Significance
The actin cytoskeleton in living cells is not static but undergoes dynamic reorganization. Actin-containing thin filaments in cardiac sarcomeres are no exception; they exhibit exchange of actin subunits at the ends within actively contracting cardiomyocytes. Fhod3, an actin organizer in cardiac sarcomeres, is implicated in regulation of actin assembly in cardiomyocytes, although the mechanism is largely unknown. We discovered a direct molecular link between Fhod3 and cMyBP-C, a thick myosin filament-associated protein that modulates myocardial contraction via cross-bridge arrangement. Because Fhod3 adversely affected cardiac function in the absence of cMyBP-C, the interaction may serve to control the Fhod3-mediated actin reorganization at the cross-bridge region. Our results provide insight into actin reorganization in cardiac sarcomeres with implications for cardiac function.
Keywords: cMyBP-C, actin, formin, cardiomyopathy, Fhod3
Abstract
Mutations in cardiac myosin-binding protein C (cMyBP-C) are a major cause of familial hypertrophic cardiomyopathy. Although cMyBP-C has been considered to regulate the cardiac function via cross-bridge arrangement at the C-zone of the myosin-containing A-band, the mechanism by which cMyBP-C functions remains unclear. We identified formin Fhod3, an actin organizer essential for the formation and maintenance of cardiac sarcomeres, as a cMyBP-C–binding protein. The cardiac-specific N-terminal Ig-like domain of cMyBP-C directly interacts with the cardiac-specific N-terminal region of Fhod3. The interaction seems to direct the localization of Fhod3 to the C-zone, since a noncardiac Fhod3 variant lacking the cMyBP-C–binding region failed to localize to the C-zone. Conversely, the cardiac variant of Fhod3 failed to localize to the C-zone in the cMyBP-C–null mice, which display a phenotype of hypertrophic cardiomyopathy. The cardiomyopathic phenotype of cMyBP-C–null mice was further exacerbated by Fhod3 overexpression with a defect of sarcomere integrity, whereas that was partially ameliorated by a reduction in the Fhod3 protein levels, suggesting that Fhod3 has a deleterious effect on cardiac function under cMyBP-C–null conditions where Fhod3 is aberrantly mislocalized. Together, these findings suggest the possibility that Fhod3 contributes to the pathogenesis of cMyBP-C–related cardiomyopathy and that Fhod3 is critically involved in cMyBP-C–mediated regulation of cardiac function via direct interaction.
Cardiac muscle contraction is driven by cyclical sliding of an array of actin-containing thin filaments into a lattice of myosin-containing thick filaments in the sarcomere. The interaction between the two filament systems, which is caused by the formation of cross-bridge structures between actin filaments and myosin heads, is controlled mainly by thin filament activation via calcium binding to the troponin–tropomyosin regulatory complex (1, 2) but also by thick filament-associated proteins, including myosin regulatory light chain (RLC) and myosin binding protein-C (MyBP-C). The fine-tuning of cross-bridge formation by these thick filament-associated proteins is absolutely required for a normal cardiac function, which is well exemplified by the fact that genetic mutations in these genes, as well as those of β-myosin heavy chain, are associated with familial hypertrophic cardiomyopathy (HCM) (3, 4).
Cardiac MyBP-C (cMyBP-C), a thick myosin filament-associated 150-kDa protein comprised of eight Ig and three fibronectin-like domains, designated C0 through C10 from the N terminus, is thought to be involved in the modulation of cardiac contractility by regulating cross-bridge formation (5–9). The functional importance of this protein is also indicated by mutations in cMyBP-C being the major leading cause of familial HCM (10). cMyBP-C differs from its two skeletal isoforms by the presence of two cardiac-specific domains in the N-terminal region: the C0 domain at the N terminus and the M domain, a phosphorylatable motif, positioned between the C1 and C2 domains. The N-terminal region (C0–C2 domains) harboring the cardiac-specific C0 and M domains appears to play a pivotal role in regulating cardiac contraction via direct binding to the myosin head or actin (9, 11, 12), although the molecular mechanism by which cMyBP-C modulates the actomyosin function is still not fully understood.
We herein report the interaction of cMyBP-C with cardiac formin Fhod3, a regulator of actin assembly in cardiac sarcomeres (13–15). Formin family proteins, which contain the formin-homology domains 1 and 2 (FH1 and FH2) in the C-terminal half of the molecule, constitute a group of actin-nucleating factors and play crucial roles in controlling actin polymerization (16–19). The FH2 domain, a central catalytic domain, directly binds to actin molecules to facilitate actin filament nucleation and promote polymerization, which is accelerated by the FH1-mediated recruitment of profilin-actin dimer (20). Formins thus direct the formation of straight actin filaments, such as stress fibers, filopodia, and contractile rings during cytokinesis (21, 22). Fhod3, a formin that is abundantly expressed in the heart, plays an essential role in the regulation of the actin assembly in cardiac myofibrils (13–15, 23). In addition, we recently showed that Fhod3 plays a role in the maintenance of the normal cardiac function of the perinatal and adult heart (24). However, the mechanism by which Fhod3 functions in the sarcomere remains largely unknown.
In the present study, we showed that the cardiac-specific N-terminal Ig-like domain C0 of cMyBP-C directly interacts with the N-terminal region of Fhod3, which is specific to the cardiac isoform of Fhod3 but absent in the short isoform expressed in the kidney and brain. The direct interaction seems to direct the localization of Fhod3 to the C-zone, since a short variant of Fhod3 lacking the region responsible for the cMyBP-C binding fails to localize to the C-zone. In contrast, the actin-binding activity of the FH2 domain, which was previously supposed to be responsible for Fhod3 targeting (14), was shown to be dispensable for Fhod3 targeting to the C-zone, because a mutant Fhod3 defective in binding to actin was also able to accumulate in the C-zone. Furthermore, homozygous cMyBP-C–null mice exhibited a diffusely distributed pattern of Fhod3 but not to the C-zone, indicating that interaction with cMyBP-C is required for Fhod3 localization. This mislocalization of Fhod3 is a clear phenotype at the sarcomere level of cMyBP-C–null mice, a well-characterized mouse model of cardiomyopathy with diastolic and systolic dysfunction. Because Fhod3 is required for the functional maintenance of the cardiac function (24), cardiac dysfunction in cMyBP-C–null mice might be associated with the mislocalization of Fhod3. Indeed, the cardiac phenotypes of cMyBP-C–null mice were exacerbated by Fhod3 overexpression but partially ameliorated by a reduction in the Fhod3 proteins. These results support the current idea, although other mechanisms besides mislocalization of Fhod3 are also possible. The present findings provide molecular insight into the mechanism underlying the cMyBP-C–mediated modulation of cardiac contractility.
Results
Colocalization of Cardiac Formin Fhod3 with cMyBP-C in the Cardiac Sarcomere.
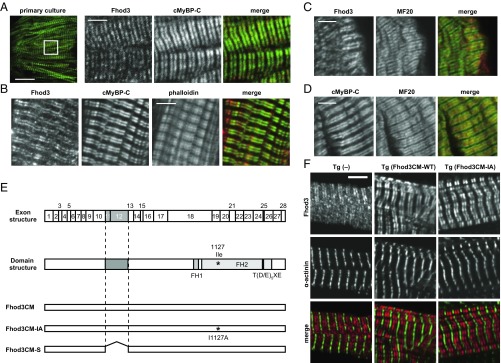
We previously showed that the cardiac formin protein Fhod3 localizes to the center of sarcomeres, specifically to the zone where thin actin filaments overlap with thick myosin filaments (13, 25, 26). Because the Fhod3 localization seems to be restricted to the C-zone, a region in the central two-thirds of the A-band (thick myosin filaments), we directly compared the localization pattern of Fhod3 with that of cMyBP-C, a structural component of the C-zone (27), in primary cultures of embryonic mouse cardiomyocytes. As shown in Fig. 1A, endogenous Fhod3 in cultured cardiomyocytes was completely colocalized with cMyBP-C at the C-zone of the sarcomere. The striking colocalization between endogenous Fhod3 and cMyBP-C was also observed in frozen sections from adult mouse hearts (Fig. 1B). In contrast, coimmunostaining with the anti-Fhod3 and anti-myosin heavy chain revealed that Fhod3 protein is not distributed throughout the A-band but strictly restricted only to the C-zone (Fig. 1C), like cMyBP-C (Fig. 1D). The sarcomeric localization of Fhod3 therefore appears to be related to the C-zone.
Fig. 1.
