Significance
The family of G-protein–coupled receptors (GPCRs) is the largest receptor family, and each member detects specific ligands, which in turn, activate selected members of one or more G-protein families. We developed a broadly applicable assay to study the specificity of GPCR–G-protein interaction in a subtype selective manner. We were able to quantitatively measure both association and dissociation rates of G proteins from agonist-activated GPCRs. We found that the stability of the receptor–G-protein complex is an inherent property of both interaction partners independent on the agonist used. Furthermore, its lifetime correlated closely with the ability of the receptor to activate the corresponding G-protein subtype.
Keywords: GPCR, G-protein selectivity, G-protein affinity, FRET, muscarinic
Abstract
G-protein–coupled receptors (GPCRs) are essential for the detection of extracellular stimuli by cells and transfer the encoded information via the activation of functionally distinct subsets of heterotrimeric G proteins into intracellular signals. Despite enormous achievements toward understanding GPCR structures, major aspects of the GPCR–G-protein selectivity mechanism remain unresolved. As this can be attributed to the lack of suitable and broadly applicable assays, we set out to develop a quantitative FRET-based assay to study kinetics and affinities of G protein binding to activated GPCRs in membranes of permeabilized cells in the absence of nucleotides. We measured the association and dissociation kinetics of agonist-induced binding of Gi/o, Gq/11, Gs, and G12/13 proteins to muscarinic M1, M2, and M3 receptors in the absence of nucleotides between fluorescently labeled G proteins and receptors expressed in mammalian cells. Our results show a strong quantitative correlation between not the on-rates of G-protein–M3–R interactions but rather the affinities of Gq and Go proteins to M3–Rs, their GPCR–G-protein lifetime and their coupling efficiencies determined in intact cells, suggesting that the G-protein subtype-specific affinity to the activated receptor in the absence of nucleotides is, in fact, a major determinant of the coupling efficiency. Our broadly applicable FRET-based assay represents a fast and reliable method to quantify the intrinsic affinity and relative coupling selectivity of GPCRs toward all G-protein subtypes.
G-protein–coupled receptors (GPCRs) are involved in many physiological functions and represent the largest receptor family, accounting for more than several hundred members. These receptors couple to four major G-protein classes (Gi, Gq/11, Gs, and G12/13) (1), which are defined by the type of Gα subunit. In total, we have a pool of 16 α, 5 β, and 12 γ subunits (2). At the same time, active G proteins are able to interact with more than 25 different effectors, such as adenylyl cyclases, ion channels (3), PLC, RhoGEFs (4, 5), and PDE (4), further enhancing the complexity of GPCR signaling. Although many classes of G proteins exhibit a high degree of selectivity by the given GPCR, it has been shown that the same receptor can couple to more than one class of G proteins (6–11). Such a large number of potential combinations gives rise to the question of how the receptor actually finds the right G protein.
Breakthroughs in GPCR crystallization gave detailed insight into the GPCR structures (12, 13) and GPCRs’ activation and interaction with downstream partners. However, the selectivity of receptor–G-protein coupling and its underlying mechanisms remain unclear (14, 15). In our study, we decided to focus on muscarinic receptors (mAChRs), which belong to class A GPCRs and constitute a family with five subtypes (16). M1– and M3–Rs are classic Gq coupling GPCRs (17–20); however, changes in the concentration of second messengers after treatment with pertussis and cholera toxins (17, 21–25) suggest a possible coupling to Gi and Gs proteins. In contrast, M2–R is a classic Gi family-coupled receptor (19, 23, 26). Despite the fact that mAChRs couple to different G-protein classes, the comparison of crystal structures of these GPCRs revealed only small differences in transmembrane helices (12, 13, 27), meaning that the current knowledge of receptor structures restricts a prediction of the coupling to a particular G-protein class or subtype (28, 29).
In accordance with ternary complex model agonist, receptor and G protein remain stably coupled until GTP binds to the Gα subunit and immediately triggers G-protein activation and its dissociation from the receptor (30, 31). We hypothesized that the mechanism underlying the coupling selectivity must be encoded in the receptor–G-protein interaction. Therefore, we set out to measure the kinetics and affinity of the GPCR–G-protein interaction in the absence of nucleotides to determine the affinity of the G proteins to the receptors in a quantitative manner. Due to the high concentration of nucleotides in the cytosol, the ternary complex has a very short lifetime (32); therefore, previous methods to measure the GPCR–G-protein affinity were mostly based on biochemical assays and required complicated protein purification steps (8, 33–35). In this study, we establish a reliable FRET-based method for quantification of the G-protein affinity to the receptor in a regular plasma membrane environment. With this method, we measured on- and off-rates as well as steady-state binding curves for the agonist-driven interaction of GPCRs with representative G proteins from all four classes.
Results
Studying Ternary Complex Formation by Means of FRET.
