Abstract
As soon as induced pluripotent stem cells (iPSCs) reprogramming of somatic cells were developed, the discovery attracted the attention of scientists, offering new perspectives for personalized medicine and providing a powerful platform for drug testing. The technology was almost immediately applied to cancer studies. As presented in this review, direct reprogramming of cancer cells with enforced expression of pluripotency factors have several basic purposes, all of which aim to explain the complex nature of cancer development and progression, therapy-resistance and relapse, and ultimately lead to the development of novel anti-cancer therapies. Here, we briefly present recent advances in reprogramming methodologies as well as commonalities between cell reprogramming and carcinogenesis and discuss recent outcomes from the implementation of induced pluripotency into cancer research.
Abbreviations: CSC, cancer stem cells; iPSCs, induced pluripotent stem cells; iCSCs, induced cancer stem-like cells; SCNT, somatic cell nuclear transfer
Keywords: Induced pluripotency, Cancer reprogramming, Cancer stem cells
1. Introduction
Induced pluripotent stem cells (iPSCs) are somatic cells that have been reprogrammed to form undifferentiated stem cells.1, 2 Direct dedifferentiation of somatic cells with enforced expression of pluripotency markers was for the first time reported by Takahashi and Yamanaka in 2006.1 Since then, the utilization of iPSC technology has grown exponentially together with advances in reprogramming methodologies using a variety of pluripotency-inducing factors and delivery systems (presented below). Induced pluripotency was used to reprogram diverse types of cells, paving the way for dedifferentiation of transformed cancer cells. Currently, most human models of cancer are based on cancer cell lines and/or xenografts of primary tumor tissues cultured in vitro, reflecting the advanced state of tumor progression from which the cells were derived.3 Due to the application of induced pluripotency, cancer research acquired a new turn. Utilization of iPSC technology in cancer studies gives the opportunity to elucidate the mechanisms which underlie the stages of cancer development. Moreover, the reprogramming of cancer cells gives rise to a population of cells that possess cancer stem cell (CSC) characteristics4 – thorough exploration of their biological properties may help in better understanding of therapy resistance and tumor relapse. Also, the application of iPSC technology to dedifferentiate cancer cells creates a powerful tool for distinguishing epigenetic and genetic alterations that occur during tumor development and progression. In this review we briefly present recent advances in reprogramming methodologies as well as commonalities between cell reprogramming and carcinogenesis and discuss recent outcomes from the implementation of induced pluripotency technology into cancer studies.
2. Somatic cell reprogramming to pluripotency
Since the first discovery that fully differentiated somatic cell could be reprogrammed to become induced pluripotent stem cell (iPSCs), numerous methods have been developed to generate iPSCs.1, 2, 5 Systematically improved strategies resulted in more efficient reprogramming (yielding a higher number of pluripotent colonies), and/or the generation of xeno-free iPSC lines lacking integration of any vector sequences into their genomes. The basal protocol established by Takahashi et al.1, 2 centers on the ectopic expression of master reprogramming factors (Oct-3/4, Sox2, Klf4 and c-Myc; OSKM) and epigenetic reactivation of endogenous pluripotency genes. In further studies reprogramming without the proto-oncogene c-Myc has been proposed either by using only 3 reprogramming factors6 or by replacing c-Myc with less critical genes such as L-Myc or Glis1.7, 8, 9 It was further demonstrated that in inducing pluripotency, the number of reprogramming factors could be reduced (to two factors Oct-3/4 and Klf4 or c-Myc) when using somatic cells that endogenously express appropriate levels of complementing factors.10, 11 Moreover, the emerging strategies of reprogramming were oriented to increase the safety of iPSC derivation; therefore, majority of them utilized transgene-free methods of reprogramming factors delivery to the host cell.12 Integration-free mouse and human iPSCs have been generated using adenoviral vectors,13, 14 Sendai viruses,15, 16, 17 Cre/loxP system,18, 19, 20 the piggyBac system,21, 22 episomal vectors,7, 23, 24 expression plasmid vectors,25 small-molecule compound distribution26, 27, 28 and direct mRNA29, 30 or protein delivery.31, 32 Examples of specific methods that have been recently utilized to reprogram mouse or human somatic cells to induced pluripotent stem cells are summarized in Table 1.
Table 1.
