Abstract
Background
As a very important coenzyme in the cell metabolism, Vitamin B12 (cobalamin, VB12) has been widely used in food and medicine fields. The complete biosynthesis of VB12 requires approximately 30 genes, but overexpression of these genes did not result in expected increase of VB12 production. High-yield VB12-producing strains are usually obtained by mutagenesis treatments, thus developing an efficient screening approach is urgently needed.
Result
By the help of engineered strains with varied capacities of VB12 production, a riboswitch library was constructed and screened, and the btuB element from Salmonella typhimurium was identified as the best regulatory device. A flow cytometry high-throughput screening system was developed based on the btuB riboswitch with high efficiency to identify positive mutants. Mutation of Sinorhizobium meliloti (S. meliloti) was optimized using the novel mutation technique of atmospheric and room temperature plasma (ARTP). Finally, the mutant S. meliloti MC5–2 was obtained and considered as a candidate for industrial applications. After 7 d’s cultivation on a rotary shaker at 30 °C, the VB12 titer of S. meliloti MC5–2 reached 156 ± 4.2 mg/L, which was 21.9% higher than that of the wild type strain S. meliloti 320 (128 ± 3.2 mg/L). The genome of S. meliloti MC5–2 was sequenced, and gene mutations were identified and analyzed.
Conclusion
To our knowledge, it is the first time that a riboswitch element was used in S. meliloti. The flow cytometry high-throughput screening system was successfully developed and a high-yield VB12 producing strain was obtained. The identified and analyzed gene mutations gave useful information for developing high-yield strains by metabolic engineering. Overall, this work provides a useful high-throughput screening method for developing high VB12-yield strains.
Electronic supplementary material
The online version of this article (10.1186/s12896-018-0441-2) contains supplementary material, which is available to authorized users.
Keywords: Vitamin B12, Cobalamin, Sinorhizobium meliloti, Riboswitch, High-throughputScreening
Background
Vitamin B12 (cobalamin, VB12) is one of the most structurally complex natural Vitamins. VB12 is only produced by some bacteria and archaea and it requires more than 30 enzymes for the de novo synthesis of VB12 [1, 2]. VB12 plays an important role as an essential co-factor in various biochemical processes such as DNA synthesis, fatty acid synthesis, amino acid metabolism and energy production [3, 4]. VB12 has been widely used in food and medicine fields [5], and the global demand for VB12 in these applications has been growing rapidly [6, 7]. A considerable number of enhancement and development of natural products mainly rely on mutagenesis and screening strategies [8], while targeted engineering strategies by gene overexpression or deletion do not always generate desired results due to the complexity of highly regulated metabolic networks.
In order to screen libraries with high genetic diversity, a key problem is developing smart screening methods. Traditional methods suffer from low dynamic range, low throughput and unwanted sensitivity to variable experimental parameters [9]. Fluorescence-activated cell sorting (FACS) allows ultrahigh throughput in cell sorting and requires a sortable signal output by biosensors in individual cells that respond to various intracellular concentrations of the target molecule with a correlation [10]. Developing a biosensor sensitive to VB12 concentration is thus very important.
Riboswitches are small structured mRNA elements that regulate gene expression and translation in response to specific small molecule ligands [11]. They act as relatively simple and direct metabolite sensors that are capable of both molecular recognition and transducing this information by altering protein expression [12–14]. Natural riboswitches are found with the highest frequency in the 5’-UTR of bacterial mRNAs, where they regulate the expression of downstream genes through structural changes undergone in response to the binding of a specific target molecule.
VB12 riboswitch has been found to widely regulate VB12 biosynthesis in bacteria [15]. Expression of cob operon in cobalamin biosynthetic pathway and transporter encoding gene btuB are repressed by the presence of vitamin B12, in particular. The VB12 riboswitch consists of a binding region (aptamer domain) and an expression platform that modulates gene expression [16, 17]. Upon ligand binding, the aptamer region undergoes a conformational change that is interpreted by the expression platform (Fig. 1), resulting in repressed gene expression [18, 19]. Comparative analysis of the genes and regulatory regions from different bacterial genomes is a powerful approach allowing the identification of regulatory patterns [20]. Using this approach, Vitreschak et al. have summarized approximately 200 VB12-elements from 67 bacterial genomes [21], providing candidates for biosens or development.
Fig. 1.
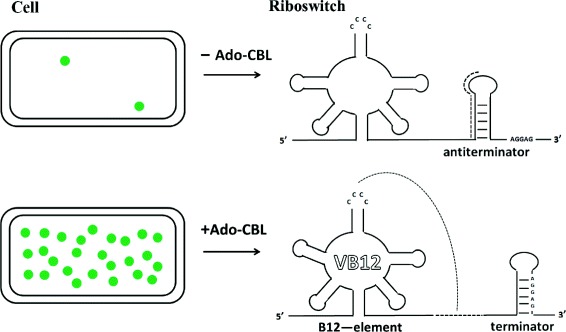
Mechanism of riboswitch based intracellular vitamin B12. In the simplified example cells containing a very low concentration or none of the VB12 (green circle), the ribosome binding site (RBS) is unstructured. Under a higher concentration of target metabolite, the aptamer domain undergoes restructuring forming a hairloop to attenuate gene expression
In this study, we screened a sensitive riboswitch to sense the amount of intracellular VB12 concentration of single cells by FACS and developed a VB12 high-throughput screening system. Using this novel VB12 screening system, high yield VB12 producing strains were screened and identified from a mutant library of S. meliloti generated by ARTP. Moreover, genomic analysis of the high yield VB12 producing strain allowed us to give insight into the mechanism of VB12 regulation pathways which would guide future study.
