Significance
Transcription by RNA Polymerase II (Pol II) is a highly dynamic process that is tightly regulated at each step of the transcription cycle. We generated GFP-RPB1 knockin cells and developed photobleaching of endogenous Pol II combined with computational modeling to study the in vivo dynamics of Pol II in real time. This approach allowed us to dissect promoter-paused Pol II from initiating and elongating Pol II and showed that initiation and promoter proximal pausing are surprisingly dynamic events, due to premature termination of Pol II. Our study provides new insights into Pol II dynamics and suggests that the iterative release and reinitiation of promoter-bound Pol II is an important component of transcriptional regulation.
Keywords: RNA Polymerase II, transcription, transcription dynamics, promoter-proximal pausing, live-cell imaging
Abstract
Initiation and promoter-proximal pausing are key regulatory steps of RNA Polymerase II (Pol II) transcription. To study the in vivo dynamics of endogenous Pol II during these steps, we generated fully functional GFP-RPB1 knockin cells. GFP-RPB1 photobleaching combined with computational modeling revealed four kinetically distinct Pol II fractions and showed that on average 7% of Pol II are freely diffusing, while 10% are chromatin-bound for 2.4 seconds during initiation, and 23% are promoter-paused for only 42 seconds. This unexpectedly high turnover of Pol II at promoters is most likely caused by premature termination of initiating and promoter-paused Pol II and is in sharp contrast to the 23 minutes that elongating Pol II resides on chromatin. Our live-cell–imaging approach provides insights into Pol II dynamics and suggests that the continuous release and reinitiation of promoter-bound Pol II is an important component of transcriptional regulation.
Eukaryotic gene expression is a highly regulated process initiated by the sequential binding of transcription factors that facilitate the recruitment of RNA polymerase II (Pol II) and the assembly of the preinitiation complex (PIC) (1). During initiation the CDK7 subunit of transcription factor (TF) IIH phosphorylates the serine (Ser) 5 of the C-terminal domain (CTD) of RPB1, the core catalytic subunit of Pol II, allowing Pol II to engage the DNA template and to start transcribing a short stretch of RNA (2, 3). This early elongation complex is paused 30–60 nucleotides downstream of the transcription start site by negative elongation factor (NELF) and DRB sensitivity-inducing factor (DSIF) (4, 5). The subsequent pause release into productive elongation is mediated by the positive transcription elongation factor b (P-TEFb), whose Cdk9 kinase converts DSIF into a positive elongation factor, facilitates the eviction of NELF, and phosphorylates the RPB1 CTD on Ser2 (2).
Traditionally it was thought that transcription is primarily regulated by Pol II recruitment and initiation, but because of the advances in genome-wide sequencing technologies, we know today that mRNA output is also controlled by the tight coordination of postinitiation steps (2, 5). For example, promoter-proximal pausing is a key rate-limiting step of RNA synthesis that serves as a checkpoint for 5′ capping of the nascent RNA and maintains an open chromatin structure near promoters (6). Originally discovered as a regulatory switch of stimulus-responsive genes in Drosophila (7), recent genome-wide studies have revealed that promoter pausing is a widespread phenomenon occurring on most metazoan genes (5, 8). However, despite this prevalence, the dynamics of promoter-paused Pol II remain under debate. The currently prevailing model suggests that Pol II pauses at promoters with a half-life of 5–15 min (8–12), serving as an integrative hub to control pause release into productive elongation, while promoter-proximal termination is infrequent. However, conflicting studies have reported that promoter-paused Pol II is less stable due to repeated premature termination and chromatin release proximal to the promoter, which is accompanied by the release of short transcription start site-associated RNAs (13–16).
