Significance
Languages evolve rapidly due to an interaction between sociocultural factors and underlying phonological processes that are influenced by genetic factors. DCDC2 has been strongly associated with core components of the phonological processing system in animal models and multiple independent studies of populations and languages. To characterize subtle language differences arising from genetic variants associated with phonological processes, we examined the relationship between READ1, a regulatory element in DCDC2, and phonemes in languages of 43 populations across five continents. Variation in READ1 was significantly correlated with the number of consonants. Our results suggest that subtle cognitive biases conferred by different READ1 alleles are amplified through cultural transmission that shape consonant use by populations over time.
Keywords: DCDC2, READ1, phoneme, language, genetics
Abstract
DCDC2 is a gene strongly associated with components of the phonological processing system in animal models and in multiple independent studies of populations and languages. We propose that it may also influence population-level variation in language component usage. To test this hypothesis, we investigated the evolution and worldwide distribution of the READ1 regulatory element within DCDC2, and compared its distribution with variation in different language properties. The mutational history of READ1 was estimated by examining primate and archaic hominin sequences. This identified duplication and expansion events, which created a large number of polymorphic alleles based on internal repeat units (RU1 and RU2). Association of READ1 alleles was studied with respect to the numbers of consonants and vowels for languages in 43 human populations distributed across five continents. Using population-based approaches with multivariate ANCOVA and linear mixed effects analyses, we found that the RU1-1 allele group of READ1 is significantly associated with the number of consonants within languages independent of genetic relatedness, geographic proximity, and language family. We propose that allelic variation in READ1 helped create a subtle cognitive bias that was amplified by cultural transmission, and ultimately shaped consonant use by different populations over time.
The main function of the phonological processing system is to translate basic word sounds called phonemes into recognizable words. Components of the phonological processing system—particularly phonological awareness and phonemic decoding—are critical for language acquisition and development (1). They are also highly heritable quantitative traits that have been associated with a limited number of genes expressed early in human brain development and implicated in neuronal migration and ciliary function (2).
Prominent among these is DCDC2, a gene strongly associated with core components of the phonological processing system in animal models and multiple independent studies of different populations and languages (3–7). Studies on genetically modified mice show that knocking out Dcdc2a decreases temporal precision of action potential firing in neurons of the neocortex (8), and impairs rapid auditory processing (9). In rats, RNAi knockdown of Dcdc2 diminishes ability to discriminate between specific speech sounds presented in continuous streams (10). Performance on a similar measure in humans, called late mismatch negativity, maps within 100,000 bps of DCDC2 on human 6p22 (11).
Associations of core language components with DCDC2 are mediated through a highly polymorphic transcriptional regulatory element called “regulatory element associated with dyslexia 1” (READ1) (12). READ1 alleles differentially alter transcription of DCDC2, which we hypothesize leads to subtle differences in phonological processing. Although READ1 was identified through clinical studies of dyslexia and specific language impairment, it is also linked to normal variation in speech and language performance within populations (13).
The evolution of language is generally discussed in terms of genetic underpinnings or sociocultural influences (14, 15). Genetic underpinnings describe the development of a general language faculty that includes the anatomical structures required for speech as well as cognitive control of these structures, which support the human capacity for language. Sociocultural influences elicit more rapid changes in language driven by factors such as linguistic, social, and population dynamics. These explanations are increasingly viewed as complementary. Theories about the evolution, development of, and changes in language have begun to incorporate the possibility that processes of cultural transmission act to amplify subtle cognitive biases conferred by genetic heterogeneity to shape differences between individual languages (16). This is supported by studies that demonstrate a correlation between frequency of certain haplotypes across two genes involved in brain growth (ASPM and Microcephalin) and the development of linguistic tones at a population level (17). Building on this work, we hypothesize that the frequency of genetic variants within a population that differentially modulate components of the phonological processing system could also influence phoneme selection and inventory size.
To test this hypothesis, we trace the evolution of READ1 beginning in nonhuman primates. Next, we compare phoneme inventory size with READ1 allele frequencies in population samples from five continental groups. Then we assess the relationship between READ1 and phonemes while adjusting for genetic relatedness between populations in a multivariate ANCOVA model. Finally, we corroborate the relationship between READ1 subunits and phonemes in a linear mixed effects model accounting for possible confounding effects due to genetic relatedness, geographic proximity, and differences in sample size and language family. These studies suggest that in addition to social and cultural factors, genetic factors likely play a role in phoneme selection and language development by different populations and cultures.
Results
Evolution of READ1.