Colocalization of Fhod3 with cMyBP-C in the cardiac sarcomere. (A) Primary culture of embryonic C57BL/6 mouse cardiomyocytes was subjected to immunofluorescent double staining for endogenous Fhod3 [anti–Fhod3-(650–802); red] and cMyBP-C (green). (Scale bar, 5 μm.) (B) Sections of adult hearts from C57BL/6 mice were subjected to immunofluorescent staining for Fhod3 [anti–Fhod3-(650–802); red] and cMyBP-C (green) followed by phalloidin staining (not depicted in merge). (Scale bar, 5 μm.) (C and D) Sections of adult hearts from C57BL/6 mice were subjected to immunofluorescent staining for myosin heavy chain (green) and Fhod3 [anti–Fhod3-(650–802)] (C) or cMyBP-C (D) (red). (Scale bars, 5 μm.) (E) Schematic presentation of exon structure (Top), domain structure (Middle), and various constructs (Bottom) of mouse Fhod3. Alternative splicing exons 11, 12, and 25 are indicated by gray boxes (Top). The alternative splicing regions are in gray boxes, and the residue responsible for actin binding (Ile1127) is indicated by asterisk (Middle). Adapted with permission from ref. 25. (F) Sarcomeric localization of endogenous and exogenous Fhod3 in the heart. Sections of adult hearts from nontransgenic mice (Left) and from transgenic mice expressing Fhod3CM-WT (Center) and Fhod3CM-IA (Right) were subjected to immunofluorescent double staining for Fhod3 [anti–Fhod3-(650–802); red] and α-actinin (green). Images were taken under the same conditions except with photomultiplier tubes (PMT) voltage (800 and 600 V for nontransgenic and transgenic mice, respectively). (Scale bar, 5 μm.)
We next evaluated the role of the actin-binding activity of the FH2 domain in the Fhod3 localization. The N-terminally tagged Fhod3 mutant proteins carrying the I1127A and K1273D substitutions, both defective in actin-binding (13), showed the C-zone pattern in neonatal rat cardiomyocytes (SI Appendix, Fig. S1), suggesting that the actin-binding activity is dispensable for the Fhod3 localization to the C-zone. To confirm the ex vivo results and to exclude the possibility that the N-terminal tag affects the proper localization, we further generated transgenic mice expressing wild-type Fhod3 of the cardiac muscle isoform (Fhod3CM-WT) and a mutant Fhod3CM carrying the I1127A substitution (Fhod3CM-IA) without epitope tag under the control of the α-MHC promoter [Fhod3Tg(α-MHC–Fhod3CM-WT) and Fhod3Tg(α-MHC–Fhod3CM-IA), respectively] (Fig. 1E). The expression of exogenous Fhod3CM-WT and Fhod3CM-IA was about 20- and 5-fold higher, respectively, than that of endogenous Fhod3 (SI Appendix, Fig. S2). Consistently, the immunofluorescent signals for Fhod3 in sections from adult hearts of both Fhod3Tg(α-MHC–Fhod3CM-WT) and Fhod3Tg(α-MHC–Fhod3CM-IA) mice were much higher than that of endogenous Fhod3, which is detected only by a long exposure (SI Appendix, Fig. S3). It should be noted that intense signals for Fhod3 were observed in all cardiomyocytes of Fhod3Tg(α-MHC–Fhod3CM-WT) mice, whereas in Fhod3Tg(α-MHC–Fhod3CM-IA) mice, some cardiomyocytes showed strong signals but others showed only weak signals comparable to endogenous levels, indicating that the Fhod3CM-IA transgene is inactivated in some cardiomyocytes (SI Appendix, Fig. S3). As shown in Fig. 1F and SI Appendix, Fig. S3, the intense signals for Fhod3, which reflect the localization of exogenously expressed Fhod3, showed a strong accumulation at the C-zone not only in Fhod3Tg(α-MHC–Fhod3CM-WT) mice but also in Fhod3Tg(α-MHC–Fhod3CM-IA) mice. The actin-binding activity of FH2 domain therefore does not seem to mediate the Fhod3 accumulation to the C-zone in the cardiac sarcomere, suggesting that unknown Fhod3-interacting proteins may mediate Fhod3 localization.
Mass Spectrometric Identification of Fhod3-Binding Proteins.
To identify Fhod3-interacting proteins that mediate Fhod3 localization, we performed immunoprecipitation experiments using a cardiac lysate of transgenic mice expressing Fhod3CM-WT with anti-Fhod3 antibodies (SI Appendix, Fig. S4). Precipitated proteins were digested with trypsin and analyzed by liquid chromatography and tandem mass spectrometry. We identified several candidate proteins that were present in all three replicates from the cardiac lysate, but had not been recovered from proteins precipitated with control IgG. Among these candidates for Fhod3-interacting proteins, we found cardiac cMyBP-C itself, suggesting that Fhod3 forms a protein complex with cMyBP-C in the cardiac sarcomere.
Fhod3 Interaction with cMyBP-C.
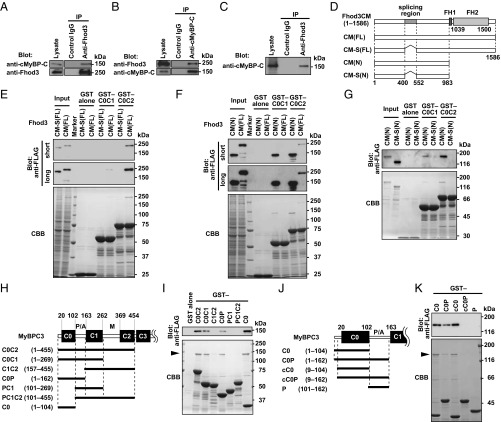
To confirm the interaction between Fhod3 and cMyBP-C, we performed a coimmunoprecipitation assay. As shown in Fig. 2A and SI Appendix, Fig. S5, anti-Fhod3 antibodies coprecipitated endogenous cMyBP-C with Fhod3 from the heart lysates of transgenic mice expressing Fhod3CM-WT. Similarly, anti–cMyBP-C antibody coprecipitated Fhod3 with endogenous cMyBP-C from the same heart lysates (Fig. 2B). To test the interaction between endogenous proteins, we further performed a coimmunoprecipitation assay using cardiac lysates of wild-type C57BL/6 mice and found that endogenous Fhod3 interacts with endogenous cMyBP-C in the heart (Fig. 2C). We next explored which region of cMyBP-C mediates Fhod3 association. cMyBP-C is a multidomain protein with 11 Ig or fibronectin-like domains, designated C0 through C10 from the N terminus (SI Appendix, Fig. S6A). The C-terminal region of cMyBP-C constitutively binds to and stabilizes the thick myosin filament through interactions with myosin rod or titin, whereas the N-terminal so-called regulatory domains (C0C2) extend from the thick filament to regulate the actin-myosin association (6). We therefore prepared the N-terminal regions of cMyBP-C as GST-fused proteins and performed an in vitro pull-down binding assay using lysates of HEK 293 cells expressing various Fhod3 proteins (Fig. 2D). As shown in Fig. 2E and SI Appendix, Fig. S7A, the full length of Fhod3 [Fhod3CM(FL)] in the lysate of HEK293 cells was effectively pulled down with GST–C0C2 and weakly pulled down with GST–C0C1, indicating that the N-terminal region of cMyBP-C is sufficient for Fhod3 association. In contrast, the full length of Fhod3CM-S [Fhod3CM-S(FL)], a short-splice variant that lacks 151 amino acids in the N-terminal region (23) (Figs. 1E and 2D) was not pulled down with either GST–C0C2 or GST–C0C1, suggesting that the spliced-out N-terminal region of Fhod3 is responsible for the binding. Consistently, the N-terminal fragment of Fhod3 [Fhod3CM(N)] was sufficiently pulled down with GST–C0C1 or GST–C0C2 (Fig. 2F), but not with GST–C3C6 or GST–C7C10 (SI Appendix, Fig. S6B). To further investigate whether or not Fhod3 directly interact with cMyBP-C, we purified the N-terminal region of Fhod3 proteins and performed an in vitro pull-down binding assay using the purified N-terminal regions of cMyBP-C. As shown in Fig. 2G, the purified Fhod3CM(N) was effectively pulled down with GST–C0C1 and GST–C0C2, but that of Fhod3CM-S [Fhod3CM-S(N)] was not. Thus, Fhod3 directly interacts with cMyBP-C, and this interaction is mediated by the N-terminal regions of both Fhod3 and cMyBP-C.
Fig. 2.
