Significance
Genetic variation of the complement factor H (CFH) gene family is associated with several complex diseases. Here, we have performed both long- and short-read sequencing of multiple humans and nonhuman primates in an effort to understand its complex evolutionary history. We find that this locus has evolved predominantly through incomplete segmental duplication and identify recurrent reuse of donor and acceptor duplications leading to CFHR fusion genes with diverse functions. Investigation of a large cohort of patients with age-related macular degeneration revealed multiple structural variation breakpoints and mutational burdens that cluster in specific domains of the CFH protein. These domains overlap sites showing signatures of natural selection, providing strong evidence for the shared role of selective pressure on diversity and disease.
Keywords: structural variation, CFH gene family, natural selection, age-related macular degeneration, AMD
Abstract
Structural variation and single-nucleotide variation of the complement factor H (CFH) gene family underlie several complex genetic diseases, including age-related macular degeneration (AMD) and atypical hemolytic uremic syndrome (AHUS). To understand its diversity and evolution, we performed high-quality sequencing of this ∼360-kbp locus in six primate lineages, including multiple human haplotypes. Comparative sequence analyses reveal two distinct periods of gene duplication leading to the emergence of four CFH-related (CFHR) gene paralogs (CFHR2 and CFHR4 ∼25–35 Mya and CFHR1 and CFHR3 ∼7–13 Mya). Remarkably, all evolutionary breakpoints share a common ∼4.8-kbp segment corresponding to an ancestral CFHR gene promoter that has expanded independently throughout primate evolution. This segment is recurrently reused and juxtaposed with a donor duplication containing exons 8 and 9 from ancestral CFH, creating four CFHR fusion genes that include lineage-specific members of the gene family. Combined analysis of >5,000 AMD cases and controls identifies a significant burden of a rare missense mutation that clusters at the N terminus of CFH [P = 5.81 × 10−8, odds ratio (OR) = 9.8 (3.67-Infinity)]. A bipolar clustering pattern of rare nonsynonymous mutations in patients with AMD (P < 10−3) and AHUS (P = 0.0079) maps to functional domains that show evidence of positive selection during primate evolution. Our structural variation analysis in >2,400 individuals reveals five recurrent rearrangement breakpoints that show variable frequency among AMD cases and controls. These data suggest a dynamic and recurrent pattern of mutation critical to the emergence of new CFHR genes but also in the predisposition to complex human genetic disease phenotypes.
The complement factor H (CFH) gene family cluster on chromosome 1q31.3 has long been recognized for its biomedical relevance to human disease. Candidate gene studies (1, 2), as well as the application of high-density single-nucleotide polymorphism microarrays (3–6) and massively (exome and targeted) parallel sequencing technologies (7, 8), have identified both common and rare mutations associated with susceptibility to complex disease [age-related macular degeneration (AMD), systemic lupus erythematosus (SLE), and atypical hemolytic uremic syndrome (AHUS)]. In particular, this locus is recognized as one of two major genetic contributors to risk of AMD (9–12), the leading cause of vision loss in the developed world.
Factor H is an abundant serum glycoprotein produced primarily in the liver, which is essential for regulating the alternative pathway of the complement system (13). Here, factor H acts at the level of C3 (14), resulting in down-regulation of alternative pathway-mediated complement activation and complement homeostasis. The N terminus displays complement regulatory activity by acting as a cofactor for factor I-mediated cleavage of C3b and facilitating the decay of C3 convertase (decay-accelerating activity) (15, 16). The C terminus of the protein mediates cell surface binding and target recognition with ligands C3b, C3d, and heparin (17, 18), and represents a critical domain for discrimination between self- and non–self-surfaces.
At the genomic sequence level, the CFH gene family comprises six genes spanning almost 360 kilobase pairs (kbp). The five CFH-related gene paralogs (CFHR3, CFHR1, CFHR4, CFHR2, and CFHR5) extend telomerically adjacent to the ancestral CFH gene, which includes four genes (CFHR1–CFHR4) embedded within a series of segmental duplications (SDs) arranged in tandem across the locus. The presence of these SD blocks renders this region genetically unstable and prone to unequal crossing over and gene conversion. Large population-based studies have identified several rare and common structural variants mediated by the SD architecture (19, 20). A common ∼84-kbp deletion that completely removes two CFH paralogs (CFHR3 and CFHR1) has been identified as one of the most population-stratified copy number variants (CNVs) in the human genome (20), with African (AFR) populations particularly enriched for the deletion allele (>50%) (21).
The common CFHR3/1 deletion haplotype associates with several complex genetic diseases, albeit with differential risk. The specific deletion, for example, is associated with protection in AMD but with risk for SLE and AHUS (6, 22, 23). In addition, there are reports of de novo rearrangements and rare disease-associated CNVs that show alternate breakpoint signatures and evidence of CFHR fusion proteins (24–26), most of which are associated with nonallelic homologous recombination (NAHR) and interlocus gene conversion (IGC) events between SD blocks (27). Moreover, the evolutionary juxtaposition of incomplete SD blocks has driven the emergence of novel CFH-like gene paralogs with overlapping, but diverse, functions distinct from their ancestral progenitors (28, 29). Incomplete SD of the CFH progenitor locus was likely critical to the neofunctionalization and subfunctionalization of CFHR genes (30).
Duplicated regions of the genome, such as the CFH gene family, are frequent sites of misassembly within reference genomes (31, 32) due to the difficulties in resolving closely related paralogous genes. In addition, studies of genetic diversity are often incomplete due to the complexity of structural variation and the limitations of having a single reference genome. For example, an examination of the most complete primate genome assemblies for this locus shows more than 93 gaps in the sequence assembly, with most corresponding to regions of recent SD. The goals of this project were to (i) reconstruct the complex evolutionary history of this locus by generating high-quality sequences from nonhuman primate (NHP) lineages (chimpanzee, gorilla, orangutan, macaque, and marmoset) and (ii) understand the complete sequence structure of this locus from a set of six diverse human haplotypes, including protective and at-risk disease haplotypes, as well as differences in their transcriptional potential. We develop specialized resources [large-insert bacterial artificial chromosome (BAC)/fosmid libraries] and apply long-read single-molecule real-time (SMRT) sequencing. We then use these data to investigate >2,400 AMD cases and controls to discover disease-associated protein-coding mutations, characterize structural variation breakpoints, and determine the IGC frequency within patients. Our results reveal a pattern of nonrandom and recurrent mutation where structural variation, disease susceptibility, and positive selection are linked.
Results
Copy Number Diversity.