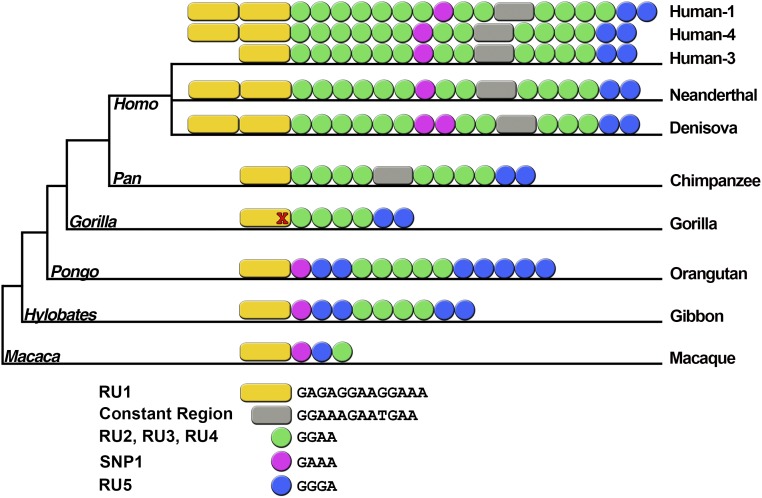
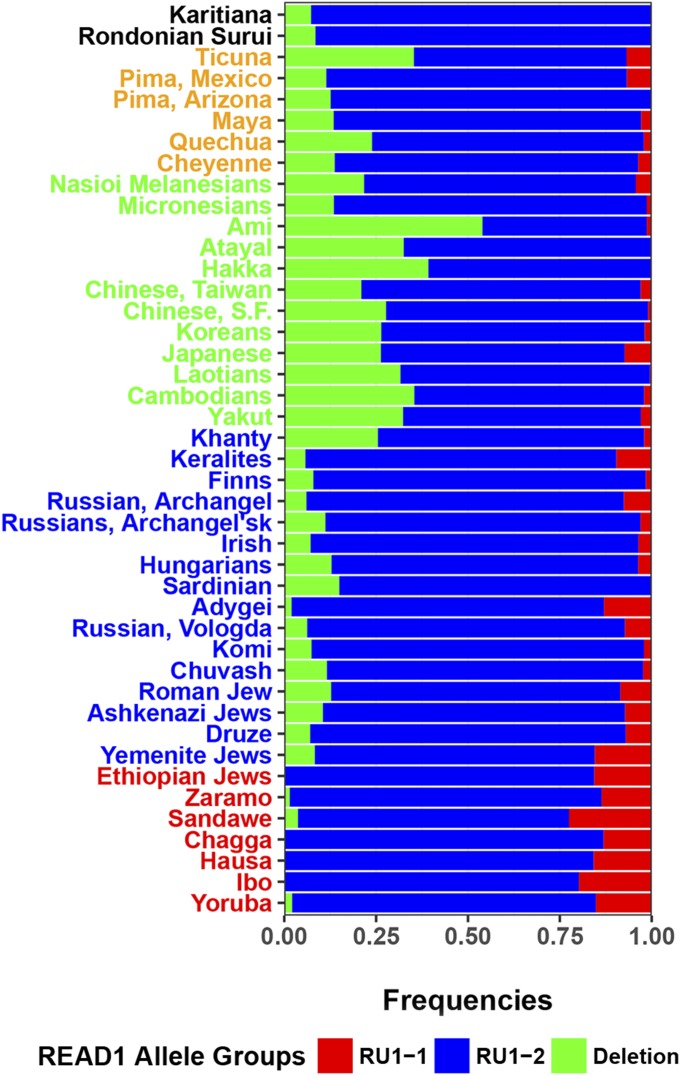
READ1 is a complex highly polymorphic transcriptional regulatory element that ranges in length from 81 to 115 base pairs (4, 12, 13, 18). It is composed of seven individual repeat units: RU1, RU2, SNP1, RU3, Constant Region, RU4, and RU5 (SI Appendix, Fig. S1). Alignment of the repeat units (Fig. 1) show that the RU1 sequence [GAGAGGAAGGAAA] is present in superfamily Hominidae (humans, chimpanzees, gorilla, orangutan, and gibbon) as well as in Old World monkey (crab-eating macaque). There is no homologous sequence present in the orthologous 2 kb location in marmoset (New World monkeys). This suggests that RU1 arose after the divergence of Old World monkeys and New World monkeys, ∼40 Mya. RU1 underwent a duplication event after the divergence of genera Pan (Pan paniscus and P. troglodytes) and Homo (4–8 Mya), but before the last common ancestors of human, Neanderthal, and Denisovan, ∼550–765 kya (19) (Fig. 1). In the few genome sequences available from archaic hominins Neanderthal and Denisovan, all have two copies of RU1. The Neanderthal sequence is identical to human allele 4. The Denisovan sequence resembles allele 4 but has two SNP1 units and only three copies of RU4. Homo sapiens has six alleles 2, 3, 9, 12, 25, and 27 that contain only one copy of RU1 (RU1-1) as well as 35 alleles with two copies of RU1 (RU1-2) (SI Appendix, Table S1). Among the 43 populations studied, the frequencies of RU1-1 alleles are low and decrease with distance from Africa (Fig. 2 and SI Appendix, Fig. S1 and Table S2).
Fig. 1.
