Significance
Nonmodel bacteria have essential roles to play in the future development of biotechnology by providing new sources of biocatalysts, antibiotics, hosts for bioproduction, and engineered “living therapies.” The characterization of such hosts can be challenging, as many are not tractable to standard molecular biology techniques. This paper presents a rapid and automated methodology for characterizing new DNA parts from a nonmodel bacterium using cell-free transcription–translation. Data analysis was performed with Bayesian parameter inference to provide an understanding of gene-expression dynamics and resource sharing. We suggest that our integrated approach is expandable to a whole range of nonmodel bacteria for the characterization of new DNA parts within a native cell-free background for new biotechnology applications.
Keywords: in vitro transcription–translation, cell-free synthetic biology, Bacillus, modeling, automation
Abstract
Native cell-free transcription–translation systems offer a rapid route to characterize the regulatory elements (promoters, transcription factors) for gene expression from nonmodel microbial hosts, which can be difficult to assess through traditional in vivo approaches. One such host, Bacillus megaterium, is a giant Gram-positive bacterium with potential biotechnology applications, although many of its regulatory elements remain uncharacterized. Here, we have developed a rapid automated platform for measuring and modeling in vitro cell-free reactions and have applied this to B. megaterium to quantify a range of ribosome binding site variants and previously uncharacterized endogenous constitutive and inducible promoters. To provide quantitative models for cell-free systems, we have also applied a Bayesian approach to infer ordinary differential equation model parameters by simultaneously using time-course data from multiple experimental conditions. Using this modeling framework, we were able to infer previously unknown transcription factor binding affinities and quantify the sharing of cell-free transcription–translation resources (energy, ribosomes, RNA polymerases, nucleotides, and amino acids) using a promoter competition experiment. This allows insights into resource limiting-factors in batch cell-free synthesis mode. Our combined automated and modeling platform allows for the rapid acquisition and model-based analysis of cell-free transcription–translation data from uncharacterized microbial cell hosts, as well as resource competition within cell-free systems, which potentially can be applied to a range of cell-free synthetic biology and biotechnology applications.
Cell-free transcription–translation systems use crude cell extracts (1–3) or purified components (4) to synthesize proteins encoded by plasmid DNA or linear PCR products. Recently, a renaissance in cell-free systems for synthetic biology has led to applications, such as prototyping genetic pathway designs (5, 6), medical viral biosensors (7), antibody production (8), and the engineering of microfluidic biochip devices (9).
One underexplored area is the use of cell-free systems to study native cellular machinery and gene-expression elements from difficult/intractable nonmodel microbial hosts; we conceptualize this approach as native cell-free (NCF) transcription–translation, hereafter simply referred to as NCF. For example, organisms adapted for growth in extreme environments that are able to produce valuable biomolecules at high yields from inexpensive substrates are of particular interest for the future engineering of cell factories (10, 11). Other potential areas of interest include the use of nonmodel organisms for novel antibiotic discovery (12, 13), while for healthcare applications there is great interest in developing new “living therapies” for the engineering of the human microbiome, which is comprised of thousands of microbial species, many of which remain uncharacterized (14). Currently, many nonmodel microbial systems are neither amenable to genetic manipulation nor fully characterized, and the range of genetic parts (e.g., promoters, transcription factors) can be limiting. Furthermore, strain engineering for bioproduction can require long design cycles (Fig. 1A). In contrast, NCF provides a rapid method to study gene expression using the endogenous components for transcription, translation, and energy regeneration (6, 15, 16), while DNA parts, such as promoters, have shown high correlation between in vivo and in vitro activity (17). In general, these studies have largely focused on the use of well-characterized Escherichia coli cell-free transcription–translation platforms and there is little work on using nonstandard microbial hosts.
Fig. 1.
Cell-free prototyping of a nonmodel microbe. (A) Testing of synthetic gene-expression plasmids in NCF using endogenous energy regeneration and transcription–translation components. (B) Parallel transcription–translation measurements with the MGapt (mRNA) aptamer and GFP in B. megaterium NCF. (C) A semiautomated workflow incorporating liquid-handling robotics for rapid screening of cell-free reactions (RXNs).
Bacillus megaterium is a giant Gram-positive bacterium with a rich history in industrial biotechnology (18), which includes the production of penicillin G amidase (19), β-amylases, and vitamin B12 (20, 21). B. megaterium is relatively uncharacterized due to a low-efficiency protoplast transformation procedure that takes 3 days to complete (22). However, in comparison with the highly characterized Bacillus subtillis, B. megaterium provides major advantages, such as stable plasmid maintenance (23), minimal neutral-alkaline protease activity (24), and the ability to metabolize low-cost substrates (25). Moreover, B. megaterium also offers a strong native Sec-dependent secretion apparatus (22), which is a desirable feature for downstream processing, while for recombinant gene expression the xylose-inducible promoter system (26, 27) produces recombinant proteins to the gram per liter scale (22), which is comparable to genetically enhanced commercial E. coli strains, such as the BL21 and Rosetta. A recent and bespoke application of its use includes characterizing the vitamin B12 pathway (21, 28) along with isolating soluble vitamin B12-dependent enzymes (29, 30). Previous attempts to isolate these proteins from E. coli, resulted in the production of inclusion bodies or inactive enzymes (21, 29, 30).
In this study, we demonstrate how readily a B. megaterium NCF transcription–translation system can be established and used as a platform to prototype gene-expression elements from this host. We have also combined this cell-free system with factorial experimental design, acoustic liquid-handling robotics with nanolitre controllability, and Bayesian model parameter inference methods to demonstrate a rapid prototyping platform to enable the rigorous characterization of gene-expression tools within hours, in comparison with weeks for in vivo analyses (Fig. 1 A and B). A significant advantage of cell-free systems is that the defined reaction conditions permit the accurate characterization of genetic parts and resource usage through mathematical modeling techniques (6, 9, 15, 31). However, such approaches have previously generally been applied to E. coli gene-expression circuits and none have used Bayesian statistical inference methods. The ordinary differential equations (ODE) models used in this work have a large number of correlated but uncertain kinetic parameters with complicated likelihood function surfaces. Bayesian methods are ideally suited to tackling these types of difficult parameter inference problems, where maximum-likelihood point estimates could be misleading, by treating the model parameters themselves as being random variables with defined probability density functions. This provides a simple and consistent framework to quantify how our knowledge of the model parameters has increased with each experimental measurement (i.e., the probability density functions become more narrowly distributed as we gain information). Robustly parameterized genetic parts are a prerequisite of computational genetic circuit design (32).
Here, we have applied a Bayesian parameter inference scheme to model the obtained experimental data, aided by quantitative proteomics, to rigorously infer unknown kinetic parameters describing the cell-free reaction. We tested a selection of Sigma A (σA) constitutive promoters from central energy pathways and also validated a xylose-inducible promoter using an automated factorial experimental design process (Fig. 1C), experimental set-up, data acquisition, and analysis. As part of this work, we have also released a general open-source C++ framework (ODE_MCMC_tools) for Bayesian parameter inference for systems of ODEs. The development of a B. megaterium NCF transcription–translation system serves as an exemplar test case for rapidly prototyping novel genetic regulatory elements from poorly characterized hosts, thus providing a basis for the future engineering of this nonmodel organism. We also suggest that this approach is expandable to a range of other important microbial chassis currently being explored for synthetic biology applications.
Results
Isolation of a Strong Constitutive Promoter and NCF Assay Optimization.
To initially test and optimize a NCF transcription–translation system in B. megaterium, we first isolated and characterized a strong constitutive σA promoter from B. megaterium. Previously, a strong σA promoter was isolated from the pyruvate dehydrogenase (pdh) operon in B. subtillis (33). For B. megaterium, we isolated the equivalent promoter from the pdhABCD (bmd_1326-1329) gene cluster by PCR along with the ribosome binding site (RBS) of pdhA (bmd_1326), and used this to control the expression of gfp (pRBBm258) or mCherry (pRBBm267). To initially validate the performance of gene expression in vivo, the optimized xylose-inducible promoter plasmid GFP variants (22) were used for comparison and tested under plate-reader conditions. As expected, the growth rate of the empty vector control and noninduced xylose promoter recombinant strains was faster than the pdh or xylose-promoter strains, with all strains reaching the stationary phase after ∼10 h of growth (SI Appendix, Figs. S3 and S4). A qualitative assessment of GFP and mCherry production was also verified by SDS/PAGE (SI Appendix, Fig. S1).
