Ocean warming and continental temperature and humidity
Ocean warming linked to continental temperature and humidity.
Changes in temperature and humidity over oceans and land are expected with changes in climate. Current climate models predict larger future increases in surface temperature and declines in relative humidity over land compared with oceans. Michael Byrne and Paul O’Gorman (pp. 4863–4868) report a direct link between ocean warming and trends in continental humidity and temperature. The authors used continental and oceanic surface-air temperature and humidity data from 1979 to 2016 to examine trends in oceanic and continental climates. Analysis of the data indicated that land temperatures increased faster than ocean temperatures, but specific humidity increased faster over oceans than over land. Moreover, the authors observed a strong decrease in relative humidity over land, consistent with climate projections. Based on a simple quantitative model incorporating atmospheric dynamics and moisture transport, the authors propose a link between ocean warming and land temperature and humidity changes. The model indicated that the amplified land temperature increases were likely due to the increased dryness of continental air relative to ocean air, in turn resulting in a decline in relative humidity over land. The findings might help interpret past and future climate change, according to the authors. — C.S.
Fossil record quality and early hominin diversity
Researchers have suggested that climate change is the primary driver of fluctuations in diversity in the hominin fossil record. However, the influence of uneven sampling on diversity patterns remains unclear. Simon Maxwell et al. (pp. 4891–4896) applied sampling-corrected methods to the hominin fossil record to examine fluctuations in species diversity from 7 million years ago to 1 million years ago. The authors compared taxic diversity, which is a measure of the number of fossil occurrences, and phylogenetic diversity, which is a measure of the number of occurrences and inferred gaps in sampling, with sampling metrics related to collection effort and the amount of exposed rock available to sample. The results revealed correlations between taxic diversity, rock exposure, and collection effort, indicating that nonuniform fossil sampling, rather than climate dynamics, might explain the pattern of early hominin taxic diversity in the fossil record. Moreover, the authors suggest that peak taxic diversity at 1.9 million years ago reflects maximal rock exposure and collection effort, rather than a major evolutionary transition. In contrast, phylogenetic diversity estimates peak at 2.4 million years ago and were unrelated to sampling metrics. According to the authors, the findings demonstrate that additional research is required to reliably determine the role of climate in hominin evolution. — C.S.
Pest moth hybridization in Brazil

H. armigera larva. Image courtesy of Cecilia Czepak (Universidade Federal de Goiás, Goiânia, Brazil).
The Old World crop pest moth Helicoverpa armigera has recently established itself in Brazil, raising concerns that the pesticide-resistant moth may spread its pestilent traits through hybridization with the native Helicoverpa zea moth. Craig Anderson et al. (pp. 5034–5039) collected specimens of Helicoverpa moths from six species and 16 countries between 2004 and 2014, including 14 individuals from Brazil, to assess the potential for hybridization. Genomic analysis of the individuals found that H. armigera contained the most genetic diversity among Helicoverpa moths, possibly contributing to its effective adaptive and invasive traits. Of the 14 individuals collected from Brazil, eight originally identified as H. armigera were found to be hybrids with H. zea, and one individual originally identified as H. zea was also shown to be a hybrid. The DNA derived from H. armigera in the H. zea hybrid included detoxification genes and gustatory receptor genes that have been associated with H. armigera’s expanded range, as well as a gene associated with insecticide resistance. Although it is still unclear whether the hybrids are being naturally selected, the results show that genetic exchange is already underway between H. armigera and H. zea, with the hybrids likely taking on the characteristics that make H. armigera a major crop pest, according to the authors. — P.G.
Characterization of a plastic-degrading enzyme
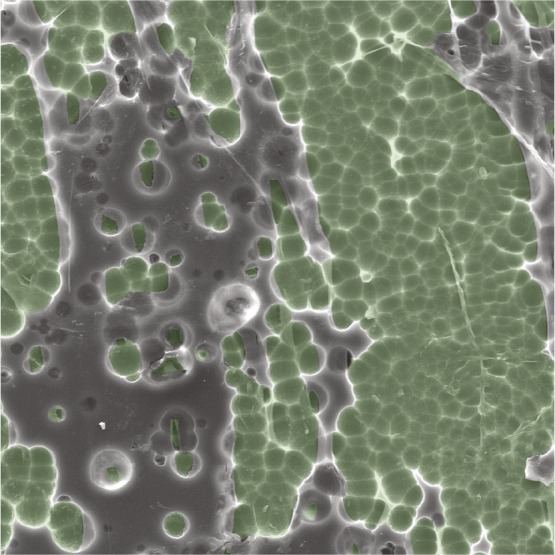
Scanning electron micrograph of a PETase-digested PET surface.
Poly(ethylene terephthalate) (PET) is an abundant polyester plastic used in everyday products. PET is a versatile raw material but is highly resistant to natural degradation. Recently, researchers reported that the bacterium Ideonella sakaiensis 201-F6 can use PET as a carbon and energy source for growth. Harry Austin et al. (pp. E4350–E4357) characterized the 3D structure of the bacterium’s PET-digesting enzyme, PETase, using X-ray crystallography. In particular, the authors identified structural features common to cutinases, lipases, and PETase. Based on the enzyme structure, the authors engineered PETase variants for PET degradation assays. An engineered double-mutant variant exhibited improved PET degradation, compared with the wild-type PETase, suggesting that PETase is not completely optimized for degrading PET despite having likely evolved in a PET-rich environment. Furthermore, the authors found that PETase degrades polyethylene-2,5-furandicarboxylate, or PEF, an emerging semiaromatic replacement for PET. Molecular dynamics simulations indicated that PETase binding is enabled by highly flexible, large aromatic side chains that induce conformational changes in PET and PEF residues. The findings might enable protein engineering efforts aimed at the biodegradation of synthetic polyesters, according to the authors. — C.S.
Explaining the deformability of red blood cells
Red blood cells (RBCs) in mammals have a characteristic biconcave disk shape and are deformable, and both attributes rely on a viscoelastic network of membrane-associated F-actin proteins interconnected with spectrin proteins, which together constitute the membrane skeleton. Alyson Smith et al. (pp. E4377–E4385) investigated the role of nonmuscle myosin II (NMII), a force-generating motor protein identified in RBCs, in controlling RBC shape. The authors found that NMIIA is the predominant isoform of NMII in RBCs and could form bipolar filaments associated with F-actin in the membrane skeleton. In vitro biochemical assays showed that the association of NMIIA with the membrane skeleton is mediated by the binding of NMIIA motor domains to F-actin. In vivo phosphorylation of the NMIIA heavy and light chains revealed that NMIIA motor activity and filament assembly are actively regulated. Inhibiting NMII motor activity in RBCs using the compound blebbistatin decreased the number of NMIIA filaments associated with membrane skeleton F-actin, caused RBCs to exhibit elongated shapes and lose membrane curvature, and enhanced RBC deformability. The latter finding suggests that NMIIA motor activity helps control the membrane tension, biconcave disk shape, and deformability of RBCs. The study identifies a mechanism by which NMIIA contractility regulates RBC shape and deformability, and the authors suggest that the same mechanism may apply to membrane skeleton networks in other cell types. — S.R.
Genetic variation tied to consonant use across populations