Alignment of READ1 repeat units in primates. Repeat units are depicted as colored dots for 4-bp motifs, and as rectangles for longer motifs, and are labeled with the same colors used in SI Appendix, Fig. S1. Human-1 (an RU1-2 allele) is the most common READ1 allele, Human-3 is the most common of the RU1-1 alleles, and Human-4 is identical to the Neanderthal allele. The Denisovan allele is similar to human alleles. Chimpanzee (Pan paniscus and P. trogdolytes) has two alleles that differ by one GGAA repeat; the longest is represented here. Gorilla gorilla lacks the final “A” in the first motif, represented by the red X at the end of the motif.
Fig. 2.
Distribution of RU1-1, RU1-2, and deletion of READ1 in 43 world populations. Populations used in this study are distributed on the y axis in an approximation of the geographic distribution out of Africa and colored by the continental group to which they were assigned in this study. The x axis shows the distribution of frequencies of three major categories of READ1 alleles: RU1-1 in red, RU1-2 in blue, and the microdeletion in green.
RU2 is a tetranucleotide tandem repeat [GGAA] that varies in length from 4 to 11 copies in humans. Only one to four RU2 copies are observed in the proto-READ1 sequences from nonhuman primates (Fig. 1). In humans, RU2 is the most polymorphic repeat unit of READ1, which likely reflects the high mutation rate of 1 in 10,000 meioses (20). Once RU2 passed a threshold of four copies, replication slippage occurred more frequently (21), and RU2 became more polymorphic and the main determinant of the large number of contemporary READ1 alleles.
There are additional changes that differentiate READ1 in humans from READ1 in chimpanzees. The shortest READ1 sequence in humans is 81 bp (allele 27), whereas the longest READ1 allele in chimpanzees is 69 bp. The expanded READ1 in humans includes a bifurcation of RU2 repeats by a GGAA to GAAA SNP (SNP1) followed by two invariant GGAAs that precede the constant region. Additionally, there is a 2.4-kb microdeletion of the entire READ1 sequence that varies in frequency in human populations. The READ1 microdeletion is rarely observed in African populations (Fig. 2). It likely arose after the migrations of humans out of Africa, with some back migrations from other continental groups accounting for sparse low frequencies.
In summary, READ1 gained Homo-specific variation sometime between 550 kya and 4 Mya, which included a duplication of RU1 seen only in the Homo genus, as well as the expansion of RU2 repeats that destabilized the locus and led to the large number of polymorphisms observed in humans today.
RU1-1 Frequency Is Associated with Number of Consonants.
Based on the results above, we divided READ1 alleles into three groups. The first group, RU1-1, contains alleles with a single copy of RU1. The second group, RU1-2, contains alleles with a duplication of RU1. The third group is the 2.4-kb microdeletion that includes the entire READ1 sequence. To examine the relationships between components of the phonological processing system and READ1, we regressed the number of consonants and vowels from 43 populations (SI Appendix, Table S3) onto the frequencies of READ1 allele groups RU1-1, RU1-2, and the 2.4-kb microdeletion (Fig. 3). This showed that consonants correlate with RU1-1 frequency (ρ = 0.45, P = 0.002), but vowels do not (ρ = −0.09, P = 0.58).
Fig. 3.
Linear regressions of consonants and vowels by RU1-1 frequencies. Log10 transformed numbers of consonants (solid dots) and vowels (open triangles) were regressed separately against the frequencies of RU1-1 alleles in 43 populations.
Association Between READ1 Allele Groups and Phonemes.
To examine whether the relationships between READ1 allele group frequencies and phonemes could be confounded by either genetic or geographic relatedness (22), we used multivariate ANCOVA to model the number of consonants and vowels with frequencies of RU1-1, RU1-2, and the microdeletion, and including the first three principal components (PCs) of the tau genetic relatedness matrix over the same 43 populations. With multivariate ANCOVA, association of RU1-1 with consonants and vowels remained significant after correcting for multiple testing over the three allele groups [F-stat = 5.51 with 37 degrees of freedom (df); false discovery rate (FDR) = 0.024; Table 1]. The analyses did not show a significant association between either the microdeletion (F-stat = 0.44 with 37 df; FDR = 0.89) or the RU1-2 allele group (F-stat = 0.12 with 37 df; FDR = 0.89) with phonemes. It remains possible that an association exists between a subset of RU1-2 alleles, but is obscured by other RU1-2 alleles with differing directions of effect in the group analysis. The RU1-1 results were robust to jackknife of populations and continental groups (SI Appendix, Table S5). Post hoc univariate analyses showed that the associations observed in the multivariate ANCOVA were driven by number of consonants (β = 1.76, P = 0.003; Table 2). RU1-1 frequency was not associated with the number of vowels (β = −0.8, P = 0.463). These analyses support the results from the univariate regressions (Fig. 3), even after accounting for possible hidden genetic and geographic relatedness between populations.
Table 1.
Multivariate ANCOVA examining the relationship between the number of consonants and vowels, compared with the frequency of RU1-1, RU1-2, and the deletion
P ≤ 0.05.