Methods
Strains and culture conditions
The initial strain, S. meliloti 320, was stored in our laboratory [22]. S. meliloti 320 was grown in LB medium supplemented with MgCl2 (2.5 mM) and CaCl2 (2.5 mM) (LB/MC medium), or minimal M9 medium supplemented with 1 μg/mL biotin and 10 ng/mL CoCl2, as well as 0.1% sucrose for liquid medium or 0.2% sucrose for solid medium (unless indicated otherwise), respectively, (named M9/sucrose) [23]. And the strain was grown at 30 °C with shaking at 200 rpm. Antibiotics were used as follows: spectinomycin (Sm; 200 μg/mL for E. coli and 600 μg/mL for S. meliloti), gentamicin (Gm; 10 μg/mL for E. coli, 20 μg/mL for S. meliloti), and kanamycin (Km; 50 μg/mL for E. coli and 100 μg/mL for S. meliloti).
Genetic techniques
For construction of Sm320-N8 strain, genetic deletions in S. meliloti 320 genome were generated by homologous recombination and allelic replacement. Two fragments of cobI-left and -right flanking DNA sequences (Additional file 1: Table S1) were ligated by overlap extension polymerase chain reaction (PCR), and the PCR product was cloned into a suicide vector pJQ200SK carrying sacB and gentamicin resistance gene. The construct was introduced into S. melilothi by triparental mating with help of the mobilizing strain MT616 [24] carrying pRK600, and the culture was plated onto LB/MC-agar containing gentamicin. A second reciprocal recombination event was selected on LB/MC-agar containing 5% sucrose, and then isolates with the desired deletions were subsequently screened by PCR verification in which primers hybridizing outside the cloned flanking sequences were used to amplify the deletion region.
Riboswitch sensor construction and validation
To construct riboswitch sensor plasmids, the promoter PmelA was amplified from pSymB megaplasmid, the approximately 200 bp VB12 riboswitch sequence containing a riboswitch and RBS from different strains were obtained by artificial synthesis (riboswitch sequences were provided in Additional file 1: Table S2), and the genes were amplified by PCR. Fragments containing PmelA promoter, VB12 riboswitch sequence and GFP reporter gene were fused by PCR. Next, the pBHR1 plasmid and the fused PCR fragments were ligated by Gibson assembly at 50 °C for 1 h, and then cloned in E. coli strain DH5α. Detailed constructions of the riboswitch-containing plasmids are shown in Additional file 1: Figure S2 and Additional file 1: Table S2. The plasmids were then separately transferred into Sm320, Sm320-N8 and Sm320-L6 (engineered mutants with different production capacities) by triparental mating. S. meliloti containing PmelA-riboswitch-gfp plasmid was grown at 30 °C for 12 h, and then transferred into shake flask fermentation with 10% (v/v) of inoculation for 3 days. The samples were harvested with centrifuging at 4000 g for 8 min, followed by washing in PBS for three times. The cells were collected to measure their GFP fluorescence by flow cytometry [9].
The flow cytometry (MoFlo XDP, Beckman, USA) was performed with a 488-nm excitation laser and FL1, respectively. The data were analyzed with Summit software. Eight riboswitches were selected based on the sequence reported by Vitreschak et al. This method was also used to characterize the other 7 strains. The strains and plasmids used in this study are presented in Table 1 and sequences of primers are listed in Table S1 in the Additional file 1.
Table 1.
Strains and plasmids used and constructed in this study
| Strain or plasmid | Relevant characteristics | Sources of reference |
|---|---|---|
| Strains | ||
| Sm320 | Sinorhizobium meliloti 320, VB12 producing strain. | [22] |
| Sm320-L6 | Sm320 derivative, carrying plasmid pBR-OhemE to reduce production capacity | This study |
| Sm320-N8 | Sm320 derivative, knocking out cobI gene | This study |
| Sm320-Mybi | Sm320 derivative, carrying plasmid pBR- SYB-lacI and plasmid pMB-gfp was used in the ARTP treat | This study |
| MT616 | MT607/pRK600; Escherichia coli helper strain for mobilizing RK2/RP4-derived plasmids | [24] |
| Plasmid | ||
| pJQ200sk | Vector for construction of deletions, sacB,GmR | [31] |
| pMB393 | Broad-host-range vector, PtauA promoter,AmpR | [32] |
| pBHR1 | Broad-host-range vector, Derivative of PBBR122,KmR | This lab |
| pBR- EC-butB | pBHR1 plasmid carrying EC-butB riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- PF-cbiB | pBHR1 plasmid carrying PF-cbiB riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- SM-bluB | pBHR1 plasmid carrying SM-bluB riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- SM-cobU | pBHR1 plasmid carrying SM-cobU riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- MLO-bluB | pBHR1 plasmid carrying MLO-bluB riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- MLO-metE | pBHR1 plasmid carrying MLO-metE riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- SY-btuB | pBHR1 plasmid carrying SY-btuB riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- BI-cbiW | pBHR1 plasmid carrying BI-cbiW riboswitch were cloned in front of a gfp reporter gene driven by the constitutive promoter PmelA | This study |
| pBR- SYB-lacI | pBHR1 plasmid carrying SY-btuB riboswitch were cloned in front of a lacI reporter | This study |
| pMB-gfp | pMB393 containing lacO and carrying gfp reporter | This study |
| pBR-OhemE | pBHR1 plasmid carrying hemE gene to reduce production capacity | This study |
Mutagenesis by ARTP and determination of optimal treatment time
Pure helium was used as the plasma working gas for ARTP mutation and the operating parameters were as follows: (1) the radio frequency power input was 100 W; (2) the distance between the plasma torch nozzle exit and the sample plate was 4 mm; and (3) the temperature of the plasma jet was below 30 °C [25]. Under these operating conditions, the ARTP mutagenesis dosage was only dependent on the treatment time.