Consonant use is tied to DCDC2 regulation across populations. Image courtesy of iStock.com/mathisworks.
Variations in READ1, a genetic sequence that regulates expression of the gene DCDC2, are tied to dyslexia and thought to influence normal speech variation, reading performance, and auditory processing of basic word sounds. Mellissa DeMille et al. (pp. 4951–4956) compared READ1 sequences in nonhuman primates, Neanderthals, Denisovans, and modern humans and found that the regulatory sequence acquired changes specific to modern humans between 550,000 and 4 million years ago. Comparison of READ1 variants across 43 modern human populations from five continents with the numbers of consonants and vowels prevalent in those populations revealed that the number of consonants—but not vowels—was positively correlated with the frequency of RU1-1, a READ1 variant; the human brain uses distinct strategies to process and encode vowels and consonants. The correlation was unaffected by the populations’ geographic proximity, genetic relatedness, or linguistic relatedness. Hence, the authors suggest, differences in READ1 sequence, and resultant differences in DCDC2 expression, may account for differences in word-sound processing across populations. Specifically, the low prevalence of RU1-1 in some populations may be associated with diminished ability to distinguish between consonants, resulting in progressive shrinkage of the populations’ consonant inventory. According to the authors, subtle but discernible genetic variations, magnified by cultural processes, might account for differences in consonant use across populations. — P.N.
Genetic adaptation to high latitudes during the Last Glacial Maximum

Mesic shrub tundra in northwestern Alaska, similar to the environment humans occupied in Beringia during the Last Glacial Maximum.
Evidence suggests that the ancestors of the first Native Americans lived in genetic isolation on the Arctic Beringian landmass during the Last Glacial Maximum, approximately 20,000–30,000 years ago. Little UV radiation, which is necessary for vitamin D biosynthesis, reaches the Earth’s surface at such high latitudes, thereby increasing the risk of vitamin D deficiency. Leslea Hlusko et al. (pp. E4426–E4432) investigated the possibility of genetic adaptation to low UV exposure in this population. The ectodysplasin A receptor (EDAR) gene influences the development of ectodermally derived structures, including teeth and mammary and sweat glands. A variant of this gene, EDAR V370A, has a highly elevated frequency in North and East Asian populations and is associated with increased sweat gland density, branching of mammary gland ducts, and incisor shoveling, among other effects. The authors found a high degree of incisor shoveling among archaeological Native American populations, suggesting that the variant was also frequent among Native Americans prior to European colonization, and therefore among their ancestral Arctic population. According to the authors, the high-latitude, low-UV environment of Beringia may have selected for this variant because of its effect on mammary ductal branching, which may amplify the transfer of vitamin D and other nutrients to offspring via breast milk. — B.D.