FDR corrected.
Table 2.
Post hoc univariate analysis of the effect of RU1-1 on vowels and consonants independently while controlling for PCs1-3
| Response | β | t | P |
| Vowels | −0.61 | −0.8 | 0.471 |
| Consonants | 1.76 | 3.2 | 0.003* |
P ≤ 0.01.
Mixed Effects Model of READ1 Allele Groups and Linguistic Traits of Worldwide Populations.
To account for possible confounding effects due to geographic proximity, language family membership, and different sample sizes, we fit a mixed effects model with RU1-1 frequency as the response variable, weighted by sample size, and consonants and vowels as fixed effects. The first three PCs of the tau genetic relatedness matrix were also included as fixed effects. Language family and continental grouping were included as random effects. The mixed effects model showed a positive association between the number of consonants and the frequency of RU1-1 [β = 0.002, 95% Bootstrap CI:(0.0008, 0.003); Table 3], corroborating the multivariate ANCOVA while controlling for sample size along with modeling genetic, geographic, and language similarities.
Table 3.
Results of a linear mixed effects model using RU1-1 as the response variable with PCs 1–3 as fixed effects, language family, and continental grouping as random effects, and using chromosome numbers as weights
| Predictor | β | t | P* | (lower, upper) |
| Vowels | −0.002 | −2.01 | 0.046 | (−0.005, 0.0002) |
| Consonants | 0.002 | 3.14 | 0.004 | (0.0008, 0.003) |
Linear mixed effects model using RU1-1 (95% CI†). Bias was less than 10−6 for both parameters.
P value under t distribution with 2 and 37 degrees of freedom.
Bootstrap estimation of error using 10,000 replicates with 95% confidence intervals.
Discussion
READ1 is a highly variable and powerful transcriptional control element embedded in a gene called DCDC2. Both READ1 and DCDC2 have been associated with reading disability and specific language impairment as well as normal variation in reading performance and phonological processing in studies of children whose primary language is English, Italian, German, Mandarin, or Cantonese (5–7, 18, 23, 24). Having established a role in processing phonological tasks in different language systems and cultures, the central question in the present study is whether READ1 alleles could have influenced variation in language components on a population level.
To address this question, we first traced the evolution of READ1 by examining genomic sequences from human populations, nonhuman primates, and Neanderthal and Denisovan genomes that are available in public databases. Proto-versions of READ1 that are missing the constant region or at least one of the five RU subunits found in human alleles, are present in chimpanzee, gorilla, orangutan, gibbon, and macaque (18). The 2.4-kb microdeletion of the entire READ1 region, duplication of RU1, and bifurcation of RU2 by SNP1 are only present in human and extinct branches of the Homo genus (Neanderthal, Denisovan) and specify three groups of READ1 alleles: the microdeletion, RU1-1 with only one copy of RU1, and RU1-2 with two copies of RU1.
RU1-1 Correlation with Consonant Numbers.
Next, we set out to determine whether READ1 could have influenced phoneme inventory size. To address this question, we used a population-based approach to assess the relationship between READ1 variants and number of phonemes in 43 populations from five continents. We examined two broad types of phonemes—consonants and vowels—because as described below, there are different neurophysiologic cues the brain uses to encode consonants versus vowels. We found a significantly positive relationship between the frequency of allele group RU1-1 and the number of consonants even after correcting for the confounding effects of geographic proximity, genetic relatedness between populations (22), and language family.
DCDC2 and Phonological Processing.
There have been a variety of animal and human studies examining the function of DCDC2 that suggest an underlying neurophysiologic effect that could account for the association between phoneme inventory and the frequency of RU1-1. Neural representation of vowels and consonants relies on distinct strategies for temporal processing of auditory information. Whereas vowel encoding is more dependent on the mean number of action potential firings per unit time (25), consonant encoding is more dependent on the precision of timing between action potential events (26). Elimination or reduction of Dcdc2 in rodent models causes a decrease in action potential temporal precision (8), impairs rapid auditory processing (9), and diminishes ability to discriminate between specific speech sounds in a continuous stream (10). In humans, decreases in late mismatch negativity (MMN) have been linked to rare genetic variants in and close to DCDC2 (11). MMN is an index of successful discrimination between speech sounds. Late MMN is mainly elicited by complex auditory stimuli like syllables and words. These independent lines of investigation in both animal models and humans suggest that variations in DCDC2 expression conferred by subtle differences in regulatory elements such as READ1 may have significant effects on phoneme encoding.
DCDC2 and Consonant Discrimination in Populations.