Thus far, genome-wide dynamics of promoter-paused Pol II have been studied by Gro-Seq (8), ChIP-Seq (10, 11), or methyltransferase footprinting (15) after inhibiting Pol II initiation. While these techniques provide gene-specific snapshots of Pol II transcription, relative abundance, or position at a given time, they do not allow measurement of steady-state Pol II kinetics (i.e., chromatin binding times) in real time. Although these studies have gained insights into the turnover of paused Pol II, most experiments have been performed after inhibiting transcription initiation by Triptolide (8, 10–12). This covalent XPB inhibitor severely affects Pol II levels (17, 18) and has been recently shown to have a slow mode of action (16), which makes it less suitable to study a potentially rapid cellular process. To overcome these limitations, we developed photobleaching of endogenously expressed GFP-RPB1 followed by computational modeling to quantitatively assess the kinetics of Pol II in unperturbed living cells.
Here we show that GFP-RPB1 knockin (KI) cells generated by CRISPR/Cas9-mediated gene targeting are fully functional and provide a promising tool to study the steady-state kinetics of endogenous Pol II. By photobleaching of GFP-RPB1, we identified three kinetically distinct fractions of chromatin-bound Pol II. Using Monte Carlo (MC) -based modeling of Pol II kinetics, we assessed the quantitative framework of the Pol II transcription cycle and elucidated its timeframe and quantitative set-up. Our findings are highly supportive of a model in which Pol II initiation and promoter pausing are highly dynamic events of iterative cycles of Pol II chromatin binding and release.
Results
Generation and Characterization of GFP-RPB1 Cells.
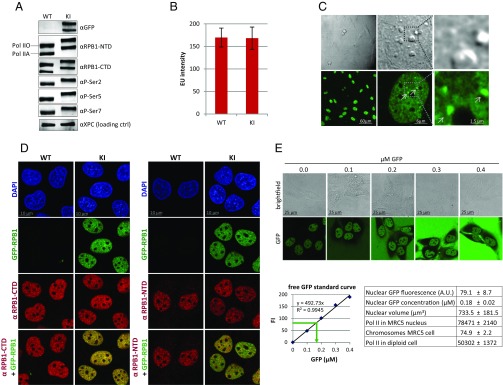
To study the in vivo kinetics of endogenous Pol II, we generated a GFP-RPB1 (POLR2A) KI cell line (MRC-5 sv40) fluorescently labeling the largest subunit of Pol II. We transiently expressed a single-guide RNA (sgRNA) to induce a CRISPR-associated protein 9 (Cas9)-mediated double-strand break (DSB) downstream of the RPB1 transcriptional start site. A repair template containing GFP cDNA flanked by homology arms comprised of the genomic RPB1 sequence (19) (Fig. S1A) was cotransfected to allow repair of the DSB by homologous recombination. GFP expressing cells were isolated by FACS and single-cell clones of homozygous KI cells were selected (Fig. S1B) in which WT RPB1 was replaced by GFP-RPB1, as shown by Western blot (Fig. 1A) and genotyping (Fig. S1C). Importantly, the GFP-RPB1 expression level and phosphorylation status in KI cells were similar to WT cells (Fig. 1A), the GFP tag did not compromise the basal transcription rate (Fig. 1B and Fig. S1D), and did not alter gene-expression profiles (Fig. S1E). Live-cell confocal imaging of KI cells revealed a nonhomogenous, nuclear distribution of GFP-RPB1 with multiple bright foci and exclusion from nucleoli (Fig. 1C). Localization of GFP-RPB1 was similar to endogenous RPB1, shown by immunofluorescent staining of the RPB1 CTD and N-terminal domain (NTD) (Fig. 1D). Taken together, these data illustrate that GFP-RPB1 is fully functional.
Fig. 1.