Interaction between Fhod3 and cMyBP-C. (A and B) Proteins in lysates of hearts from transgenic mice expressing Fhod3CM-WT were immunoprecipitated with the anti–Fhod3-C-20 (A) or the anti–cMyBP-C (B) antibodies, and then analyzed by immunoblot with the indicated antibodies. Uncropped images are shown in SI Appendix, Fig. S5. (C) Proteins in lysates of hearts from nontransgenic C57BL/6 mice were immunoprecipitated with the anti–Fhod3-C-20 or control IgG, and then analyzed by immunoblot with the anti–cMyBP-C antibody. Uncropped images are shown in SI Appendix, Fig. S5. (D) Schematic structures of various Fhod3 constructs used in the present pull-down binding assay (E–K). (E) FLAG-tagged Fhod3CM(FL) or Fhod3CM-S(FL) in the lysate of HEK-293F cells (input) was incubated with GST–cMyBP-C (C0C1 or C0C2) or GST alone and pulled down with glutathione-Sepharose 4B beads. Precipitated proteins were subjected to SDS/PAGE and analyzed by immunoblot with the anti-FLAG antibody (Upper) or stained with Coomassie Brilliant Blue (CBB) (Lower). Positions for marker proteins are indicated in kilodaltons. (F) FLAG-tagged full-length (FL) or N terminus (N) of Fhod3CM in the lysate of HEK-293F cells (input) was incubated with GST–cMyBP-C (C0C1 or C0C2) or GST alone and pulled down with glutathione-Sepharose 4B beads. Precipitated proteins were analyzed as in E. (G) Purified N-terminal region of Fhod3 proteins [Fhod3CM(N) or Fhod3CM-S(N)] tagged with FLAG was incubated with GST–cMyBP-C (C0C1 or C0C2) or GST alone and pulled down with glutathione-Sepharose 4B beads. Precipitated proteins were analyzed as in E. (H and J) Schematic structures of various cMyBP-C constructs used in I and J, respectively. (I and K) FLAG-tagged Fhod3CM(N) in the lysate of HEK-293F cells was incubated with indicated GST-fused fragments of cMyBP-C and pulled down with glutathione-Sepharose 4B beads. Precipitated proteins were analyzed as in E. The arrowheads indicate the position of Fhod3CM(N).
To identify the domains in the N-terminal cMyBP-C responsible for the interaction, we further prepared a series of smaller fragments of cMyBP-C (Fig. 2H). C0C2 bound to Fhod3CM(N) more strongly than C0C1 did (Fig. 2I and SI Appendix, Fig. S7B). Deletion of the C0 domain resulted in a markedly reduced interaction (Fig. 2I and SI Appendix, Fig. S8), whereas the C0 domain alone was sufficient for strong binding to Fhod3 (Fig. 2I). Compared with the human cMyBP-C, the mouse homolog has a unique N-terminal extension of eight amino acids (GenBank accession no. NM_008653). To exclude the possibility that the interaction is mediated via the mouse-specific N-terminal sequence, we further prepared the N-terminally truncated core C0 domain (cC0), which corresponds to the human C0 domain (Fig. 2J). As shown in Fig. 2K, cC0 sufficiently bound to Fhod3CM(N), indicating that the N-terminal extension specific to mice is dispensable for the interaction. In contrast, the addition of the P/A region to C0 or to cC0 attenuates the interaction, indicating that the P/A region seems to negatively regulate the binding. The extent of the inhibitory effect of the P/A region seems higher in the case of cC0P than that of C0P (SI Appendix, Fig. S7C), possibly suggesting that the inhibitory effect of the P/A region is attenuated by the presence of the N-terminal extension. The M and C2 domains might also attenuate the inhibitory effect. These findings indicate that the core C0 domain of cMyBP-C is sufficient for the interaction with Fhod3, whereas the binding may be regulated by other domains in a complex manner and differently in mice and human.
Mode of Interaction of the C0 Domain of cMyBP-C with the N-Terminal Region of Fhod3.
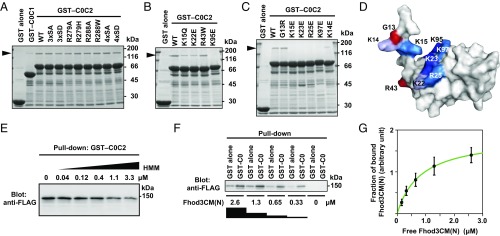
Phosphorylation of the M domain of cMyBP-C is known to modulate the interaction of cMyBP-C with the thin and thick filaments, thereby affecting the cross-bridge cycle and cardiac hemodynamics (5). In addition, familial HCM missense mutations have been identified in the M domain (10). To determine whether phosphorylation or a mutation in the M domain affects the interaction between Fhod3 and cMyBP-C, we prepared various mutant fragments of cMyBP-C and examined the interaction with Fhod3 (Fig. 3A). Nonphosphorylatable alanine substitutions (3SA and 4SA) or phosphorylation mimetic substitutions (3SD and 4SD) for serines in the M domain had no effect on the binding. The HCM missense mutations in the M domain (R279H and R288W: corresponding to R273H and R282W in human cMyBP-C, respectively) also did not cause any effects. Thus, no substitutions in the M domain examined showed any effects on the interaction with Fhod3.
Fig. 3.
Specific interaction of the C0 domain of cMyBP-C with Fhod3. (A–C) Effect of amino acid substitutions on the interaction of C0C2 with the N-terminal region of Fhod3. FLAG-tagged Fhod3CM(N) in the lysate of HEK-293F cells was incubated with GST-fused C0C2 fragment carrying the indicated amino acid substitution: 3xSA, the S281A/S290A/S310A substitution; 4xSA, the S281A/S290A/S310A/S315A substitution. Proteins were pulled down with glutathione-Sepharose 4B beads. Precipitated proteins were subjected to SDS/PAGE and stained with CBB. The arrowheads indicate the position of Fhod3CM(N). (D) Mapping of the residues responsible for the Fhod3 interaction on the surface of the C0 domain. The figure is drawn using PyMOL from the structure at PDB ID code 2K1M (28). Amino acid residues responsible for the interaction with Fhod3 in B and C are shown in blue, and residues associated with cardiomyopathy are shown in red. (E) Competitive effects of heavy meromyosin on Fhod3 binding to cMyBP-C–C0C2. GST–cMyBP-C–C0C2 was incubated with the lysate of HEK-293F cells expressing FLAG-tagged Fhod3CM(N) (final concentration of 2.0 μM) in the presence of various concentration of heavy meromyosin. (F) Representative pull-down assay for quantification of binding between cMyBP-C–C0 and Fhod3CM(N). GST–cMyBP-C–C0 or GST alone immobilized to glutathione particles was incubated with the indicated concentrations of the lysate of HEK-293F cells expressing FLAG-tagged Fhod3CM(N). To avoid disturbing the binding equilibrium, bound Fhod3CM(N) were analyzed directly without washing by immunoblot with the anti-FLAG antibodies followed by fluorescence measurement. (G) Quantitative analysis for binding of cMyBP-C–C0 to Fhod3CM(N). Fractions of specifically bound Fhod3-N were determined as fraction bound to GST–C0 minus fraction to GST alone in F.
We further investigated the effect of substitutions in the C0 domain. Similar to the HCM mutations in the M domain, those in the C0 domain, G13R and R43W (corresponding to G5R and R35W in human cMyBP-C, respectively), did not affect the interaction (Fig. 3 B and C). In contrast, substitutions of lysine or arginine residues positioned on the surface of the C0 domain (28) to glutamate effectively abolished the interaction. Because the electrophoretic mobility of mutant proteins is not markedly altered, these substitutions of surface residues are expected not to cause major structural changes. Intriguingly, the residues responsible for Fhod3 binding (Fig. 3D) partially overlapped with those for binding to the RLC of myosin (28), suggesting that the interaction between Fhod3 and cMyBP-C is affected by the presence of myosin. Indeed, as shown in Fig. 3E, heavy meromyosin attenuated the interaction in a dose-dependent manner, suggesting the possibility that the C0C2-mediated bindings to Fhod3 and myosin are mutually exclusive. Furthermore, we titrated in different concentrations of Fhod3CM(N) to estimate a dissociation constant (Fig. 3F) and found that Fhod3CM(N) bound to the C0 domain in a concentration-dependent manner with an estimated Kd value of about 0.8 μM (Fig. 3G). Although the binding affinity of cMyBP-C for Fhod3 is higher than that for the RLC of myosin (28) or for the S2 segment of myosin (29), cumulative contribution of the RLC and S2 segment may affect the overall affinity of myosin head. Thus, the cardiac formin Fhod3 interacts with cMyBP-C via a charge-dependent interaction between the N-terminal region of Fhod3 and the cardiac-specific C0 domain of cMyBP-C.