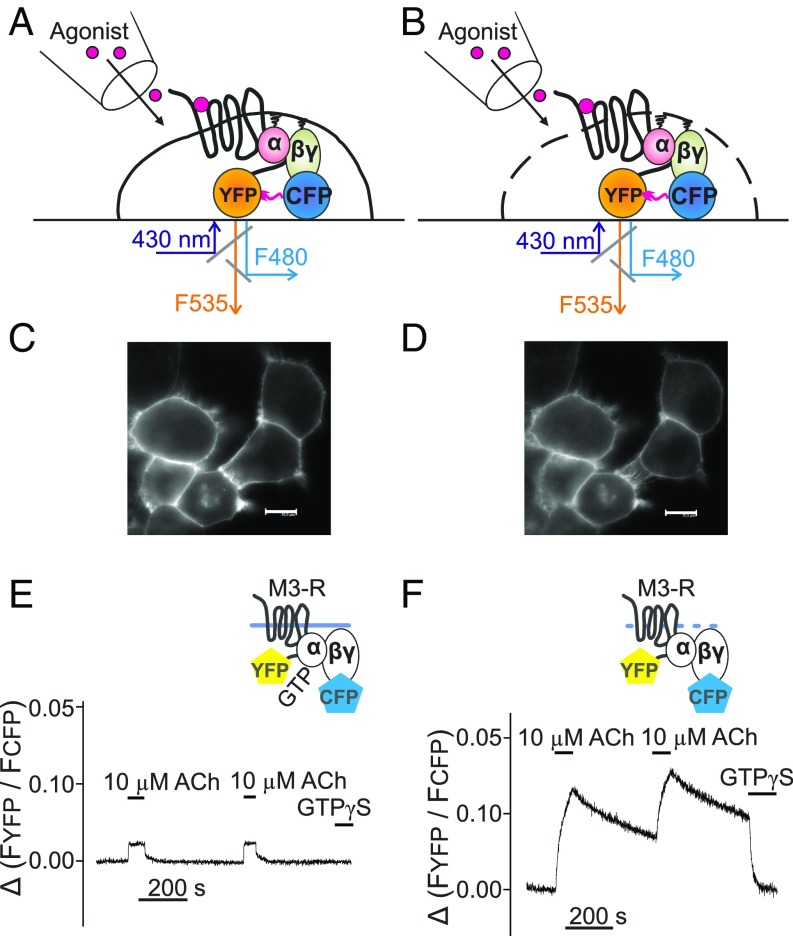
To investigate the affinity of receptor–G-protein interaction, the dynamics of G proteins binding to activated GPCRs and their subsequent dissociation were analyzed by FRET under conditions of GTP depletion. HEK293T cells expressing M3–R–YFP, Gαq, Gβ1 subunits, and CFP-Gγ2 were subjected to single-cell FRET imaging (Fig. 1A), similar to the process described previously (36). In intact non–GTP-depleted cells, we observed a small and rapidly reversible increase in FRET as also shown previously for other receptors (Fig. 1 C and E) (37). We then permeabilized cells by 2-min exposure to 0.05% saponin (Fig. 1 B, D, and F) to deplete membranes from nucleotides, thus allowing the development of relatively stable agonist–receptor–G-protein complexes. The amplitude of FRET signal in permeabilized cells was substantially higher, indicating a largely increased occupancy of receptors with G proteins (Fig. 1F). Most importantly, after withdrawal of agonist, we observed much slower dissociation kinetics of Gq from M3–R in absence of nucleotides (Fig. 1E vs. Fig. 1F), which reflects the high intrinsic affinity of G proteins to active receptors under nucleotide-depleted conditions. The rapid drop of the FRET signal in response to GTPγS indicates nucleotide-dependent fast dissociation of the remaining receptor–G-protein complexes (Fig. 1F).
Fig. 1.
Assay to measure G-protein–receptor interaction. Gq-protein binding to the M3–R was measured either in intact (A, C, and E) or permeabilized (B, D, and F) cells by means of brief exposure to saponin as schematically illustrated in B. HEK293T cells transfected with M3–R C-terminally labeled with YFP, Gαq-WT, Gβ1-WT, and Gγ2 N-terminally labeled with CFP (C–F) were subjected to confocal microscopy and imaged for the CFP fluorescence before (C) and after membrane permeabilization (D). (Scale bars, 10 μm.) For FRET measurements, cells were excited at 430 nm, and YFP and CFP emission was simultaneously imaged using a dual-emission fluorescence microscope. The YFP/CFP emission ratio derived from a single cell was calculated and plotted over time (E and F), and the superfusion of cells with agonists or GTPγS occurred as indicated.
Affinity of G Protein to mAChRs Determines GPCR–G-Protein Selectivity.
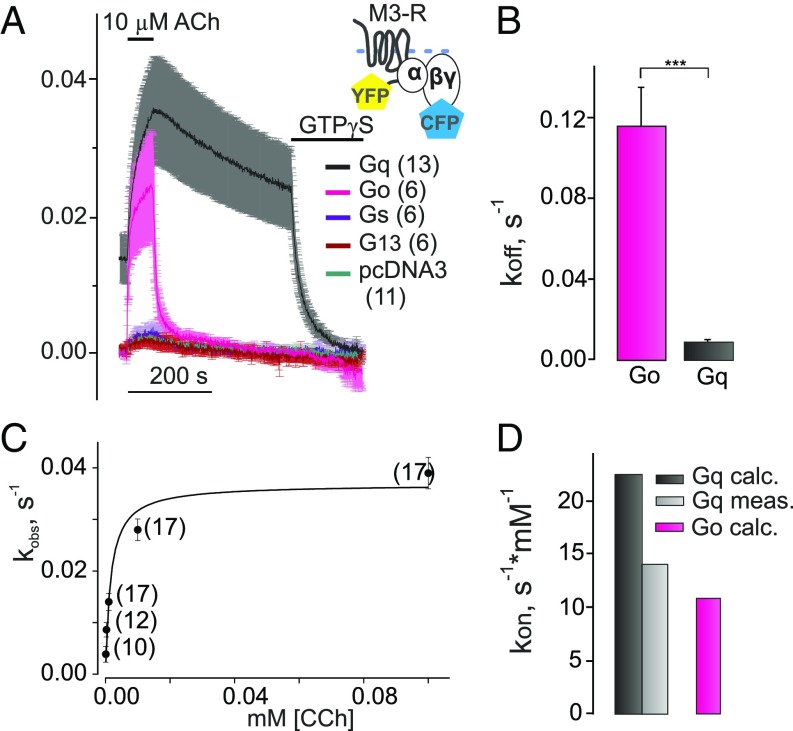
Based on reports that M3–Rs may also couple to Gi/o and even Gs proteins (38), we wanted to quantify the selectivity of M3–Rs binding to four different classes of G proteins under conditions of nucleotide depletion. Therefore, we compared the agonist [10 µM acetylcholine (ACh)]-evoked FRET signal between fluorescent M3–R and fluorescent Gβγ subunits when Gαq, Gαo, Gαs, Gα13, or pcDNA3 instead of Gα was transfected (Fig. 2A). As depicted in Fig. 2A, there were no significant differences in the small agonist-evoked FRET signal in cells transfected with Gαs or Gα13 compared with those that were transfected with empty vector instead of cDNA encoding for Gα subunits, indicating no detectable interactions. Although the binding of Gαs and Gα13 to M3–R was not detected, these G proteins did bind with high affinity to receptors, such as β-andrenergic receptors (β1-AR and β2-AR) and thromboxane TxA2 receptor (TP-R) (SI Appendix, Fig. S1), in accordance with the literature data. Correspondingly, we could not detect G13 or Gs activation via stimulation of M3–R (SI Appendix, Fig. S2). However, we observed a robust agonist-induced FRET signal in cells transfected with Gαo, which was comparable in amplitude with signals obtained in Gαq-expressing cells (Fig. 2A, pink vs. black). In experiments studying M3–R interaction with either Gq or Go proteins in the presence of high or low concentrations of GDP or GTP, we verified accurate control of nucleotides in permeabilized cells (SI Appendix, Fig. S3) and also verified the absence of nucleotide-sensitive preassociation of receptors and G proteins (SI Appendix, Fig. S4).