Mouse and human iPSC lines derived with different methods of reprogramming factor delivery.
| Delivery method | Cell origin | Cell type | Reprogramming factors | Reference |
|---|---|---|---|---|
| Adenoviruses | Mouse | Tail tip fibroblasts, fetal liver, hepatocytes | OSKM | 13 |
| Human | Embryonic fibroblasts | OSKM | 14 | |
| Sendai virus | Human | Neonatal foreskin, dermal fibroblasts | OSKM | 15 |
| Human | Fibroblasts | OSKM | 16 | |
| Human | Nasal epithelial cells | OSKM | 17 | |
| Cre/loxP | Mouse | Primary fibroblast | OSKM | 18 |
| Human | Extraembryonic fetal cells | OSKM | 19 | |
| Human | Fibroblast | OSKM | 20 | |
| PiggyBac transposon | Mouse | Fibroblast | OSKM | 21 |
| Mouse/human | Embryonic fibroblasts | OSKM | 22 | |
| Episomal vector | Human | Fibroblast | OSKM and NANOG, LIN28 + SV40LT | 23 |
| Human | Neonatal foreskin, adult scar tissue, amniotic fluid, urine | OSKM and LIN28 + mutated TP53 | 24 | |
| Human | Dermal fibroblasts | OSKM and LIN28 + TP53 knockdown | 7 | |
| Plasmid DNA transfection | Mouse | Embryonic fibroblasts | OSK + M | 25 |
| Small-molecule compounds | Mouse | Embryonic fibroblasts | None | 26 |
| Mouse | Embryonic fibroblasts | Oct-3/4, Klf4 + BIX-01294 and BayK8644 | 27 | |
| Mouse | Embryonic fibroblasts | Oct-3/4 + VPA, tranylcypromine, CHIR99021 and 616452 (Tgfbeta inhibitor) | 28 | |
| mRNA | Human | ES-derived dH1f fibroblasts | OSKM | 29 |
| Human | Fibroblast | OSKM, Lin28 | 30 | |
| Protein delivery | Mouse | Embryonic fibroblasts | OSKM | 31 |
| Human | Newborn fibroblasts | OSKM | 32 | |
OSKM – Oct-3/4, Sox2, Klf4, c-Myc.
3. Reprogramming and carcinogenesis
The connection between oncogenesis and induced pluripotency is commonly discussed by the fact that during the reprogramming, somatic cells acquire potential for unlimited proliferation and ability to self-renew – both are well known features of transformed cancer cells.33 Also, induced pluripotent stem cells lack contact inhibition of proliferation34 and exhibit high telomerase activity and telomere elongation.35, 36 These are also two essential characteristics of cancer cells that facilitate tumor growth. Moreover, iPSC and cancer cell metabolism are overtly similar, with metabolite levels directly influencing chromatin organization and transcription.37 To promote rapid cell proliferation and duplication, induced pluripotent stem cells balance their energy with biosynthetic requirements, which results in a metabolic shift from oxidative state to a glycolytic state in pluripotency,38 a feature shared with highly proliferative cancer cells. Furthermore, the core pluripotency genes involved in the reprogramming process also play a central role in tumorigenicity.39, 40 The cocktail of Yamanaka's factors1, 2 that enables the dedifferentiation of somatic cells to a stem-like state is composed of well-known oncogenes, such as c-Myc and Klf4,41, 42 or genes that exhibit high expression in various types of cancer, such as Oct-3/4 and Sox2.43, 44, 45, 46 Oct-3/4 was demonstrated to promote tumorigenesis and inhibit apoptosis of cervical cancer cells,44 support drug-resistance of prostate cancer,47 and play a crucial role in maintaining cancer stem-like cells in lung,48 liver,49 breast,50, 51 brain52 and other cancers.53, 54 Similarly, Sox2 enhances the tumorigenicity and chemoresistance of cancer stem-like cells derived from gastric carcinoma,55 and is required to maintain cancer stem cells in the breast,46 bladder,56 ovarian,57 and other cancers.58, 59, 60 The importance of the thorough understanding of somatic cell reprogramming for enforced expression of “pluripotency factors” is evident when we consider the proposed theory of cancer evolution from dedifferentiated cancer cell that acquire stem cell traits (so called, cancer stem cell).61 Advanced analyses of this multistep process will be extremely helpful in the recognition of specific molecular changes that reprogrammed cells and, in parallel, cancer cells need to make to achieve stem cell state. These molecular or cellular modifications may serve as therapeutic targets in anti-cancer treatment.62, 63
Similarities between iPSCs and cancer cells also encompass the overall gene expression pattern64 and epigenetic status.65 Reprogramming of somatic cells to iPSCs requires profound alterations in the epigenetic landscape. During the reprogramming, pluripotent stem cells acquire a unique epigenetic profile enriched for active chromatin modifications (including H3K4me3, H3K36me3, histone acetylation, and hypomethylated DNA)66 which are frequently found within the regions of pluripotency-associated genes. Also, alterations in DNA methylation within cancer-specific gene promoters as well as aberrant responses to epigenetic-modifying drugs resembling those for cancer cells65 were observed in induced pluripotent stem cells, suggesting that thorough exploration of mechanisms governing induced reprogramming may provide a significant insight into the origins of epigenetic gene silencing associated with human carcinogenesis.67
4. Generation of induced cancer stem cells (iCSCs)
Generation of induced pluripotent stem cells from both normal and malignant patient tissue could be used to model carcinogenesis and elucidate the mechanisms which underlie the stages of cancer development. Also, reprogramming cancer cells could offer a platform for studying tumor heterogeneity and origin of cancer stem cells and a source for cancer type-specific drug discovery studies. Moreover, induced cancer stem cells (iCSCs) would be beneficial to explore the possibility to normalize in vivo the malignant phenotype of cancer stem cells, as an alternative to conventional therapeutic protocols. In vitro generation of iCSCs may yield a large number of stem-like cells, which would be available for experimental manipulation and for exploration of their biological properties that ultimately may help in better understanding of therapy-resistant tumors. Specific techniques developed so far for in vitro cancer cell reprogramming are presented below.