Role of the Interaction with cMyBP-C in Fhod3 Localization.
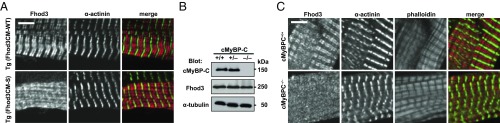
We next investigated whether or not the interaction between Fhod3 and cMyBP-C is required for the in situ localization of Fhod3 in the sarcomeres. To investigate this, we generated transgenic mice expressing the short form (Fhod3CM-S) (Fig. 1E), defective in binding to cMyBP-C (Fig. 2E), under the control of the α-MHC promoter [Fhod3Tg(α-MHC–Fhod3CM-S)]. From over 200 injections, we obtained only one transgenic line expressing Fhod3CM-S protein at a low level (SI Appendix, Fig. S9 A and B). In the Fhod3CM-S–expressing cells shown in Fig. 4A and SI Appendix, Fig. S3, the intense signals for Fhod3, which reflect the localization of exogenously expressed Fhod3CM-S, distributed in a diffuse pattern and failed to show the characteristic localization pattern to the C-zones. Consistent with this observation, the N-terminally tagged short variant of Fhod3 also failed to localize at the C-zone in neonatal rat cardiomyocytes (SI Appendix, Fig. S1) (13). Thus, the binding with cMyBP-C seems to be necessary for the in situ localization of Fhod3 in the sarcomeres. It should be noted that Fhod3CM-S was expressed only in a small subpopulation of cardiomyocytes in the cardiac section (SI Appendix, Fig. S3), indicating that, similar to Fhod3CM-IA, the Fhod3CM-S transgene is inactivated in some cardiomyocytes. Although the precise mechanism for this is presently unknown, a toxicity of the aberrant Fhos3CM-S protein dislocated from the C-zone may explain the reason. Full expression of the toxic peptide in the cardiomyocytes is supposed to impair normal cardiac development and result in lethality. If the exogenous expression of the toxic peptide is suppressed by inactivation of the transgene, the lethality could be overcome. Indeed, the obtained transgenic line expressing Fhod3CM-S protein at a very low level appears phenotypically normal (SI Appendix, Fig. S9 C and D).
Fig. 4.
Interaction between Fhod3 and cMyBP-C is required for Fhod3 localization to the C-zone. (A) Sarcomeric localization of Fhod3CM-S in the heart. Sections of adult hearts from transgenic mice expressing Fhod3CM-S were subjected to immunofluorescent double staining for Fhod3 [anti–Fhod3-(650–802); red] and α-actinin (green). (Scale bar, 5 μm.) (B) Immunoblot analysis of total proteins from the heart of wild-type (+/+), cMyBP-C+/− (+/−), and cMyBP-C−/− (−/−) mice using antibodies against Fhod3 (anti–Fhod3-C-20), cMyBP-C, or α-tubulin. (C) Sarcomeric localization of endogenous Fhod3 in the heart from cMyBP-C–null mice. Sections of adult hearts from cMyBP-C–null mice were subjected to immunofluorescent double staining for Fhod3 [anti–Fhod3-(650–802); red] and α-actinin (green) followed by phalloidin staining (not depicted in merge). (Scale bar, 5 μm.)
To confirm the crucial role of the interaction with cMyBP-C on Fhod3 localization in the physiological state, we prepared cMyBP-C–null mice from cMyBP-C mutant mice carrying the targeted trap allele (SI Appendix, Fig. S10). The cMyBP-C–null mice showed cardiomyopathic changes at morphological and histological levels (Fig. 5 D and F and SI Appendix, Figs. S10 C and D and S14) but did not display any significant alterations in the sarcomere structure (SI Appendix, Fig. S13), which is in line with previous reports (30–33). Consistent with our hypothesis, in the cardiac section of the cMyBP-C–null mice shown in Fig. 4C and SI Appendix, Fig. S11, Fhod3 did not show a periodic accumulation pattern, but instead demonstrated a diffuse distribution, although the expression level of Fhod3 was not affected in the cMyBP-C–null mice (Fig. 4B). Thus, cMyBP-C is required for the characteristic localization of Fhod3 to the C-zone in the sarcomere. Furthermore, these findings raise the intriguing question of whether or not the mislocalization of Fhod3 is responsible for the pathogenesis of cardiomyopathy in cMyBP-C–null mice.
Fig. 5.
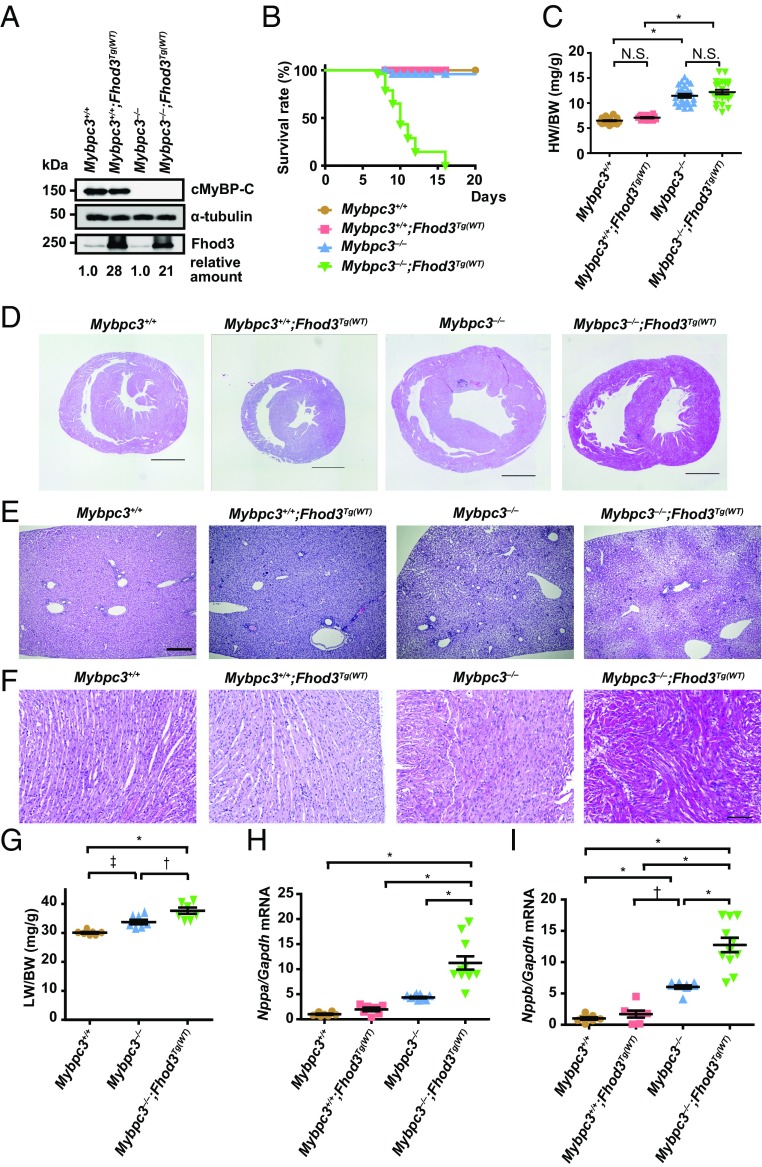
Physiological effects of transgenic overexpression of Fhod3 in cMyBP-C–null mice. (A) Immunoblot analysis of Fhod3, cMyBP-C, and α-tubulin using whole heart tissue lysates from wild-type (Mybpc3+/+), Fhod3-Tg [Mybpc3+/+;Fhod3Tg(WT)], cMyBP-C–null (Mybpc3−/−), and cMyBP-C–null mice overexpressing Fhod3 [Mybpc3−/−;Fhod3Tg(WT)] at P9. (B) Survival curves of mice of the indicated genotypes [Mybpc3+/+, n = 52; Mybpc3+/+;Fhod3Tg(WT), n = 38; Mybpc3−/−, n = 50; Mybpc3−/−;Fhod3Tg(WT), n = 52]. (C) Heart-to-body weight ratio of cMyBP-C–null mice overexpressing Fhod3 [Mybpc3−/−;Fhod3Tg(WT), n = 23] and control mice [Mybpc3+/+, n = 30; Mybpc3+/+;Fhod3Tg(WT), n = 14; Mybpc3−/−, n = 24] at P9. Values are means (long bars) ± SEM (short bars). *P < 0.001; N.S., not significant. (D–F) Histological analysis of hearts and livers of cMyBP-C–null mice overexpressing Fhod3 [Mybpc3−/−;Fhod3Tg(WT)] and control mice at P9. Short-axial sections of hearts (D), liver tissues (E), and short-axial–sectioned lateral wall of the left ventricles (F) were stained with H&E. (Scale bars: 1 mm in D and E and 100 μm in F.) (G) Liver-to-body weight ratio of cMyBP-C–null mice overexpressing Fhod3 [Mybpc3−/−;Fhod3Tg(WT), n = 7] and control mice (Mybpc3+/+, n = 7; Mybpc3−/−, n = 7) at P9. Values are means (long bars) ± SEM (short bars). *P < 0.001, †P < 0.01, ‡P < 0.05. (H and I) Quantitative real-time PCR analysis of fetal cardiac gene expression in cMyBP-C–null mice overexpressing Fhod3 [Mybpc3−/−;Fhod3Tg(WT), n = 11] and control mice [Mybpc3+/+, n = 8; Mybpc3+/+;Fhod3Tg(WT), n = 7; Mybpc3−/−, n = 9] at P9. Values are means (long bars) ± SEM (short bars). Nppa, encoding ANF; Gapdh, encoding GAPDH; Nppb, encoding BNP. *P < 0.001, †P < 0.01.