Fig. 2.
Selectivity of G protein binding to M3–R. (A) Agonist-dependent association and dissociation of M3–R with Go, Gq, Gs, or G13 proteins were measured after nucleotide depletion by means of FRET similar to that described in Fig. 1F. Average traces of YFP/CFP emission ratio (normalized to initial values) reflecting FRET between YFP-labeled M3–R and CFP-labeled Gγ2 subunit are illustrated when Gαo-WT (pink curve), Gαq-WT (black), Gs (purple), G13 (red), or empty pcDNA3 (dark green) was cotransfected. A much faster decay of the FRET signal after withdrawal of agonist was detected for Go proteins compared with Gq. (B) The constants of dissociation (koff; s−1) for Go and Gq proteins from M3–R are shown (magenta, n = 14; black, n = 30, respectively). (C) Observed association kinetics (kobs; s−1) of Gq protein with M3–R under GTP-depleted conditions were plotted over different CCh concentrations and fitted by a hyperbolic function. (D) Measured kon value of M3–R–Gq association kinetics (light gray, Gq meas.) obtained from fitted hyperbolic function using Eq. 2 compared with of Gq (black, Gq calc.) and Go (magenta, Go calc.), which were calculated based on steady-state experiments data as shown in Eq. 3. All data are plotted as means ± SEM for each condition; n of each experiment is shown in parentheses if not indicated. Statistical anlysis was performed using one-way ANOVA followed by Student’s t test (***P <0.001).
The comparison of Gq and Go dissociation rates from M3–R during the withdrawal of agonist in the absence of nucleotides revealed a significant 13-fold difference (0.009 ± 0.002 and 0.116 ± 0.019 s−1, respectively) (Fig. 2B and Fig. 4A, exponential fit example), which indicates a lower affinity of the M3–R–Go complex. Moreover, since we actually measured interaction of fluorescent Gβγ with fluorescent receptors using overexpressed native Gα subunits for a more accurate comparison between receptor–G-protein coupling, we also performed experiments with fluorescently labeled versions of both Gαq and Gαo as a control. No labeling-associated differences in the dissociation kinetics for both G proteins were detected (SI Appendix, Fig. S5). Similarly, long lifetimes of receptor–G-protein complexes were observed for other receptors, such as β1-, β2-, and α2A-ARs as well as TP-R, only for G-protein subtypes, which are known to be activated by the respective receptor (SI Appendix, Fig. S1). Furthermore, we measured the association kinetics of M3–R with Go and Gq proteins under nucleotide-depleted conditions in dependence of the agonist concentration (Fig. 2 C and D). Based on generally accepted agonist–receptor occupancy models (39, 40), the rates of receptor–G-protein interaction (kobs) should increase with an exponential correlation (Eq. 1) to the agonist concentration applied. Taking into account that the exponential function is a subcase of a particular hyperbolic sector, kon was calculated as a first derivative of kobs described as hyperbolic function (Eq. 2):
| [1] |
| [2] |
| [3] |
Moreover, the kon values for the Gq, Go, and Gi2 were also calculated according to Eq. 3 based on the determined koff values and the apparent EC50 values measured under steady-state conditions in permeabilized cells (Eq. 3, Fig. 3A, and SI Appendix, Fig. S6). Both kon and values for Gq (Fig. 2D) seemed to be relatively similar (22.40 and 14.04 s−1·mM−1, respectively). We also attempted to experimentally determine the on-rate for the M3–R–Go interaction (SI Appendix, Fig. S7); however, in this case, the apparent on-kinetics were faster, and if plotted against the agonist concentration, we failed to fit the data to a simple one-component hyperbolic function (SI Appendix, Table S1). Obviously, the measured and calculated association kinetics did not reflect the differences in coupling preference for Gq over Go proteins. This suggests—at least for the M3–R—that the lifetimes of the ternary complexes play a major role in the determination of receptor–G-protein selectivity.
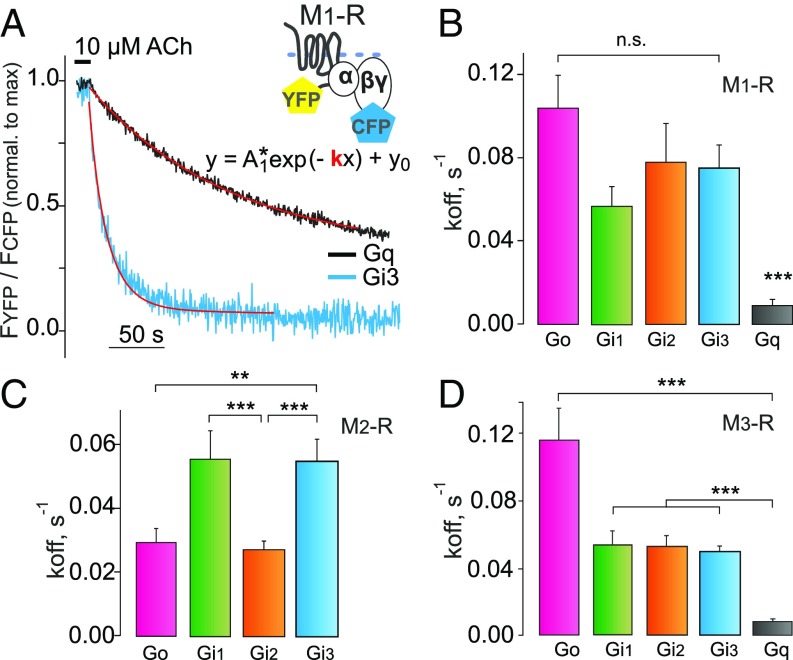
Fig. 4.
Comparison of the stability of G-protein–receptor complexes. Decay kinetics of receptor–G-protein complexes were determined in response to agonist withdrawal for M1–, M2–, and M3–Rs and different members of the Gi/o- and Gq-protein family in experiments similar to those shown in Fig. 1F. (A) Examples of exponential curve fittings to the decline in the YFP/CFP ratio after agonist withdrawal for M1–R and Gi3 (blue) or Gq (black) protein. The constants of dissociation (koff; s−1) of Gi1, Gi2, Gi3, Go, and Gq proteins (green, orange, blue, magenta, and black, respectively, in all bar graphs) were calculated for M1– (B), M2– (C), and M3–Rs (D). Calculated koff (s−1) values, including the replication numbers (n), are given in SI Appendix, Table S2. The data are represented as means ± SEM. (Data for Go and Gq are taken from Fig. 2B for comparison.) Normalization of alterations in FRET was performed as described in Fig. 2. Statistical analysis was performed using one-way ANOVA followed by Dunnett’s multiple comparison test (**P < 0.01; ***P < 0.001; n.s., P ≥ 0.05).