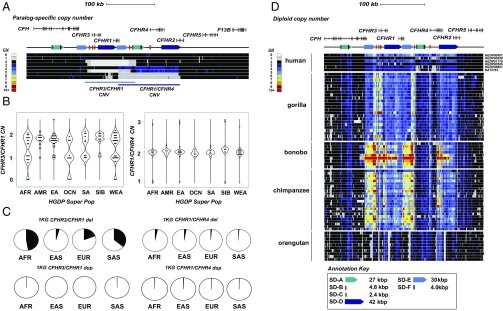
We initially assessed copy number variation at the CFH 1q31.3 locus by assessing read depth from genome sequence data mapped back to the human reference genome from 2,367 human [224 from Human Genome Diversity Project (HGDP) (33) and 2,143 from 1000 Genomes Project (1KG) (19)] and 86 NHP (34) genomes. Among humans, we readily distinguish two large copy number polymorphisms that are ∼84 kbp and ∼120 kbp in length and include three paralogous CFH-related genes (CFHR3, CFHR1, and CFHR4) (Fig. 1 A and B and SI Appendix, Fig. S1). The 84-kbp CFHR3/CFHR1 deletion is highly differentiated (maximum Vst = 0.28) between AFR and East Asian (EAS) populations and shows the highest allele frequency among AFR and South Asian populations (0.36 and 0.47, respectively) (Fig. 1C and Datasets S1 and S2). In particular, >57% of Yoruban and 41% of Gujarati individuals contain at least one copy of the deletion allele. The ∼120-kbp deletion of CFHR1 and CFHR4 is rarer but twofold more frequent among AFR and EAS populations than Europeans (Fig. 1C and Dataset S1). By contrast, the apparent reciprocal duplications of these deletion events occur at low frequency across all human populations.
Fig. 1.
CFHR copy number diversity in humans and great apes. (A) Schematic of the organization of the CFHR family (208 kbp) with respect to common copy number (CN) polymorphisms. Deletions (gray) and duplications (blue) are shown in the context of a heat map for nine human genomes [CN estimated based on mapping sequence reads to singly unique nucleotide k-mers (SUNKs)]. Gene models are shown in the context of SD orientation (colored arrows). (B) Scatter plots depicting CN estimates obtained using whole-genome sequencing data from 224 diverse humans from the HGDP. CN is estimated by sequence read depth using SUNK identifiers over CFHR3 and CFHR1 and CFHR1 and CFHR4. Frequency and Vst analyses are described in Datasets S1 and S2. (C) Pie charts per population group estimate the deletion/duplication allele frequency for CFHR3/CFHR1 and CFHR1/CFHR4 CNVs. AFR and South Asian (SAS) populations show enrichment of the CFHR3/CFHR1 deletion allele (0.35 and 0.47) relative to East Asian (EA/EAS) and European (EUR) populations (0.04 and 0.19). The rarer CFHR1/CFHR4 deletion is twice as frequent in AFR and EA/EAS (0.027 and 0.035) populations compared with EUR and SAS populations (0.008 and 0.01). (D) Diploid aggregate CN heat maps over CFHR SDs from 86 NHP genomes. Regions of CN expansion include the 3′ regions of CFHR4 and CFH. The chimpanzee and bonobo genomes demonstrate increased CN (six to 10 copies) for an ∼28-kbp segment containing the last three exons of the ancestral CFH. This segment is subject to independent and recurrent expansions in chimpanzees and gorillas.
Analysis among NHP species (chimpanzee, bonobo, gorilla, and orangutan) reveals distinct lineage-specific patterns of copy number variation. The sampled orangutan genomes show little copy number variation at 1q31.3, indicating a fairly static genome organization where many of the large-scale duplications occurred in the common ancestor of humans-gorillas-chimpanzees and bonobos (Fig. 1D). Among NHPs, we identified several regions of copy number expansion, including an ∼15-kbp region containing the 3′ exons of CFHR4 and an ∼28-kbp region containing the last three exons of CFH that had expanded to high copy among chimpanzees and bonobos (e.g., some bonobos carried between six and 10 copies of this duplicated segment). The most unstable region among NHPs maps to a 30-kbp segment (SD-E) that appears to have been subject to independent and recurrent expansions in both chimpanzees and gorillas (Fig. 1D). This sequence element also mediates the recurrent rearrangement associated with the CFHR1/CFHR3 deletion in humans, which is protective against AMD.
Sequence Assembly of Human Haplotypes and Breakpoint Analyses.
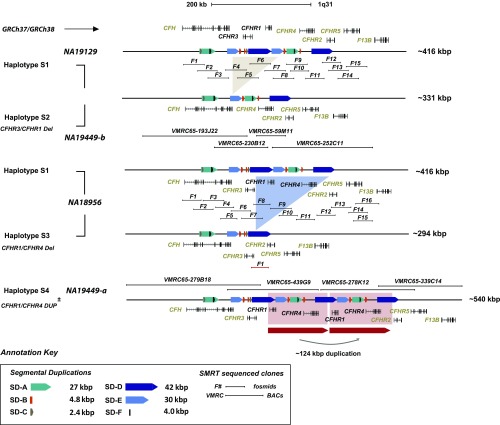
To gain insight into the structural diversity of the 1q31.3 CFHR locus, we sequenced and assembled 99 large-insert clones (BACs and fosmids) from 12 diverse human genome libraries using SMRT sequencing (SI Appendix, SI Materials and Methods and Datasets S3 and S4). We generated and deposited >7.2 Mbp of high-quality finished sequence, producing data from six alternative reference haplotypes, four of which contain alternate structural configurations (Fig. 2 and Dataset S5). To avoid confusion with previously defined single-nucleotide variant haplotypes (H1–H5) (1), we designate these alternative haplotype structures as S1–S4 (Fig. 2). The haplotypes range in size from 294 kbp (S3) to 540 kbp (S4), and their length varies almost exclusively due to unequal crossover between tandem SDs. We designate the human reference genome (GRCh37 and GRCh38) as S1 and identify two genomes [NA19129 (Yoruban) and CHM1 (hydatidiform mole)] with organizations consistent to the reference. S1 carries all five CFHR paralogs (CFHR1–CFHR5) and a series of SDs, the largest of which, SD-D (∼42 kbp) and SD-E (∼30 kbp), correspond to the breakpoints associated with common deletion CNVs (Dataset S6).
Fig. 2.
Alternative human structural haplotypes at the 1q31.3 locus. The sequence organization of four common structural haplotypes at 1q31.3 is compared. Annotations include the location of breakpoints associated with flanking SDs (colored arrows). Regions of CFHR gene gain and gene loss are represented by colored shading. Large-insert clone tiling paths [BAC (VMRC) and fosmid clone inserts (F designation)] used to assemble each haplotype are shown.
Using these high-quality sequences, we refine the breakpoint intervals associated with each structural haplotype, taking advantage of paralogous sequence variants (PSVs) (20) that distinguish SDs mediating the rearrangements (SI Appendix, SI Materials and Methods). Based on the S2 structural haplotype (SI Appendix, Figs. S2 and S3A) obtained from AFR samples NA18517 and NA19449, we initially define a region of 6.4 kbp within the duplication SD-E as the location of the breakpoints associated with the common AMD-protective 84.68-kbp CFHR1–CFHR3 deletion. HMMSeg (35) further refines the breakpoint to a 489-bp sequence interval mapping to a dense cluster of long interspersed nuclear element (LINE)/L1 repeat elements embedded within SD-E (SI Appendix, Fig. S4 and Dataset S7). It is interesting that this predicted breakpoint is flanked by several kilobases of perfect sequence identity [including the breakpoints defined previously (22)]. Sequence analysis shows that the proposed breakpoints map to an ∼15-kbp IGC hotspot between the SD-E paralogs, complicating precise localization of the breakpoints (SI Appendix, Fig. S5 and Dataset S8).