To determine the optimal treatment time, S. meliloti was cultured for 72 h (mid-log phase) and then subjected to ARTP treatment. Briefly, 10 μL culture (1 × 108 cells/mL) was treated with the helium-based ARTP for 15, 30, 60, 75, 90 and 120 s. Lethality rate was calculated for determining the appropriate treatment time. The culture without treatment was used as a control. After each treatment, the steel plate was eluted with sterile distilled water into new tubes and properly diluted, incubated on fermentation medium at 30 °C for 1 h, and then cultured on solid medium with 600 μg/mL spectinomycin at 30 °C for 4 days. The lethality rate was determined as follows:
Whole genome sequencing
The genomic DNA of strain S. meliloti 320 was extracted from bacterial cells grown overnight in LB/MC medium at 30 °C and 200 rpm. The DNA was extracted using OMEGA Bacterial DNA kits (Omega bio-tek, Feyou Biotechnology, China) following the manufacturer’s protocol [26, 27]. To verify DNA quality, PCR amplifications were performed in a final volume of 50 μL, containing 10 μM each primer, 1.25 units of Q5 High-Fidelity DNA polymerase (New England Biolabs, USA), dNTP mixture (250 μM each dNTP), and approximately 50 ng of template DNA. The PCR amplification conditions consisted of an initial denaturation for 10 min at 94 °C, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 1 min with a final extension of 10 min at 72 °C. The quality and quantity of DNA were analyzed using 1% agarose gel electrophoresis and spectrophotometric method, respectively. The verified genomic DNA was selected for further analysis.
Analytical methods
Biomass was determined with a spectrophotometer at 600 nm. The sample for measuring cell concentrations was obtained as follows: 1 mL of broth was centrifuged at 10,000×g for 10 min, the supernatant was discarded, and the cell pellets were resuspended in 1 mL of 0.85% NaCl buffer. The concentrations of the principal fermentation products were determined by high performance liquid chromatography (HPLC) [28]. The sample (25 mL) was mixed with 2.5 mL of NaNO2 8% (w/v) and 2.5 mL of glacial acetic acid. The mixtures were boiled for 30 min, and then centrifuged. The supernatants were filtrated through a 0.22-μm membrane filter. Then, 20 μl of NaCN 10% (w/v) was added to 1 mL of the aqueous phase. 20 μL of the final sample was injected into an Agilent 1260 HPLC with a 5C18-250A column (Agilent, 4.6 mm id × 250 mm, 5 μm) thermostated at 35 °C. The mobile phase consists of water and acetonitrile (70:30 [v/v]) at 0.8 mL/min [22]. Samples were detected at 361 nm. All values presented in this study were averages of at least three independent trials.
Preparation of cell extracts and detection of the amount of intracellular VB12
Centrifugation was used to sediment 500 μL of cell suspension through 100 μL silicone oil on the bottom of the tube. Cell pellets were then resuspended by 200 μL perchloric acid (20%, v/v) by gentle vortexing after disruption by sonication. After breaking up the cells, extracts were neutralized by addition of 200 μL Na2CO3 and the membrane materials and denatured proteins were removed by centrifugation. Then, 100 μL of NaNO2 8% (w/v) and 100 μL of glacial acetic acid were added to 1 mL of aqueous phase, and the mixture was boiled for 30 min, centrifuged and the supernatant was filtrated through a filter membrane (0.22 μm), and then 2 μl of NaCN 10% (w/v) was added to 100 μl of the aqueous phase. The samples were stored in − 20 °C for VB12 analysis by HPLC. The amount of intracellular VB12 was calculated as the mill grams of intracellular VB12 per liter of culture.
Fermentations
The fermentation medium contained (per liter of deionized water): sucrose 60 g/L; corn steep liquor (CSL) 20 g/L; betaine 6 g/L; ammonium sulfate 2 g/L; potassium dihydrogen phosphate 2.6 g/L; cobalt chloride 0.06 g/L; and 0.1 g/L DMB was added to the medium. The pH before autoclaving was adjusted to 7.0–7.4. The stock sugar solution was autoclaved separately before mixing with the rest of the medium.
For cultivating VB12-producing strains in microtiter plates, colonies were cultivated in 100 μL LB/MC medium overnight using 96-well microtiter plates (Corning Costar 3930, square U-bottom, 350 μL) and then 10% (v/v) inocula was transferred to 2 mL fermentation medium using 24-well deep microtiter plates (Corning Costar 3524, square U-bottom, 5 mL) at 30 °C, 700 rpm, 80% humidity in a Microtron shaker (Infors, China).
For shake flask fermentation, the seed culture was grown at 30 °C for 12 h and was used to provide 10% (v/v) inocula for fermentation. Experiments were conducted in 250 mL Erlenmeyer flasks containing 30 mL of each medium at 30 °C at an agitation rate of 200 rpm in rotary shakers for 7 days. The samples were used to determine cell mass (OD600 nm), residual sugars every 24 h, and product at 7 days.