Exacerbation of Cardiac Dysfunction by Overexpression of Fhod3 in cMyBP-C–Null Mice.
To investigate the physiological significance of Fhod3 anchoring to the C-zone and to explore the consequences of Fhod3 mislocalization, we examined the effect of Fhod3 overexpression in cMyBP-C–null mice. If the cardiomyopathic phenotype of cMyBP-C–null mice worsens by overexpression of Fhod3, the aberrant Fhod3 protein is likely toxic, possibly in a similar manner to that of the Fhos3CM-S protein. By crossing cMyBP-C–null mice with transgenic mice expressing wild-type Fhod3 (Fhod3CM) under the control of the α-MHC promoter [Fhod3Tg(α-MHC–Fhod3CM)], we generated cMyBP-C–null mice overexpressing Fhod3 in the heart [Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM)] (Fig. 5A). As shown in Fig. 5B, these Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) pups all died around 2 wk after birth. Although the heart-to-body weight ratio at postnatal day 9 (P9) did not differ significantly between Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) mice and Mybpc3−/− mice (Fig. 5C), dilatation of the right ventricle with severe liver congestion was evident (Fig. 5 D, E, and G), suggesting that Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) mice died due to cardiac failure. In contrast, lung congestion was not apparent (SI Appendix, Fig. S12). Disarray of cardiomyocytes, which is a characteristic change for cMyBP-C–null heart (31, 32), was also more prominent in Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) mice (Fig. 5F). Consistent with these findings, the expression of the cardiac remodeling-associated fetal genes was significantly elevated in Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) mice compared with Mybpc3−/− mice (Fig. 5 H and I). Thus, the overexpression of Fhod3 seems to induce exacerbation of cardiac dysfunction in cMyBP-C–null mice, although it does not affect the cardiac function of wild-type mice.
Disorganization of the Sarcomeric Structure Due to Overexpression of Fhod3 in the cMyBP-C–Null Heart.
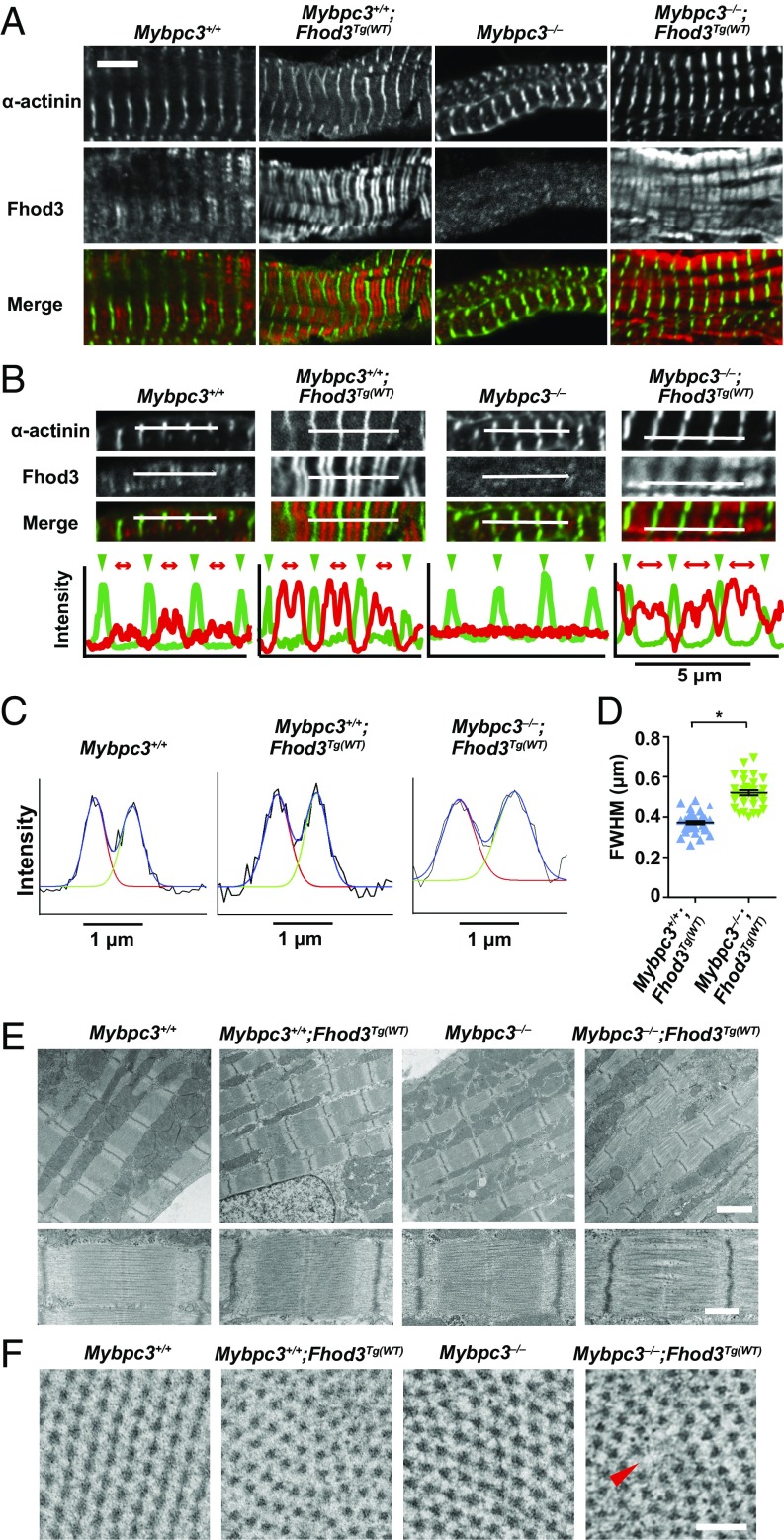
To clarify the mechanism by which Fhod3 overexpression induces the exacerbation of cardiac dysfunction only in cMyBP-C–null mice, we examined the sarcomere structure in the heart of Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) mice. There were no significant differences in sarcomeric structures comprising the Z-line marked with the antibody to α-actinin and thin actin filaments stained with phalloidin (SI Appendix, Fig. S13). However, somewhat to our surprise, the localization pattern of Fhod3 showed striking differences; the intense signals for overexpressed Fhod3 in Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) mice distributed in a periodic pattern despite the absence of cMyBP-C, although weak signals for endogenous Fhod3 in Mybpc3−/− mice distributed diffusely in the absence of cMyBP-C (Fig. 6A), as already shown in Fig. 4C. Intriguingly, the periodic pattern of overexpressed Fhod3 in cMyBP-C–null mice was apparently different from the characteristic C-zone pattern of endogenous Fhod3 or that of overexpressed Fhod3 in wild-type mice; the accumulation of overexpressed Fhod3 in cMyBP-C–null mice was not restricted to the central C-zone, but it expanded to the outer peripheral region of the A-band. The difference in localization pattern was evident in line scan graphs of fluorescent intensity along the long axis of sarcomeres (Fig. 6B). The width of the Fhod3 distribution pattern (full-width at half-maximum, FWHM) was significantly larger in Mybpc3−/−;Fhod3Tg(α-MHC–Fhod3CM) mice than in Fhod3Tg(α-MHC–Fhod3CM) mice (Fig. 6 C and D). Thus, in the absence of cMyBP-C, overexpressed Fhod3 protein, seems to distribute along the entire region of the A-band.
Fig. 6.