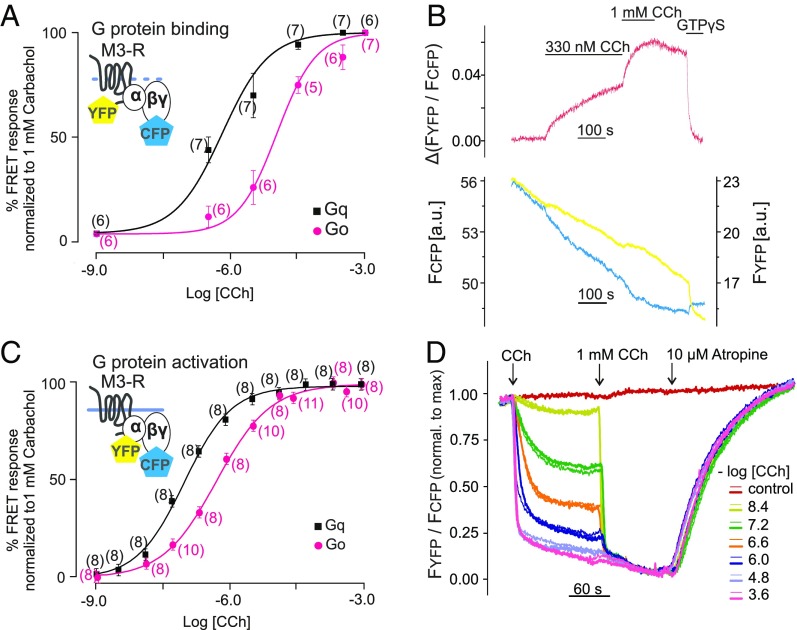
Fig. 3.
Comparison of receptor–G-protein binding and activation of G proteins. (A) Concentration–response curves of Gq (black) and Go (pink) binding to M3–R under GTP-free conditions. FRET imaging in single permeabilized cells was performed as described in Fig. 1F. Increasing concentrations of CCh were applied as shown in a representative trace of a Gq-expressing cell (B). The change of YFP/CFP emission ratio was normalized to the amplitude observed on application of saturating CCh concentration (1 mM; maximum response) and the YFP/CFP ratio measured under the application of GTPγS (minimum response). To achieve steady-state conditions at low CCh concentrations (under 100 nM), cells were incubated for 10 min. (C and D) Concentration–response curves of Gq and Go activation by M3–R were determined in intact cells using established G-protein FRET assays (61). (D) Representative traces of FRET between YFP-labeled Gα subunit and CFP-labeled Gγ2 subunit were measured in duplicates (overflow technique) simultaneously in 12 individual wells derived from a 96-well plate and normalized to the maximal response (1 mM CCh). All concentration–response data are represented as means ± SEM for each condition; n of each experiment is shown in parentheses. Statistical analysis was performed by one-way ANOVA with Bonferroni post hoc test (P < 0.05).
The Difference in Go and Gq Affinity to M3–R Reflects Coupling Efficiency.
To experimentally verify whether the measured kon and koff were realistic, we next determined steady-state binding curves for the carbachol (CCh)-induced nucleotide-free complex formation of M3–R with Gαq- or Gαo-containing G proteins in dependence of agonist concentration by determining the stable plateau reached after agonist application (Fig. 3 A, black vs. pink and B). The concentration–response curve of Go binding to M3–R was found to be 16.5-fold (right shifted in comparison with Gq; EC50 = 10.68 ± 0.32 and 0.64 ± 0.04 µM, respectively), which indicates a higher affinity of binding of Gq to M3–R compared with Go. Under these conditions, we determined the G-protein/receptor expression ratio to be two- to sixfold (SI Appendix, Fig. S8) and also detected no major alterations of Gβγ expression in dependence of the coexpressed Gα subtype (SI Appendix, Fig. S9).
To test whether the stability of the ternary complex correlates with the efficiency of G-protein activation, we determined Go- and Gq-protein coupling efficiency to M3–R (Fig. 3 C and D) by measuring FRET between YFP-labeled Gα and CFP-labeled Gβγ subunits (41) in thousands of nonpermeabilized adherent transiently transfected cells (Fig. 3D) in a 96-well format. Similar to the binding data, the concentration–response curve of Gαo activation (EC50 = 0.79 ± 0.01 µM) by M3–R was 6.6-fold right shifted in comparison with Gαq activation (EC50 = 0.12 ± 0.01 µM) (Fig. 3C, pink vs. black). Considering our findings regarding the on-kinetics of Go and Gq binding to activated M3–R, this result shows that the affinity of the mAChRs–G-protein interaction underlying G-protein selectivity is primarily reflected by the stability (lifetime) of the complex and correlates closely with the coupling efficiency. Moreover, we measured dissociation kinetics of M3–R–Gi/o proteins complexes in dependence of Gαi subtypes and found no significant differences for Gαi1, Gαi2, and Gαi3 (Fig. 4D and SI Appendix, Fig. S10A). Similar results were obtained for M1–R (Fig. 4 A and B and SI Appendix, Fig. S10 B and E).
To test for the Gi/o-subtype selectivity of M2–R, we repeated similar experiments for M2–R. Here, we observed a higher amplitude and slightly enhanced complex stability for Gαo and Gαi2 compared with Gαi1 and Gαi3, as the calculated koff values of Gαo and Gαi2 from M2–R were twofold smaller in comparison with Gαi1 and Gαi3, a result that correlates with the G-protein homology within the Gi family (Fig. 4C and SI Appendix, Fig. S10 D and F). Also, as expected, no binding of Gαq to M2–R in the absence of nucleotides was detected (SI Appendix, Fig. S10C). A summary of all koff values is shown in SI Appendix, Table S2.
Agonist Affinity Is Not Crucial for Gq- and Go-Protein Affinity to M3–R.