Protein-damaging truncations and missense mutations of DCDC2 can cause heritable deafness (27) and other congenital ciliopathies (28–30). However, variations in READ1 alleles do not alter DCDC2 protein and would have less severe and potentially targeted effects on temporal processing of phonemes, sparing broader cognitive functions. READ1, which appeared later in primate evolutionary history, may be critical for fine-tuning transcription of DCDC2 in specific neurons that constitute key language circuits by modifying temporal precision of action potentials, which is critical for discriminating consonants. Depending on the distribution of READ1 allele frequencies and their neurophysiologic effects on DCDC2 expression, we hypothesize that low RU1-1 prevalence in some populations could lead to diminished ability to discriminate consonants, and over time reduce consonant inventory. This is compatible with theories of sound change and linguistic typology that highlight the role of speech perception and language changes. For example, Ohala (31) hypothesized that listeners contribute to the mechanism of sound change by failing to reconstruct intended speech as produced by a speaker, and then in turn produce the reconstructed speech sound when acting as the speaker. A recurrent error could therefore become established as the new norm. This hypothesis has been examined by comparing patterns of sound change with patterns of perceptual errors. The directionality of the cross-linguistic sound change mirrors the directionality of perception errors in both laboratory-induced speech perception errors (32) as well as naturally occurring speech perception errors (33).
The effect of speech perception on language change is highlighted in Australian Aboriginal children (34). As reported, 80% of these children have a significant conductive hearing loss due to chronic otitis media (COM). COM typically affects both ends of the hearing scale, below 500 Hz, and above 4,000 Hz. Interestingly, the Aboriginal phonemic inventory lacks contrasts that depend on low-frequency acoustic cues such as high vowels and voiced obstruents, as well as contrasts that depend on high-frequency cues such as fricatives and aspirated stops (35). This suggests that for Australian Aboriginal language, a constrained phonemic inventory was an adaptation in response to the effect of COM.
The coding regions of DCDC2 are highly conserved, but there is no evidence that READ1 confers either a selective reproductive advantage or disadvantage. However, in instances of transcriptional pleiotropy, regulatory elements can confer tissue-specific expression of the same gene in different contexts of tissue type and developmental stages (36). DCDC2 is expressed in many different tissue types and is observed at high levels in the kidney cortex. While mutations in the protein coding regions can cause autosomal recessive ciliopathies involving the kidney (28), the noncoding regulatory element READ1 has not been linked to kidney disease. Human studies mostly focus on protein-coding exons, concentrating on high effect protein truncations and missense mutations. It is therefore possible that subtle selective advantages conferred by READ1 for controlling the expression of DCDC2 in the kidney in different environments could be underappreciated. As whole genome sequencing becomes more widespread for molecular genetic description of variation in biological processes, future studies could test for possible transcriptional pleiotropy and selection.
We present converging lines of evidence to suggest that READ1, a transcriptional regulator of DCDC2, is important for distinguishing among consonants. This is consistent with the neurobiological mechanisms influenced by DCDC2, and with linguistic theories of language change. While differences in ability to discriminate between consonants conferred by READ1 are likely subtle, even weak biases can have a cumulative effect on linguistic structures (16). For READ1, a highly polymorphic transcriptional control element with more than 40 alleles in various frequency distributions throughout worldwide populations, the reduction of RU1-1 alleles dates back at least 90,000 years, before anatomically modern humans began migration out of Africa. As human cultures and languages have changed over that time, the differences in the distributions of READ1 allele frequencies may have modified consonant perception and influenced language change through cultural transmission of subtle cognitive biases. Although conventional theories mostly attribute language changes to random fluctuations, historical conquests, and migrations, these results suggest that genetic variants affecting auditory processing may also be important.
Methods
READ1 Genotyping.
Genomic DNA from 2,138 individuals representing 43 populations was extracted from transformed lymphoblastoid cell lines that were kindly provided from the curated collection of Dr. Kenneth Kidd and Dr. Judith Kidd (SI Appendix, Table S2) (37). These cell lines were generated from normal, apparently healthy adults who provided informed consent under protocols approved by their respective governmental and institutional review boards. More complete descriptions of all of the population samples are in Allele Frequency Database (ALFRED) (38). Genomic DNA for chimpanzee (Pan troglodytes; n = 3), bonobo (Pan paniscus; n = 3), gorilla (Gorilla gorilla; n = 3), orangutan (Pongo pygmaeus; n = 3), and gibbon (Hyoblates.; n = 3) were also obtained from the Kidd laboratory. Allele identifications for READ1 (GenBank: BV677278) and a 2,445-bp READ1 microdeletion (dbVar: esv3608367) were determined through Sanger sequencing and allele-specific PCR, respectively. Primers and amplification protocols have been described previously (18). Hardy-Weinberg analysis was performed using the function hw.test from pegas, an R package (39). Primate sequences were compared with those found in the University of California, Santa Cruz (UCSC) genome browser (40). Sequences for the Old World monkeys crab-eating macaque (Macaca fascicularis), green monkey (Chlorocebus sabaeus), and baboon (Papio sp.), as well as the New World monkey marmoset (Callithrix jacchus), were inspected in the UCSC Genome browser to identify READ1 sequences. Archaic hominin alleles were obtained from Denisovan and Neanderthal sequences available on the UCSC Genome Browser and from the Neanderthal Genome Project (41).