The concept of cancer cell reprogramming to pluripotency is not new and before Yamanaka's discovery of enforced expression of ectopic master reprogramming factors in somatic cell reprogramming, the Somatic Cell Nuclear Transfer (SCNT) was adapted in cancer studies.68, 69 In 2004, Hochedlinger et al. reversed malignant melanoma cells to a pluripotent state by nuclear transplantation to oocytes.68 However, not all cancer types could be reprogrammed with this technique. Hochedlinger et al.68 have observed that leukemia, lymphoma, and breast cancer cell nuclei transplanted into the oocyte could support normal preimplantation development to the blastocyst stage but failed to produce embryonic stem (ES) cells. Blelloch et al.69 have observed a strong correlation between the differentiation state of the donor cancer nucleus and the efficiency of producing stem cell lines after nuclear transfer, suggesting that cancer cells that initially express high levels of pluripotency markers, including Oct-3/4, Sox2 and SSEA, are more prone to reprogramming that differentiated cancer cells. They have found that the derivation of stem cells after NT of a differentiated tumor model occurs at a low efficiency if at all.
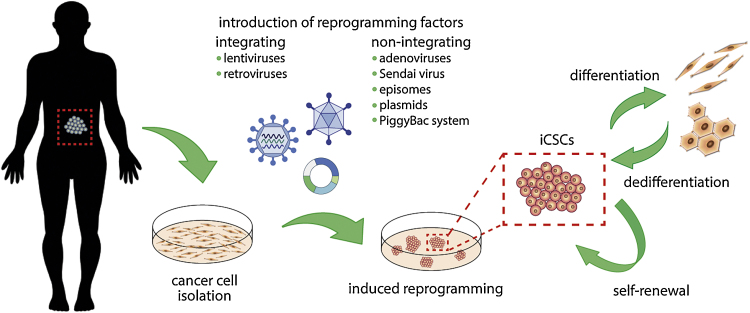
As previously mentioned, the reprogramming of cancer cells seems to be a valuable approach to analyze cancer-associated genes and the interaction between the genes and cell environment before and after reprogramming, to explain the mechanisms regulating cancer development. Fortunately, utilization of new protocols to obtain induced cancer stem cells (iCSCs) resulted in the higher efficiency of reprogramming (Fig. 1).70
Fig. 1.
Generation of induced cancer stem cells (iCSCs) with defined reprogramming factors. Patient derived cancer cells could be dedifferentiated into cancer-stem like population upon introduction of reprogramming factors either with integrating or non-integrating methods. Emerging iCSCs colonies possess the ability to both self-renew and give rise of more differentiated progeny.
For the first time, core Yamanaka's reprogramming factors were adapted to dedifferentiate cancer cells into induced cancer stem-like cells with lentiviral vector in 2009 by Utikal et al.71 They demonstrated that mouse melanoma cells do not require Sox2 transcription factor to acquire pluripotency fully. Emerging induced cancer stem-like cells phenotypically resemble embryonic stem cell-like colonies and show demethylation of the endogenous OCT-3/4 and NANOG gene promoters. They also formed teratomas and gave rise to chimeras after blastocyst injection. Lentiviral reprogramming method was further used by Mathieu et al.72 who observed iPSC-like colonies generated from lung cancer cell line. However, relatively low level of endogenous OCT-3/4 and NANOG mRNA and partially unmethylated status of OCT-3/4 gene promoter suggested not fully re-programmed phenotype. Zhang et al.73 demonstrated direct reprogramming of several sarcoma cell lines and resulting iCSCs were further utilized to explore the relationship between cell reprogramming and the epigenetics of oncogenes. Interestingly, they observed that reprogramming of cancer cells affects the ability of the cancer cells to re-engage and terminally execute normal cellular differentiation pathways with consequent loss of tumorigenicity. Later, Kim et al.74 have established iPSC-like cell lines from patient-derived pancreatic ductal adenocarcinoma as well as matched healthy tissue. However, they were successful in deriving pluripotent cell lines from only one patient, which demonstrates the difficulty of reprogramming primary cancer cells to iCSCs. Recently, Bernhardt et al.75 have obtained metastable melanoma induced cancer stem-like cells with the ability to differentiate into non-tumorigenic lineages terminally. Moreover, reprogrammed cells, as well as their differentiated progeny, were more resistant to targeted therapies, although the cells harbor the same oncogenic mutations and signaling activity as the parental melanoma cells.75
Also, other approaches of reprogramming factor delivery that include retroviruses76, 77, 78, 79 and Sendai virus,80 as well as non-integrating methods: episomes,81 PiggyBac transposon82 or plasmid transfection,70 were used to dedifferentiate several cancer types to induced pluripotency.