Next, to test constitutive and inducible promoter systems in vitro, a B. megaterium cell-free extract was prepared using the E. coli S30 protocol (34) with some optimization (SI Appendix, Extended Materials and Methods and Figs. S5–S7). Importantly, we have now found this protocol to be generally adaptable to a selection of Gram-positive bacterial species, including B. megaterium (35, 36). Using this protocol, up to 4.96 μM of GFP was synthesized within a 4-h batch reaction using the strong PDH (pRBBm258) plasmid.
LC-MS/MS Quantitation of NCF Transcription–Translation Enzymes.
To initially parameterize the B. megaterium NCF extract, we next quantified a selection of proteins from transcription–translation. For E. coli cell-free, this is well-characterized (1, 37). However, for nonmodel microbes, such as B. megaterium, this is generally not the case. From E. coli studies, cell-free protein synthesis is restricted by a combination of limiting factors, such as active 70S ribosomes, RNA polymerase (RNAP) holoenzyme, energy, and substrate resources, with NTP regeneration and inorganic phosphate accumulation thought to be major limiting factors under batch-synthesis mode (2, 38). Transcription and translation processes require ∼36 proteins and 70S ribosomes to undertake these reactions (4). In this work, we selected key marker proteins for quantitation including the ribosomal proteins (30S and 50S subunits), RNAP, and the σA factor, using targeted LC-MS/MS (SI Appendix, Figs. S25–S27). We also measured the methionyl-tRNA (Met-tRNA) and alanyl-tRNA (Ala-tRNA) synthetases because they encode for both low- and high-frequency amino acids, respectively (39). In comparison with E. coli measurements (1, 40), the values we obtained were similar (SI Appendix, Fig. S25 and Table S5). For example, the average 30S and 50S ribosome content (S4, S7, L2, and L4 ribosomal proteins) was determined at 2.18 ± 0.86 μM across three biological replicates in a standard 10 mg·mL−1 cell-extract reaction. For E. coli cell-free systems, active ribosomes range from 1.6 to 2.3 μM (1, 40) under the same cell-extract concentration (10 mg·mL−1). Our total protein quantitation is likely to provide an overestimate, since a fraction (∼20–30%) of ribosomes are dissociated as 50S and 30S subunits (41). In addition, we calculated the RNAP β-subunit at 0.41 ± 0.15 μM and the σA factor at 53 ± 30 nM. The two aminoacyl tRNA synthetases (methionyl- and alanyl-) we selected ranged from 0.13 to 0.22 μM.
Experimental Characterization of the Model Xylose-Inducible Promoter System by Acoustic Liquid-Handling Robotics.
Before characterizing unknown gene-expression elements, we first investigated the relatively well-characterized model xylose-repressor system native to B. megaterium. Here we analyzed its regulatory interactions using liquid-handling robotic-assisted experiments and model-based analyses with Bayesian parameter inference. We inferred parameters using quantitative time-course data for both mRNA levels (with the malachite green aptamer) and protein levels (using the GFP fluorescent reporter), as well as incorporating the proteomic parameters obtained earlier by LC-MS/MS. We then carried out two separate experiments with the xylose-inducible promoter (described below) to obtain both quantitative mRNA/protein data and xylose-repressor system behavior for our modeling analyses.
The xylose-inducible promoter is derived from the native B. megaterium xylABT operon, which encodes the xylose isomerase (xylA), xylulokinase (xylB), and permease (xylT) genes (42, 43). To characterize the kinetics of the xylose promoter, we used our cell-free system to simultaneously monitor transcription and translation in vitro. To monitor mRNA expression, we modified the 3′UTR of the xylose-inducible promoter plasmid (pKMMBm5) to provide real-time fluorescent measurement of mRNA synthesis as previously described (5, 16) (SI Appendix, Figs. S11–S16). First, using an in vitro-synthesized GFP-MGapt transcript, we measured the fluorescence for this aptamer in a B. megaterium cell-free reaction for 2 h to generate a calibration curve and observed that mRNA degradation followed a single-phase exponential decay rate (SI Appendix, Figs. S13 and S14). The half-life for this transcript was estimated at 15.6 min, which is similar (18 min) to previously observed E. coli cell-free measurements (6). In a parallel time-course measurement, qRT-PCR confirmed that actual mRNA levels correlated well with real-time fluorescence measurement (SI Appendix, Fig. S15).
We next tested a range of DNA concentrations in cell-free reactions with simultaneous measurement of MGapt and GFP fluorescence (Fig. 2). With an increasing DNA concentration of pKMMBm5-MGapt, the rate of MGapt fluorescence rose rapidly to a peak concentration of ∼235 nM of GFP-MGapt transcript at 25–27 min after the start of incubation (Fig. 2). Thereafter, the signal decays suggesting that NTP levels become limiting for further mRNA synthesis. In fact, spiking the reaction with a purified GFP-MGapt transcript, shows that mRNA concentrations decay more slowly throughout the assay when expressed from plasmid DNA, suggesting that continuous transcription provides mRNA substrate for translation throughout the reaction time period (SI Appendix, Fig. S16). Additionally, we also independently verified that the extracts displayed undetectable interference from native XylR (SI Appendix, SI Text and Fig. S28).
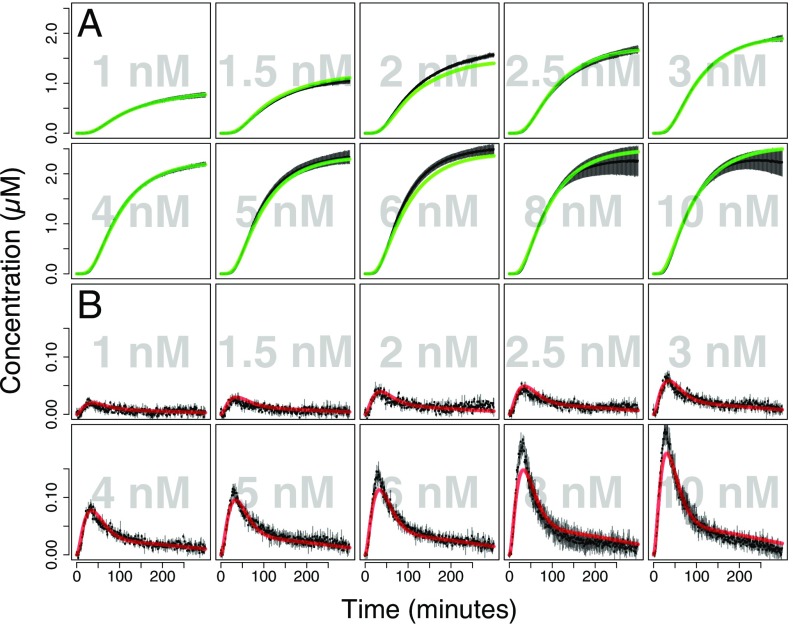
Fig. 2.
Transcription and translation of the xylose-inducible promoter. Cell-free extracts (10 mg·mL−1) were incubated at 30 °C for 6 h with a range of DNA concentrations (1–10 nM) of the pKMMBm5-MGapt plasmid. Fluorescence data were collected every 60 s for (A) GFP (protein) and (B) MGapt (mRNA) signals and displayed as black points in the plot with gray bars representing SE. Experimental data were modeled using ordinary differential equations with a system of 14 species and 26 parameters (SBML model available in the GitHub software repository, see Methods), and parameters were inferred using MCMC (SI Appendix, Fig. S18 and Table S9). Simulated trajectories using these inferred parameters are shown with green (for GFP) and red (mRNA) lines.