Continental Grouping and Accounting for Genetic Relatedness.
To quantify overall genetic relatedness, we calculated the genetic pairwise distances (tau) for all populations using 165 SNPs (SI Appendix, Table S4) that have been shown to robustly separate populations by genetic variation (38, 42). The first three principal components (PCs) of these tau scores account for most (99.4%) of the genetic variation between populations (SI Appendix, Fig. S3). Assignment of populations to five continental groups (Africa, Middle East plus Europe, Asia, two groups from the Americas) was achieved using hclust centroid clustering of tau scores from the basic stats package in R (43) (SI Appendix, Figs S4 and S5). The first three PCs of the tau matrix explained geographic variation between the population group centers, whose latitude and longitude coordinates were calculated as the average of the two opposite corners of a rectangle encompassing the area where the respective populations live, as specified by the ALFRED database (44). The association between genetic and geographic variation was quantified by correlating, for each pair of populations, the geographic distance (in kilometers) between their respective centers and Euclidean distance between the first three PCs of the tau matrix using Pearson’s product-moment correlation (SI Appendix, Fig. S6A), with the latter serving as a proxy for genetic distance between each population pair. Pairwise geographic distances between populations were evaluated using both great circle distances computed using the rdist function in R package fields (45) and distances via migratory waypoints (SI Appendix, SI Methods). Pairwise genetic distances had significant positive correlations with pairwise geographic distances based on both great circle calculations [ρ = 0.69, 95% CI = (0.66, 0.72)] and via migratory waypoints [ρ = 0.71, 95% CI = (0.68, 0.74); SI Appendix, Fig. S6B]. These observations provide evidence that the first three PCs of the tau matrix accounted for geographic proximity as well as genetic relatedness between the populations in our study. Finally, we observed that distances between each population center and a putative location of human origins in South Africa, via five migratory waypoints, had a strong negative correlation (ρ = −0.94, P < 10−15) with PC1 of the tau matrix (SI Appendix, Fig. S6C).
Language Metrics.
Populations were assigned International Organization for Standardization (ISO) 639–3 codes that were used for comprehensive representation of language names, corresponding to the language(s) used by that population (SI Appendix, Table S3). For populations with multiple languages listed, we used the most ancient language available. Populations were only included if there were more than 20 DNA samples, and if there was adequate historical documentation to support assignment of language. Furthermore, pidgin or creole languages were excluded because they are strongly associated with language contact and borrowing (46, 47). Counts for consonants and vowels for ISO codes were retrieved from the Phonetics Information Base and Lexicon (PHOIBLE) database (48). If PHOIBLE counts were not available, we averaged counts for phonemes, consonants, and vowels from available reports or retrieved the data manually from alternative sources (SI Appendix, Table S3). Top-level language families were used for the analyses (49).
Statistical Analyses.
To assess the relationship between population frequencies of READ1 alleles with linguistic variables, we restricted our analysis to the 43 populations with valid language data available for the number of vowels and consonants. We conducted regressions with log10 numbers of consonants and numbers of vowels as dependent variables and the frequency of RU1-1 as the independent variable using the lm function in R (50).
To account for genetic relatedness between populations we used multivariate ANCOVA to model SNP genotypes and phoneme inventory size. Each model included the first three PCs of the tau genetic relatedness matrix as covariates. For each of three groups of READ1 alleles (RU1-1, RU1-2, and the microdeletion of READ1) (g), we tested whether the observed frequency of g in the sample of population i, denoted by , had a significant effect on the log10 of number of vowels () and consonants () appearing in the associated language. We estimated the multivariate linear regression models
where , , and are covariates for scores on the first three PCs of the tau matrix. Model parameters are intercepts , effects from the three covariates , and the effect of the gth allele . We assessed significance of genetic effects on vowels and consonants using multivariate ANCOVA through Pillai’s test of the null hypothesis
Linear mixed effect analyses were conducted using lmer, part of the lme4 R package (50), to estimate the association between RU1-1 and numbers of consonants and vowels of each language, while controlling for geographic distances and language family membership. The first three PCs were used as fixed effects representing genetic and geographic similarity among populations. Continental grouping (1–5) and top-level language family were modeled as random effects. Chromosome count was also included as a weighting variable to reflect the varying precision of the RU1-1 proportion estimates associated with the different populations. Model criticism showed that the residuals of this regression were normally distributed with none being greater than three SDs from the mean. We evaluated 95% confidence intervals for parameter estimates in this model through 10,000 bootstraps. Sensitivity analyses were also conducted using jackknifing to characterize the dependence of the observed pattern in our results on each of the 43 individual populations in our sample as well as regional groupings of populations in Africa, Europe, Asia, and the Americas. Further descriptions are provided in the SI Appendix and SI Appendix, Table S5.