To activate mAChRs, we used three different agonists exhibiting different affinities and efficacies toward binding and activation: ACh, its synthetic analog CCh, and the partial agonist arecoline (Are) (42). Thus, we also addressed the question of whether the type of the ligand can affect the affinity of the G protein to the receptor. HEK293T cells were transiently transfected and permeabilized as mentioned above. Single cells were first stimulated with ACh and after its withdrawal, stimulated a second time with CCh or Are (SI Appendix, Fig. S11A). This procedure allows a comparison of the dissociation kinetics of G-protein–receptor complexes in the same cell, meaning equal expression levels of interacting proteins. We observed similar dissociation rates of Gq from ACh-, CCh-, and Are-activated M3–R (SI Appendix, Fig. S11 A and B). Relatedly, the dissociation kinetics of Go from M3–R were not significantly different for all three agonists (SI Appendix, Fig. S11 C and D). Thus, we could conclude that, at least in the case of agonists with moderate to low affinity, the stability of the G-protein–receptor complex is an intrinsic property of the mAChR–G-protein pair and not influenced by the agonist affinity.
Discussion
Understanding the molecular mechanism of selectivity and efficiency of receptor-mediated G-protein activation is one of the key research topics in the field of GPCR physiology and pharmacology. Based on the fact that agonist–receptor–G-protein complexes exhibit the highest stability in absence of nucleotides (43, 44), we quantified the affinity of different G proteins to M1–, M2–, and M3–Rs (22, 28, 45) by means of FRET imaging on permeabilized membranes of single cells. By laminar superfusion of these membranes, we ensured excellent control of agonist and nucleotides (SI Appendix, Fig. S3) and resolved the dynamics of interactions between nucleotide-free G proteins with several different receptors. The lifetime of receptor–G-protein complexes was determined by measuring complex dissociation in response to withdrawal of agonist and was longest for those complexes that contained G-protein subtypes that are known to be activated best by the respective receptor (Figs. 2 and 4 and SI Appendix, Figs. S1 and S10). Our detailed analysis (Fig. 2B vs. Fig. 2 C and D and SI Appendix, Fig. S7) of M3–R revealed that the lifetime of the nucleotide-free GPCR–G-protein complex (rather than the association kinetics) is the major determinant for the differences in affinity of these complexes (Fig. 3A and SI Appendix, Fig. S6).
Unlike the dissociation kinetics of the nucleotide-free receptor–G-protein complex, the steady-state and on-rate measurements might not exclusively be restricted to complexes after GDP release but also include initially GDP-bound G proteins, although these do not contribute a lot to the overall signal (SI Appendix, Fig. S3 B and D). GDP release is much faster for Go (46) than for Gq (47), which needs to be considered. We attempted to calculate the kon of GPCR–G-protein interactions in native cell membranes. The correlation of apparent M3–R–Gq association kinetics and agonist concentration is consistent with a hyperbolic function (Fig. 2C), allowing us to determine the kon directly. The resulting value of kon = 22.4 s−1·mM−1 was remarkably close to the kon = 14.1 s−1·mM−1 calculated by division of koff (Fig. 2 A and B) by the EC50 value (Fig. 3A and SI Appendix, Table S1), clearly indicating the applicability of our method. Based on the much faster equilibration kinetics of agonist binding to non-G-protein–bound muscarinic receptors (48), the kinetics of agonist-induced receptor–G-protein complex formation under nucleotide-free conditions is probably a close approximation to the optimal situation of G proteins binding to equilibrated agonist–receptor complexes. In the case of M3–R–Go interaction, the correlation of apparent association kinetics of Go proteins and M3–R with the agonist concentration is best explained by a two-component hyperbolic function (SI Appendix, Fig. S7). A comparison of the correspondingly determined on-rates with kon (calculated using Eq. 3) revealed, even with the slower on-rate, a fivefold deviation from the calculated kon. Possible reasons for this could be effects due to the above-mentioned multistep binding reaction. This might affect the apparent association kinetics of receptors and G proteins and complicates interpretation of the measured kon. Therefore, at the current research stage, we propose that measuring the off-rates of the complex together with steady-state association gives a more robust quantitative correlate of the affinity of the subtype-specific receptor–G-protein complex.
Our results show that both the steady-state curves of G protein binding to the M3–Rs as well as the dissociation kinetics of this complex quantitatively reflect M3–R–G-protein selectivity (Fig. 3). Specifically, the dissociation kinetics of Go protein from M3– and M1–Rs were 13-fold faster in comparison with Gq (Figs. 2 A and B and 4 B and D and SI Appendix, Fig. S10 A, B, and E). Similarly, we observed an approximate 16.5-fold right shift of the concentration–response curves of Go proteins binding to M3–R in comparison with Gq (Fig. 3A). A quantitatively similar 6.6-fold difference in coupling efficiency of Go and Gq to M3–R, measured in an FRET-based G-protein activation assay in intact cells (Fig. 3C), suggests that the efficacy of coupling to a certain G-protein subtype is indeed reflected in the relative M3–R–G-protein affinity and can thus be detected by measurement of the lifetime of the G-protein–receptor complex. For moderate- to low-affinity agonists with different efficacy, we could so far not detect differences in the stability of M3–R–Gq or Go complexes. An important finding was that the Gq-coupled and evolutionary close M1– and M3–Rs exhibited very close ternary complex stabilities, with a very similar pattern for the different G-protein subtypes (Fig. 4 A, B, and D and SI Appendix, Fig. S10 A, B, and E). Remarkably, the stability of the complex of different GPCRs under investigation with their best coupling G protein varied over at least two orders of magnitude, being lowest for M2–R–Go protein complexes (lifetime only a few seconds) (SI Appendix, Table S2) and highest for TP-R–G13 and β-AR–Gs complexes (lifetime >200 s) (SI Appendix, Fig. S1 A, C, and E), which likely reflect the parallel evolution of the coupling mechanism (49). In light of the recent advances in understanding the allosteric effect of G protein binding (or mimicking nanobodies) to the M2–R–agonist interaction, specifically the closure of a tyrosine lid on the extracellular side of the agonist exit path (13, 50, 51), our results that M2–R–Go interaction is very short lived may be somewhat unexpected. However, they are not contradictory to published results, as the lifetime of M2–R–Go complexes was not addressed in previous studies. The reliability of the method that we developed was also confirmed by testing other G-protein classes. For instance, we were unable to detect Gs or G13 binding to M3–R or M1–R under nucleotide-depleted conditions (Fig. 2A and SI Appendix, Fig. S10B) or to observe activation of these G proteins measured by means of FRET (SI Appendix, Fig. S2).