A second, larger-scale experiment was carried out to separately characterize the XylR promoter in cell-free reactions. This experiment was designed to simultaneously monitor GFP translation in 108 unique conditions in triplicate (324 total reactions) made up of an augmented full factorial combination of different concentrations of purified recombinant XylR repressor protein (0–1,000 nM) (SI Appendix, Figs. S18–S22), d-xylose (0–1,000 μM), and pKMMBm5-MGapt DNA template (1, 2, and 5 nM) in a 384-well microtiter plate using an acoustic liquid-handling robot to provide nanolitre control of liquid transfers (Fig. 1C). Factorial experimental design and transfer instructions for the robot were automatically generated using a Python script (available in the GitHub repository, see Methods). In summary, when XylRHis is titrated in the absence of d-xylose, a repression effect on GFP expression is observed, as expected (Fig. 3A). Following the titration of d-xylose in the presence of 0.1–1.0 μM XylRHis, GFP synthesis is recovered, which is consistent with the release of the XylRHis repressor from the promoter region upon binding of d-xylose.
Fig. 3.
Cell-free quantitative characterization of the xylose-inducible promoter system. d-xylose (mM), purified XylRHis (µM), and pKMMBm5 plasmid DNA (nM) was titrated into a cell-free reaction using an acoustic liquid-handling robot with full factorial experimental design and transfer instructions automatically generated using a Python script (SI Appendix, SI Text), giving a total of 108 unique conditions in triplicate (324 reactions). The full experimental time-course data (SI Appendix, Figs. S19–S21) from the xylose experiment were simultaneously used to infer ODE model parameters (SI Appendix, Fig. S22 and Table S9), but for simplicity only end-point values are shown in this figure. (A) Experimentally measured end-point GFP concentrations are shown as black points, the green surface contour map represents simulated end-point GFP values as a function of XylRHis and xylose concentrations. Differences between the experimental values and the simulated values are displayed as vertical black lines. (B) The inferred univariate and bivariate marginal posterior distributions (the diagonal and off-diagonal plots, respectively) over the KD and Hill-coefficients for binding of XylR and xylose (see SI Appendix, Fig. S22 for the full posterior over all model parameters). The posterior distribution is the inferred probability density function of the model parameters given the experimental data. The points were sampled from the posterior using MCMC and are colored by local point density.
Mathematical Modeling of the Xylose-Repressor System.
To date, the binding affinities and Hill coefficients of the XylR repressor protein to DNA and the inducer (xylose) to the repressor protein haven’t been experimentally determined. To estimate these and other unknown parameters, we modeled the system using a coarse-grained system of ODEs describing cell-free transcription–translation (Methods and SI Appendix), with 18 chemical reactions, 14 species, and 26 parameters (SI Appendix, Table S9). For the xylose-inducible promoter, the initial concentration of free DNA was approximated using the Hill equation. The mRNA degradation reaction was assumed to be a first-order reaction. The translation reaction produced nonfluorescent immature GFP and this was assumed to mature in a first-order reaction into the detectable fluorescent species, GFPmat.
Initial concentrations for all species other than NTP, the secondary energy source, ribosome, and RNAP were set to zero. The LC-MS/MS determined concentrations for RNAP:σA holoenzyme and the ribosome 70S complex were incorporated as upper bounds for initial values during parameter inference (0.08 and 2.5 μM, respectively). Although initial NTP concentrations in the reaction are known, the overall NTP capacity of the system was unknown as the efficiency of NTP regeneration from the secondary energy source was unknown. For this reason, the initial concentrations of NTP and the secondary energy source were inferred. In all experimental conditions, the mature GFP concentration was measured during the course of the reaction. However, where the mRNA concentrations were also experimentally determined, this was also included in the log-likelihood function.
Model parameters were first inferred from the DNA titration dataset from both GFP and mRNA time-course observations using our ODE_MCMC_tools software (Methods). The posterior distribution from the first experiment (SI Appendix, Fig. S18 and Table S9) was then used as a prior distribution for parameter inference on the full factorial dataset. Multiple Markov chain Monte Carlo (MCMC) runs were generated from different parameter set starting points and convergence was confirmed for all parameters using the Gelman–Rubin diagnostic (44). Simulated trajectories from the model closely matched the experimental time-course data for both GFP and mRNA concentration at different DNA template concentrations (Fig. 2), while the posterior distribution was found to be unimodal (SI Appendix, Fig. S18). The translational machinery appeared to be saturated at around 5-nM template DNA with no further increase in total protein produced; however, mRNA expression continued increasing with higher DNA concentrations. This implied that the ribosomes were saturated with mRNAs but the host RNAPs were not saturated with DNA template.
Over the diverse range of conditions in the full factorial XylR/xylose experiment, both the simulated end-point GFP concentrations (Fig. 3A) and the time-course trajectories were found to fit the experimental data remarkably well (SI Appendix, Figs. S19–S21). The posterior distribution for these experiments was also found to be unimodal and confidence intervals were determined (Fig. 3B and SI Appendix, Fig. S22 and Table S9). The maximal translation rate was inferred to be 5.4–6.3 amino acids min−1 (95% confidence intervals), which is lower than previously estimated values for E. coli cell-free reactions (45), while the maximal transcription rate was inferred at between 488 and 688 bp min−1, similar to previously estimated values for E. coli cell-free (300–600 bp min−1) (1). Previously, the maturation rate of GFP+ (a synonym for GFPuv3) was measured to be 0.066 min−1 (46). Here, we inferred the maturation rate to be between 0.058 and 0.063 min−1 without taking into account any prior knowledge. The difference may reflect experimental inaccuracies, simplicities in the model, or indicate that a lower oxygen concentration was limiting maturation in the reaction mixture. Moreover, the XylR repressor was found to cooperatively bind to its operator sequence (dissociation constant 12.9–14.2 nM and Hill coefficient 1.74–1.8), while xylose appeared to cooperatively bind XylR (dissociation constant 12.2–13.5 μM and Hill coefficient 1.44–1.49). In summary, our modeling approach provides a detailed kinetic analyses of the XylR promoter and repressor without any prior knowledge, illustrating the potential of integrating Bayesian inference modeling into NCF part characterization.
Resource Competition and Energy Usage in Our NCF System.
We next wanted to identify the strengths and limitations within our new NCF system by focusing on RNAP/ribosome availability and energy consumption. We anticipated that by providing a rigorous characterization of our NCF system, we would be able to properly assess the limits of quantifying new DNA parts in B. megaterium NCF extracts. To achieve this, we designed a competition experiment described below.
To gain insight into the sharing of resources in our NCF system, we investigated the simultaneous expression of two different proteins, both under the control of a strong constitutive promoter, inspired by previous studies (47). Our plasmid constructs allowed the simultaneous real-time measurement of GFP, RFP, and the GFP mRNA transcript concentrations. The two plasmids were then titrated together into cell-free reactions at different plasmid concentrations (from 0 to 40 nM) (Fig. 4). It was found to be difficult to avoid leakage from the mCherry fluorescence signal into the MGapt signal (SI Appendix, Fig. S30); however, a peak for the mRNA signal was observed at around 50 min, before significant mature mCherry was detected around 90 min. In reactions where no mCherry plasmid was present, the full mRNA time-course signal was used in parameter inference. Where the mCherry plasmid was present, the mRNA MGapt data up to 90 min was used.
Fig. 4.
Competition for cell-free shared resources. GFP (pdh-RiboJ-RBS-GFP-MGapt-Bba_B0015) and mCherry (pdh-RiboJ-RBS-mCherry-Bba_B0015) encoding plasmid DNA were simultaneously titrated into cell-free reactions at a range of concentrations from 0 to 40 nM. The light green points (with light green error bars indicating SEs) represent the experimentally measured GFP concentrations, while the dark green lines represent the simulated trajectories. The light red points (with light red error bars indicating SE) represent the experimentally measured mCherry concentrations, while the dark red lines represent the simulated trajectories. In reactions where no mCherry plasmid was present, the full mRNA time course signal was used in parameter inference. Where the mCherry plasmid was present, the mRNA MGapt data up to 90 min was used. The system was modeled using an ODE model with 29 species and 31 parameters (an SBML model is available in the GitHub software repository, see Methods). Model parameters were simultaneously inferred from all experimental data points (GFP mRNA time-course data are shown in SI Appendix, Fig. S30) using MCMC (SI Appendix, Fig. S29 and Table S10).