Retroviral reprogramming approach was used by Miyoshi et al.79 who dedifferentiated several types of gastrointestinal cancer cells with combination of Yamanaka's factors as well as oncogenes (BCL2 and KRAS) and shRNA targeting tumor suppressor genes (TP53, CDKN2A, PTEN, FHIT, RB1). Surprisingly, Miyoshi et al.79 demonstrated higher sensitivity to chemotherapeutic agents and differentiation-inducing treatment in induced cancer stem cells when compared to parental cell lines which contradicts previous results showing cancer stem cell resistance to chemotherapy.4, 63 However, retroviral reprogramming of cancer cells applied to other cancer types, such as chronic myeloid leukemia78, 83 or breast cancer,84 resulted in the acquisition of stem-like drug-resistant phenotype by reprogrammed cells. Recently, direct reprogramming of lung cancer cells with retrovirus-mediated enforced expression of pluripotency factors resulted in the acquisition of stem-like phenotype by cancer cells which were characterized with a distinct epigenetic pattern when compared to parental cells.85 Mahalingam et al.85 have suggested that direct reprogramming was able to perturb the epigenetics of lung cancer cells by causing the reversal of the aberrantly methylated promoters resulting in some instances in activation of gene transcription. They propose that direct cell dedifferentiation amends the aberrant epigenetic signatures of cancer cells which may contribute to the abrogation of their malignant properties.85 Surprisingly, further studies with lung cancer dedifferentiation enabled Lai et al.86 to observe that enforced cell reprogramming not only perturb the epigenetic state of the cell when compared to parental cell but also impacts the genetic mutation status. However, it is clear that a cell is not capable of recovering a mutated gene during reprogramming87, 88; therefore, Lai et al.86 observation suggests that enforced reprogramming discriminates within the heterogeneous cancer cell population of varying degrees of genetic insult and enriches a minor subpopulation with not affected genetic background.
Generation of iCSCs still requires improvement to increase the efficiency of direct reprogramming. Recently, Zhao et al.70 have reported a highly optimized protocol for reprogramming cancer cells to pluripotency using transfection with non-viral plasmid vectors. Transfection of cancer cells with specific vectors encoding Yamanaka's reprogramming factors resulted in extremely high differentiation effectiveness, reaching more than 50% of transfected cells emerging with stem-like phenotype. Surprisingly, the critical parameter in optimizing reprogramming effectiveness was the cell density initially seeded in the cultures. They believe that parallel principles established in their study can be used to generate virus-free iCSCs from other cancer types, thereby providing a general protocol with widely applicable potential for future studies.70
5. Application of induced reprogramming in cancer research
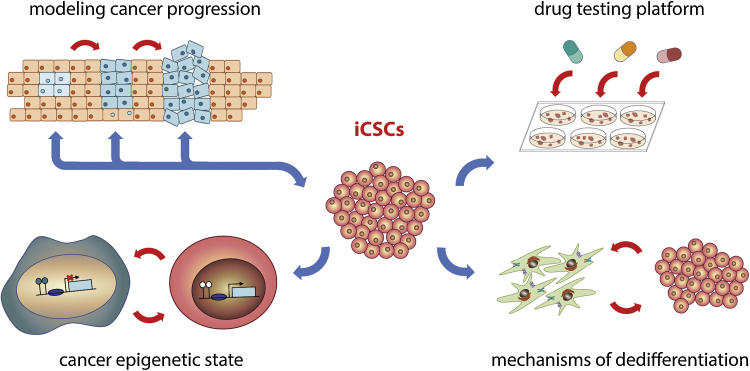
Primary tumors resected from patients and patient-derived cancer cell lines allow to understand late-stage markers and cellular phenotypes; however, they are inadequate models to study the early stages of tumor progression. Utilization of induced reprogramming technology gives the opportunity to elucidate the mechanisms which underlie the stages of cancer development (Fig. 2). In their work Kim et al.74 have reprogrammed human patient-derived pancreatic ductal adenocarcinoma to induced pluripotent state to test whether, upon differentiation, a subset of reprogrammed cells would undergo early developmental stages of human cancer. They observed very low efficiency of dedifferentiation, being successful only once in reprogramming a cell from a recurrent, late stage human pancreatic cancer to a near-pluripotent state. Emerging induced cancer stem-like cells when injected into immunodeficient mice, consistently generate pancreatic intraepithelial neoplasia lesions that progress to the invasive stage. Established protocol allowed identification of specific proteins secreted by tumor cells at distinct stages of tumor progression. Kim et al.74 proposed that the combined detection of released proteins that are the products of the HNF4α, TGFβ, and integrin networks within pancreatic intraepithelial neoplasia and invasive ductal adenocarcinoma cells may provide the best means for noninvasively detecting the progression of pancreatic cancer in humans.