Similarly, using the S3 sequence assembly, we refine the CFHR1/CFHR4 (121,898 bp) deletion breakpoints to a 59-bp sequence interval completely contained within a LINE/L2 repeat element inside the SD-D duplication (SI Appendix, Fig. S3B and Dataset S7). Interestingly, sequence analysis of the supposed reciprocal duplication (S4) of the CFHR1–CFHR4 deletion event predicts that breakpoints differ. The S4 haplotype creates a 124-kbp tandem duplication block of 99.7% sequence identity. A multiple sequence alignment and HMMSeg refine the breakpoint to a 1,179-bp region corresponding to a cluster of LINEs and short interspersed nuclear elements (SINEs) (SI Appendix, Fig. S3C and Dataset S7). This repeat cluster maps 4.3 kbp downstream of the breakpoint associated with the AMD-protective CFHR1/CFHR4 deletion haplotype, and therefore does not represent the reciprocal product of an unequal crossover event.
Evolution of CFHR Duplications.
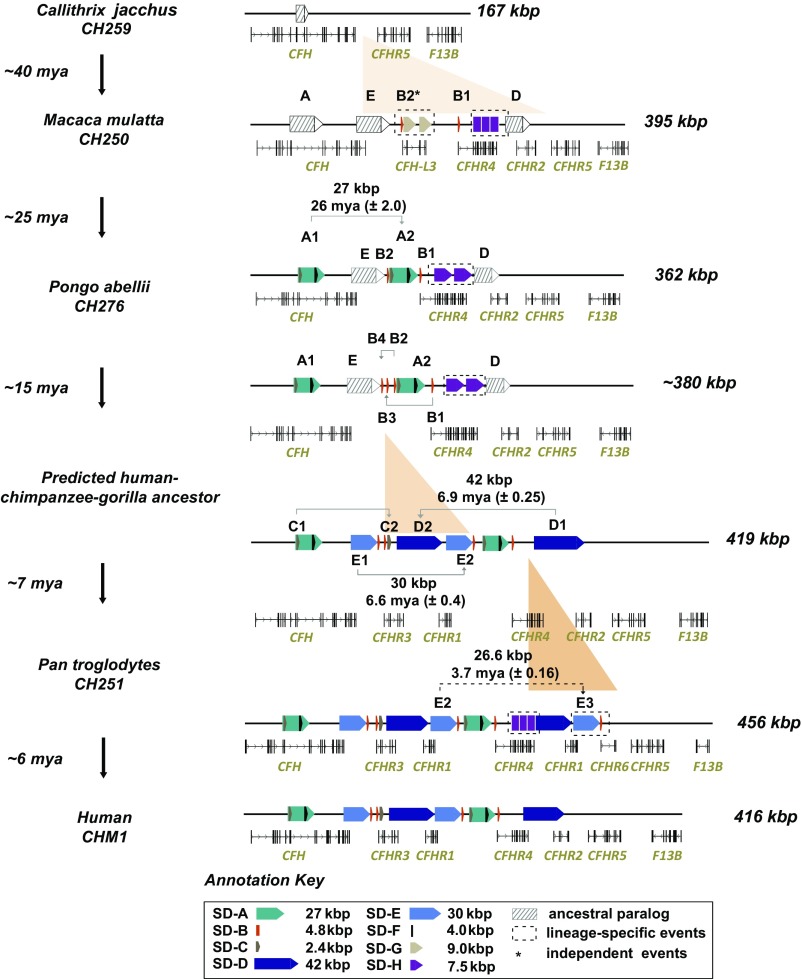
To build a model for the evolution of this locus, we sequenced and assembled 20 additional large-insert BAC clones from three NHP species (chimpanzee, gorilla, and orangutan) and representatives of the Old World (macaque) and New World (marmoset) monkey lineages (Fig. 3 and Dataset S3). To estimate the evolutionary age of the duplications, we construct a series of phylogenetic trees for each SD and estimate the divergence of the corresponding haplotypes (SI Appendix, Fig. S6). A comparison of these assembled sequences (∼1.92 Mbp) with current NHP genome assemblies showed that all existing reference sequences were incomplete and/or misassembled (SI Appendix, Figs. S7 and S8). In total, we resolve 93 euchromatic gaps, adding 218.4 kbp of sequence to these reference assemblies. This included missing and lineage-specific CFHR genes, SDs, and common repeat annotations (Datasets S9–S11). Over the last 40 My of primate evolution, we estimate that this locus has expanded about threefold almost entirely as a result of SD (Fig. 3). Based on the human CFH gene structure, we note that all SD events were incomplete but harbored protein-encoding exons with respect to the ancestral gene model. All duplications occurred in close proximity to their ancestral source (<100 kbp) (Fig. 3 and Dataset S6).
Fig. 3.
CFHR gene family evolution. Sequence structure and organization of the chromosome 1q31.3 CFHR locus are compared among primates (marmoset, macaque, orangutan, chimpanzee, and human) based on BAC clone sequencing. Gene models show CFHR gain and loss at different stages with respect to changes in the SD architecture (colored and shaded arrows represent recent and ancestral SDs, respectively). Sequence alignment and phylogenetic analyses are used to predict the timing and size of various duplications over 40 My of primate evolution (SI Appendix, SI Materials and Methods).
We predict at least seven evolutionary structural changes (ranging from 1.6 to 40.2 kbp in size) to reconcile the organization of the CFHR locus between modern humans and other primates (Fig. 3). Using the common S1 haplotype as a point of reference for human genome organization, we observe that CFHR4 and CFHR2 arose after divergence of the Old World and New World monkey lineages (∼35 Mya) as a result of independent duplications of CFH. In the case of CFHR4, exons 7, 8, and 9 formed a cassette that was subsequently tandemized, underwent exon exaptation, and became juxtaposed to a promoter element mapping to a 4.8-kbp segment (SD-B). The origin of the SD-B promoter element is complicated as no homology can be identified within the progenitor CFH locus, but we do identify it in association with other CFHR expansions in different mammalian lineages. For example, the mouse contains four copies of the promoter element associated with two rodent-specific CFHR paralogs. We also note that SD-B is present twice in the macaque in association with CFHR4 and a derived CFHR homolog, CFH-L3, which appears specific to Old World monkey lineages, and it is also part of a gorilla-specific duplication that could not be sequence-resolved at the level of clone-based assembly (SI Appendix, Fig. S9).
Next, a 26.9-kbp incomplete segment (SD-A) duplicatively transposed exons 8, 9, and 10 from CFH to a region immediately adjacent to the promoter of CFHR4 in the common ape ancestor (estimated 26 ± 2.0 Mya) (Fig. 3 and SI Appendix, Fig. S10). The breakpoints of this event map precisely between two direct SD-B elements (SD-B1 and SD-B2) that contain the promoter and first exon of CFHR4 (SI Appendix, Fig. S11). Three additional smaller duplication events likely occurred in the human-chimpanzee-gorilla ancestor. These included two independent duplications of the 4.8-kbp CFHR4 promoter (SD-B1 to SD-B3 and SD-B2 to SD-B4) and a 1.6-kbp duplication (SD-C) from CFH (exons 8 and 9) to the telomeric end of SD-B3 (Fig. 3 and SI Appendix, Fig. S11). The presence of a complete LINE/L1 element at the boundary of the SD-C duplication is suggestive of L1-mediated transduction as previously proposed for the origin of this smaller duplication (36).