Sarcomeric effects of transgenic overexpression of Fhod3 in cMyBP-Cvnull mice. (A) Sarcomeric localization of endogenous and exogenous Fhod3 in the mouse heart at P9. Sections of hearts from mice of the indicated genotypes were subjected to immunofluorescent double staining for Fhod3 [anti–Fhod3-(650–802); red] and α-actinin (green). (Scale bar, 5 μm.) (B) Fluorescence intensity profiles in a line scan of sarcomeres. Line scan profiles of fluorescence intensities for the anti–α-actinin antibody (green) and the anti–Fhod3-(650–802) (red) are generated from immunofluorescent images of Upper panels. Green arrowheads and red two-headed arrows indicate the positions of Z-lines and the range of Fhod3 staining, respectively. (C) Representative fitting curves for Fhod3 intensity profiles in a line scan of sarcomeres from mice with the indicated genotypes. (D) The FWHM of Fhod3 intensity profiles in cMyBP-C–null mice overexpressing Fhod3 [Mybpc3−/−;Fhod3Tg(WT), n = 32] and control mice [Mybpc3+/+;Fhod3Tg(WT), n = 36]. *P < 0.001. (E) Electron micrographs of longitudinal sarcomere sections of the mouse heart at P9. [Scale Bar: 2 μm (Upper) and 500 nm (Lower).] (F) Electron micrographs of cross-sections of sarcomeres at the level of M-line or of nonoverlap region. The red arrowhead indicates the area where thick filaments are missing. (Scale bar, 100 nm.)
To determine the consequences of the abnormal distribution of Fhod3 throughout the entire A-band, we further examined the ultrastructure of sarcomeres using transmission electron microscopy. Fhod3 overexpression in the presence of cMyBP-C did not affect the ultrastructure of sarcomeres: intact sarcomeres with Z-lines, M-lines, and A-bands were observed (Fig. 6E). Depletion of cMyBP-C per se also did not alter the sarcomere organization (Fig. 6E) as previously reported (30–32). In contrast, in Fhod3-overexpressing cMyBP-C–null mice, disorder of the array of myosin filaments was observed: gaps between thick filaments are frequently detected at the center of the sarcomere (Fig. 6E). In some sarcomeres, thick filaments appear to be bundled or clumped at the level of the M-line, suggesting the disordering of hexagonal myosin filament lattice. Consistent with this, cross-sections of sarcomeres showed disorder of the hexagonal lattice of myosin filaments in Fhod3-overexpressing cMyBP-C–null mice at the level of the M-line or of nonoverlapping regions that contained only thick filaments (Fig. 6F). Thus, an overexpression of Fhod3 in the cMyBP-C–null heart induces the abnormal accumulation of Fhod3 to the entire A-band, thereby leading to a loss of sarcomeric integrity, which may be responsible for the cardiac dysfunction and death of those mice. These findings suggest that an excess amount of aberrant Fhod3 functions deleteriously as a toxic peptide in cMyBP-C–null mice.
Modulation of the Cardiac Function in cMyBP-C–Null Mice by Reduced Fhod3 Protein Levels.
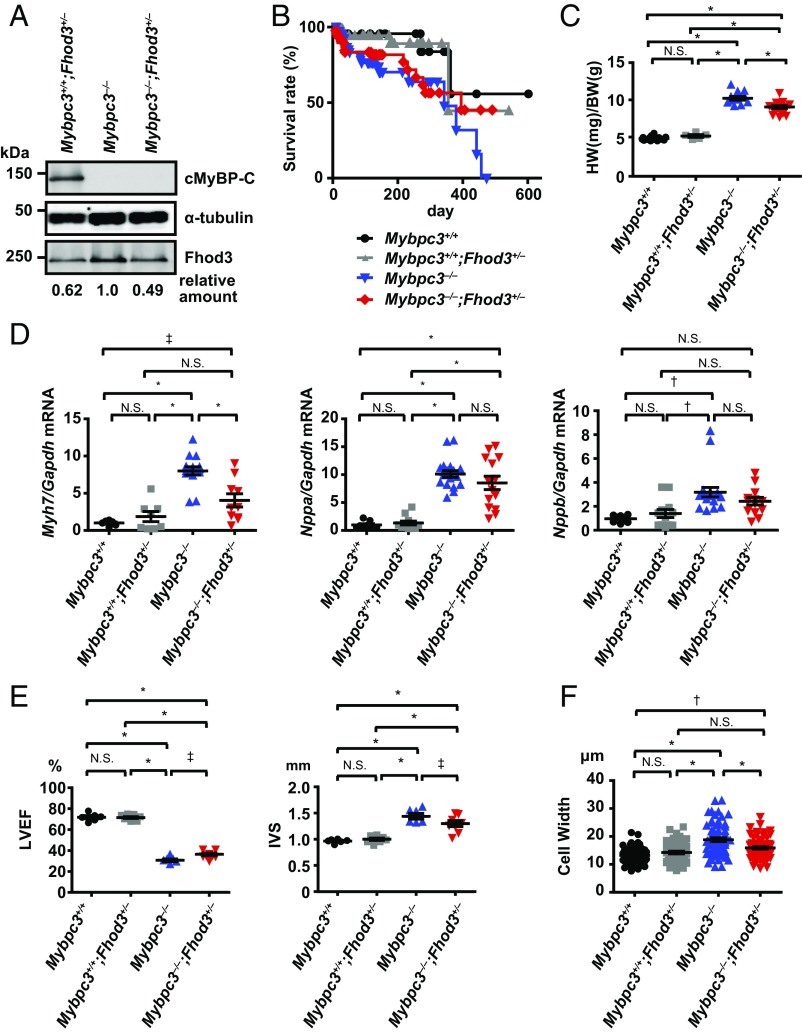
To evaluate the pathogenic role of the aberrant Fhod3 in cMyBP-C–null mice, we finally investigated the effect of the reduction of Fhod3 protein levels in cMyBP-C–null mice. By crossing cMyBP-C–null mice with heterozygous Fhod3 knockout mice (Fhod3+/−) (15), we generated cMyBP-C–null mice expressing half the normal amount of Fhod3 protein (Mybpc3−/−;Fhod3+/−) (Fig. 7A). Although no significant difference in the long-term survival over a year was observed between Mybpc3−/−;Fhod3+/− mice and Mybpc3−/−;Fhod3+/+ mice, about 50% of Mybpc3−/−;Fhod3+/− mice were alive at the time point when all Mybpc3−/−;Fhod3+/+ mice died (Fig. 7B). The heart-to-body weight ratio of Mybpc3−/−;Fhod3+/− mice at 16 wk was significantly lower than that in Mybpc3−/−;Fhod3+/+ mice (Fig. 7C). The expression of the cardiac remodeling-associated fetal gene β-MHC was consistently decreased in Mybpc3−/−;Fhod3+/− mice compared with Mybpc3−/−;Fhod3+/+ mice, although differences in the expressions of ANF and BNP were not statistically significant (Fig. 7D). An echocardiographic analysis revealed that a reduction of the ventricular ejection fraction observed in cMyBP-C–null mice was slightly but significantly recovered in Mybpc3−/−;Fhod3+/− mice (Fig. 7E). Although the changes in ventricular dimensions were not significant, the ventricular wall thickening was significantly recovered in Mybpc3−/−;Fhod3+/− mice (Fig. 7E). Consistent with this, the width of individual cardiomyocytes was significantly reduced in Mybpc3−/−;Fhod3+/− mice (Fig. 7F). Such improvement was also observed at 36 wk of age (SI Appendix, Fig. S14). Thus, reductions in protein levels of Fhod3 in cMyBP-C–null mice appeared to partially antagonize the hypertrophic changes in cMyBP–C-null mice.
Fig. 7.