Fig. 2.
Potential application of induced cancer stem cells in biomedical studies. Utilization of induced reprogramming technology in cancer studies (1) gives the opportunity to elucidate the mechanisms which underlie the stages of cancer development, (2) creates a powerful tool for distinguishing epigenetic (and genetic) alterations that occur during tumor development and progression, (3) allows a better understanding of dedifferentiation mechanism that occurs within heterogeneous cancer population and (4) serves as suitable platforms for the identification and validation of therapeutic targets that directly eradicate stem-cell like population within tumor cells.
So far, the reprogramming efficiency is extremely low in many types of cancer cells, suggesting that some properties of these cells impede induced dedifferentiation. During the acquisition of a pluripotent state, the genome of reprogrammed cells undergoes dynamic epigenetic changes.89 The fact that cancer cells are highly resistant to enforced dedifferentiation might be associated with relatively stable epigenetic regulations that maintain cancer cell integrity.90 Therefore, application of induced reprogramming into cancer studies may help to determine the epigenetic state of cancer cell and to evaluate the relative contribution of reversible epigenetic and irreversible genetic changes that occur during carcinogenesis.67, 71, 85, 86, 91 Moreover, uncovering the epigenetic stability of cancer cells may ultimately contribute to the development of therapeutics that effectively target epigenetic modifications in cancer cells.
Nowadays, standard high-throughput cell viability assays applied to bulk populations of cancer cells are ineffective in identifying agents with CSC-specific toxicity due to the low frequency of CSCs within cancer cell populations. Respectively, screening for agents that preferentially target CSCs depends on the ability to propagate stable, highly-enriched populations of CSCs in vitro. Therefore, human induced cancer stem-like cells could empower translational-research areas, serving as suitable platforms for the identification and validation of therapeutic targets and more relevant models for compound screening and drug repurposing. Recently, Nishi et al.92 using a model of iCSCs have developed a phenotypic drug assay to identify agents that inhibit the stemness and self-renewal properties of CSCs. They determined the selectivity of tested agents with three distinct assays characterized by cell viability, cellular stemness and tumor sphere formation. With this approach, the withaferin A was identified as a potent inhibitor of cancer stemness leading to cellular senescence primarily via the induction of p21Cip1 expression.
Observation that induced reprogramming of cancer cells could be achieved with enforced expression of reprogramming factors reveals the possibility that stem cell-like phenotypes could be gained and maintained by cancer cells during tumor progression in response to other exogenous stimuli in vitro or in vivo (i.e. hypoxia, therapeutic agents, heterogeneous stromal cells). Recent studies have demonstrated that cancer stem cells could be induced from non-stem cancer cells by radio- or chemotherapy, constituting the subpopulation of iCSCs that is characterized by increased sphere-forming ability, enriched tumorigenicity, and expression of genes related to stemness.93, 94, 95 It is, therefore, of the highest importance to identify direct mechanisms that govern dedifferentiation of cancer cells to stem-like populations to more precisely select therapeutic strategies that would not only destroy non-stem cancer cells, but also eradicate cancer stem cell population as well as disrupt the machinery of stemness acquisition.
6. Conclusion
Direct reprogramming of cancer cells with enforced expression of pluripotency factors have several primary purposes, all of which aim to explain the complex nature of cancer development and progression, therapy-resistance and relapse, and ultimately lead to the development of novel anti-cancer therapies. Generation of a large number of iCSCs that resemble cancer stem cell population in vivo may give the opportunity to explore biological properties of these cells. Moreover, since the prevalent in vitro cancer models miss the heterogeneous tumor structure and profoundly express the late-stage markers of cancer progression, the induced pluripotent stem cell technology could provide an alternative model to explore the early stages of carcinogenesis. Reprogramming of cancer cells into induced cancer stem-like cells provides the opportunity to convert malignant cells into any cell type, however, the effectiveness of cancer cell reprogramming remains to be a barrier. The difficulties in cancer cell dedifferentiation are thought to stem (at least partially) from the accumulation of DNA damage and cancer-specific mutations, epigenetic modifications and reprogramming-triggered cellular senescence. Therefore, development of more effective reprogramming approaches that would overcome current technological and biological challenges is essential to fully exploit the potential of induced pluripotency stem cell technology in cancer research.
Conflict of interest
None declared.
Financial disclosure
This work was supported by Greater Poland Cancer Centre intramural grants (Nos. 17/2016 (132) and 22/2016 (137)) and Poznań University of Medical Sciences intramural grant (No. 502-14-02233381-11055) to Patrycja Czerwińska.
Authors’ contributions
PC was a major contributor in writing the manuscript. MW coordinated the manuscript preparation. All authors read and approved the final manuscript.
Contributor Information
Patrycja Czerwińska, Email: patrycja.czerwinska@wco.pl.
Sylwia Mazurek, Email: syl.mazurek@gmail.com.