The final series of SDs occurred concurrently or within a very narrow evolutionary window during the separation of human and African great ape lineages (5–8 Mya). These included a 40.2-kbp distal duplication (SD-D) in direct orientation (6.9 ± 0.25 Mya), creating truncated versions of CFHR4 (exons 7–10) and CFHR2 (exons 1–3). This was followed by a 28.6-kbp duplication (SD-E) containing C-terminal exons from CFH to the 3′ end of SD-D (6.6 ± 0.4 Mya). The arrangement of SD-E and SD-D in a “head-to-tail” configuration, in combination with the juxtaposition of SD-C and SD-B, established the most recent of the CFHR paralogs, CFHR3 and CFHR1, which are exclusive to the African great ape lineage. The last two duplications also create the genomic architecture necessary for the common/rare deletions associated with AMD protection. Interestingly, we mapped the breakpoints of these last duplications to the same 4.8-kbp CFHR4 promoter segment that defined the breakpoints for one of the initial duplication events (SD-A) (SI Appendix, Fig. S12). We note the other SDs that resulted in lineage-specific CFHR genes also show the SD-B element at the duplication breakpoints, including CFHR6 in the chimpanzee and CFH-L3 in the macaque (SI Appendix, Fig. S11). In total, these data argue that the promoter duplication SD-B served as a preferential target for the majority of SDs that led to the emergence of paralogous and lineage-specific CFHR genes.
Gene Innovation, Transcript Diversity, and Selection.
We identify seven distinct CFHR paralogs that arose as a result of duplication and juxtaposition of nine SDs in the primate lineage (Dataset S6). Notably, we sequence-resolved 25 gene-intersecting structural variants >50 bp in size among great apes (Table 1). The largest predicted ORF results from a 7.6-kbp tandem duplication of CFHR4 observed in all primates with the exception of humans and gorillas (Dataset S10). There are some general trends regarding the evolution of the gene family. First, we have determined that CFH exons 8 and 9 have been reused at least five times during the construction of these genes, suggesting that this particular domain has been a preferential donor of duplicated sequences. Notably, exon 9 is the same region where the common AMD risk variant (Y402H) has been mapped (1). Sequence analysis shows that the H402 variant can be identified among lesser apes, indicating it is at least ∼20 My old and may have arisen at the root of the ape lineage. Second, we find that SD-B defines the breakpoints of most primate duplication events, suggesting it has served as a preferential acceptor. Importantly, this segment corresponds to the CFHR4 promoter, and we determine that it has served as a 5′ transcript initiator for at least four CFHR gene fusions in the primate lineage that maintain an ORF (SI Appendix, Fig. S11). As an example, a chimpanzee-specific duplication of CFHR1 resulting from an additional copy of SD-E (SD-E3 estimated to have occurred 3.7 ± 0.16 Mya) contains this promoter duplication, defining the telomeric boundary of this event (SI Appendix, Fig. S13). The juxtaposition of the CFHR4 promoter duplication with two unique exons from CFHR2 (exons 4 and 5) results in a chimpanzee-specific 146-aa ORF, which we designate CFHR6. We amplified by RT-PCR the putative full-length ORF spanning both the predicted 5′ and 3′ untranslated regions (UTRs) using chimpanzee RNA generated from liver source material and confirmed expression of CFHR6 in the chimpanzee liver.
Table 1.
Gene-intersecting structural variation (>50 bp) detected among humans and NHPs
| Gene | GRCh37 coordinates | Primates | Size, bp | SV type |
| CFH | Chr1:196699022–196700021 | Orangutan | 999 | Deletion |
| CFH | Chr1:196681710–196682216 | Orangutan | 506 | Deletion |
| CFH | Chr1:196628274–196628275 | Chimpanzee, gorilla, orangutan | 50 | Insertion |
| CFH | Chr1:196714624–196714625 | Chimpanzee | 56 | Insertion |
| CFH | Chr1:196652722–196652723 | Gorilla | 323 | Insertion |
| CFH | Chr1:196706208–196706209 | Orangutan | 304 | Insertion |
| CFH | Chr1:196662187–196662188 | Orangutan | 5,586 | Insertion |
| CFH | Chr1:196636697–196636698 | Orangutan | 6,132 | Insertion |
| CFHR3 | Chr1:196761302–196761358 | Chimpanzee, gorilla | 327 | Deletion |
| CFHR3 | Chr1:196758756–196758764 | Chimpanzee, gorilla | 369 | Insertion |
| CFHR3* | Chr1:196734114–196748223 | Human | ∼14,110 | Deletion |
| CFHR1 | Chr1:196920353–196920890 | Chimpanzee | 26,596 | Duplication |
| CFHR3, CFHR1 | Chr1:196726714–196811885 | Human | 84,683 | Deletion |
| CFHR1, CFHR4 | Chr1:196782714–196904670 | Human | 121,898 | Deletion |
| CFHR1, CFHR4 | Chr1:196787137–196912332 | Human | 124,017 | Deletion |
| CFHR1, CFHR4† | Chr1:196763623–196763901 | Human | 125,164 | Deletion |
| CFHR4 | Chr1:196872498–196872844 | Orangutan | 346 | Deletion |
| CFHR4 | Chr1:196872237–196872238 | Orangutan | 79 | Insertion |
| CFHR4 | Chr1:196878738–196878739 | Orangutan | 177 | Insertion |
| CFHR4 | Chr1:196880789–196880844 | Chimpanzee | 7,630 | Insertion |
| CFHR4 | Chr1:196880789–196880844 | Orangutan | 15,584 | Insertion |
| CFHR2 | Chr1:196920020–196920095 | Gorilla | 75 | Deletion |
| CFHR5 | Chr1:196976072–196976443 | Gorilla | 371 | Deletion |
| CFHR5 | chr1:196961355–196961491 | Orangutan | 136 | Deletion |
| CFHR5 | chr1:196977326–196977327 | Orangutan | 208 | Insertion |
SV, structural variant.
Unable to estimate breakpoints by MIP resequencing.
Breakpoint size estimated by MIP resequencing.
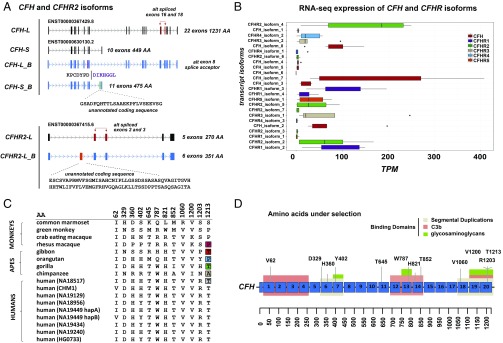
To understand the protein-coding potential and levels of transcript diversity for this gene family, we focused on the human organization and mapped isoform sequencing (Iso-Seq) data generated from liver source material to identify the gene models based on the most common European CFH structural haplotype S1. The nearly full-length cDNA data from Iso-Seq allows paralogs and isoforms to be more readily distinguished. In total, we identify 12 isoform transcripts (supported by at least one full-length read and more than one non–full-length read) that were previously unannotated by the GENCODE or RefSeq database (Dataset S12). We find no sequence support for seven of 12 GENCODE-predicted isoforms. We find that the most abundant patterns of alternative splicing occur at CFH, defining four short and four long isoforms (Fig. 4A). In contrast, CFHR5 is completely devoid of alternatively spliced transcripts, with all full-length reads representing the canonical 10-exon, 569-aa ORF. We identify unannotated exons for three spliced isoforms for CFH and CFHR2, with the former yielding an increase of 26 amino acids to the ORF of the canonical short CFH isoform (Fig. 4A). Additionally, we identify an unannotated exon 2 within the long isoform of CFHR2 that increases the ORF length by 81 amino acids. We also extend the 5′ and 3′ UTRs, and in five cases, we identify alternative splice donor/acceptor sites (Dataset S12). To quantitate expression differences between CFHR isoforms, we next analyze RNA-sequencing (RNA-seq) data generated from liver source material (Genotype-Tissue Expression project) (37) and map these data back to the isoform models. Of the 25 isoforms used in the analysis, we show that 56% (14 of 25) are highly expressed in the liver (Fig. 4B), with 67% (eight of 12) of the unannotated isoforms identified by long-read sequencing showing the highest expression patterns among CFH and the gene paralogs (Fig. 4B). While this analysis does not take into account haplotypic or intrinsic regulatory differences that would likely impact gene expression levels, it is interesting that four genes (CFH, CFHR1, CFHR4, and CFHR2) show the highest expression patterns for the noncanonical isoforms as annotated by GENCODE. Such data suggest that long-read Iso-Seq will have a substantial impact in annotating complex/duplication-rich regions of the genome.