Effect of heterozygous deletion of Fhod3 in cMyBP-C–null mice. (A) Cardiac lysates from mice of the indicated genotypes at 16 wk were analyzed by immunoblot with antibodies against Fhod3 (anti–Fhod3-C-20), cMyBP-C, and α-tubulin. (B) Survival curves of mice of the indicated genotypes (Mybpc3+/+, n = 21; Mybpc3+/+;Fhod3+/−, n = 69; Mybpc3−/−, n = 71; Mybpc3−/−;Fhod3+/−, n = 52). (C) Heart-to-body weight ratio of mice of the indicated genotypes (Mybpc3+/+, n = 17; Mybpc3+/+;Fhod3+/−, n = 5; Mybpc3−/−, n = 12; Mybpc3−/−;Fhod3+/−, n = 17) at 16 wk. Values are means (long bars) ± SEM (short bars). *P < 0.001; N.S., not significant. (D) Quantitative real-time PCR analysis of fetal cardiac gene expression in mice with the indicated genotypes (Mybpc3+/+, n = 8; Mybpc3+/+;Fhod3+/−, n = 12; Mybpc3−/−, n = 9; Mybpc3−/−;Fhod3+/−, n = 14) at 16 wk. Values are means (long bars) ± SEM (short bars). Myh7, encoding β-MHC; Gapdh, encoding GAPDH; Nppa, encoding ANF; Nppb, encoding BNP. *P < 0.001, †P < 0.01, ‡P < 0.05; N.S., not significant. (E) Echocardiography analysis of hearts of mice with the indicated genotypes (Mybpc3+/+, n = 6; Mybpc3+/+;Fhod3+/−, n = 9; Mybpc3−/−, n = 6; Mybpc3−/−;Fhod3+/−, n = 8) at 16 wk. Values are means (long bars) ± SEM (short bars). IVS, interventricular septum thickness; LVEF, left ventricular ejection fraction. *P < 0.001, †P < 0.01, ‡P < 0.05; N.S., not significant. (F) The width of cardiomyocyte at the nuclei level by wheat-germ agglutinin staining of left ventricle septum from mice of the indicated genotypes (Mybpc3+/+, n = 58; Mybpc3+/+;Fhod3+/−, n = 58; Mybpc3−/−, n = 68; Mybpc3−/−;Fhod3+/−, n = 67) at 16 wk was estimated by wheat-germ agglutinin staining. Values are means (long bars) ± SEM (short bars). *P < 0.001, †P < 0.01; N.S., not significant.
Discussion
In the present study, we show that the cardiac formin Fhod3 directly interacts with the cardiac isoform of MyBP-C; the interaction is mediated via cardiac isoform-specific regions of both proteins. It seems that cMyBP-C serves as a Fhod3-anchoring protein, because homozygous cMyBP-C–null mice exhibit the diffusely distributed pattern of Fhod3 instead of showing Fhod3 accumulation to the C-zone. Furthermore, a short variant of Fhod3 lacking the region responsible for the cMyBP-C binding fails to localize to the C-zone, indicating that this binding is required for the precise localization of Fhod3. Given that Fhod3 is required for functional maintenance of the heart (24), cardiac dysfunction caused by cMyBP-C depletion may be associated with the mislocalization of Fhod3. Consistent with this idea, the cardiomyopathic changes observed in cMyBP-C–null mice are exacerbated by Fhod3 overexpression, whereas the cardiac phenotypes are partially improved by the reduction of Fhod3 protein levels. The present results offer a molecular and functional link between Fhod3 and cMyBP-C in the cardiac sarcomere.
Fhod3, a formin that is abundantly expressed in the heart, plays an essential role in the regulation of the actin assembly in cardiac myofibrils (13–15). In addition, we recently showed that Fhod3 plays a role in the maintenance of the normal cardiac function of the adult heart (24). Interestingly, despite its ability to associate with the barbed end of actin filaments, Fhod3 does not actually accumulate at the Z-line where the barbed ends of thin actin filaments are anchored, instead exhibiting a unique localization pattern at the C-zone (13, 25, 26). The mechanism underlying this paradoxical localization of Fhod3 has been unclear, but the present study shows that the sarcomeric accumulation of Fhod3 at the C-zone is not determined by the actin binding activity of the FH2 domain but by the direct interaction with cMyBP-C. Although Iskratsch et al. (14, 34) have previously shown that CK2-mediated phosphorylation of the FH2 domain of Fhod3 is required for its localization, the relationship between Fhod3 phosphorylation and cMyBP-C binding is presently unknown.
The physiological role of Fhod3 at the C-zone is still largely unclear. One intriguing speculation is that Fhod3 is involved in the reorganization of actin filaments, possibly through the end-to-end annealing to the pointed end of thin filaments, as discussed in our previous papers (13, 15, 26), because thin actin filaments at the C-zone, where the cross-bridge occurs, are expected to be damaged by repetitive myosin-driven forces. Indeed, the contractility-dependent reorganization of actin filaments has been observed in cardiomyocytes (35, 36). Fhod3 is also expected to participate in the regulation of cross-bridge kinetics because it interacts directly with the N-terminal C0 domain of cMyBP-C, which mediates cross-bridge modulation via direct interaction with actin and myosin (11, 12, 37). Although the functional link between actin dynamics and cross-bridge regulation in the sarcomere is presently unknown, the interaction of Fhod3 with cMyBP-C may represent a novel mode of regulation of cardiac contractility.
Because the N-terminal fragment of Fhod3 interacts with cMyBP-C more strongly than the full-length Fhod3 does (Fig. 2F and SI Appendix, Fig. S7A), it seems likely that the binding of Fhod3 to cMyBP-C is regulated by a conformational change in Fhod3. It has been shown that Fhod1, a nonmuscle homolog of Fhod3, is usually kept in an inactive form through intramolecular interaction and can be activated by phosphorylation of the C-terminal region (38). Because the C-terminal region of Fhod3 is also phosphorylated and can be inactivated via intramolecular interaction (34), the phosphorylation of Fhod3 at the C-terminal region may induce a conformational change in Fhod3, thereby regulating its interaction with cMyBP-C. In addition, the binding also may be affected by a conformational change in the N-terminal region of cMyBP-C. Because phosphorylation alters the conformation of the N-terminal domains C0–C2 (39, 40), it is possible that the interaction between Fhod3 and cMyBP-C undergoes phosphorylation-dependent regulation, albeit substitutions for serine residues within the M domain had no effect on the interaction (Fig. 3A).
Although cMyBP-C–null mice show impairment of diastolic and systolic function with disarray of cardiomyocytes, no specific changes in the sarcomeric structure, including the striation of myosin heavy chain and titin (32) or ultrastructure of Z-lines, M-lines, and A-bands (31), have been detected. In the present study, we found that, in the sarcomere of cMyBP-C–null mice, Fhod3 fails to localize to the C-zone and distributes diffusely (Fig. 4C). The aberrant mislocalization of Fhod3 detected in the present study is a major structural change in the sarcomeres of cMyBP-C–null mice. Since a reduction in the protein levels of Fhod3 partially antagonizes the hypertrophic changes in cMyBP-C–null mice (Fig. 7), cardiac dysfunction caused by cMyBP-C depletion may be associated with adverse effects of mislocalized Fhod3. This hypothesis is supported by the finding that Fhod3 overexpression exacerbates cardiomyopathic changes in cMyBP-C–null mice (Figs. 5 and 6), although we cannot exclude the possibility that Fhod3 overexpression acts as a nonspecific second stressor. In addition, it may be possible that mislocalization of Fhod3 is involved specifically in cMyBP-C–related cardiomyopathy, since Fhod3 localization is not affected in the hearts of some cardiomyopathy patients (25). On the other hand, the majority of human patients with cMyBP-C–related cardiomyopathy are heterozygous, and those patients have a decreased amount of normal cMyBP-C protein. It should be investigated in the future whether such a decrease in the amount of cMyBP-C protein induces mislocalization of Fhod3, as discussed below.
Somewhat unexpectedly, overexpressed Fhod3 is not diffusely distributed, but instead accumulates at the A-band in the cMyBP-C–null heart. Although the reason for this accumulation is presently unknown, Fhod3 may have an intrinsic activity to bind myosin thick filaments besides cMyBP-C. In contrast to the accumulation of overexpressed Fhod3 at the A-band in the cMyBP-C–null heart, overexpressed Fhod3 in the presence of cMyBP-C was strictly colocalized with cMyBP-C at the C-zone (Fig. 1F) like endogenous Fhod3, despite high amounts of Fhod3 protein. Because cMyBP-C is a major sarcomeric protein constituting 2% of myofibrillar mass (41), sarcomeres may thus have a sufficient capacity for tethering an excess of Fhod3 protein to the C-zone. This seems to be the reason why overexpression of Fhod3 per se does not affect the cardiac function despite the toxicity of aberrant Fhod3. One important role of cMyBP-C may therefore be to tether Fhod3 to the C-zone to avoid the adverse effects of aberrant Fhod3.
The mislocalization of Fhod3 to the A-band in cMyBP-C–null mice seems to be harmful for the cardiac function, although the precise mechanism by which aberrant Fhod3 functions as a toxic factor at the A-band is presently unknown. Judging from the ultrastructural changes in the alignment of thick myosin filaments (Fig. 6 E and F), accumulated Fhod3 protein may affect the cross-bridge formation. The detailed mechanism should therefore be elucidated in future studies.