Maciej Wiznerowicz, Email: maciej.wiznerowicz@gmail.com.
References
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K., Tanabe K., Ohnuki M. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Katt M.E., Placone A.L., Wong A.D., Xu Z.S., Searson P.C. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front Bioeng Biotechnol. 2016;4:12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czerwinska P., Kaminska B. Review: Regulation of breast cancer stem cell features. Contemp Oncol. 2015;1A:7–15. doi: 10.5114/wo.2014.47126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa M., Koyanagi M., Tanabe K. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 7.Okita K., Matsumura Y., Sato Y. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 8.Maekawa M., Yamaguchi K., Nakamura T. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474(7350):225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 9.Maekawa M., Yamanaka S. Glis1, a unique pro-reprogramming factor, may facilitate clinical applications of iPSC technology. Cell Cycle. 2011;10(21):3613–3614. doi: 10.4161/cc.10.21.17834. [DOI] [PubMed] [Google Scholar]
- 10.Kim J.B., Zaehres H., Wu G. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454(7204):646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 11.Hester M.E., Song S., Miranda C.J., Eagle A., Schwartz P.H., Kaspar B.K. Two factor reprogramming of human neural stem cells into pluripotency. PLoS ONE. 2009;4(9):e7044. doi: 10.1371/journal.pone.0007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okano H., Nakamura M., Yoshida K. Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res. 2013;112(3):523–533. doi: 10.1161/CIRCRESAHA.111.256149. [DOI] [PubMed] [Google Scholar]
- 13.Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322(5903):945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W., Freed C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27(11):2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- 15.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ban H., Nishishita N., Fusaki N. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. PNAS. 2011;108(34):14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono M., Hamada Y., Horiuchi Y. Generation of induced pluripotent stem cells from human nasal epithelial cells using a Sendai virus vector. PLOS ONE. 2012;7(8):e42855. doi: 10.1371/journal.pone.0042855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varga E., Nemes C., Davis R.P. Generation of transgene-free mouse induced pluripotent stem cells using an excisable lentiviral system. Exp Cell Res. 2014;322(2):335–344. doi: 10.1016/j.yexcr.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Spitalieri P., Talarico R.V., Botta A. Generation of human induced pluripotent stem cells from extraembryonic tissues of fetuses affected by monogenic diseases. Cell Reprogram. 2015;17(4):275–287. doi: 10.1089/cell.2015.0003. [DOI] [PubMed] [Google Scholar]
- 20.Loh Y.-H., Yang J.C., De Los Angeles A. Excision of a viral reprogramming cassette by delivery of synthetic Cre mRNA. Curr Protoc Stem Cell Biol. 2012 doi: 10.1002/9780470151808.sc04a05s21. Chapter 4: Unit 4A.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458(7239):771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woltjen K., Michael I.P., Mohseni P. PiggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J., Hu K., Smuga-Otto K. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drozd A.M., Walczak M.P., Piaskowski S., Stoczynska-Fidelus E., Rieske P., Grzela D.P. Generation of human iPSCs from cells of fibroblastic and epithelial origin by means of the oriP/EBNA-1 episomal reprogramming system. Stem Cell Res Ther. 2015;6:122. doi: 10.1186/s13287-015-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322(5903):949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 26.Hou P., Li Y., Zhang X. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341(6146):651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y., Desponts C., Do J.T., Hahm H.S., Scholer H.R., Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3(5):568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Zhang Q., Yin X. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21(1):196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren L., Manos P.D., Ahfeldt T. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandal P.K., Rossi D.J. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat Protocols. 2013;8(3):568–582. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H., Wu S., Joo J.Y. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4(5):381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D., Kim C.-H., Moon J.-I. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedmann-Morvinski D., Verma I.M. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15(3):244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Zheng Y.-L., Qiu D.-B. An extracellular matrix culture system for induced pluripotent stem cells derived from human dental pulp cells. Eur Rev Med Pharmacol Sci. 2015;19(21):4035–4046. [PubMed] [Google Scholar]