Results
Engineered mutants with different production capacities of VB12
To evaluate the dose-response of riboswitches on transcriptional regulation under variable the amount of intracellular VB12 levels, we engineered mutants of the original S. meliloti production strain Sm320 with different production capacities. The complete VB12 biosynthetic pathway contains approximately 30 genes (Fig. 2a) and deleting any of them will prevent VB12 biosynthesis. Strain Sm320-N8 with an in-frame deletion of essential VB12 biosynthetic gene cobI was constructed as a negative control strain that does not produce VB12. To construct mutant with low production capacity, we overexpressed gene hemE encoding the key enzyme in the heme branch pathway to reduce VB12 biosynthesis. This strain was named Sm320-L6. The VB12 concentration of the original strain Sm320 and the two derivatives, Sm320-N8 and Sm320-L6, were measured by HPLC. The total amounts of VB12 produced by strains Sm320-N8, Sm320-L6 and Sm320 were 0, 65.21 and 126.28 mg/L with 7d’s cultivation, respectively, the amounts of intracellular VB12 of strains Sm320-N8, Sm320-L6 and Sm320 were 0, 9.65 and 18.5 mg/L (Fig. 2b, c). The details of cell growth are provided in Additional file 1: Figure S1. It was important to note that the amounts of intracellular VB12 was used to indicate the intracellular VB12 level, and but was not the real intracellular VB12 concentration.
Fig. 2.
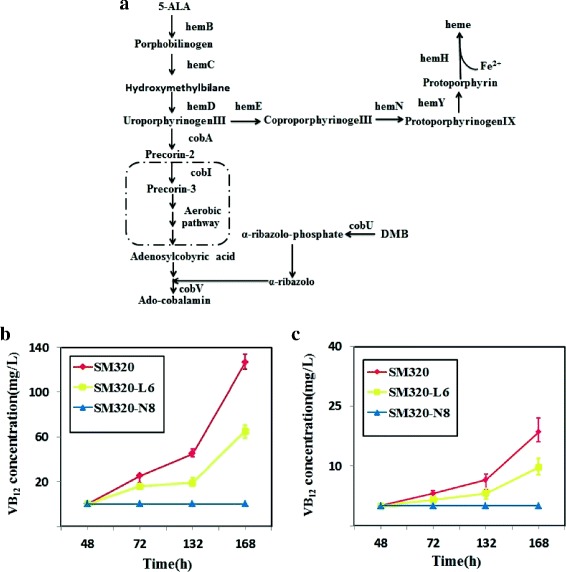
Constructed engineered mutants with different production capacities. a Oxygen-dependent VB12 biosynthetic pathway in S. meliloti. The branched biosynthesis of heme is also depicted. b The concentration of VB12 in producer strains in fermentation broth was determined by HPLC. Different colors represent the different strains. Values shown are HPLC-determined and are the means of three independent fermentation cultures for each strain. c the amount of intracellular VB12 in recombinant strains in (b)
Construction of riboswitch-based VB12 sensors
To identify a sensitive element that allows efficient screening of positive mutants with high VB12 production, eight different potential riboswitches (Table 2) were selected according to the criteria described above, plasmids containing VB12-dependent riboswitches coupled to a GFP reporter were separately transformed into Sm320, Sm320-L6 and Sm320-N8, then the expression levels of GFP under the control of the eight riboswitches were compared. The strains containing the plasmids were grown at 30 °C for 12 h and were used to provide shake flask fermentation for 3 days with 10% (v/v) inoculant and then tested for GFP expression by flow cytometry (Fig. 3).
Table 2.
Conserved RNA elements upstream of some B12-regulated genes
| Genome abbreviations | The start codon | Proximal downstream genes | Coding of enzyme protein |
|---|---|---|---|
| CBL biosynthesis | |||
| Propionibacterim freudenreichii (PF) | 285 | cbiB | VB12 biosynthesis protein |
| Bacillus megaterium(BI) | 266 | cbiW | VB12 biosynthesis protein |
| Mesorhizobium loti(MLO) | 233 | bluB | 5,6-dimethylbenzimidazole synthase |
| Sinorhizobium meliloti(SM) | 251 | bluB | 5,6-dimethylbenzimidazole synthase |
| Sinorhizobium meliloti(SM) | 227 | cobU | adenosylcobinamide kinase |
| Vitamin B12 transport | |||
| Escherichia coli(EC) | 248 | btuB | VB12 transporter BtuB |
| Salmonella typhimurium(SY) | 252 | btuB | VB12 transporter BtuB |
| B12-dependent or alternative metabolic pathways | |||
| Mesorhizobium loti(MLO) | 309 | metE | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase |
Fig. 3.