Materials and Methods
Transgenic mice expressing wild-type Fhod3CM [Fhod3Tg(α-MHC–Fhod3CM)], a mutant Fhod3CM-IA [Fhod3Tg(α-MHC–Fhod3CM-IA)], or a short variant Fhod3CM-S [Fhod3Tg(α-MHC–Fhod3CM-S)] under the control of α-MHC promoter were also generated on a C57BL/6 background. Mice carrying the Mybpc3tm1a(EUCOMM)Hmgu allele [B6Dnk;B6N-Mybpc3tm1a(EUCOMM)Hmgu/H] were obtained from the European Mouse Mutant Archive. All of the experimental protocol was approved by the Animal Care and Use Committee of Kyushu University (Permit no. A26-102) and the Animal Care and Use Committee of University of Miyazaki (Permit no. 2014-526-3). All mice were housed and maintained in a specific pathogen-free animal facility at Kyushu University or at University of Miyazaki, and all efforts were made to minimize the number of animals used and their suffering. A detailed description of the materials and methods used in this study is provided in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jeffery Robbins (Cincinnati Children’s Hospital Medical Center) for providing the α-MHC promoter; Dr. Takuo Yasunaga (Kyusyu Institute of Technology) for helpful discussion; Dr. Hideki Nishitoh (University of Miyazaki) for providing reagents; Mizuho Oda and Emiko Koba (Kyushu University) for the mass spectrometric analysis; Masato Tanaka and Kaori Nagatoshi (Kyushu University) for generation of transgenic mice; Sayo Yamamoto and Haruka Yagita (Kyushu University) for manipulation of mouse embryos; Kanako Motomura (Kyusyu University) and Ritsuko Sotomura (University of Miyazaki) for histological analysis; Yoshiteru Goto (University of Miyazaki) for transmission electron microscopic analysis; and Shoko Miura (Kyushu University), Ami Inayoshi (University of Miyazaki), and Asami Akiyama (University of Miyazaki) for secretarial assistance. For their technical support, we thank the Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences; the Laboratory for Technical Support, Medical Institute of Bioregulation, Kyushu University; and the Frontier Science Research Center, University of Miyazaki. This work was supported in part by the Japan Society for the Promotion of Science through a Grant-in-Aid for Scientific Research on Innovative Areas “Oxygen Biology: a new criterion for integrated understanding of life” Grant 26111009 (to H.S.), a Grant-in-Aid for Scientific Research (C) Grant 26460371 (to R.T.), a Grant-in-Aid for Scientific Research on Innovative Areas “Harmonized supramolecular motility machinery and its diversity” Grant 25117515 (to R.T.), and a Grant-in-Aid for Scientific Research on Innovative Areas–Platforms for Advanced Technologies and Research Resources “Advanced Bioimaging Support” Grant JP16H06280 (to R.T.). This work was also supported in part by grants from the Takeda Science Foundation (to R.T.) and the Institute of Seizon and Life Sciences (to R.T.); Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University (R.T.); the joint research program of Biosignal Research Center, Kobe University (R.T.); and President Strategic Priority Budget of University of Miyazaki (R.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716498115/-/DCSupplemental.
References
- 1.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 2.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: Clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 4.Kimura A. Molecular genetics and pathogenesis of cardiomyopathy. J Hum Genet. 2016;61:41–50. doi: 10.1038/jhg.2015.83. [DOI] [PubMed] [Google Scholar]
- 5.James J, Robbins J. Signaling and myosin-binding protein C. J Biol Chem. 2011;286:9913–9919. doi: 10.1074/jbc.R110.171801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadayappan S, de Tombe PP. Cardiac myosin binding protein-C as a central target of cardiac sarcomere signaling: A special mini review series. Pflugers Arch. 2014;466:195–200. doi: 10.1007/s00424-013-1396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrier L, Mearini G, Stathopoulou K, Cuello F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene. 2015;573:188–197. doi: 10.1016/j.gene.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moss RL, Fitzsimons DP, Ralphe JC. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ Res. 2015;116:183–192. doi: 10.1161/CIRCRESAHA.116.300561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Previs MJ, Michalek AJ, Warshaw DM. Molecular modulation of actomyosin function by cardiac myosin-binding protein C. Pflugers Arch. 2014;466:439–444. doi: 10.1007/s00424-013-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris SP, Lyons RG, Bezold KL. In the thick of it: HCM-causing mutations in myosin binding proteins of the thick filament. Circ Res. 2011;108:751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfuhl M, Gautel M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): Who does what, with what, and to whom? J Muscle Res Cell Motil. 2012;33:83–94. doi: 10.1007/s10974-012-9291-z. [DOI] [PubMed] [Google Scholar]
- 12.Craig R, Lee KH, Mun JY, Torre I, Luther PK. Structure, sarcomeric organization, and thin filament binding of cardiac myosin-binding protein-C. Pflugers Arch. 2014;466:425–431. doi: 10.1007/s00424-013-1426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi K, et al. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J Biol Chem. 2009;284:29873–29881. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iskratsch T, et al. Formin follows function: A muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J Cell Biol. 2010;191:1159–1172. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan-O M, et al. Mammalian formin Fhod3 plays an essential role in cardiogenesis by organizing myofibrillogenesis. Biol Open. 2012;1:889–896. doi: 10.1242/bio.20121370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 18.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shekhar S, Pernier J, Carlier M-F. Regulators of actin filament barbed ends at a glance. J Cell Sci. 2016;129:1085–1091. doi: 10.1242/jcs.179994. [DOI] [PubMed] [Google Scholar]
- 20.Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 22.Skau CT, Waterman CM. Specification of architecture and function of actin structures by actin nucleation factors. Annu Rev Biophys. 2015;44:285–310. doi: 10.1146/annurev-biophys-060414-034308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaya H, et al. Fhos2, a novel formin-related actin-organizing protein, probably associates with the nestin intermediate filament. Genes Cells. 2005;10:665–678. doi: 10.1111/j.1365-2443.2005.00867.x. [DOI] [PubMed] [Google Scholar]
- 24.Ushijima T, et al. The actin-organizing formin protein Fhod3 is required for postnatal development and functional maintenance of the adult heart in mice. J Biol Chem. 2018;293:148–162. doi: 10.1074/jbc.M117.813931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kan-o M, et al. Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One. 2012;7:e34765. doi: 10.1371/journal.pone.0034765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto N, et al. Transgenic expression of the formin protein Fhod3 selectively in the embryonic heart: Role of actin-binding activity of Fhod3 and its sarcomeric localization during myofibrillogenesis. PLoS One. 2016;11:e0148472. doi: 10.1371/journal.pone.0148472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig R, Offer G. The location of C-protein in rabbit skeletal muscle. Proc R Soc Lond B Biol Sci. 1976;192:451–461. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- 28.Ratti J, Rostkova E, Gautel M, Pfuhl M. Structure and interactions of myosin-binding protein C domain C0: Cardiac-specific regulation of myosin at its neck? J Biol Chem. 2011;286:12650–12658. doi: 10.1074/jbc.M110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruen M, Gautel M. Mutations in β-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J Mol Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 30.McConnell BK, et al. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1235–1244, and erratum (1999) 104:1771. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris SP, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 32.Carrier L, et al. Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc Res. 2004;63:293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Nixon BR, et al. Alterations in sarcomere function modify the hyperplastic to hypertrophic transition phase of mammalian cardiomyocyte development. JCI Insight. 2017;2:e90656. doi: 10.1172/jci.insight.90656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iskratsch T, et al. Two distinct phosphorylation events govern the function of muscle FHOD3. Cell Mol Life Sci. 2013;70:893–908. doi: 10.1007/s00018-012-1154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skwarek-Maruszewska A, Hotulainen P, Mattila PK, Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- 36.Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 37.Finley NL, Cuperman TI. Cardiac myosin binding protein-C: A structurally dynamic regulator of myocardial contractility. Pflugers Arch. 2014;466:433–438. doi: 10.1007/s00424-014-1451-0. [DOI] [PubMed] [Google Scholar]
- 38.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 2008;27:618–628. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colson BA, Thompson AR, Espinoza-Fonseca LM, Thomas DD. Site-directed spectroscopy of cardiac myosin-binding protein C reveals effects of phosphorylation on protein structural dynamics. Proc Natl Acad Sci USA. 2016;113:3233–3238. doi: 10.1073/pnas.1521281113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Previs MJ, et al. Phosphorylation and calcium antagonistically tune myosin-binding protein C’s structure and function. Proc Natl Acad Sci USA. 2016;113:3239–3244. doi: 10.1073/pnas.1522236113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.