- 35.Hiyama E., Hiyama K. Telomere and telomerase in stem cells. Br J Cancer. 2007;96(7):1020–1024. doi: 10.1038/sj.bjc.6603671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y., Liang P., Liu D., Huang J., Songyang Z. Telomere regulation in pluripotent stem cells. Protein Cell. 2014;5(3):194–202. doi: 10.1007/s13238-014-0028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J., Nuebel E., Daley G.Q., Koehler C.M., Teitell M.A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11(5):589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panopoulos A.D., Yanes O., Ruiz S. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22(1):168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J., Orkin S.H. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med. 2011;3(11):75. doi: 10.1186/gm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadjimichael C., Chanoumidou K., Papadopoulou N., Arampatzi P., Papamatheakis J., Kretsovali A. Common stemness regulators of embryonic and cancer stem cells. World J Stem Cells. 2015;7(9):1150–1184. doi: 10.4252/wjsc.v7.i9.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Wang H., Li Z. c-Myc is required for maintenance of glioma cancer stem cells. PLoS ONE. 2008;3(11):e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu F., Li J., Chen H. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30(18):2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Marzoqee F.Y., Khoder G., Al-Awadhi H. Upregulation and inhibition of the nuclear translocation of Oct4 during multistep gastric carcinogenesis. Int J Oncol. 2012;41(5):1733–1743. doi: 10.3892/ijo.2012.1608. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y.-D., Cai N., Wu X.-L., Cao H.-Z., Xie L.-L., Zheng P.-S. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013;4:e760. doi: 10.1038/cddis.2013.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y., Futtner C., Rock J.R. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE. 2010;5(6):e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leis O., Eguiara A., Lopez-Arribillaga E. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31(11):1354–1365. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 47.Linn D.E., Yang X., Sun F. A role for OCT4 in tumor initiation of drug-resistant prostate cancer cells. Genes Cancer. 2010;1(9):908–916. doi: 10.1177/1947601910388271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y.-C., Hsu H.-S., Chen Y.-W. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS ONE. 2008;3(7):e2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami S., Ninomiya W., Sakamoto E., Shibata T., Akiyama H., Tashiro F. SRY and OCT4 are required for the acquisition of cancer stem cell-like properties and are potential differentiation therapy targets. Stem Cells. 2015;33(9):2652–2663. doi: 10.1002/stem.2059. [DOI] [PubMed] [Google Scholar]
- 50.Xun J., Wang D., Shen L. JMJD3 suppresses stem cell-like characteristics in breast cancer cells by downregulation of Oct4 independently of its demethylase activity. Oncotarget. 2017;8(13):21918–21929. doi: 10.18632/oncotarget.15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim R.-J., Nam J.-S. OCT4 expression enhances features of cancer stem cells in a mouse model of breast cancer. Lab Anim Res. 2011;27(2):147–152. doi: 10.5625/lar.2011.27.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Q.-W., Zhou Y.-W., Li W.-X. Akt-mediated phosphorylation of Oct4 is associated with the proliferation of stemlike cancer cells. Oncol Rep. 2015;33(4):1621–1629. doi: 10.3892/or.2015.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiou S.-H., Yu C.-C., Huang C.-Y. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14(13):4085–4095. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- 54.Kumar S.M., Liu S., Lu H. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31(47):4898–4911. doi: 10.1038/onc.2011.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tian T., Zhang Y., Wang S., Zhou J., Xu S. Sox2 enhances the tumorigenicity and chemoresistance of cancer stem-like cells derived from gastric cancer. J Biomed Res. 2012;26(5):336–345. doi: 10.7555/JBR.26.20120045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu F., Qian W., Zhang H. SOX2 is a marker for stem-like tumor cells in bladder cancer. Stem Cell Rep. 2017;9(2):429–437. doi: 10.1016/j.stemcr.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen Y., Hou Y., Huang Z., Cai J., Wang Z. SOX2 is required to maintain cancer stem cells in ovarian cancer. Cancer Sci. 2017;108(4):719–731. doi: 10.1111/cas.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lundberg I.V., Edin S., Eklof V., Oberg A., Palmqvist R., Wikberg M.L. SOX2 expression is associated with a cancer stem cell state and down-regulation of CDX2 in colorectal cancer. BMC Cancer. 2016;16:471. doi: 10.1186/s12885-016-2509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boumahdi S., Driessens G., Lapouge G. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511(7508):246–250. doi: 10.1038/nature13305. [DOI] [PubMed] [Google Scholar]
- 60.Keysar S.B., Le P.N., Miller B. Regulation of head and neck squamous cancer stem cells by PI3K and SOX2. J Natl Cancer Inst. 2017;109(1) doi: 10.1093/jnci/djw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dalerba P., Cho R.W., Clarke M.F. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 62.Tang C., Ang B.T., Pervaiz S. Cancer stem cell: target for anti-cancer therapy. FASEB J. 2007;21(14):3777–3785. doi: 10.1096/fj.07-8560rev. [DOI] [PubMed] [Google Scholar]