As in the previous XylR experiment, our ODE_MCMC_tools software was used to infer model parameters. Based on the reaction scheme described in Methods, a new ODE model was created to account for the simultaneous expression of two proteins from two different promoters in the same reaction. This model included accounting for the use of shared resources (NTPs, amino acids, RNAP, and ribosomes) and resulted in a model with 31 parameters and 29 species. The LC-MS/MS determined concentrations for the RNAP:σA holoenzyme and the ribosome 70S complex were incorporated as upper bounds for initial values during parameter inference (0.08 and 2.5 μM, respectively). Model parameters were inferred simultaneously from the full time-course datasets using MCMC. The two-promoter model with full resource accounting, was able to simultaneously fit the 36 different experimental conditions with time-course data for mature GFP, mature mCherry, and GFP mRNA species (Fig. 4 and SI Appendix, Fig. S30). The maturation kinetics of mCherry are not as well understood as GFP, and the fluorescence curves can be seen to have a significantly longer lag-time. For this reason, mCherry maturation was modeled to have a three-step maturation process similar to its progenitor, dsRed (48). The mCherry mRNA species were unobserved and were assumed to have the same degradation rate as the GFP mRNA species. Transcription elongation rate was assumed to be the same for both transcripts, but other parameters related to transcription and translation were not assumed to be identical. Nevertheless, as would be expected given that the same RiboJ-insulated pdh promoter was used to express both proteins, transcriptional parameters (promoter escape rate and promoter binding constant) were inferred to fall within similar ranges (SI Appendix, Table S10).
The effect of competition for both transcription and translational resources can be seen experimentally. Where both promoters were competing for RNAPs, the levels of GFP mRNA decreased, implying that the transcriptional machinery is fully saturated (SI Appendix, Fig. S30). Taking the most transcriptionally burdened condition (40 nM pdh-RiboJ-GFP-MGapt and 40 nM pdh-RiboJ-mCherry plasmid concentrations), we find that our model predicts that increasing the transcription elongation rate and increasing the RNAP concentration can increase the total amount of translated amino acids, suggesting there is some spare translational capacity in this condition and that, in some circumstances, the transcriptional capacity of the system can be the limiting factor (SI Appendix, Fig. S32).
Parameters related to protein translation were found to vary a lot more widely, with an order-of-magnitude difference in translation elongation rate and almost two orders-of-magnitude difference in the RBS binding constant (SI Appendix, Table S10). The maximal final level of mCherry synthesis was ∼0.4 μM for a Bacillus codon-optimized variant (low G + C%), with a high G + C% variant providing up to 0.09 μM of mCherry for comparison (SI Appendix, Fig. S7). In vivo synthesis of mCherry was also comparatively lower than GFP (SI Appendix, Fig. S1). This suggests that mCherry translation elongation is rate-limiting and therefore creating more burden to the translational machinery. To investigate this further, 500 random parameter sets were drawn from the posterior distribution and simulated free-ribosome concentration trajectories were plotted as a function of the 36 different experimental conditions (SI Appendix, Fig. S33). In all experimental conditions, the model predicts a minimum in the concentration of free ribosomes at around 50 min into the reaction after an initial lag-phase where mRNAs are increasing in concentration and while there is still a high NTP concentration available for translation. It can be seen that the pdh-RiboJ-mCherry plasmid exerts a greater burden on the ribosomes, even though total protein expression from this plasmid is much lower than for the pdh-RiboJ-GFP-MGapt plasmid. Our model suggests that this is due to the longer ribosome residency time on the mCherry mRNA during the elongation process, predicting a much higher mean number of ribosomes per polysome for the mCherry mRNA than for the GFP mRNA (SI Appendix, Figs. S34 and S35). We also find that only around 6–7% of the total theoretical energy input was accessible to the system, with only 0.6–0.7% of the theoretical energy input directed to protein translation. A detailed analysis of the energy usage can be found in SI Appendix, SI Text, Fig. S36, and Table S10. In summary, our detailed characterization illustrates how important a full kinetic understanding of a particular NCF system and reporter protein is for robust measurements of new parts in a high-throughput context.
Characterization of B. megaterium Promoter Parts in Vitro and in Vivo.
After testing the transcriptional, translational, and energy resource limitations of the B. megaterium NCF system, we next sought to use our platform to identify new DNA parts for synthetic biology applications. The primary advantage of developing a NCF platform in a nonmodel host is to characterize new gene-expression elements using the endogenous polymerase and σ factors from the native host. Additionally, we also measured the promoter activities in vivo. Previously, we have demonstrated that E. coli promoter activity correlates between in vivo and in cell-free–based measurement (17).
To provide a proof-of-concept as to whether this trend is reproducible in another cell-free host, a small selection of uncharacterized B. megaterium glycolysis and gluconeogenesis promoters were chosen for promoter testing (SI Appendix, SI Text). Unlike some better-characterized microbes, no prior proteomic or RNAseq information was available to guide this selection process. However, we selected a set of promoters from central metabolism, which share some partial conservation to the −10 and −35 consensus (SI Appendix, SI Text) sequence (−35TTGACA and −10TATAAT). This represents a characteristic recognition motif for transcription initiation by the σA protein, the major growth regulatory factor of Bacillus species (49). This indicated that the B. megaterium promoters are putative σA promoters. In addition, the pdh promoter used in the competition experiments, was used as a positive control for a strong σA constitutive promoter.
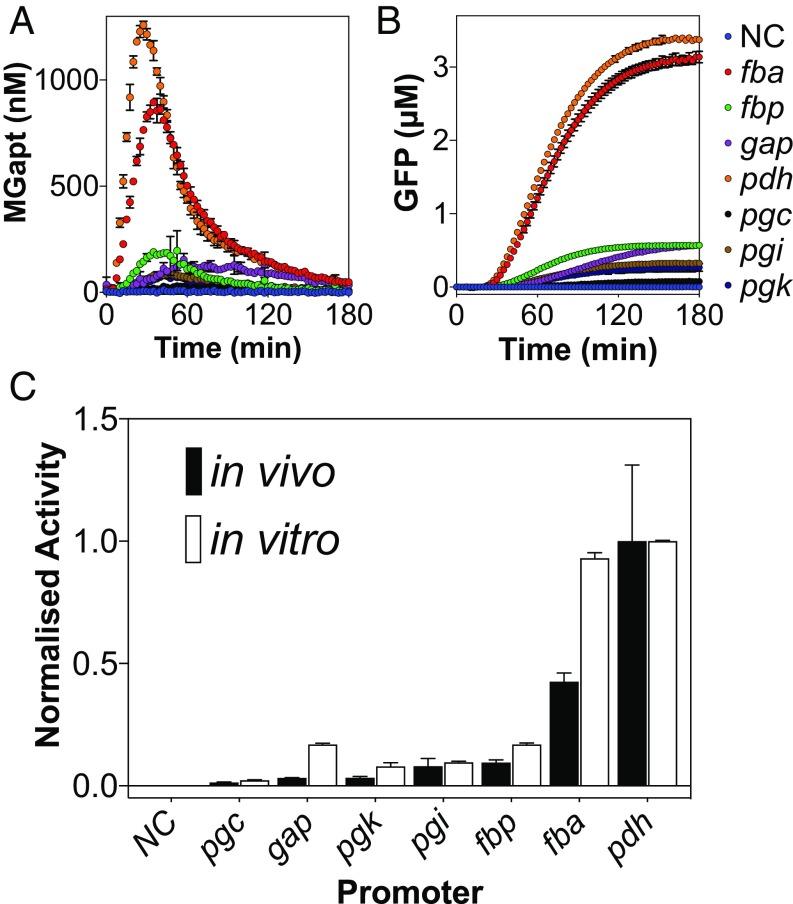
To test these promoters under the same context, the well-characterized RiboJ insulator (50) was introduced and assembled by EcoFlex (51) with an RBS (derived from pKMMBm5), GFP, MGapt, Bba_B0015 terminator, and Bacillus shuttle vector backbone (tetA-rebB). For cell-free, the strongest levels of mRNA and GFP were produced with the pdh and fba promoters (SI Appendix, Figs. S8, S9, and S37), with pdh achieving a maximum GFP yield of 3.40 μM with 20 nM of plasmid DNA (Fig. 5 A and B). The other promoters, gap, fbp, pgi, and pgk, displayed medium strength activities with an end-point range of 0.20–0.61 μM GFP synthesized. The pgc promoter produced the weakest end-point yield of 0.08 μM GFP, while the activities of the pyk and tpi promoters were not detectable. These observations were also proportional to the mRNA signal (SI Appendix, Figs. S8 and S9). However, while pdh and fba demonstrated similar levels of GFP expression, pdh showed stronger mRNA expression (Fig. 5A and SI Appendix, Fig. S8), suggesting that cell-free translation is saturated with the strongest promoter, pdh.