Characterization of GFP-RPB1 KI cells. (A) Western blot of MRC-5 WT and GFP-RPB1 KI cells. Pol IIA, hypophosphorylated Pol II; Pol IIO, hyperphosphorylated Pol II; P-Ser, phosphoserine. (B) EU incorporation levels of WT and KI cells. Mean total nuclear fluorescence intensity (FI) ± SD, n = 60 cells, two independent experiments. (C) Live-cell images of KI cells. Arrows indicate foci of locally enriched Pol II. (D) Immunofluorescent stainings of RPB1-CTD (Left) and RPB1-NTD (Right) in WT and KI cells. DNA is stained with DAPI, GFP fluorescence from endogenously expressed GFP-RPB1. (E) Representative images of KI cells with increasing concentrations of free GFP in the culture medium. Extracellular GFP fluorescence was plotted against GFP concentration to make a standard curve (graph, Lower Left) and nuclear Pol II concentration was calculated based on the standard curve (indicated with the green arrow). Numbers used to calculate the amount of Pol II in a diploid nucleus are summarized in the table (Lower Right). Table shows mean ± SD, n = 40 cells of two independent experiments.
Because GFP-RPB1 is expressed from its endogenous gene loci and RPB1 only translocates to the nucleus as part of the fully assembled Pol II complex (20), nuclear GFP fluorescence can be used as a direct readout for endogenous Pol II localization and concentration in living cells. To estimate the number of Pol II complexes, we compared the nuclear GFP intensity of KI cells to the extracellular fluorescence of known, increasing concentrations of recombinant fluorescent GFP added to the culture medium (Fig. 1E). This direct comparison of fluorescence on the same microscope slide revealed a nuclear Pol II concentration of 0.18 µM. Based on the determined average nuclear volume of the KI cells of 734 µm3, this equals ∼50,000 Pol II per diploid genome (Fig. 1E), which is within the range of previously determined Pol II quantities (21).
Initiation, Promoter Pausing, and Productive Elongation Are Characterized by Distinct Pol II Kinetics.
This multitude of Pol II complexes is divided over the different stages of the transcription cycle, including initiation, promoter pausing, and productive elongation. Previous studies have indicated that Pol II mobility is linked to its state of engagement during transcription (22–24); however, thus far promoter-proximally paused Pol II could not be discriminated and quantified in living cells. To test whether this fraction could be identified in GFP-RPB1 KI cells expressing Pol II at endogenous levels, we determined the kinetic framework of the Pol II transcription cycle by fluorescence recovery after photobleaching (FRAP) (25, 26).
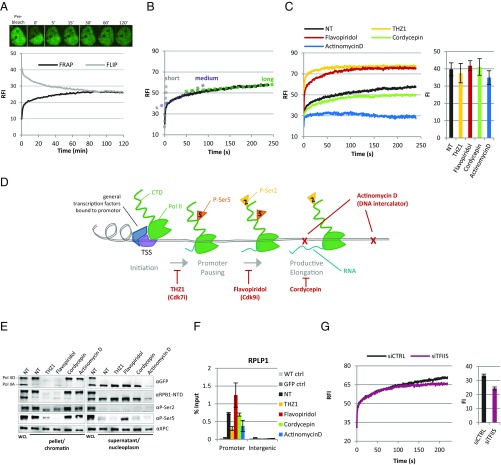
First, we measured the redistribution of Pol II after photobleaching in half the nucleus (Fig. 2A). Half-nucleus FRAP revealed an initial, fast redistribution of Pol II followed by a slow recovery of apparently less mobile Pol II. In line with previous studies (22–24), we assumed that the latter fraction represents long-term chromatin-bound, elongating Pol II that are released from chromatin over time when transcription terminates. Pol II fluorescence was fully recovered 80 min after the bleach pulse, likely reflecting the engagement of a small fraction of Pol II with very long or slowly transcribed genes. (Fig. 2A)
Fig. 2.