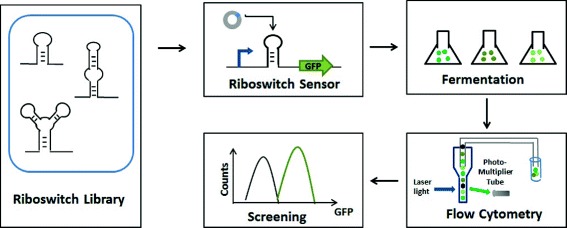
Flow diagram of constructed VB12 riboswitches. The ideal phenotypes were selected from a VB12 riboswitch library. VB12 sensor constructs were made that contained a predicted riboswitch and a GFP reporter. The fermentation broth was collected and analyzed by Fluorescence Activated Cell Sorting
Figure 4 shows the GFP fluorescence of different S. meliloti strains carrying different riboswitches. The strains containing riboswitches shown in Fig. 4-b, c, d, e and h exhibited a similar fluorescence intensity, although they could produce different levels of VB12. Fig. 4-g shows that the fluorescence intensity of different S. meliloti strains containing SY-btuB-gfp plasmid were sensitive to the increasing amount of intracellular VB12, while the other strains in Fig. 4-a, f, g exhibited a negative correlation between their fluorescence intensity and the amounts of intracellular VB12. The results indicated that some of these predicted riboswitches indeed responded to the concentrations of VB12 in S. meliloti. Thus, we further examined the feasibility of using the SY-btuB riboswitch as a sensitive sensor for VB12 concentration.
Fig. 4.
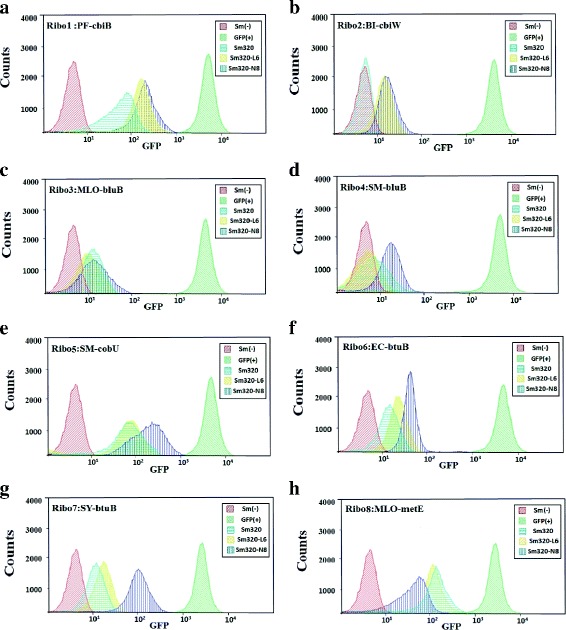
Validation and selection of a sensitive element that allows efficient identification of positive mutants. Fluorescence-activated cell sorting results of engineered VB12 producing strains with different production capacity (Sm320-N8, Sm320-L6, and Sm320) were tested separately with eight riboswitches. Fig. a-h exhibited S. meliloti strains carrying different riboswitches, and the genome abbreviations of eight different potential riboswitches were marked in the top left corner of the figures
Examination of the SY-btuB riboswitch sensor
We attempted to demonstrate the feasibility of the sensor for screening hyperproducing strains from a library. As SY-btuB VB12 riboswitch is a repressive type of selector, the single gfp reporter gene was substituted with a “dual-repression” mode that contained a lacI repressor protein and a gfp reporter gene (Fig. 5a). We tested this reconstructed sensor in strains Sm320, Sm320-L6 and Sm320-N8. A positive correlation between the GFP fluorescence and the VB12 productivity of the strains was showed by flow cytometry (Fig. 5c).
Fig. 5.
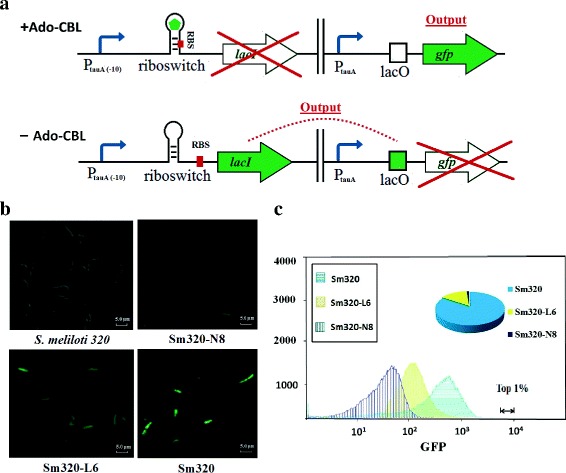
Validation of feasibility of riboswitch SY-btuB in different performances. a Mechanism of VB12 dependent riboswitch SY-btuB double plasmid work mode. VB12 directly binds to the riboswitch region, producing a conformational change in the secondary structure of mRNA, and thus inhibiting lacI gene expression and causing the gfp reporter to show a fluorescent signal. For cells containing a very low concentration of VB12, the ribosome binding site (RBS) is unstructured. This allows for efficient translation of lacI gene and then lacI and can bind lacO and repress gfp expression. b The microscopic image of S. meliloti 320 and fluorescence microscopic images of Sm320, Sm320-L6 and Sm320-N8 cells carrying SY-btuB sensor plasmid, respectively. c FACS (Fluorescence Activated Cell Sorting) results of three strains mixed with equal optical density. Strains exhibiting top 1% of the fluorescence values were selected by FCM. The screened strains were then distinguished the selected colonies by Summit 5.2 analysis
In order to verify the response behavior of the new sensor system to the amount of intracellular VB12 level, GFP fluorescence was observed by fluorescence microscope (DM5000B, Leica, Germany). A very weak fluorescence value was detected for the samples without VB12, and there was a corresponding increase in GFP fluorescence value along with the increase amount of intracellular VB12 (Fig. 5b). This result indicated that this sensor could work in different strains to sense the different levels of the amount of intracellular VB12 by producing corresponding levels of fluorescence intensity. Moreover, a corresponding rise in GFP fluorescence values of Sm320-N8 strains carrying SY-btuB sensor double plasmids could be observed by increasing VB12 concentration in the medium (Additional file 1: Figure S3).