Fig. 4.
Gene family selection and expression. (A) Iso-Seq identifies two main canonical isoforms of CFH: a long isoform (22 exons) and a short isoform (10 exons) (annotated in black), with unannotated isoforms depicting alternatively spliced exons and their coding potential. Similarly, Iso-Seq identifies two major CFHR2 transcripts, including unannotated isoforms containing alternatively spliced exons 2 and 3 (annotated in red) and an unannotated exon 2 (annotated in orange). (B) RNA-seq reads from liver source tissue [GTEx Consortium (37)] were used to estimate expression [transcripts per million (TPM)] of full-length PacBio-sequenced isoforms from CFH and its duplicate gene paralogs. CFH demonstrates the highest expression level in the liver; however, three of the youngest CFH gene paralogs (CFHR1, CFHR3, and CFHR2) are highly expressed relative to evolutionary older gene paralogs (CFHR5 and CFHR4). Box plots indicate median and interquartile range (IQRs) with outliers shown beyond 1.5 × IQR. (C) Twelve sites of positive selection (P < 0.05) are shown for CFH based on the ratio of nonsynonymous to synonymous changes (dN/dS) for the canonical ORF (1,231 amino acids). (D) Sites of positive selection are projected onto the CFH protein model with short consensus repeat domains annotated in blue, SDs annotated in gray, and binding domains annotated in green and orange.
Next, using sequence data generated as part of the 1KG, we searched for signatures of positive selection, restricting our analysis to unique regions of 1q31.3. Applying the extended haplotype homozygosity metric, we found no evidence of recent selective sweeps for any of the CFH/CFHR loci (CFH, CFHR5, and F13B) in the human population (SI Appendix, Figs. S14 and S15). Finally, using a high-quality sequence generated from both primate and multiple human haplotypes, we also investigated the ratio of nonsynonymous to synonymous variation (omega = ω) as a measure of selective pressure on CFH and its gene paralogs. We incorporated data generated from SMRT sequencing of large-insert clones (14 sequenced haplotypes and 21 fosmid inserts) and phased whole-genome sequencing short-read Illumina data from the HGDP (76 haplotypes) (Dataset S13). Only at the ancestral CFH locus did we observe an elevated nonsynonymous change (dN)/synonymous change (dS) value consistent with signals of positive selection (P = 1.14 × 10−17, dN/dS ω = 7.6) (280.66 kbp of coding sequence). At the protein level, we estimate that ∼3.5% of amino acid sites show signatures of positive selection and identify 12 unique amino acid sites that remain significant after correction (P < 0.05) (Fig. 4C and Dataset S13). Notably, these sites intersect sites of SD (seven of 12 of the positively selected amino acid sites) or functional binding domains such as C3b (seven of 12 domains) or glycosaminoglycan domains (five of 12 domains) (Fig. 4D). For example, we identify a cluster of three positively selected amino acids (1,200, 1,203, and 1,213) located in short consensus repeat (SCR) domain 20, a domain important for discriminating between self- and non–self-complement activation.
Disease Association.
We designed 405 molecular inversion probe (MIP) assays to distinguish and sequence CFH and CHFR paralogs (including F13B) in humans, taking advantage of singly unique nucleotide identifiers that distinguish paralogous copies (Dataset S14). We sequenced the coding portion of these genes in 1,574 AMD cases and 855 controls from three separate cohorts. Analysis of these cohorts replicated previous findings that showed an excess of common, private, and damaging missense mutations of the ancestral CFH in association with AMD (38) (Datasets S15–S17). Interestingly, we also found four significant AMD associations with coding mutations in CFHR paralogs, including a rare putative loss-of-function mutation [P = 0.003, odds ratio (OR) = 3.6, 95% confidence interval (CI) (1.7–9.8) adjusted for age and gender] and a severe missense mutation [P = 6.88 × 10−6, OR = 0.26, 0.95 CI (0.14–0.40) adjusted for age and gender] [combined annotation-dependent depletion (CADD) score = 23] in CFHR2 (Datasets S18 and S19).