Fig. 5.
σA Constitutive promoter activity correlates between in vivo and cell-free transcription–translation characterization. (A and B) RNA and GFP cell-free time-course of σA constitutive promoters. (C) Normalized cell-free and in vivo promoter characterization data. SD is representative of three measurements in cell-free, and four biological replicates in vivo. NC, negative control.
In comparison, in vivo promoter activities displayed a similar trend to the cell-free data (Fig. 5B). The promoters fbp, gap, pgc, pgi, and pgk were found to be relatively low strength (Fig. 5 and SI Appendix, Figs. S8 and S9), while pyk and tpi were below the limit of detection (see SI Appendix, SI Text for further discussion). Overall, the expression levels of the tested promoters (pdh, fba, fbp, gap, pgc, pgi, and pgk) showed a statistically significant correlation coefficient of 0.916 (P = 0.0001) (SI Appendix, Fig. S10), providing a further proof-of-concept of a relationship between in vivo and cell-free measured promoter activities in a prokaryotic system.
Semiautomated Screening of a Synthetic RBS Library.
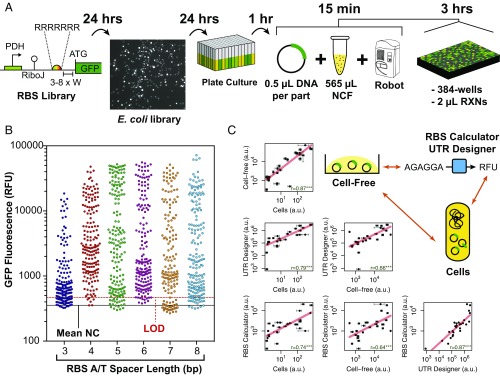
To demonstrate robustness and scalability of our NCF system, we developed a semiautomated workflow for screening libraries of biological parts using an acoustic liquid-handling robot (Fig. 6A). For E. coli and B. subtilis, the RBS is well-characterized, unlike B. megaterium, where only a handful have been tested for recombinant protein expression, providing an ideal test case for our platform (18). The RBS is composed of a semiconserved polypurine motif (5′-AGGAGGA-3′) Shine–Dalgarno sequence, followed typically by an A/T-rich spacer of 2–10 nucleotides. In addition, a spacer range of 5–10 nucleotides for E. coli and B. subtilis was previously determined as being optimal for translation initiation (52).
Fig. 6.
Rapid quantification of RBS library parts in B. megaterium NCF. (A) A semiautomated workflow and time-scale from library generation to cell-free screening of RBS activities. [The image in the second place in A was taken with a Gel Doc XR (Bio-Rad Laboratories, Hercules, CA) at ∼10× magnification.] (B) Distribution of library groups (RBS-3, -4, -5, -6, -7, and -8) rapidly screened in 2-μL reactions in 384-well plates. Solid black line represents mean negative control (NC) signal and dotted red line represents limit of detection (3σ from mean NC signal). Four measurement repeats per RBS variant (44 per group) were randomly distributed within the plate by the Echo PickList software. (C) Correlation of cell-free, in vivo, and in silico prediction (54, 55) activities of 26 RBS-6 variants with correlation coefficients and statistical significance shown within each individual plot. For full data, please see SI Appendix, Figs. S38 and S39 and Tables S12 and S13.
To generate a synthetic RBS library, a degenerate poly-R (A/G) Shine–Dalgarno sequence and poly-W (A/T) spacer varying from 3 to 8 nucleotides was designed and combined with the strong PDH promoter described earlier in this paper. Libraries with varying spacer sequence lengths (3–8 bases, termed RBS-3, -4, -5, -6, -7, and -8) were generated using PCR and individually screened in NCF. The upper library sizes for the RBS-3 and RBS-8 libraries were 1,024 (210) and 32,768 (215) combinations, respectively. These libraries were transformed into E. coli and 44 colonies were randomly sampled from each RBS spacer variant library category (n = 264) and isolated using 96-well DNA purification. The library was screened in two stages, with an initial screen to find active hits, followed by a more detailed analysis of selected variants.
To minimize reagent usage and reduce automated assay set-up time, we scaled down our cell-free reactions to 2-μL reaction volumes using 0.5 μL of DNA and 1.5 μL of NCF mix. This gave a mean final DNA concentration of 7.5 ± 1.8 nM within the NCF reaction with a range of 1.4–11.0 nM from the preliminary DNA screened (SI Appendix, Fig. S38). DNA concentrations were not normalized in this initial screen, as we were only concerned with selecting active variants at this stage. This reaction scale gave typical z-factor scores of between 0.517 and 0.744 with the positive control plasmid (PDH-GFP) versus a buffer negative control, indicating a sufficient statistical effect size for this screening assay. For comparison, a 10-μL volume reaction gave a stronger z-factor score of 0.90, although this was offset by increased transfer time (30 min) and reagent cost (2,880 μL NCF mix) for a 384-well plate. In contrast, despite increased variability at a 2-μL scale, this has the advantage of rapid transfer (10 min), minimal reagent cost (576 μL NCF mix), and low dead volume (200 μL for a six-well source plate). At the 2-μL reaction scale, ∼1 L of cell culture (∼6 mL cell-extract) provides enough NCF mix (∼18 mL) for 12,000 reactions, enabling scalable cell-free–based screening assays. Moreover, to account for transfer delay differences using the acoustic liquid-handling robot and positional effects, four technical repeats for each RBS variant and controls were randomly distributed in the plate.
Aside from the RBS-3 library, which displayed minimal hits, all libraries provided a range of positive hits, which is consistent with previous E. coli and B. subtilis RBS studies (52, 53). Fifteen variants above the limit of detection (Fig. 6B) from each of the RBS-4, -5, -7, and -8 libraries, along with 30 from the RBS-6 library, were selected for sequencing and more detailed analysis. After elimination of duplication events (four in RBS-6 group and two in RBS-4 to -7), each RBS variant was characterized in our NCF system with normalized DNA concentrations of 10 nM as four technical repeats and is presented as a ranked RBS activity library in SI Appendix, Fig. S39 and Tables S12 and S13.
Focusing on the RBS-6 library, additional in vivo and in silico data provided a comparison of methods. Here 26 RBS-6 variants were transformed and relative RBS activity was determined as a steady-state measurement with cell-free, in vivo, and through in silico prediction, using the RBS Calculator (54) and UTR Designer (55) models. Interestingly, two variants (pSJM1211 - 5′-GAGGAGGTATTAT-3′ and pSJM1223 - 5′-AAGGGGGTATTAT-3′) identified from the cell-free screen displayed a twofold enhanced activity in vivo in comparison with the positive control RBS (5′-AGGAGGTGAATGTCAT-3′). This was also consistent with in silico predictions (SI Appendix, Tables S12 and S13). Statistically significant (P < 0.001) correlations were found between cell-free, in vivo, and in silico datasets (Fig. 6C).