Next, we simulated a screening process to verify the reliability of the sensor-reporter. The three strains with different production capacities were mixed together at equal optical density and the mixture of strains was used for screening with FCAS. Colonies exhibiting the top 1% of fluorescence values were analyzed to evaluate the sorting performance of the SY-btuB sensor double plasmid (Summit 5.2 analysis). As expected, nearly 84% of sorted cells were identified as the high-producing Sm320 strain (Fig. 5c). Overall, Sm320-Mybi represented the best regulatory device, and the strain Sm320-Mybi was used in the subsequent experiments.
High throughput screening of mutants using riboswitch
ARTP-irradiation was used to induce random mutations, and high VB12-producing mutants were selected. Growing cells were treated with helium-based ARTP for 15, 30, 60, 75, 90 and 120 s, respectively. The lethality rate was over 80% when the exposure time exceeded 60 s, and there was no cell survival after 120 s treatment (Fig. S4). Therefore, the optimal exposure time used in subsequent studies was set at 60 s with lethality ranged from 80 to 85%.
We then attempted to screen hyperproducing strains from a library of variant strains using the engineered riboswitch. After treatment of ARTP, selection was carried out using the “dual-repression” SY-btuB riboswitch. Colonies exhibiting the top 0.1% of fluorescence signals were selected from the genetic library. The wild type strain and the mutants were cultured in LB/MC medium in a 96-well microtiter plate at 30 °C for 12 h and then transferred to fresh medium for fermentation in 24-well deep plate at 30 °C with shaking for 7 days.
After 3 rounds of mutation and screening, a total of 144 colonies were selected from the genetic library containing roughly 6 × 107 mutants. After fermentation in 24-well plates, 58.3% (84/144) of the mutants produced higher VB12 levels than the initial strains (Fig. 6). Six mutants (MD1–6, MB3–5, MD3–2 MC4–5, MB5–1, and MC5–2) showed a significant enhancement of 10% in their VB12 content compared with the initial strains. These mutants were then cultured in 250 mL flasks at 30 °C with shaking for 7 days. The mutant MC5–2 showed VB12 production of 156 ± 4.2 mg/L, which was 21.82% higher than the wild-type strain. After 7-generation cultivation, mutant MC5–2 still maintained high VB12 yield reaching to 154 ± 3.2 mg/L. The VB12 titer of MD3–2 was the second highest among the mutants with an increase of 20.31% than that of the wild-type strain.
Fig. 6.
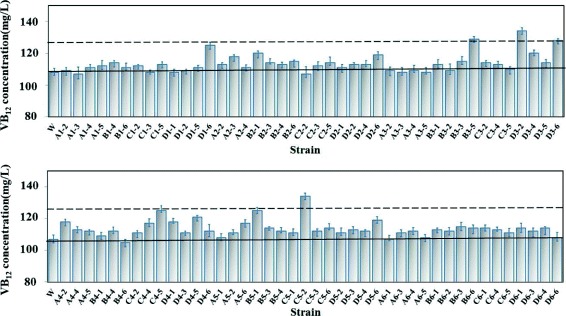
VB12 concentration of Sm320 mutants obtained with ARTP treatments. “W” indicates the wild-type strain of S. meliloti with a VB12 content of 108 mg/L in a 24-well deep plate at 30 °C for 7 d. The solid line indicates the original strain, and the dotted line indicates the position where the VB12 content increased 10% compared with the original strain. Error bars show the means of three independent fermentation cultures for each strain by HPLC analysis
Whole genome sequencing analyses
To identify the gene mutations contributing to the high VB12 production, we sequenced the genomic DNA of the mutant Sm320MC5–2. The gene mutations were identified from the sequence data. A total of 17 SNP (single nucleotide polymorphism) and 1 Indel (insertion and deletion) were identified in the Sm320MC5–2 genome (Additional file 1: Table S3). Genome-scale gene expression analysis of Sm320 and Sm320MC5–2 revealed that the mutant genes were located in the VB12 metabolic pathway, heme branch pathway, ABC transporters, VB12 precursor synthesis pathway, tricarboxylic acid cycle and other metabolic pathways. These SNPs functioned through altering the transcription level or enzyme activities and may have resulted in enhanced metabolic flux towards production of VB12 or by weakening the competing branches. This study demonstrates a new paradigm of revealing unexpected gene mutations to guide metabolic engineering for increasing VB12 yield.
Discussion
In the past few years, various mRNA elements containing engineered riboswitches and reporter genes have been described and applied in modulating genetic switches, discovering the genetic pathway and optimizing cellular behaviors. However, none of simple and direct metabolite sensors has been applied in screening to increase the yield and productivity of target molecules. In this study, we reported successful application of a riboswitch sensor as a sensitive and novel detection method for VB12 screening. Moreover, we identified a VB12 riboswitch element that is responsible for expression of the VB12 transporter, btuB, in the genome of Salmonella typhimurium [29]. We validated the applicability of this riboswitch by screening higher VB12 producers from a genetic DNA library.