Supplementary Material
Acknowledgments
We are grateful for the advice and DNA samples provided by Judy and Ken Kidd. We are also grateful to Andrew Pakstis for generating the tau genetic relatedness matrix and William Speed for retrieving SNPs from the Kidd database. We thank Derek Nurse, Malin Petzell, and Gerard Philippson for their help with locating the phonology of Zaramo, and Claire Bowern for her discussion regarding relevant literatures. Support for J.R.G., M.M.C.D., C.M.M., J.G.M., N.R.P., D.T.T., and J.C.F. was provided by The Manton Foundation. Support for J.R.G., M.M.C.D., and D.T.T. was provided by NIH/Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) P50 HD027802. Support for B.M.B. was provided by NIH/NICHD T32 HD007149. Support for J.R.G. and N.R.P. was provided by NIH/National Institute of Neurological Disorders and Stroke (NINDS) R01 NS43530. Support for D.T.T. was provided by the Lambert Fund.
Footnotes
Conflict of interest statement: Yale University has applied for a patent covering READ1. J.R.G. is an inventor named on the patent application.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710472115/-/DCSupplemental.
References
- 1.Schulte-Körne G, Deimel W, Bartling J, Remschmidt H. The role of phonological awareness, speech perception, and auditory temporal processing for dyslexia. Eur Child Adolesc Psychiatry. 1999;8:28–34. doi: 10.1007/pl00010690. [DOI] [PubMed] [Google Scholar]
- 2.Peterson RL, Pennington BF. Developmental dyslexia. Lancet. 2012;379:1997–2007. doi: 10.1016/S0140-6736(12)60198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardon LR, et al. Quantitative trait locus for reading disability on chromosome 6. Science. 1994;266:276–279. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- 4.Meng H, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA. 2005;102:17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludwig KU, et al. Investigation of the DCDC2 intron 2 deletion/compound short tandem repeat polymorphism in a large German dyslexia sample. Psychiatr Genet. 2008;18:310–312. doi: 10.1097/YPG.0b013e3283063a78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scerri TS, et al. DCDC2, KIAA0319 and CMIP are associated with reading-related traits. Biol Psychiatry. 2011;70:237–245. doi: 10.1016/j.biopsych.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, et al. Association study of developmental dyslexia candidate genes DCDC2 and KIAA0319 in Chinese population. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:627–634. doi: 10.1002/ajmg.b.32267. [DOI] [PubMed] [Google Scholar]
- 8.Che A, Girgenti MJ, LoTurco J. The dyslexia-associated gene DCDC2 is required for spike-timing precision in mouse neocortex. Biol Psychiatry. 2014;76:387–396. doi: 10.1016/j.biopsych.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truong DT, et al. Mutation of Dcdc2 in mice leads to impairments in auditory processing and memory ability. Genes Brain Behav. 2014;13:802–811. doi: 10.1111/gbb.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centanni TM, et al. Knockdown of dyslexia-gene Dcdc2 interferes with speech sound discrimination in continuous streams. J Neurosci. 2016;36:4895–4906. doi: 10.1523/JNEUROSCI.4202-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czamara D, et al. Association of a rare variant with mismatch negativity in a region between KIAA0319 and DCDC2 in dyslexia. Behav Genet. 2011;41:110–119. doi: 10.1007/s10519-010-9413-6. [DOI] [PubMed] [Google Scholar]
- 12.Meng H, et al. A dyslexia-associated variant in DCDC2 changes gene expression. Behav Genet. 2011;41:58–66. doi: 10.1007/s10519-010-9408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers NR, et al. The regulatory element READ1 epistatically influences reading and language, with both deleterious and protective alleles. J Med Genet. 2016;53:163–171. doi: 10.1136/jmedgenet-2015-103418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitch WT. Empirical approaches to the study of language evolution. Psychon Bull Rev. 2017;24:3–33. doi: 10.3758/s13423-017-1236-5. [DOI] [PubMed] [Google Scholar]
- 15.Kirby S. Culture and biology in the origins of linguistic structure. Psychon Bull Rev. 2017;24:118–137. doi: 10.3758/s13423-016-1166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson B, Kirby S, Smith K. Culture shapes the evolution of cognition. Proc Natl Acad Sci USA. 2016;113:4530–4535. doi: 10.1073/pnas.1523631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dediu D, Ladd DR. Linguistic tone is related to the population frequency of the adaptive haplogroups of two brain size genes, ASPM and Microcephalin. Proc Natl Acad Sci USA. 2007;104:10944–10949. doi: 10.1073/pnas.0610848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powers NR, et al. Alleles of a polymorphic ETV6 binding site in DCDC2 confer risk of reading and language impairment. Am J Hum Genet. 2013;93:19–28. doi: 10.1016/j.ajhg.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun JX, et al. A direct characterization of human mutation based on microsatellites. Nat Genet. 2012;44:1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, Strassmann JE, Queller DC. Insertions, substitutions, and the origin of microsatellites. Genet Res. 2000;76:227–236. doi: 10.1017/s001667230000478x. [DOI] [PubMed] [Google Scholar]