Importantly, the strongest correlation was found to be between cell-free and in vivo cell data (r = 0.87, P < 0.001). This not only provides further support to the earlier proof-of-concept findings with the natural B. megaterium promoters, but also complements in silico prediction methods and demonstrates the speed and precision of using liquid-handling robotics in a semiautomated cell-free screening workflow.
Discussion
E. coli-based cell-free platforms have become an established cornerstone of cell-free synthetic biology for prototyping gene expression (6, 15, 16) and optimizing metabolic pathway designs (1, 5, 56). Led by the early developments of Swartz and colleagues (57), cell-free has demonstrated the potential for scalable industrial protein manufacturing to provide an alternative and controlled environment for engineering valuable biomolecules outside of the living cell (58). However, in depth studies of cell-free systems have generally focused on the well-characterized synthetic biology chassis, such as E. coli. To address this shortcoming, we have developed the concept of NCF. Prerequisites for a minimum NCF proof-of-concept study include the ability to harvest a sufficient concentration of active cell-extracts (∼15–30 mg·mL−1), which can be limiting for slow-growing bacteria, and an initial handful of gene-expression tools. If energy regeneration, transcription, or translation present a bottleneck, this can be alleviated by the addition of purified components, such as creatine kinase, T7 polymerase, or 70S ribosomes, respectively. We have found the 3-PGA S30 energy mix (34) to be active in a selection of Gram-positive bacteria species (35, 36), including B. megaterium. This energy mix may be found to be more generally active in new NCF systems in the future. In this paper, we have extended this work to establish an integrated NCF platform that uses acoustic liquid-handling robotics (nanolitre transfers) combined with a Bayesian model parameter inference approach, providing a quantitative model-based parameterization of cell-free transcription–translation dynamics. We also further apply a semiautomated workflow for characterizing new library parts and show that reaction volume can be scaled down to minimize reagent usage at scale, while still permitting the acquisition of precise and statistically significant data. We believe this is particularly useful for cell-free screening applications where reagent cost or availability (e.g., mammalian cell-free) is a limiting factor for automation.
Our choice of B. megaterium is in part due to the important use of industrial microbes for the production of valuable biomolecules, ranging from vitamins (20), amino acids (59), and commodity fine chemicals (60). B. megaterium has industrial potential for the overproduction of proteins but suffers from a lack of genetic tools, poor genetic tractability, and an uncharacterized metabolome (18). Indeed many industrial microbial hosts have been established through strain evolution using random mutagenesis coupled with screening to optimize growth rates, improve resistance to toxic metabolites, and elevate product titers of a chosen biomolecule (20). Given the plethora of microbial species that have not been characterized, there is a need to develop new approaches that would enable such microbial systems to be explored rapidly for biotechnology applications, including and beyond bioprocessing. One such application is in the engineering of the human gut microbiome, which comprises thousands of poorly characterized bacterial species (14). For such an application, there is a need for novel bacterial strains to colonize and persist in the human gut for extended periods of time and such strains need to be genetically engineered to perform specific tasks, like sensing and memory (61).
Using our NCF platform we were able to characterize and model the native B. megaterium xylose-inducible promoter system, as well as a selection of B. megaterium σA constitutive promoters and RBS library members, which significantly expands the available toolset for this organism. Our NCF platform uses endogenous RNAP, translational apparatus, and energy regeneration enzymes for coupled protein synthesis and does not require the addition of exogenous components, such as T7 RNAP or enzymes for ATP synthesis (e.g., pyruvate kinase, creatine kinase) and thus provides, in part, a native environment for measurement of endogenous components. Cell-free systems also have the general advantages of permitting direct access to the reaction and providing a simple fluorescent read-out of complex assays. Here, we have used these properties to rapidly infer previously unknown XylR transcription factor binding affinities, which would be difficult or impossible to access using live-cell experiments. Other inferred model parameters, such as GFP maturation rates and transcription elongation rates, were found to closely match previously known values (SI Appendix, Tables S9 and S10).
For B. megaterium we found that in vivo RBS and promoter activities show strong correlation with cell-free activity, providing further evidence to our and other earlier findings for E. coli cell-free (17, 62). However, cell-free does present certain limitations. For example, cell-free is unable to fully capture biological perturbations resulting from growth-associated stress (63, 64). In addition, while two new strong RBS variants were identified in vivo, these displayed similar activity to the standard RBS (pRBBm258) in cell-free. From our prior resource competition and modeling experiments, we suggest that in cell-free, ribosomes are rate-limiting, therefore presenting a bottleneck for identifying stronger RBS or promoter elements. Therefore, while NCF provides a fast tool to initially test uncharacterized gene-expression elements from nonmodel organisms, secondary verification in the in vivo platform will also further inform this design process. We anticipate that NCF can open up the possibility of designing in vivo genetic circuits for less well-characterized organisms.
While the quantitative parameterization of individual genetic elements in vitro is important, their behavior in living cells is coupled to other genetic elements by the use of shared resources, such as free ribosomes. A combination of robustly parameterized genetic components, together with models capable of accounting for resource competition, could help address the particularly challenging problem of complex dynamical circuit design (65, 66). To analyze how such resources are utilized in cell-free reactions under conditions that potentially mimic cellular stress, we have used multiplexed absolute MS quantification of transcription–translation components together with cell-free reactions with multiple promoters. By titrating varying quantities of two competing promoters into our cell-free reactions, we were able to estimate resource sharing using mathematical modeling with Bayesian parameter inference (Fig. 4 and SI Appendix, Figs. S29–S36). Interestingly, for two different genes (one expressing GFP and one mCherry), we observed strikingly different propensities to exert translational burden on the system. The mCherry mRNA was inferred to have a much slower translation elongation rate, depleting the concentration of free ribosomes at much lower promoter concentrations than the corresponding GFP-expressing promoter (SI Appendix, Fig. S33). The reasons for this are unclear, but it could be due to the codon usage being suboptimal or it could be an intrinsic property of the mCherry protein sequence itself.
In comparison with a fully optimized E. coli cell-free reaction (38), our initial B. megaterium system is approximately one order-of-magnitude less active, even though the protein synthesis capacity of wild-type B. megaterium in vivo is comparable (gram per liter scale) to commercial E. coli strains (22). From the LC-MS analysis, this confirmed that the level of core transcription–translation components is comparable to previous summarized E. coli cell-free literature and therefore is unlikely to be rate-limiting (1). Taking into account the combined experimental and modeling analysis, we suggest that a number of factors are currently rate-limiting for our B. megaterium cell-free transcription–translation system. These include inefficient NTP regeneration, energy wastage, suboptimal codon usage, and ribosome elongation rates. Surprisingly our model, with parameters inferred from the two-promoter resource competition experiment, suggests that only ∼6% of the total theoretical energy input into the system is accessible in practice. Our model predicts that improving the metabolism of the secondary energy source would increase the available energy and, therefore, substantially increase the overall protein yield (SI Appendix, SI Text). We find that of the total accessible energy, a large fraction (>74%) is typically diverted to competing nonspecific phosphorylase activity as previously discussed (67). In the future, we will look to improve energy regeneration in B. megaterium NCF to assess its potential use for scaled-up recombinant protein production processes in vitro.
In summary, we have established an integrated experimental platform that combines automation, factorial experimental design, and modeling to study NCF extracts from different organisms (35, 36). Our approach opens up the possibility of systematically exploring gene-expression tools, cell-free transcription–translation, and energy regeneration machinery from understudied host cells for new synthetic biology and biotechnology applications.
Methods
Strains and Plasmids.
B. megaterium wild-type strain DSM319 was used for preparing the cell-free extract (68), E. coli strain BL21 (DE3) Star-pLysS (LifeTechnologies) for the recombinant production of the His6-tagged B. megaterium DSM319 repressor protein XylR (SI Appendix, SI Text), and E. coli strain DH10B (Invitrogen) for routine cloning. All of the plasmids and oligos used in this study are listed in SI Appendix, Tables S1–S4, respectively. For construction of plasmids, please see SI Appendix, SI Text.