Real-time measurements of Pol II kinetics at different transcription cycle stages. (A) FRAP of the bleached and fluorescence loss in photobleaching (FLIP) of the nonbleached half of nuclei are plotted in time. Images are snapshots of a representative cell at indicated time points. RFI, Relative fluorescence intensity, Mean of n = 20 cells of two independent experiments. Image size is 15 × 18 μm. (B) Strip-FRAP analysis of GFP-RPB1 in NT cells. GFP-RPB1 was bleached in a narrow strip spanning the nucleus. Fluorescence recovery was measured every 0.4 s for 4 min, background-corrected, and normalized to prebleach fluorescence intensity. The dotted lines indicate kinetically distinct Pol II fractions, which mainly represent Pol II complexes that are short-term (gray), medium-term (purple), or long-term (green). Mean ± SD, n = 20 cells of two independent experiments. (C) Strip-FRAP of GFP-RPB1 in NT cells and after 1 h of treatment with the indicated transcription inhibitors. Column chart (Right) shows average GFP-RPB1 prebleach fluorescence intensities (FI) of cells analyzed by FRAP as a measure for Pol II protein levels. n > 16 cells per condition measured in two independent experiments. FI chart shows mean ± SD. (D) Schematic representation of Pol II transcription cycle and points of action of the used transcription inhibitors. THZ1 inhibits the phosphorylation of Ser5 by Cdk7. Flavopiridol inhibits the phosphorylation of Ser2 by Cdk9. Cordycepin is a 3′deoxy adenosine analog that stalls chain elongation when incorporated into the mRNA. Actinomycin D is a DNA intercalator. (E) Western blot of GFP-RPB1 KI whole-cell lysates (WCL) and chromatin and nucleoplasm fractions of NT cells and cells treated for 90 min with the indicated transcription inhibitors. NTD, N-terminal domain of RPB1. (F) Pol II binding to the RPLP1 promoter measured by chromatin-immunoprecipitation. WT and cells expressing free GFP (GFP) were analyzed as controls (ctrl). Mean ± SEM of n = 3 independent experiments. (G) FRAP of GFP-RPB1 after transfection with a nontargeting control siRNA (siCTRL) or siRNAs targeting the TFIIS paralogues TCEA1 and TCEA2 (siTFIIS). Mean of n = 20 cells of two independent experiments.
For a more detailed analysis of Pol II kinetics, we performed FRAP in a narrow strip spanning the nucleus (Strip-FRAP) (26), allowing fluorescence measurements every 0.4 s. In line with half-nucleus FRAP, Strip-FRAP of GFP-RPB1 in nontreated (NT) cells showed a long-term immobilization of a large fraction of Pol II (Fig. 2B), hereafter referred to as long-bound fraction (for clarification, approximated with a green dotted line in Fig. 2B). To investigate the nature of this long-bound fraction, we measured Pol II mobility after treatment for 1 h with 100 μM Cordycepin, a 3′-deoxy nucleoside that inhibits transcript elongation when incorporated into RNA (27) (Fig. 2D). Indeed, Cordycepin reduced 5′ethynyl uridine (EU) incorporation (Fig. S2 A and B) without affecting Pol II protein levels, determined by quantifying GFP fluorescence either before FRAP (Fig. 2C, Right), by immunofluorescence (Fig. S2B, Lower), or Western blot (Fig. 2E). The chromatin release of specifically the long-bound fraction was markedly decreased after Cordycepin (Fig. 2C), indicating that the long-bound fraction mainly represents elongating Pol II. Cordycepin-mediated inhibition of elongation also markedly delayed the redistribution of Pol II after half-nucleus FRAP (Fig. S2C), corroborating that the long-bound fraction mainly reflects the behavior of productively elongating Pol II. This allocation was further validated by the depletion of TFIIS (Fig. 2G and Fig. S2D). TFIIS stimulates the intrinsic cleavage activity of Pol II needed to reactivate complexes that have been arrested during productive elongation (28). Similar to that after Cordycepin (Fig. 2C), simultaneous siRNA-mediated knockdown of the TFIIS paralogues TCEA1 and TCEA2 (Fig. 2F and Fig. S2D) decreased the chromatin release of long-bound Pol II (Fig. 2G).
In addition to the large fraction of elongating Pol II with long-term chromatin binding, Strip-FRAP revealed a fraction of short-term chromatin-bound Pol II and a fraction with intermediate chromatin binding, hereafter referred to as the short-bound and the medium-bound fractions (approximated with gray and purple dotted lines, respectively, in Fig. 2B). To investigate the nature of these fractions, we performed Strip-FRAP using inhibitors that specifically arrest Pol II at defined, consecutive stages of the transcription cycle (Fig. 2D).