- 22.Creanza N, et al. A comparison of worldwide phonemic and genetic variation in human populations. Proc Natl Acad Sci USA. 2015;112:1265–1272. doi: 10.1073/pnas.1424033112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lind PA, et al. Dyslexia and DCDC2: Normal variation in reading and spelling is associated with DCDC2 polymorphisms in an Australian population sample. Eur J Hum Genet. 2010;18:668–673. doi: 10.1038/ejhg.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Association of DCDC2 polymorphisms with normal variations in reading abilities in a Chinese population. PLoS One. 2016;11:e0153603. doi: 10.1371/journal.pone.0153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez CA, et al. Different timescales for the neural coding of consonant and vowel sounds. Cereb Cortex. 2013;23:670–683. doi: 10.1093/cercor/bhs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engineer CT, et al. Cortical activity patterns predict speech discrimination ability. Nat Neurosci. 2008;11:603–608. doi: 10.1038/nn.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grati M, et al. A missense mutation in DCDC2 causes human recessive deafness DFNB66, likely by interfering with sensory hair cell and supporting cell cilia length regulation. Hum Mol Genet. 2015;24:2482–2491. doi: 10.1093/hmg/ddv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schueler M, et al. DCDC2 mutations cause a renal-hepatic ciliopathy by disrupting Wnt signaling. Am J Hum Genet. 2015;96:81–92. doi: 10.1016/j.ajhg.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girard M, et al. DCDC2 mutations cause neonatal sclerosing cholangitis. Hum Mutat. 2016;37:1025–1029. doi: 10.1002/humu.23031. [DOI] [PubMed] [Google Scholar]
- 30.Grammatikopoulos T, et al. University of Washington Center for Mendelian Genomics Mutations in DCDC2 (doublecortin domain containing protein 2) in neonatal sclerosing cholangitis. J Hepatol. 2016;65:1179–1187. doi: 10.1016/j.jhep.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohala JJ. Historical linguistics: Problems and Perspectives. Routledge; New York: 1993. The phonetics of sound change; pp. 237–278. [Google Scholar]
- 32.Chang S, Plauché MC, Ohala JJ. The Role of Speech Perception in Phonology. Academic; London: 2001. Markedness and consonant confusion asymmetries; pp. 79–101. [Google Scholar]
- 33.Tang K. Naturalistic speech misperception. PhD dissertation. University College London; London: 2015. [Google Scholar]
- 34.Coates HL, Morris PS, Leach AJ, Couzos S. Otitis media in aboriginal children: Tackling a major health problem. Med J Aust. 2002;177:177–178. doi: 10.5694/j.1326-5377.2002.tb04727.x. [DOI] [PubMed] [Google Scholar]
- 35.Butcher A. Australian aboriginal languages: Consonant-salient phonologies and the “place-of-articulation imperative”. In: Harrington J, Tabain M, editors. Speech Production: Models, Phonetic Processes, and Techniques. Psychology Press; New York: 2013. pp. 187–210. [Google Scholar]
- 36.Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science. 2014;346:1004–1006. doi: 10.1126/science.1257493. [DOI] [PubMed] [Google Scholar]
- 37.Cherni L, et al. Genetic variation in Tunisia in the context of human diversity worldwide. Am J Phys Anthropol. 2016;161:62–71. doi: 10.1002/ajpa.23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kidd KK, Cavalli-Sforza LL. The role of genetic drift in the differentiation of Icelandic and Norwegian cattle. Evolution. 1974;28:381–395. doi: 10.1111/j.1558-5646.1974.tb00759.x. [DOI] [PubMed] [Google Scholar]
- 39.Paradis E. pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 40.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kidd JR, et al. Single nucleotide polymorphisms and haplotypes in Native American populations. Am J Phys Anthropol. 2011;146:495–502. doi: 10.1002/ajpa.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Core Team 2016. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing Vienna), Version 3.2.4.
- 44.Rajeevan H, et al. ALFRED: The ALelle FREquency database. Update. Nucleic Acids Res. 2003;31:270–271. doi: 10.1093/nar/gkg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nychka D, Furrer R, Sain S. 2015. Fields: Tools for Spatial Data (University Corporation for Atmospheric Research, Boulder, CO), R Package Version 8.2-1.
- 46.Bickerton D. Pidgin and Creole Linguistics. Indiana University; Bloomington, IN: 1977. Pidginization and creolization: Language acquisition and language universals; pp. 49–69. [Google Scholar]
- 47.McWhorter J. Language Change and Language Contact in Pidgins and Creoles. John Benjamins Publishing; Amsterdam: 2000. [Google Scholar]
- 48.Moran S, McCloy D, Wright R. PHOIBLE Online. Max Planck Institute for Evolutionary Anthropology; Leipzig, Germany: 2014. [Google Scholar]
- 49.Hammarström H, Forkel R, Haspelmath M, Bank S. 2015 Glottolog 2.3. (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany). Available at glottolog.org/. Accessed February 27, 2017.
- 50.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.