Growth and Recombinant Protein Production in B. megaterium.
We used a fluorescence variant (GFP+-F64L/S65T/Q80R/F99S/M153T/V163A) of the wild-type GFP, herein simply referred to as GFP, to monitor protein production in B. megaterium cell-free transcription–translation, along with mCherry protein. B. megaterium cells were individually transformed with the corresponding plasmids and cultivated as described previously (22).
B. megaterium Cell-Extract Preparation and Transcription–Translation Reaction Conditions.
A B. megaterium DSM319 cell-free transcription–translation system was developed based on the E. coli method of Sun et al. (34). Full details and modifications are outlined in SI Appendix, SI Text and Table S6. This method yielded between 27–34 mg·mL−1 of cell-free crude lysate. All cell-free transcription–translation experiments were repeated with at least two biological repeats and three repeat measurements. Calibration standards of purified GFP-MGapt mRNA, His6-tagged GFP and mCherry were prepared in the standard reaction mixture (SI Appendix, Figs. S2 and S14).
Acoustic Liquid-Handling Robotics and Experimental Set-Up.
Full factorial experimental design and Echo525 liquid handler (Labcyte) pick list transfer instructions were automatically generated using a Python script (available at https://github.com/jmacdona/ODE_MCMC_tools) or Labcyte PickList software. Liquid droplets were transferred as multiples of 25 nL to a final volume of 10 μL as three triplicate replicates. Plates were sealed with Breathe-Easy sealing membrane (Sigma) for 10-μL reactions and a sealed aluminum foil for 2-μL reactions, and briefly centrifuged at 1,000 × g for 10 s. A CLARIOStar plate reader (BMG Labtech) was used for cell-free incubations and fluorescence measurements. Standard measurements were recorded every 10 min for 40 cycles at 30 °C with 10 s of 250 rpm orbital shaking before measurement. Further details and fluorescence settings are provided in SI Appendix, SI Text. For details of XylR experiments, please see SI Appendix, SI Text and Fig. S17.
Multiplexed Protein Quantification by LC-MS/MS.
A proteomics LC-MS/MS protocol based on previous studies was developed to quantitate cell-free transcription–translation enzymes from a triplicate biological replicates of B. megaterium cell extracts. Full information is provided in SI Appendix, SI Text, Figs. S23–S26, and Tables S7, S8, and S11.
Mathematical Modeling of Cell-Free Transcription–Translation.
A general ODE model scheme was created to account for the expression of single or multiple genes in the same reaction. This model included accounting for the use of shared resources (NTPs, amino acids, RNAP, and ribosomes). Further details are provided in SI Appendix, SI Text. SBML models are available from Github, as linked below.
Bayesian Parameter Inference.
A general C++ framework, called ODE_MCMC_tools, for inferring model parameters and initial values from experimental datasets for systems of ODEs using adaptive MCMC and multithreaded parallel tempering MCMC (PT-MCMC) was developed. Further details are in SI Appendix, SI Text. Source code and precompiled binaries are available from https://github.com/jmacdona/ODE_MCMC_tools.
Supplementary Material
Acknowledgments
S.W. and R.B. would like to thank Dieter Jahn for support and discussion. This work was supported by the Engineering and Physical Sciences Research Council [Grants EP/K038648/1 (to S.J.M.) and EP/L011573/1 (to J.T.M., A.I., A.T., D.J.B., D.W.M., and K.J.)]; Biotechnology and Biological Sciences Research Council Case Studentship DTG BB/F017324/1 (to N.K.); and Deutsche Forschungsgemeinschaft priority Program SPP1617 (to S.W. and R.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The plasmids reported in this paper have been deposited in the AddGene database (accession nos. 107576, 107577, 107578, 107581, and 107582). SBML models, source code, and precompiled binaries are available from https://github.com/jmacdona/ODE_MCMC_tools.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715806115/-/DCSupplemental.
References
- 1.Garamella J, Marshall R, Rustad M, Noireaux V. The all E. coli TX-TL Toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth Biol. 2016;5:344–355. doi: 10.1021/acssynbio.5b00296. [DOI] [PubMed] [Google Scholar]
- 2.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 3.Swartz JR. Universal cell-free protein synthesis. Nat Biotechnol. 2009;27:731–732. doi: 10.1038/nbt0809-731. [DOI] [PubMed] [Google Scholar]
- 4.Kuruma Y, Ueda T. The PURE system for the cell-free synthesis of membrane proteins. Nat Protoc. 2015;10:1328–1344. doi: 10.1038/nprot.2015.082. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi MK, et al. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth Biol. 2015;4:503–515. doi: 10.1021/sb400206c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegal-Gaskins D, Tuza ZA, Kim J, Noireaux V, Murray RM. Gene circuit performance characterization and resource usage in a cell-free “breadboard”. ACS Synth Biol. 2014;3:416–425. doi: 10.1021/sb400203p. [DOI] [PubMed] [Google Scholar]
- 7.Pardee K, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 8.Groff D, et al. Engineering toward a bacterial “endoplasmic reticulum” for the rapid expression of immunoglobulin proteins. MAbs. 2014;6:671–678. doi: 10.4161/mabs.28172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geertz M, Shore D, Maerkl SJ. Massively parallel measurements of molecular interaction kinetics on a microfluidic platform. Proc Natl Acad Sci USA. 2012;109:16540–16545. doi: 10.1073/pnas.1206011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenstein M. Living factories of the future. Nature. 2016;531:401–403. doi: 10.1038/531401a. [DOI] [PubMed] [Google Scholar]
- 11.Florea M, et al. Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose-producing strain. Proc Natl Acad Sci USA. 2016;113:E3431–E3440. doi: 10.1073/pnas.1522985113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Escribano JP, Bibb MJ. Streptomyces coelicolor as an expression host for heterologous gene clusters. Methods Enzymol. 2012;517:279–300. doi: 10.1016/B978-0-12-404634-4.00014-0. [DOI] [PubMed] [Google Scholar]
- 14.Huttenhower C, et al. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuza ZA, Singhal V, Kim J, Murray RM. Proceedings of the 52nd IEEE Conference on Decision and Control. IEEE; Firenze, Italy: 2013. An in silico modeling toolbox for rapid prototyping of circuits in a biomolecular “breadboard” system; pp. 1404–1410. [Google Scholar]
- 16.Sun ZZ, Yeung E, Hayes CA, Noireaux V, Murray RM. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth Biol. 2014;3:387–397. doi: 10.1021/sb400131a. [DOI] [PubMed] [Google Scholar]
- 17.Chappell J, Jensen K, Freemont PS. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 2013;41:3471–3481. doi: 10.1093/nar/gkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korneli C, David F, Biedendieck R, Jahn D, Wittmann C. Getting the big beast to work—Systems biotechnology of Bacillus megaterium for novel high-value proteins. J Biotechnol. 2013;163:87–96. doi: 10.1016/j.jbiotec.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, et al. High yield recombinant penicillin G amidase production and export into the growth medium using Bacillus megaterium. Microb Cell Fact. 2006;5:36. doi: 10.1186/1475-2859-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens JH, Barg H, Warren MJ, Jahn D. Microbial production of vitamin B12. Appl Microbiol Biotechnol. 2002;58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 21.Moore SJ, et al. Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12) Proc Natl Acad Sci USA. 2013;110:14906–14911. doi: 10.1073/pnas.1308098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stammen S, et al. High-yield intra- and extracellular protein production using Bacillus megaterium. Appl Environ Microbiol. 2010;76:4037–4046. doi: 10.1128/AEM.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G, et al. Expression of recombinant Clostridium difficile toxin A and B in Bacillus megaterium. BMC Microbiol. 2008;8:192. doi: 10.1186/1471-2180-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittchen KD, Meinhardt F. Inactivation of the major extracellular protease from Bacillus megaterium DSM319 by gene replacement. Appl Microbiol Biotechnol. 1995;42:871–877. doi: 10.1007/BF00191184. [DOI] [PubMed] [Google Scholar]
