kumamoto K, Wang XW, Hickson ID, Maizels N, Monnat RJ Jr, Harris CC (2014) Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc Natl Acad Sci USA 111:9905–9910. 10.1073/pnas.1404807111 [DOI] [PMC free article] [PubMed]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA. DNA replication through G-Quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Serero A, Boule JB, Legoix-Ne P, Lopes J, Nicolas A. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet. 2012;8:e1003033. doi: 10.1371/journal.pgen.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Cui X, Adrian M, Samazan F, Heddi B, Phan AT, Nicolas AG (2017) Non-Canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae. eLife 610.7554/eLife.26884 [DOI] [PMC free article] [PubMed]
- Ribeyre C, Lopes J, Boulé J, Piazza P, Guédin A, Zakian V, Mergny J-L, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado GF, Cazenave C, Kerkour A, Mergny JL. G-quadruplex DNA and ligand interaction in living cells using NMR spectroscopy. Chem Sci. 2015;6:3314–3320. doi: 10.1039/C4SC03853C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Paeschke K (2017) G-quadruplex unwinding helicases and their function in vivo Biochem Soc Trans [DOI] [PubMed]
- Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA 99:11593–11598. 10.1073/pnas.182256799 [DOI] [PMC free article] [PubMed]
- Sun H, Bennett R, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucl Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Robles AI, Beyer RP, Gray LT, Nguyen GH, Oshima J, Maizels N, Harris CC, Monnat RJ., Jr The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Hum Mol Genet. 2016;25:2060–2069. doi: 10.1093/hmg/ddw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Tijsterman M. Genomes and G-quadruplexes: for better or for worse. J Mol Biol. 2013;425:4782–4789. doi: 10.1016/j.jmb.2013.09.026. [DOI] [PubMed] [Google Scholar]
- Vaisica JA, Baryshnikova A, Costanzo M, Boone C, Brown GW. Mms1 and Mms22 stabilize the replisome during replication stress. Mol Biol Cell. 2011;22:2396–2408. doi: 10.1091/mbc.e10-10-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton AL, Prioleau MN. G-Quadruplexes in DNA replication: a problem or a necessity? Trends Genet. 2016;32:697–706. doi: 10.1016/j.tig.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintome C, Riou JF, Prioleau MN. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kregten M, Tijsterman M. The repair of G-quadruplex-induced DNA damage. Exp Cell Res. 2014;329:178–183. doi: 10.1016/j.yexcr.2014.08.038. [DOI] [PubMed] [Google Scholar]
- Van Dyke MW, Nelson LD, Weilbaecher RG, Mehta DV. Stm1p, a G4 quadruplex and purine motif triplex nucleic acid-binding protein, interacts with ribosomes and subtelomeric Y’ DNA in Saccharomyces cerevisiae. J Biol Chem. 2004;279:24323–24333. doi: 10.1074/jbc.M401981200. [DOI] [PubMed] [Google Scholar]
- Wanzek K, Schwindt E, Capra JA, Paeschke K. Mms1 binds to G-rich regions in Saccharomyces cerevisiae and influences replication and genome stability. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CG, Spies M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmstrom J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008;9:1034–1040. doi: 10.1038/embor.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]