- 63.Dragu D.L., Necula L.G., Bleotu C., Diaconu C.C., Chivu-Economescu M. Therapies targeting cancer stem cells: current trends and future challenges. World J Stem Cells. 2015;7(9):1185–1201. doi: 10.4252/wjsc.v7.i9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ben-Porath I., Thomson M.W., Carey V.J. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohm J.E., Mali P., Van Neste L. Cancer-related epigenome changes associated with reprogramming to induced pluripotent stem cells. Cancer Res. 2010;70(19):7662–7673. doi: 10.1158/0008-5472.CAN-10-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gladych M., Andrzejewska A., Oleksiewicz U., Estecio M.R.H. Epigenetic mechanisms of induced pluripotency. Contemp Oncol. 2015;19(1A):A30–A38. doi: 10.5114/wo.2014.47135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stricker S., Pollard S. Reprogramming cancer cells to pluripotency: an experimental tool for exploring cancer epigenetics. Epigenetics. 2014;9(6):798–802. doi: 10.4161/epi.28600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hochedlinger K., Blelloch R., Brennan C. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18(15):1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blelloch R.H., Hochedlinger K., Yamada Y. Nuclear cloning of embryonal carcinoma cells. PNAS. 2004;101(39):13985–13990. doi: 10.1073/pnas.0405015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao H., Davies T.J., Ning J. A highly optimized protocol for reprogramming cancer cells to pluripotency using nonviral plasmid vectors. Cell Reprogram. 2015;17(1):7–18. doi: 10.1089/cell.2014.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Utikal J., Maherali N., Kulalert W., Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122(Pt 19):3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathieu J., Zhang Z., Zhou W. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71(13):4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X., Cruz F.D., Terry M., Remotti F., Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32(18) doi: 10.1038/onc.2012.237. 2249–60, 2260.e1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J., Hoffman J.P., Alpaugh R.K. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3(6):2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernhardt M., Novak D., Assenov Y. Melanoma-derived iPCCs show differential tumorigenicity and therapy response. Stem Cell Rep. 2017;8(5):1379–1391. doi: 10.1016/j.stemcr.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim H.J., Jeong J., Park S. Establishment of hepatocellular cancer induced pluripotent stem cells using a reprogramming technique. Gut Liver. 2017;11(2):261–269. doi: 10.5009/gnl15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choong P.F., Teh H.X., Teoh H.K. Heterogeneity of osteosarcoma cell lines led to variable responses in reprogramming. Int J Med Sci. 2014;11(11):1154–1160. doi: 10.7150/ijms.8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumano K., Arai S., Hosoi M. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119(26):6234–6242. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- 79.Miyoshi N., Ishii H., Nagai K. Defined factors induce reprogramming of gastrointestinal cancer cells. PNAS. 2010;107(1):40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iskender B., Izgi K., Canatan H. Reprogramming bladder cancer cells for studying cancer initiation and progression. Tumor Biol. 2016;37(10):13237–13245. doi: 10.1007/s13277-016-5226-4. [DOI] [PubMed] [Google Scholar]
- 81.Harada K., Ferdous T., Cui D. Induction of artificial cancer stem cells from tongue cancer cells by defined reprogramming factors. BMC Cancer. 2016;16:548. doi: 10.1186/s12885-016-2416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yin J., Fan Y., Qin D., Xiaocui Bian X., Bi X. Generation and characterization of virus-free reprogrammed melanoma cells by the piggyBac transposon. J Cancer Res Clin Oncol. 2013;139(9):1591–1599. doi: 10.1007/s00432-013-1431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carette J.E., Pruszak J., Varadarajan M. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115(20):4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Corominas-Faja B., Cufi S., Oliveras-Ferraros C. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell Cycle. 2013;12(18):3109–3124. doi: 10.4161/cc.26173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahalingam D., Kong C.M., Lai J., Tay L.L., Yang H., Wang X. Reversal of aberrant cancer methylome and transcriptome upon direct reprogramming of lung cancer cells. Sci Rep. 2012;2:592. doi: 10.1038/srep00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lai J., Kong C.M., Mahalingam D., Xie X., Wang X. Elite model for the generation of induced pluripotent cancer cells (iPCs) PLOS ONE. 2013;8(2):e56702. doi: 10.1371/journal.pone.0056702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H., Collado M., Villasante A. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marion R.M., Strati K., Li H. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mattout A., Biran A., Meshorer E. Global epigenetic changes during somatic cell reprogramming to iPS cells. J Mol Cell Biol. 2011;3(6):341–350. doi: 10.1093/jmcb/mjr028. [DOI] [PubMed] [Google Scholar]
- 90.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moore J.B., Loeb D.M., Hong K.U. Epigenetic reprogramming and re-differentiation of a Ewing sarcoma cell line. Front Cell Dev Biol. 2015;3:15. doi: 10.3389/fcell.2015.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nishi M., Akutsu H., Kudoh A. Induced cancer stem-like cells as a model for biological screening and discovery of agents targeting phenotypic traits of cancer stem cell. Oncotarget. 2014;5(18):8665–8680. doi: 10.18632/oncotarget.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang L., Huang X., Zheng X. Enrichment of prostate cancer stem-like cells from human prostate cancer cell lines by culture in serum-free medium and chemoradiotherapy. Int J Biol Sci. 2013;9:472–479. doi: 10.7150/ijbs.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y., Li W., Patel S.S. Blocking the formation of radiation-induced breast cancer stem cells. Oncotarget. 2014;5:3743–3755. doi: 10.18632/oncotarget.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Debeb B.G., Lacerda L., Xu W. Histone deacetylase inhibitors stimulate dedifferentiation of human breast cancer cells through WNT/beta-catenin signaling. Stem Cells. 2012;30:2366–2377. doi: 10.1002/stem.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]