THZ1 inhibits CDK7 (29), the TFIIH subunit that phosphorylates Ser5 of the RPB1 CTD, and thereby prevents promoter pausing, mRNA capping, and subsequently Pol II transition into productive elongation (30). As expected, treatment with 1 μM of THZ1 for 90 min resulted in a loss of hyperphosphorylated Pol II (Pol IIo), P-Ser2 (Fig. 2E), and loss of EU incorporation (Fig. S2 A and B, Upper), indicating a run-off of most engaged, elongating Pol II, without affecting Pol II protein levels (Fig. 2C, Right and Fig. S2 A and B, Lower) or the cell cycle (Fig. S2E). In addition to the loss of promoter-paused Pol II (30), THZ1-treated cells showed a loss of phosphorylated Ser5 (P-Ser5) (Fig. 2E) and reduced Pol II promoter-binding (Fig. 2F). Accordingly, FRAP after THZ1 showed a decreased slope of the curve compared with NT cells at time points >100 s (Fig. 2C, yellow line): that is, a loss of elongating Pol II. In addition, THZ1 extensively enlarged the short-bound fraction of Pol II, indicating that Pol II is not stably chromatin-bound. This was confirmed by the increased detection of nonphosphorylated Pol II (Pol IIo) in the nucleoplasm after cellular fractionation (Fig. 2E), concomitant with a loss of Pol II in the pellet fraction. Because Pol II can still be incorporated into the PIC in the absence of CDK7 activity (30), this indicated that the short-bound fraction mainly represents free and initiating Pol II. Remarkably, the very fast replacement of bleached Pol II by new and fluorescent Pol II in THZ1-treated cells suggests that initiation is a highly dynamic process of continuous cycles of Pol II release and rebinding of new Pol II. In addition to the increase of short-bound Pol II, THZ1 treatment resulted in the loss of both the long- and the medium-bound Pol II fractions (Fig. 2C). As THZ1 prevents both Pol II promoter pausing and productive elongation (Fig. 2 D and F and Fig. S2 A and B) (30), and we assigned the long-bound fraction to productively elongating Pol II, we hypothesized that the medium-bound fraction might represent promoter-paused Pol II.
To test this, we inhibited the transition of Pol II from pausing to productive elongation with the CDK9 inhibitor Flavopiridol. The CDK9 activity of the PTEF-b complex licenses Pol II pause release by phosphorylating DSIF, NELF, and Ser2 of the RPB1 CTD (2, 8, 31, 32). As expected, inhibition of Pol II pause release by incubation with 1 µM Flavopiridol for 90 min increased Pol II promoter binding (Fig. 2F), and inhibited productive elongation (Fig. 2E and Fig. S2 A and B, Upper) (8, 31, 32). Similar to THZ1, Flavopiridol markedly enlarged the fraction of short-bound Pol II (Fig. 2C) and resulted in a loss of productively elongating Pol II, as shown by the horizontal slope of the slow fraction at time points >100 s. Most importantly and in striking contrast to THZ1, the medium-bound fraction of Pol II remained after Flavopiridol (Fig. 2C, yellow vs. red curve), strongly suggesting that Pol II with intermediate kinetics represent mostly promoter-paused Pol II. Similar results were obtained with another CDK9 inhibitor, DRB. Intriguingly, the fluorescence recovery after photobleaching in Flavopiridol-treated cells was complete after ∼100 s (Fig. 2C), which implied that within this short time frame all Pol II that were paused during the bleach pulse were already replaced by new, fluorescent Pol II. This Flavopiridol-induced accumulation of a fraction of Pol II that is only transiently chromatin-bound was corroborated by an accumulation of P-Ser5 modified Pol II in the nucleoplasm after cellular fractionation (Fig. 2E). Taken together, these results suggested that Pol II promoter pausing is surprisingly dynamic and presumably consists of iterative rounds of pause entry and promoter-proximal termination (i.e., release from chromatin and not release into productive elongation). Importantly, the expression of noncoding genomic loci, such as enhancer RNAs and upstream antisense RNAs, which would result in short Pol II chromatin binding times, has been shown to be dependent on CDK9 activity (33–35), and hence we conclude that their contribution to the medium-bound fraction, which is not reduced but rather increased after Flavopiridol (Fig. 2 C and F), is limited.