Furthermore, we screened other Gi family proteins, and although we could not observe significant differences in the affinities of Gi proteins to M3– or M1–Rs, M2–R did show a higher specificity for Go and Gi2 over Gi1 and Gi3 (Fig. 4C and SI Appendix, Fig. S10 D and F). This distinct affinity profile of GPCR is also supported by recently delineated GPCR “fingerprints” determined as the efficiency of G-protein activation by different GPCRs (52). Moreover, recent computational and evolutionary studies assume that the selectivity mechanism of GPCRs is likely disclosed on the G-protein level (15, 49). Thus, we conclude that receptor–G-protein affinity represents a major determinant for receptor–G-protein subtype selectivity. We could essentially attribute the observed differences in the affinity of Go versus Gq toward active M3–Rs to differences in the lifetime of the complex, suggesting that the apparent on-rate of complex formation is less important for defining coupling selectivity.
The potential and distinctive advantage of our FRET-based nucleotide-free method over biochemical assays is the ability to perform experiments in a regular membrane environment. By excluding the steps of protein purification and protein reconstitution (53, 54), a remarkably high degree of signal specificity is still exhibited. This is particularly important, as G proteins are well-known to be very sensitive to detergents (55). Furthermore, this method is easily accessible to all GPCRs and G proteins and can be used as a reliable way to quantify GPCR–G-protein specificity.
Materials and Methods
Chemicals.
DMEM, FCS, PBS, penicillin/streptomycin, l-glutamine, and trypsin-EDTA were from Biochrom. Saponin was purchased from AppliChem. All other substances were purchased from Sigma-Aldrich.
Plasmids.
cDNAs for Gαq; Gαq-YFP (56); Gβ1-WT; Gγ2-WT; Gαo-YFP (C351I) (57); Gαi1 (C351I)-, Gαi2-, Gαi3- (58), and Gαo-WT (59); CFP-Gγ2 (37); and M3–R–YFP (48) were described previously. The M3–R was obtained from the Missouri S&T cDNA Resource Center. M3–R–mTurquoise fluorescent protein was cloned analogously to M3–R–YFP.
Cell Culture and Transfection.
All experiments were carried out in HEK293T cells (a gift from Dr. Martin Lohse, Institute of Pharmacology, Würzburg, Germany). Cells were cultured in high-glucose DMEM supplemented with 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, and 0.1 mg/mL streptomycin at 37 °C and 5% CO2. Cells were transiently transfected with 0.5 µg DNA (6-cm dish) of each particular mAChR-YFP, 1.5 µg DNA Gα-WT of interest (if not otherwise indicated), 0.5 Gβ1-WT, 0.2 CFP-Gγ2, and 0.3 pcDNA3 (if not otherwise indicated) using Effectene Transfection Reagent according to the manufacturer’s instructions (Qiagen). For the G-protein activation, HEK293T cells were transfected with (micrograms DNA, 6-cm dish) 0.5 µg M3–R–WT, 0.8 µg Gα-YFP of interest, 0.5 µg Gβ1-WT, and 0.2 µg CFP-Gγ2. Experiments were performed 48 h after transfection at room temperature on cells, which were plated into six-well plates with 25-mm coverslips (cover glasses were preincubated 30 min with poly-l-lysine) 1 d after transfection.
Permeabilization Procedure.
Coverslips with transiently transfected HEK293T cells were fixed in a microscope chamber and washed once with external buffer (137 mM NaCl, 5.4 mM KCl, 10 mM Hepes, 2 mM CaCl2, 1 mM MgCl2, pH 7.3). To permeabilize cells, coverslips were incubated for 2 min with 0.05% saponin and afterward washed five times with internal buffer (100 mM K+-aspartate, 30 mM KCl, 10 mM Hepes, 5 mM EGTA, 1 mM MgCl2, 10 mM NaCl, pH 7.35.), inducing the depletion of GTP and GDP (60).
Single-Cell FRET Imaging.
The FRET measurements were performed on single cells selected for membrane staining of YFP and CFP fluorescence. The round shape of cells was taken as an indicator of proper permeabilization. Dual-emission imaging of YFP and CFP of a single cell (or its membrane) was performed as previously described in the work by Milde et al. (36) and modified as described in SI Appendix, SI Methods. Individual traces are shown as either absolute or relative YFP/CFP emission ratio changes. Absolute changes in YFP/CFP emission ratio were calculated as the differences in average values of the last 10 s before or after an event. The relative change of FYFP/FCFP was calculated by normalization to the maximum peak after application of saturating agonist concentration relative to the G-protein– state determined by application of GTPγS. As a measure of the affinity of the G-protein–receptor complex, dissociation kinetics of the complex were measured under nucleotide-free conditions in response to agonist withdrawal. Resulting data of the offset kinetics (decrease in FRET) were fitted by a monoexponential function.
Multiple-Cell FRET Imaging.
The efficiency of G-protein activation by the receptor of interest was determined by means of a multiple-cell FRET assay in intact cells transiently transfected as previously described in the work by Frank et al. (59). Transfected HEK293T cells cultivated in 12-well strips (TC; 96-well format; Greiner Bio-One) were washed with and maintained in external buffer A (137 mM NaCl, 5.4 mM KCl, 1.0 mM MgCl2, 2.0 mM CaCl2, 10 mM Hepes, 2% Brilliant Black) at 32.5 °C. The final volume was 275 µL per well. Injections of test compounds (agonists and antagonists) were made with a volume of 22 µL added gradually with gentle up and down movements of the 12-channel injecting unit equipped with 50-mm needles (1.2-mm diameter), resulting in a 13.5-fold dilution of stock solutions. Alternatively, cells were stimulated with an overflow apparatus, allowing almost immediate (<3 s) change of the standard buffer to induce receptor stimulation (Fig. 3D). Details for the optical equipment are given in SI Appendix, SI Methods.
Data Processing.
Fluorescence intensities were acquired using the imaging software NIS-Elements advanced research (Nikon Corporation). Values are given as means ± SEM of n experiments. Other programs used for the analysis are given in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Cornelius Krasel for the fruitful discussions and helpful comments. This work was partially funded by the Deutscher Academischer Austauschdienst.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715751115/-/DCSupplemental.