It is necessary to emphasize that the titer of VB12 was heavily dependent on the culture conditions. In our previous study, we did some research to select the nitrogen source, considering the sediment of CSL may interfere the accuracy of the experiments, we replaced the nitrogen sources “2% CSL” by “2% AYP, 1% peptone” in the fermentation medium when the strains were screened by FACS, and the comparison was shown in Additional file 1: Figure S5. When the strains were cultivated on shake flasks, VB12 was produced at millimolar concentrations. But when the strains were screened by FACS, the amount of intracellular VB12 was only about 1.06 μg/mL (micromolar levels of B12). So, the levels of VB12 produced could be detected by riboswitch normally.
In this study, 156 ± 4.2 mg/L of VB12 was produced by the selected mutant MC5–2. In order to identify gene mutations that contributed to the high production of VB12, we sequenced the genomic DNA of the mutant Sm320MC5–2 Many gene mutations in Sm320MC5–2 genome may play key roles in the VB12 hyperproduction. The SNPs in hemin, hemN, sdhA, betB, ribH, and metH were not reported in S. meliloti. These SNPs may alter transcription levels and enzyme activities to enhance metabolic flux towards producing VB12 or weakening the competing branches to improve productivity.
There were two changes identified in the heme branch pathway, mutation of hemin (c.C1228T: p.H410Y) and hemN (c.C632T: p.T211I). The mutation of hemN is speculated to decrease urinary porphyrins original III metabolic flow to the synthesis of hemoglobin leading to increasing productivity of VB12. The gene hemin encodes an iron complex transport system substrate-binding protein, and its mutation may decrease the active efflux of hemoglobin, reducing the heme anabolic flow to maintain the cell intracellular requirement.
In addition, we speculate that the mutation of sdhA (c.G1846A:p.A616T) in the tricarboxylic acid cycle conversion of succinate as carbon sources is necessary but insufficient for the VB12 metabolic pathways. The mutant of betB (c.T301C:p.W101R) gene may enhance the utilization of betaine to improve VB12 productivity. The ribH (c.T95C:p.L32P) encodes riboflavin kinase in riboflavin metabolism and may play a key role in 5, 6-dimethylbenzimidazo-le (DMB) synthesis, a precursor of VB12 biosynthesis.
Finally, one cobalamin-dependent enzyme in S. meliloti, methionine synthase (MetH c.G733A:p.G245S), catalyzes the final step in methionine biosynthesis and is required for growth [30]. In future work, the contribution of these enzymes to VB12 production needs to be determined, which may help elucidate the underlying mechanism and analyze metabolic circuits for further improvement of VB12 production in S. meliloti.
Conclusions
We developed a Vitamin B12 high-throughput screening system by riboswitch sensor for the selection of VB12hyperproducing strains. The S. meliloti mutant library was generated by ARTP. Numerous mutants were obtained, and Sm320MC5–2 exhibited 21.82% higher VB12 yield than the wild type strain. The genomic DNA of the hyperproducing mutant was subjected to whole genome sequencing, and the potential contributions of the gene mutations were discussed. This study provides a new paradigm in high throughput screening and elucidating the unexpected gene mutations which would be helpful for rewiring metabolic pathway to raise the yield of VB12.
Additional file
Figure S1. Growth conditions of Sm320-N8, Sm320-L6 and Sm320 producing strains. Figure S2. Cloning scheme for riboswitch sensor constructs showing the relevant plasmid features. Figure S3. Characterization of our biosensor plasmids in response to addition of increasing VB12 into the medium. Figure S4. Lethal rate of S. meliloti irradiated by atmospheric and room temperature plasma (ARTP). Figure S5. Concentration of VB12 in producer strains during different growth conditions. Table S1. Oligonucleotide primers used in in this study. Table S2. Sequences of eight cobalamin riboswitches. Table S3. Analysis of mutations of mutant Sm320MC5–2 with original strain. Supplementary experimental procedures: VB12 feeding assay. (DOC 322 kb)
Funding
Tianjin Science Fund for Distinguished Young Scholars (17JCJQJC45300), Nature Science Foundation of Tianjin (CN) (16JCYBJC23500, 15JCQNJC09500), Tianjin Science and Technology project (15PTCYSY00020), the Key Projects in the.
Tianjin Science & Technology Pillar Program (14ZCZDSY00058), and the Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences. All the findings mentioned above have no roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Authors’ contributions
YYC and DZ conceived of the manuscript. YYC, MMX, HD and YQ performed the experiment. YYC, MMX, HD, TCZ, BWZ, JCW and DZ wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12896-018-0441-2) contains supplementary material, which is available to authorized users.
Contributor Information
Yingying Cai, Email: yy_cai@163.com.
Miaomiao Xia, Email: xia_mm@tib.cas.cn.
Huina Dong, Email: dong_hn@tib.cas.cn.
Yuan Qian, Email: qian_yuan@tib.cas.cn.
Tongcun Zhang, Email: tony@tust.edu.cn.
Beiwei Zhu, Email: zhubeiwei@163.com.
Jinchuan Wu, Email: wu_jinchuan@ices.a-star.edu.eg.
Dawei Zhang, Phone: +86-22-24828749, Email: zhang_dw@tib.cas.cn.