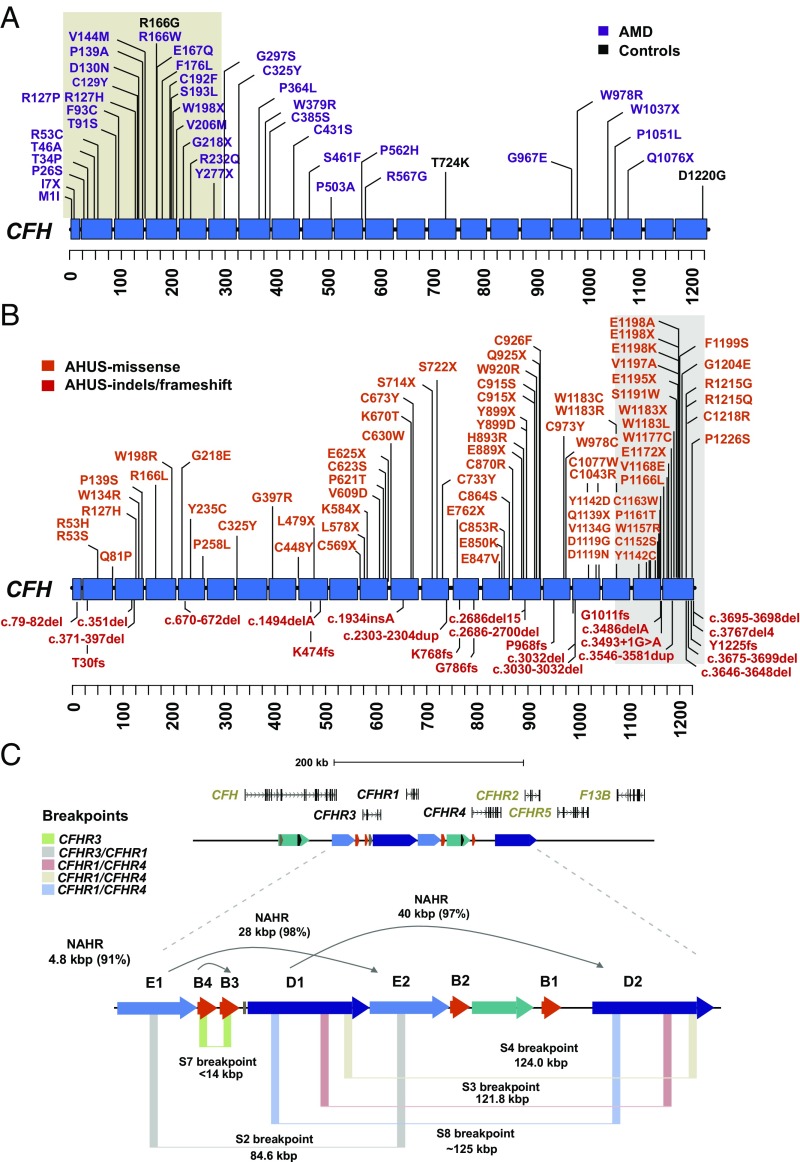
To create a more refined map of potential pathogenic mutations in the ancestral CFH, we combined our data with variant calls from five resequencing studies (38–41), which were limited to the CFH locus. In the combined set, we identify a total of 101 CFH missense mutations in >5,000 individuals (3,452 cases and 1,645 controls). Among this nonredundant set of missense mutations, 48.5% (49 of 101) have not been observed in Exome Aggregation Consortium (ExAC), with a further 42.6% (43 of 101) of mutations demonstrating a CADD score of >20 (Dataset S20). Likely gene-disruptive (LGD) mutations were identified solely in AMD cases at a frequency of 0.28% versus 0% in controls [P = 0.01987, OR = (1.37-Inf), Fisher’s exact test where Inf = Infinity]. Similarly, we identify 37 private (absent from ExAC) deleterious missense mutations (CADD score > 20) in AMD cases and one mutation in controls [1.07% vs. 0.06%; P = 6.29 × 10−6, OR = 17.9 (3.58-Inf), Fisher’s exact test]. This association became stronger when we restrict on frequency (<0.001), identifying 61 in AMD cases and three in controls [P = 5.81 × 10−8, OR = 9.8 (3.67-Inf), Fisher’s exact test] (Fig. 5A and Dataset S16). Notably, we identify four amino acids that have increased burden (more than four mutations) in AMD cases, with no occurrences identified in controls (R53C, R127P/H, D130N, and P562H). These are likely candidates for high-impact mutations among AMD patients; however, they did not reach statistical significance.
Fig. 5.
Recurrent and clustered mutations associated with disease. (A) Pattern of missense variation in >5,000 AMD cases (purple; n = 3,452) versus controls (black; n = 1,645) is plotted against a schematic of the CFH protein with SCR domains (blue). Missense mutations in cases are significantly clustered (tan highlighted region) around SCR domains 1–5 as determined by CLUMP analysis. (B) Pattern of missense, splice-site, and indel variation (152 total mutations) in AHUS cases (www.fh-hus.org/fullList.php?protein=FH) is similarly plotted. AHUS missense mutations significantly cluster around SCR domains 18–20 (highlighted). (C) Refinement of five NAHR-mediated breakpoints (colored vertical boxes) with respect to SD (colored horizontal arrows) and CFHR gene family organization indicates recurrent rearrangements. The size and percent sequence identity of the SDs that mediate the rearrangements are shown.
Previous studies have observed a nonrandom pattern of pathogenic mutations with respect to CFH protein domains (38, 42), namely, N terminus for patients with AMD and C terminus for patients with AHUS. Among the private deleterious missense mutations, we find that 59% (23 of 39) map to SCR domains 1–5 (Fig. 5A). Similarly, an analysis of 106 private missense mutations identified in the AHUS factor H database (43) shows that 43% (46 of 106) of mutations are located within SCR domains 19 and 20 (Fig. 5B). To assess the significance of these observations, we apply an unbiased clustering approach using CLUstering by Mutation Position (CLUMP) (44) to first compare the pattern of missense variation in AMD cases versus controls and then separately compared with a population control of 503 European individuals from the 1KG. In both cases, we identify significant clustering around medoids (CLUMP score = 1.19; P < 10−3) for AMD-associated missense mutations (Fig. 5A). As AHUS mutations are typically more deleterious than those described in AMD, we compare our list of mutations with 182 private missense alleles detected in ExAC. Once again, we observe significant clustering of AHUS mutations (CLUMP score = 3.1; P = 0.0079), but to the opposite end of the protein (Fig. 5B). Interestingly, we find that ∼53% (56 of 106) of private AHUS mutations and 28.6% (16 of 56) of AMD mutations map to SCR domains where we also report evidence of positive selection at the amino acid level, including the two missense mutations (Y402H and V62I), which tag as the reported risk (H1) and protective (H2) haplotypes in AMD. Using a random distribution of AMD and AHUS mutations across the CFH protein, we tested by simulation (10,000 permutations) whether AMD and AHUS variants were significantly more likely to fall in protein domains under selection. Notably, we find that AHUS variants are significantly enriched in these domains (P = 0.002); however, this did not reach statistical significance for AMD under this model (P = 0.084).
Recurrent CNVs, Mutational Burden, and Disease Risk.
CNVs of the CFHR gene cluster have been associated with three immune diseases (AHUS, SLE, and AMD); however, the breakpoints of these events, with a few exceptions (22), have not typically been resolved in patients beyond those afforded by microarrays. We took advantage of our diversity panel of sequence-resolved haplotypes coupled to the targeted resequencing of duplicate loci to identify and refine both rare and common CNVs in 2,427 AMD cases and controls. Since our sequencing assays focused on sequence differences among the duplicated CFHR genes, the assays allowed us to distinguish canonical from alternate breakpoint signatures mapping within the mediating SDs (SD-D and SD-E) (SI Appendix, Fig. S17). In total, we identify eight structurally distinct haplotypes with variable frequency in this cohort (Fig. 5C and Datasets S7 and S21), seven of which correspond to deletions and duplications of CFHR3, CFHR1, and CFHR4. Focusing on the structural variants thought to be protective against AMD, we identify five distinct NAHR-mediated structural variants: three corresponding to the CFHR1–CFHR4 and two corresponding to the CFHR1–CFHR3. We designate the nonreciprocal CFHR1–CFHR4 structural variant haplotype as S8 (breakpoints estimated to map ∼700 bp upstream of the 3′ UTR of CFHR4) and the nonreciprocal CFHR3 deletion haplotype as S7 (Fig. 5C). Due to the lack of informative PSVs that distinguish the S3 and S4 CFHR1–CFHR4 breakpoints (4.3 kbp apart), we could not distinguish these two haplotypes with certainty using our MIP genotyping assay (SI Appendix, Fig. S3 B and C).
Consistent with previous reports that deletions are protective (21, 22, 45, 46), we observe that control individuals are enriched for haplotypes carrying the deletion of CFHR3-1 [P = 2.2 × 10−16, OR = 0.35, 0.95 CI (0.30–0.41)] or CFHR1-4 [P = 0.01, OR = 0.43, 0.95 CI (0.22–0.83) adjusted for age and gender]. While we could not stratify all CFHR1-4 structural variants by haplotype (36 deletion and 4 duplication carriers), it is interesting that only the S4 duplication haplotype breakpoint maps within 565 bp of the 5′ UTR of CFHR2, while the other two deletion haplotypes (S3 and S8) have their breakpoints at least 5,000 bp away from this gene. This suggests that expression of CFHR2 may also be disrupted if a reciprocal deletion occurs on this haplotype. Similarly, for CFHR1–CFHR3 deletions, we find the protective association only with haplotype S2 but note that S7 could only be confidently assigned in one individual (and we are thus underpowered to assess its effect on AMD disease risk). The latter structural haplotype is interesting because the breakpoints map precisely to the SD-B “promoter” duplication, which was critical to the evolution of the gene cluster.