- 25.Korneli C, Biedendieck R, David F, Jahn D, Wittmann C. High yield production of extracellular recombinant levansucrase by Bacillus megaterium. Appl Microbiol Biotechnol. 2013;97:3343–3353. doi: 10.1007/s00253-012-4567-1. [DOI] [PubMed] [Google Scholar]
- 26.Rygus T, Hillen W. Catabolite repression of the xyl operon in Bacillus megaterium. J Bacteriol. 1992;174:3049–3055. doi: 10.1128/jb.174.9.3049-3055.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biedendieck R, Yang Y, Deckwer W-D, Malten M, Jahn D. Plasmid system for the intracellular production and purification of affinity-tagged proteins in Bacillus megaterium. Biotechnol Bioeng. 2007;96:525–537. doi: 10.1002/bit.21145. [DOI] [PubMed] [Google Scholar]
- 28.Moore SJ, et al. Characterization of the enzyme CbiH60 involved in anaerobic ring contraction of the cobalamin (vitamin B12) biosynthetic pathway. J Biol Chem. 2013;288:297–305. doi: 10.1074/jbc.M112.422535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payne KAP, et al. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature. 2015;517:513–516. doi: 10.1038/nature13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bridwell-Rabb J, Zhong A, Sun HG, Drennan CL, Liu HW. A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis. Nature. 2017;544:322–326. doi: 10.1038/nature21689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niederholtmeyer H, Xu L, Maerkl SJ. Real-time mRNA measurement during an in vitro transcription and translation reaction using binary probes. ACS Synth Biol. 2013;2:411–417. doi: 10.1021/sb300104f. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen AAK, et al. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 33.Tojo S, Kumamoto K, Hirooka K, Fujita Y. Heavy involvement of stringent transcription control depending on the adenine or guanine species of the transcription initiation site in glucose and pyruvate metabolism in Bacillus subtilis. J Bacteriol. 2010;192:1573–1585. doi: 10.1128/JB.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun ZZ, et al. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J Vis Exp. 2013:e50762. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore SJ, Lai H-E, Needham H, Polizzi KM, Freemont PS. Streptomyces venezuelae TX-TL—A next generation cell-free synthetic biology tool. Biotechnol J. 2017;12:1600678. doi: 10.1002/biot.201600678. [DOI] [PubMed] [Google Scholar]
- 36.Kelwick R, Webb AJ, MacDonald JT, Freemont PS. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab Eng. 2016;38:370–381. doi: 10.1016/j.ymben.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus. 2008;3 doi: 10.1128/ecosal.5.2.3. [DOI] [PubMed] [Google Scholar]
- 38.Caschera F, Noireaux V. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie. 2014;99:162–168. doi: 10.1016/j.biochi.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 39.Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Gu L, Aach J, Church GM. Improved cell-free RNA and protein synthesis system. PLoS One. 2014;9:e106232. doi: 10.1371/journal.pone.0106232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varricchio F, Monier R. Ribosome patterns in Escherichia coli growing at various rates. J Bacteriol. 1971;108:105–110. doi: 10.1128/jb.108.1.105-110.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rygus T, Hillen W. Inducible high-level expression of heterologous genes in Bacillus megaterium using the regulatory elements of the xylose-utilization operon. Appl Microbiol Biotechnol. 1991;35:594–599. doi: 10.1007/BF00169622. [DOI] [PubMed] [Google Scholar]
- 43.Rygus T, Scheler A, Allmansberger R, Hillen W. Molecular cloning, structure, promoters and regulatory elements for transcription of the Bacillus megaterium encoded regulon for xylose utilization. Arch Microbiol. 1991;155:535–542. doi: 10.1007/BF00245346. [DOI] [PubMed] [Google Scholar]
- 44.Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7:457–472. [Google Scholar]
- 45.Karzbrun E, Shin J, Bar-Ziv RH, Noireaux V. Coarse-grained dynamics of protein synthesis in a cell-free system. Phys Rev Lett. 2011;106:048104. doi: 10.1103/PhysRevLett.106.048104. [DOI] [PubMed] [Google Scholar]
- 46.Iizuka R, Yamagishi-Shirasaki M, Funatsu T. Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal Biochem. 2011;414:173–178. doi: 10.1016/j.ab.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 47.Gyorgy A, Murray RM. 2016 IEEE 55th Conference on Decision and Control (CDC) IEEE; Piscataway, NJ: 2016. Quantifying resource competition and its effects in the TX-TL system; pp. 3363–3368. [Google Scholar]
- 48.Verkhusha VV, Akovbian NA, Efremenko EN, Varfolomeyev SD, Vrzheshch PV. Kinetic analysis of maturation and denaturation of DsRed, a coral-derived red fluorescent protein. Biochemistry (Mosc) 2001;66:1342–1351. doi: 10.1023/a:1013325627378. [DOI] [PubMed] [Google Scholar]
- 49.Haldenwang WG. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lou C, Stanton B, Chen Y-J, Munsky B, Voigt CA. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore SJ, et al. EcoFlex: A multifunctional MoClo kit for E. coli synthetic biology. ACS Synth Biol. 2016;5:1059–1069. doi: 10.1021/acssynbio.6b00031. [DOI] [PubMed] [Google Scholar]
- 52.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guiziou S, et al. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res. 2016;44:7495–7508. doi: 10.1093/nar/gkw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salis HM. The ribosome binding site calculator. Methods Enzymol. 2011;498:19–42. doi: 10.1016/B978-0-12-385120-8.00002-4. [DOI] [PubMed] [Google Scholar]
- 55.Seo SW, et al. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab Eng. 2013;15:67–74. doi: 10.1016/j.ymben.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Karim AS, Jewett MC. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab Eng. 2016;36:116–126. doi: 10.1016/j.ymben.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Liu DV, Zawada JF, Swartz JR. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol Prog. 2005;21:460–465. doi: 10.1021/bp049789y. [DOI] [PubMed] [Google Scholar]
- 58.Zawada JF, et al. Microscale to manufacturing scale-up of cell-free cytokine production—A new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krömer JO, Sorgenfrei O, Klopprogge K, Heinzle E, Wittmann C. In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J Bacteriol. 2004;186:1769–1784. doi: 10.1128/JB.186.6.1769-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000;54:287–301. doi: 10.1007/s002530000403. [DOI] [PubMed] [Google Scholar]
- 61.Riglar DT, et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol. 2017;35:653–658. doi: 10.1038/nbt.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noireaux V, Bar-Ziv R, Libchaber A. Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci USA. 2003;100:12672–12677. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Münch KM, et al. Polar fixation of plasmids during recombinant protein production in Bacillus megaterium results in population heterogeneity. Appl Environ Microbiol. 2015;81:5976–5986. doi: 10.1128/AEM.00807-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ceroni F, Algar R, Stan G-B, Ellis T. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods. 2015;12:415–418. doi: 10.1038/nmeth.3339. [DOI] [PubMed] [Google Scholar]
- 65.Barnes CP, Silk D, Sheng X, Stumpf MPH. Bayesian design of synthetic biological systems. Proc Natl Acad Sci USA. 2011;108:15190–15195. doi: 10.1073/pnas.1017972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J, Khetarpal I, Sen S, Murray RM. Synthetic circuit for exact adaptation and fold-change detection. Nucleic Acids Res. 2014;42:6078–6089. doi: 10.1093/nar/gku233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calhoun KA, Swartz JR. Energy systems for ATP regeneration in cell-free protein synthesis reactions. In: Tymms MJ, editor. In Vitro Transcription and Translation Protocols. Humana Press; Totowa, NJ: 2007. pp. 3–17. [DOI] [PubMed] [Google Scholar]
- 68.Eppinger M, et al. Genome sequences of the biotechnologically important Bacillus megaterium strains QM B1551 and DSM319. J Bacteriol. 2011;193:4199–4213. doi: 10.1128/JB.00449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.