References
- 1.Cheng Z, Li K, Hammad LA, et al. Vitamin B12 regulates photosystem gene expression via the CrtJ antirepressor AerR in Rhodobacter capsulatus. Mol Microbiol. 2014;91(4):649–664. doi: 10.1111/mmi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escalante-Semerena JC. Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J Bacteriol. 2007;189:4555–4560. doi: 10.1128/JB.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eschenmoser A. Vitamin B12: experiments concerning the origin of its molecular structure. Angew Chem Int Ed. 1988;27(1):5–39. doi: 10.1002/anie.198800051. [DOI] [Google Scholar]
- 4.Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72(1):209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, Zhang Z, et al. Improved propionic acid and 5, 6-dimethylbenzimidazole control strategy for vitamin B12 fermentation by Propionibacterium freudenreichii. J Biotechnol. 2015;193:123–129. doi: 10.1016/j.jbiotec.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Survase SA, Singhal RS. Biotechnological production of vitamins. Food Technol Biotechnol. 2006;44:381–396. [Google Scholar]
- 7.Cheng X, Chen W, Peng WF. Improved vitamin B12 fermentation process by adding rotenone to regulate the metabolism of Pseudomonas denitrificans. Appl Biochem Biotechnol. 2014;173:673–681. doi: 10.1007/s12010-014-0878-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Zhang C, Zhang QQ, Xing XH. Quantitative evaluation of DNA damage and mutation rate by atmospheric and room-temperature plasma (ARTP) and conventional mutagenesis. Appl Microbiol Biotechnol. 2015;99:5639–5646. doi: 10.1007/s00253-015-6678-y. [DOI] [PubMed] [Google Scholar]
- 9.Jha RK, Kern TL, Fox DT, M Strauss CE. Engineering an Acinetobacter regulon for biosensing and high-throughput enzyme screening in E. coli via flow cytometry. Nucleic Acids Res. 2014;42:8150–8160. doi: 10.1093/nar/gku444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang G, Withers SG. Ultrahigh-throughput FACS-based screening for directed enzyme evolution. Chembiochem. 2009;10:2704–2015. doi: 10.1002/cbic.200900384. [DOI] [PubMed] [Google Scholar]
- 11.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler CC, Brown ED, Li Y. Using a riboswitch sensor to examine coenzyme B12 metabolism and transport in E. coli. Chem Biol. 2010;17:756–765. doi: 10.1016/j.chembiol.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 13.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 15.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4:441–1. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Geneti. 2004;20:44–50. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Watson PY, Fedor MJ. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol. 2011;18:359–363. doi: 10.1038/nsmb.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelfand MS, Novichkov PS, Novichkova ES. Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform. 2000;1:357–371. doi: 10.1093/bib/1.4.357. [DOI] [PubMed] [Google Scholar]
- 21.Vitreschak AG. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong H, Li S, Fang H, et al. A newly isolated and identified vitamin B12 producing strain: Sinorhizobium meliloti 320. Bioprocess Biosyst Eng. 2016;39:1527–1537. doi: 10.1007/s00449-016-1628-3. [DOI] [PubMed] [Google Scholar]
- 23.MacLean AM, MacPherson G, Aneja P, et al. Characterization of the β-ketoadipate pathway in Sinorhizobium meliloti. Appl Environ Microbiol. 2006;72(8):5403–5413. doi: 10.1128/AEM.00580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bringhurst RM, Gage DJ. Control of inducer accumulation plays a key role in succinate-mediated catabolite repression in Sinorhizobium meliloti. J Bacteriol. 2002;184:5385–5392. doi: 10.1128/JB.184.19.5385-5392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Liu R, Li J, Chang M. Enhanced arachidonic acid production from Mortierella alpina combining atmospheric and room temperature plasma (ARTP) and diethyl sulfate treatments. Bioresour Technol. 2015;177:134–140. doi: 10.1016/j.biortech.2014.11.051. [DOI] [PubMed] [Google Scholar]
- 26.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe KH. Comparative genomics and genome evolution in yeasts. Philos Trans R Soc Lond B Bio sci. 2006;361:403–412. doi: 10.1098/rstb.2005.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia W, Chen W, Peng W, Li K. Industrial vitamin B12 production by Pseudomonas denitrificans using maltose syrup and corn steep liquor as the cost-effective fermentation substrates. Bioprocess Biosyst Eng. 2015;38:1065–1073. doi: 10.1007/s00449-014-1348-5. [DOI] [PubMed] [Google Scholar]
- 29.Campos-Gimenez E, Fontannaz P, Trisconi M. Determination of vitamin B12 in infant formula and adult nutritionals by liquid chromatography/UV detection with immunoaffinity extraction: first action 2011.08. J AOAC Int. 2012;95:307–312. doi: 10.5740/jaoacint.CS2011_08. [DOI] [PubMed] [Google Scholar]
- 30.Taga ME, Walker GC. Sinorhizobium meliloti requires a cobalamin-dependent ribonucleotide reductase for symbiosis with its plant host. Mol Plant-Microbe Interact. 2010;23:1643–1654. doi: 10.1094/MPMI-07-10-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 32.Kovach ME, Elzer PH, Hill DS, Robertson GT. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Growth conditions of Sm320-N8, Sm320-L6 and Sm320 producing strains. Figure S2. Cloning scheme for riboswitch sensor constructs showing the relevant plasmid features. Figure S3. Characterization of our biosensor plasmids in response to addition of increasing VB12 into the medium. Figure S4. Lethal rate of S. meliloti irradiated by atmospheric and room temperature plasma (ARTP). Figure S5. Concentration of VB12 in producer strains during different growth conditions. Table S1. Oligonucleotide primers used in in this study. Table S2. Sequences of eight cobalamin riboswitches. Table S3. Analysis of mutations of mutant Sm320MC5–2 with original strain. Supplementary experimental procedures: VB12 feeding assay. (DOC 322 kb)