Discussion
Here, we performed targeted long-read sequencing of the CFH family in multiple humans and NHPs in an effort to understand the evolution, genetic diversity, and transcript potential of this locus as it relates to complex diseases, such as AMD and AHUS. Because recent SDs are enriched for misassembly, the high-quality sequence resource (10.6 Mbp) closes an estimated 93 gaps in primate genomes, discovers new paralogous genes, and significantly improves gene and genome annotation. Our evolutionary reconstruction and disease analysis highlight several important features.
First, incomplete gene duplication has been the predominant mechanism for evolutionary change at the CFHR locus. Specifically, we find no evidence of a complete duplication of the CFH locus; rather, the most common human genome organization arose as a result of at least 12 incomplete SDs, where each duplication event harbored a few protein-coding exons. This mechanism has the potential for rapid neofunctionalization (as well as pseudogenization) because the new duplicate genes at the time of their inception lack a complete ORF but differ radically in structure from the progenitor loci (47). Recent functional data for several human CFHR proteins support this observation (28, 29, 48, 49). For example, a recent study reported that CFHR3 functions as an inhibitor protein capable of blocking C3d-mediated B cell activation (48). The juxtaposition of three incomplete SDs at this position effectively rescued these partial gene duplications from pseudogenization, and CFHR3 thus acquired novel B cell regulatory function.
Second, the process of duplication appears highly nonrandom in that we observe preferential donor and acceptor sequences for all duplicative transpositions. Specifically, during primate evolution, an SD cassette corresponding to CFH exons 8 and 9 was duplicated at least five times, leading to the evolution of three of the CFHR family members. Similarly, we identify reuse of the CFHR gene promoter duplication (SD-B) as a preferential acceptor region. Our phylogenetic analysis shows that this ∼4.8-kbp sequence expanded on at least four occasions independently (copy number ranging from four to 12 copies), preceding and defining the breakpoints of larger evolutionary duplications in most primate lineages. For example, SD-B copy number expansion in ancestral African apes occurred before and defines the boundaries of the >85-kbp duplicative transposition that predisposes to common rearrangements associated with immune disease (6, 22, 23). Remarkably, this same sequence acts as a 5′ transcript initiator for at least four CFHR fusion genes, including a lineage-specific CFHR gene, CFHR6, in chimpanzees. The results suggest a particularly prolific and unstable genomic element driving transcription akin to the “core” duplicons that demarcate the boundaries of African ape SD expansion genome-wide (50, 51). While the evolutionary origin of the promoter duplication is uncertain, it is noteworthy that the 5′ UTR and exon of this cassette are conserved in the mouse, where they are associated with two rodent-specific CFHR fusion genes. In conjunction with CFH exons 8 and 9 donor duplications, this suggests broader reuse of this element during mammalian evolution.
Third, rare missense/LGD mutations associated with disease are nonrandomly distributed and cluster in distinct protein domains of CFH (N terminus for AMD and C terminus for AHUS). Consistent with previous observations, low-frequency deleterious mutations (CADD score > 20) are enriched among AMD cases and exhibit a distinct N-terminal clustering pattern (38). We observe a fivefold enrichment of LGD mutation and a tenfold enrichment of rare mutation (<0.001 frequency based on ExAC). This effect becomes stronger when restricting to private mutations [P = 6.29 × 10−6, OR = 17.9 (3.58-Inf)] (Dataset S16). Putative pathogenic missense/LGD mutations frequently map to canonically spliced exons (SI Appendix, Fig. S18), despite CFH exhibiting complex patterns of alternative splicing (Dataset S12). Importantly, missense and LGD mutations cluster in protein domains that also show signals of selection. In some cases, positive selection occurs at sites that are themselves susceptibility alleles for AMD and AHUS (V62, Y402, R1203, and V1200) (1, 52, 53). These protein domains are also binding sites for several pathogenic microbes (54–56), suggesting that natural selection has shaped a portion of the genetic variation at CFH underlying susceptibility to immune-associated diseases.
Finally, we provide evidence of recurrent and nonreciprocal deletions and duplications of the cluster. While previous studies have shown that both deletions CFHR3/1 (21, 22) and CFHR1/4 (45) are protective in AMD, our copy number analysis shows that these rearrangements occur as a result of five distinct classes of breakpoints with variable frequency in this cohort (Fig. 5C and Dataset S21). The S4 breakpoint, for example, occurs in close proximity to the promoter of CFHR2 and may also result in disruption of its expression. Notably, we identify a structural haplotype (S7) that likely removes the promoter and first exon of CFHR3. While this haplotype appears to be rare, it is interesting that the deletion is mediated by the same duplications (SD-B) critical in restructuring the locus throughout primate evolution. In total, these observations suggest that fine-scale mapping of CNV deletion breakpoints in combination with enhanced variant detection (particularly among duplicate gene paralogs) will be increasingly important for discriminating disease and at-risk haplotypes more broadly for this locus.
Materials and Methods
Detailed methods, including copy number genotyping, PacBio sequence and assembly, phylogenetic and evolutionary analysis, PacBio Iso-Seq, RNA-seq expression analysis, patient data, and Illumina-based short-read sequencing, are provided in SI Appendix. All groups collected data according to Declaration of Helsinki principles. At the Columbia center, the study was reviewed and approved by Columbia University Human Research Protection Office Institutional Review Board. In Melbourne, the study was approved by the Human Research and Ethics Committee of the Royal Victorian Eye and Ear Hospital. In Regensburg, the study was approved by the Ethics Committees at the University Eye Clinics of Würzburg (Study 78/01) and München (Study 226/02). Written informed consent was obtained for all study participants before participation.
Supplementary Material
Acknowledgments
We thank T. Brown for assistance with manuscript preparation. This work was supported, in part, by grants from the US NIH (Grant R01HG002385 to E.E.E. and Grant U41HG007635 to R.K.W. and E.E.E.). S.C. was supported by a National Health and Medical Research Council (NHMRC) C. J. Martin Biomedical Fellowship (1073726). P.N.B. was supported by an NHMRC Senior Research Fellowship (APP1138585). The Centre for Eye Research Australia receives operational infrastructure support from the Victorian Government. R.A. was supported, in part, by NIH Grants R01-EY013435 and P30-EY019007 and by an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology, Columbia University. E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: E.E.E. is on the scientific advisory board of DNAnexus, Inc.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited as a National Center for Biotechnology Information BioProject (accession no. PRJNA401648).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717600115/-/DCSupplemental.