References
- 1.Simon MI, Strathmann MP, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 2.Hillenbrand M, Schori C, Schöppe J, Plückthun A. Comprehensive analysis of heterotrimeric G-protein complex diversity and their interactions with GPCRs in solution. Proc Natl Acad Sci USA. 2015;112:E1181–E1190. doi: 10.1073/pnas.1417573112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schultz G, Rosenthal W, Hescheler J, Trautwein W. Role of G proteins in calcium channel modulation. Annu Rev Physiol. 1990;52:275–292. doi: 10.1146/annurev.ph.52.030190.001423. [DOI] [PubMed] [Google Scholar]
- 4.Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel M, et al. Gα13/PDZ-RhoGEF/RhoA signaling is essential for gastrin-releasing peptide receptor-mediated colon cancer cell migration. Mol Pharmacol. 2014;86:252–262. doi: 10.1124/mol.114.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allgeier A, et al. The human thyrotropin receptor activates G-proteins Gs and Gq/11. J Biol Chem. 1994;269:13733–13735. [PubMed] [Google Scholar]
- 7.Xiao R-P, Ji X, Lakatta EG. Functional coupling of the β 2-adrenoceptor to a pertussis toxin-sensitive G protein in cardiac myocytes. Mol Pharmacol. 1995;47:322–329. [PubMed] [Google Scholar]
- 8.Eason MG, Kurose H, Holt BD, Raymond JR, Liggett SB. Simultaneous coupling of α 2-adrenergic receptors to two G-proteins with opposing effects. Subtype-selective coupling of α 2C10, α 2C4, and α 2C2 adrenergic receptors to Gi and Gs. J Biol Chem. 1992;267:15795–15801. [PubMed] [Google Scholar]
- 9.Kilts JD, et al. β(2)-adrenergic and several other G protein-coupled receptors in human atrial membranes activate both G(s) and G(i) Circ Res. 2000;87:705–709. doi: 10.1161/01.res.87.8.705. [DOI] [PubMed] [Google Scholar]
- 10.Hermans E. Biochemical and pharmacological control of the multiplicity of coupling at G-protein-coupled receptors. Pharmacol Ther. 2003;99:25–44. doi: 10.1016/s0163-7258(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 11.Herrlich A, et al. Involvement of Gs and Gi proteins in dual coupling of the luteinizing hormone receptor to adenylyl cyclase and phospholipase C. J Biol Chem. 1996;271:16764–16772. doi: 10.1074/jbc.271.28.16764. [DOI] [PubMed] [Google Scholar]
- 12.Thal DM, et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Zhou X, Dai Z, Zou X. Classification of G proteins and prediction of GPCRs-G proteins coupling specificity using continuous wavelet transform and information theory. Amino Acids. 2012;43:793–804. doi: 10.1007/s00726-011-1133-6. [DOI] [PubMed] [Google Scholar]
- 15.Flock T, et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudermann T, Kalkbrenner F, Schultz G. Diversity and selectivity of receptor-G protein interaction. Annu Rev Pharmacol Toxicol. 1996;36:429–459. doi: 10.1146/annurev.pa.36.040196.002241. [DOI] [PubMed] [Google Scholar]
- 17.Dippel E, Kalkbrenner F, Wittig B, Schultz G. A heterotrimeric G protein complex couples the muscarinic m1 receptor to phospholipase C-β. Proc Natl Acad Sci USA. 1996;93:1391–1396. doi: 10.1073/pnas.93.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berstein G, et al. Reconstitution of agonist-stimulated phosphatidylinositol 4,5-bisphosphate hydrolysis using purified m1 muscarinic receptor, Gq/11, and phospholipase C-β 1. J Biol Chem. 1992;267:8081–8088. [PubMed] [Google Scholar]
- 19.Leach K, Simms J, Sexton PM, Christopoulos A. Structure-function studies of muscarinic acetylcholine receptors. Handb Exp Pharmacol. 2012;208:29–48. doi: 10.1007/978-3-642-23274-9_2. [DOI] [PubMed] [Google Scholar]
- 20.Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Regulation of polyphosphoinositide-specific phospholipase C activity by purified Gq. Science. 1991;251:804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Activation of M3 muscarinic receptors inhibits T-type Ca(2+) channel currents via pertussis toxin-sensitive novel protein kinase C pathway in small dorsal root ganglion neurons. Cell Signal. 2011;23:1057–1067. doi: 10.1016/j.cellsig.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Burford NT, Tobin AB, Nahorski SR. Differential coupling of m1, m2 and m3 muscarinic receptor subtypes to inositol 1,4,5-trisphosphate and adenosine 3′,5′-cyclic monophosphate accumulation in Chinese hamster ovary cells. J Pharmacol Exp Ther. 1995;274:134–142. [PubMed] [Google Scholar]
- 23.Wess J. Molecular basis of muscarinic acetylcholine receptor function. Trends Pharmacol Sci. 1993;14:308–313. doi: 10.1016/0165-6147(93)90049-p. [DOI] [PubMed] [Google Scholar]
- 24.Burford NT, Nahorski SR. Muscarinic m1 receptor-stimulated adenylate cyclase activity in Chinese hamster ovary cells is mediated by Gs α and is not a consequence of phosphoinositidase C activation. Biochem J. 1996;315:883–888. doi: 10.1042/bj3150883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramachandran J, et al. Structural and functional diversity of muscarinic acetylcholine receptor subtypes. Prog Clin Biol Res. 1989;289:327–339. [PubMed] [Google Scholar]
- 26.Dell’Acqua ML, Carroll RC, Peralta EG. Transfected m2 muscarinic acetylcholine receptors couple to G α i2 and G α i3 in Chinese hamster ovary cells. Activation and desensitization of the phospholipase C signaling pathway. J Biol Chem. 1993;268:5676–5685. [PubMed] [Google Scholar]
- 27.Albert PR, Robillard L. G protein specificity: Traffic direction required. Cell Signal. 2002;14:407–418. doi: 10.1016/s0898-6568(01)00259-5. [DOI] [PubMed] [Google Scholar]
- 28.Wess J. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol Ther. 1998;80:231–264. doi: 10.1016/s0163-7258(98)00030-8. [DOI] [PubMed] [Google Scholar]
- 29.Wong SK-F. G protein selectivity is regulated by multiple intracellular regions of GPCRs. Neurosignals. 2003;12:1–12. doi: 10.1159/000068914. [DOI] [PubMed] [Google Scholar]
- 30.Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Model systems for the study of seven-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- 31.De Lean A, Stadel JM, Lefkowitz RJ. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled β-adrenergic receptor. J Biol Chem. 1980;255:7108–7117. [PubMed] [Google Scholar]