References
- 1.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ying L, et al. Complement factor H gene mutation associated with autosomal recessive atypical hemolytic uremic syndrome. Am J Hum Genet. 1999;65:1538–1546. doi: 10.1086/302673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 4.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 5.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J, et al. BIOLUPUS Network; GENLES Network Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet. 2011;7:e1002079. doi: 10.1371/journal.pgen.1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raychaudhuri S, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seddon JM, et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013;45:1366–1370. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritsche LG, et al. AMD Gene Consortium Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–439, 439e1–439e2. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakobsdottir J, et al. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera A, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez de Córdoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sánchez-Corral P. The human complement factor H: Functional roles, genetic variations and disease associations. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Józsi M, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008;29:380–387. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Zipfel PF, et al. Factor H family proteins: On complement, microbes and human diseases. Biochem Soc Trans. 2002;30:971–978. doi: 10.1042/bst0300971. [DOI] [PubMed] [Google Scholar]
- 16.Zipfel PF, Heinen S, Józsi M, Skerka C. Complement and diseases: Defective alternative pathway control results in kidney and eye diseases. Mol Immunol. 2006;43:97–106. doi: 10.1016/j.molimm.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Oppermann M, et al. The C-terminus of complement regulator factor H mediates target recognition: Evidence for a compact conformation of the native protein. Clin Exp Immunol. 2006;144:342–352. doi: 10.1111/j.1365-2249.2006.03071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira VP, Herbert AP, Hocking HG, Barlow PN, Pangburn MK. Critical role of the C-terminal domains of factor H in regulating complement activation at cell surfaces. J Immunol. 2006;177:6308–6316. doi: 10.4049/jimmunol.177.9.6308. [DOI] [PubMed] [Google Scholar]
- 19.Sudmant PH, et al. 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudmant PH, et al. 1000 Genomes Project Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hageman GS, et al. Extended haplotypes in the complement factor H (CFH) and CFH‐related (CFHR) family of genes protect against age‐related macular degeneration: Characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38:592–604. [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes AE, et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 23.Zipfel PF, et al. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3:e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Challis RC, et al. A de novo deletion in the regulators of complement activation cluster producing a hybrid complement factor H/complement factor H-related 3 gene in atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2016;27:1617–1624. doi: 10.1681/ASN.2015010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyler SJ, et al. A novel hybrid CFHR1/CFH gene causes atypical hemolytic uremic syndrome. Pediatr Nephrol. 2013;28:2221–2225. doi: 10.1007/s00467-013-2560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis NJ, et al. A novel hybrid CFH/CFHR3 gene generated by a microhomology-mediated deletion in familial atypical hemolytic uremic syndrome. Blood. 2012;119:591–601. doi: 10.1182/blood-2011-03-339903. [DOI] [PubMed] [Google Scholar]
- 27.Heinen S, et al. De novo gene conversion in the RCA gene cluster (1q32) causes mutations in complement factor H associated with atypical hemolytic uremic syndrome. Hum Mutat. 2006;27:292–293. doi: 10.1002/humu.9408. [DOI] [PubMed] [Google Scholar]
- 28.Eberhardt HU, et al. Human factor H-related protein 2 (CFHR2) regulates complement activation. PLoS One. 2013;8:e78617. doi: 10.1371/journal.pone.0078617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinen S, et al. Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood. 2009;114:2439–2447. doi: 10.1182/blood-2009-02-205641. [DOI] [PubMed] [Google Scholar]
- 30.Dennis MY, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichler EE. Segmental duplications: What’s missing, misassigned, and misassembled–And should we care? Genome Res. 2001;11:653–656. doi: 10.1101/gr.188901. [DOI] [PubMed] [Google Scholar]
- 32.Chaisson MJP, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sudmant PH, et al. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Day N, Hemmaplardh A, Thurman RE, Stamatoyannopoulos JA, Noble WS. Unsupervised segmentation of continuous genomic data. Bioinformatics. 2007;23:1424–1426. doi: 10.1093/bioinformatics/btm096. [DOI] [PubMed] [Google Scholar]
- 36.Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL. Complement factor H: Sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Mol Immunol. 2000;37:41–52. doi: 10.1016/s0161-5890(00)00024-9. [DOI] [PubMed] [Google Scholar]
- 37.Lonsdale J, et al. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triebwasser MP, et al. Rare variants in the functional domains of complement factor H are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:6873–6878. doi: 10.1167/iovs.15-17432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duvvari MR, et al. Analysis of rare variants in the CFH gene in patients with the cuticular drusen subtype of age-related macular degeneration. Mol Vis. 2015;21:285–292. [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes AE, Meng W, Bridgett S, Bradley DT. Rare CFH mutations and early-onset age-related macular degeneration. Acta Ophthalmol. 2016;94:e247–e248. doi: 10.1111/aos.12822. [DOI] [PubMed] [Google Scholar]
- 41.Wagner EK, et al. Mapping rare, deleterious mutations in Factor H: Association with early onset, drusen burden, and lower antigenic levels in familial AMD. Sci Rep. 2016;6:31531. doi: 10.1038/srep31531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pérez-Caballero D, et al. Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet. 2001;68:478–484. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders RE, Goodship THJ, Zipfel PF, Perkins SJ. An interactive web database of factor H-associated hemolytic uremic syndrome mutations: Insights into the structural consequences of disease-associated mutations. Hum Mutat. 2006;27:21–30. doi: 10.1002/humu.20268. [DOI] [PubMed] [Google Scholar]
- 44.Turner TN, et al. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum Mol Genet. 2015;24:5995–6002. doi: 10.1093/hmg/ddv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivakumaran TA, et al. A 32 kb critical region excluding Y402H in CFH mediates risk for age-related macular degeneration. PLoS One. 2011;6:e25598. doi: 10.1371/journal.pone.0025598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritsche LG, et al. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD) Hum Mol Genet. 2010;19:4694–4704. doi: 10.1093/hmg/ddq399. [DOI] [PubMed] [Google Scholar]
- 47.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 48.Buhlmann D, et al. FHR3 blocks C3d-mediated coactivation of human B cells. J Immunol. 2016;197:620–629. doi: 10.4049/jimmunol.1600053. [DOI] [PubMed] [Google Scholar]
- 49.Goicoechea de Jorge E, et al. Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc Natl Acad Sci USA. 2013;110:4685–4690. doi: 10.1073/pnas.1219260110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antonacci F, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat Genet. 2014;46:1293–1302. doi: 10.1038/ng.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohajeri K, et al. Interchromosomal core duplicons drive both evolutionary instability and disease susceptibility of the Chromosome 8p23.1 region. Genome Res. 2016;26:1453–1467. doi: 10.1101/gr.211284.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bresin E, et al. European Working Party on Complement Genetics in Renal Diseases Combined complement gene mutations in atypical hemolytic uremic syndrome influence clinical phenotype. J Am Soc Nephrol. 2013;24:475–486. doi: 10.1681/ASN.2012090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westra D, et al. Genetic disorders in complement (regulating) genes in patients with atypical haemolytic uraemic syndrome (aHUS) Nephrol Dial Transplant. 2010;25:2195–2202. doi: 10.1093/ndt/gfq010. [DOI] [PubMed] [Google Scholar]
- 54.Blackmore TK, Fischetti VA, Sadlon TA, Ward HM, Gordon DL. M protein of the group A Streptococcus binds to the seventh short consensus repeat of human complement factor H. Infect Immun. 1998;66:1427–1431. doi: 10.1128/iai.66.4.1427-1431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meri T, et al. Microbes bind complement inhibitor factor H via a common site. PLoS Pathog. 2013;9:e1003308. doi: 10.1371/journal.ppat.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.