- 32.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 33.Cerione RA, et al. The mammalian β 2-adrenergic receptor: Reconstitution of functional interactions between pure receptor and pure stimulatory nucleotide binding protein of the adenylate cyclase system. Biochemistry. 1984;23:4519–4525. doi: 10.1021/bi00315a003. [DOI] [PubMed] [Google Scholar]
- 34.Cerione RA, et al. Specificity of the functional interactions of the β-adrenergic receptor and rhodopsin with guanine nucleotide regulatory proteins reconstituted in phospholipid vesicles. J Biol Chem. 1985;260:1493–1500. [PubMed] [Google Scholar]
- 35.Rubenstein RC, Linder ME, Ross EM. Selectivity of the β-adrenergic receptor among Gs, Gi’s, and Go: Assay using recombinant alpha subunits in reconstituted phospholipid vesicles. Biochemistry. 1991;30:10769–10777. doi: 10.1021/bi00108a023. [DOI] [PubMed] [Google Scholar]
- 36.Milde M, Rinne A, Wunder F, Engelhardt S, Bünemann M. Dynamics of Gαi1 interaction with type 5 adenylate cyclase reveal the molecular basis for high sensitivity of Gi-mediated inhibition of cAMP production. Biochem J. 2013;454:515–523. doi: 10.1042/BJ20130554. [DOI] [PubMed] [Google Scholar]
- 37.Hein P, Bünemann M. Coupling mode of receptors and G proteins. Naunyn Schmiedebergs Arch Pharmacol. 2009;379:435–443. doi: 10.1007/s00210-008-0383-7. [DOI] [PubMed] [Google Scholar]
- 38.Jones SV, Heilman CJ, Brann MR. Functional responses of cloned muscarinic receptors expressed in CHO-K1 cells. Mol Pharmacol. 1991;40:242–247. [PubMed] [Google Scholar]
- 39.Kenakin T. Receptor theory. Curr Protoc Pharmacol. 2008;1:1.2. doi: 10.1002/0471141755.ph0102s41. [DOI] [PubMed] [Google Scholar]
- 40.Pierre N. Determination of association (kon) and dissociation (koff) rates of spiperone on the dopamine D2 receptor using a platform for GPCR applications. Am Lab. 2011;2013:2–4. [Google Scholar]
- 41.Vilardaga J-P, Bünemann M, Krasel C, Castro M, Lohse MJ. Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat Biotechnol. 2003;21:807–812. doi: 10.1038/nbt838. [DOI] [PubMed] [Google Scholar]
- 42.Stamatiou R, et al. Long-term exposure to muscarinic agonists decreases expression of contractile proteins and responsiveness of rabbit tracheal smooth muscle cells. BMC Pulm Med. 2014;14:39. doi: 10.1186/1471-2466-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- 44.Roberts DJ, Waelbroeck M. G protein activation by G protein coupled receptors: Ternary complex formation or catalyzed reaction? Biochem Pharmacol. 2004;68:799–806. doi: 10.1016/j.bcp.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 45.Kostenis E, Zeng FY, Wess J. Structure-function analysis of muscarinic acetylcholine receptors. J Physiol Paris. 1998;92:265–268. doi: 10.1016/s0928-4257(98)80030-2. [DOI] [PubMed] [Google Scholar]
- 46.Higashijima T, Ferguson KM, Sternweis PC, Smigel MD, Gilman AG. Effects of Mg2+ and the β γ-subunit complex on the interactions of guanine nucleotides with G proteins. J Biol Chem. 1987;262:762–766. [PubMed] [Google Scholar]
- 47.Mukhopadhyay S, Ross EM. Rapid GTP binding and hydrolysis by G(q) promoted by receptor and GTPase-activating proteins. Proc Natl Acad Sci USA. 1999;96:9539–9544. doi: 10.1073/pnas.96.17.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann C, et al. Comparison of the activation kinetics of the M3 acetylcholine receptor and a constitutively active mutant receptor in living cells. Mol Pharmacol. 2012;82:236–245. doi: 10.1124/mol.112.077578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flock T, et al. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–322. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeVree BT, et al. Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature. 2016;535:182–186. doi: 10.1038/nature18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carr R, 3rd, et al. Interdicting Gq activation in airway disease by receptor-dependent and receptor-independent mechanisms. Mol Pharmacol. 2016;89:94–104. doi: 10.1124/mol.115.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee E, Linder ME, Gilman AG. Expression of G-protein α subunits in Escherichia coli. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- 54.Linder ME, Ewald DA, Miller RJ, Gilman AG. Purification and characterization of Go α and three types of Gi α after expression in Escherichia coli. J Biol Chem. 1990;265:8243–8251. [PubMed] [Google Scholar]
- 55.Sýkora J, Bouřová L, Hof M, Svoboda P. The effect of detergents on trimeric G-protein activity in isolated plasma membranes from rat brain cortex: Correlation with studies of DPH and Laurdan fluorescence. Biochim Biophys Acta. 2009;1788:324–332. doi: 10.1016/j.bbamem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 56.Hughes TE, Zhang H, Logothetis DE, Berlot CH. Visualization of a functional Gαq-green fluorescent protein fusion in living cells. J Biol Chem. 2001;276:4227–4235. doi: 10.1074/jbc.M007608200. [DOI] [PubMed] [Google Scholar]
- 57.Bünemann M, Frank M, Lohse MJ. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci USA. 2003;100:16077–16082. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wise A, Watson-Koken M-A, Rees S, Lee M, Milligan G. Interactions of the α2A-adrenoceptor with multiple Gi-family g-proteins: Studies with pertussis toxin-resistant g-protein mutants. Biochem J. 1997;321:721–728. doi: 10.1042/bj3210721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frank M, Thümer L, Lohse MJ, Bünemann M. G Protein activation without subunit dissociation depends on a Gα(i)-specific region. J Biol Chem. 2005;280:24584–24590. doi: 10.1074/jbc.M414630200. [DOI] [PubMed] [Google Scholar]
- 60.Hommers LG, Klenk C, Dees C, Bünemann M. G proteins in reverse mode: Receptor-mediated GTP release inhibits G protein and effector function. J Biol Chem. 2010;285:8227–8233. doi: 10.1074/jbc.M109.015388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolters V, Krasel C, Brockmann J, Bünemann M. Influence of Gαq on the dynamics of m3-acetylcholine receptor-G-protein-coupled receptor kinase 2 interaction. Mol Pharmacol. 2015;87:9–17. doi: 10.1124/mol.114.094722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.