Treatment with 1 μg/mL of Actinomycin D, a DNA intercalator (36), for 90 min, completely inhibited transcription (Fig. S2 A and B, Upper). In line with the loss of nucleoplasmic Pol II after Actinomycin D (Fig. 2E), both the short and the medium-bound fractions were completely lost (Fig. 2C). Most Pol II remained chromatin-bound after Actinomycin D, resembling the Pol II FRAP curve in PFA-fixed cells, suggesting that after Actinomycin D most Pol II are stably trapped on DNA.
Computational Model of Steady-State Pol II Kinetics.
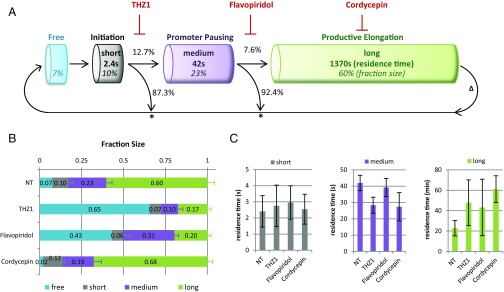
By inhibiting specific transitions in the Pol II transcription cycle, we demonstrated that Pol II engaged in initiating, promoter pausing, and productive elongation are characterized by significantly distinct kinetics. While the use of transcription inhibitors allowed us to assign kinetically distinct Pol II fractions to specific stages of the transcription cycle, we cannot exclude possible additional effects of these inhibitors on Pol II dynamics and stability. For example, it has recently been shown that Cdk9 inhibition by Flavopiridol also affects Pol II initiation (11). It is therefore important to extract information about Pol II kinetics from unperturbed cells. To do so, we used MC modeling to computationally simulate Pol II kinetics and fitted the simulated curves to the Pol II Strip-FRAP curves. MC modeling is based on the assumption that (molecular) events have a certain probability to occur within a specific short period of time (in a living cell) (37). As every transcription cycle starts with Pol II binding to the promoter, we defined one single binding step upon initiation (Fig. 3A). Supported by our analysis with specific transcription inhibitors (Fig. 2C), we proposed, in addition to freely diffusing Pol II, three interconnected Pol II fractions with three possible exit points: Pol II release after initiation, after promoter pausing, and after transcription termination (13–15, 24, 38) (Fig. 3A). The best simulations obtained from this model fit the experimental FRAP curves very well (Fig. S3 A–I). From the average of the 10 best-fitted MC simulations (Fig. S3 J–M), we extracted quantitative information about the relative size of the kinetically distinct Pol II fractions, their respective residence times on chromatin, and their binding constants (kon and koff). This approach favored a model in which 60% of all nuclear Pol II are productively elongating with a residence time on chromatin for ∼23 min on average (Fig. 3 B and C, Right). These results were in line with the large fraction of long-term chromatin-bound Pol II observed during FRAP experiments (Fig. 2 A and B). When combined with the number and length of expressed genes, these results allowed us to deduce the average Pol II elongation speed in unperturbed conditions. Therefore, we determined the number and average length of all actively transcribed genes in immortalized human fibroblasts using existing nascent RNA-Seq data (39) (Dataset S1). We included all genes with >0.2 reads per kilobase of transcript per million mapped reads (RPKM) and a gene length >300 bp in the analysis, which resulted in ∼13,600 actively transcribed genes with an average, expression frequency-corrected gene length of 46 kb (Dataset S1, sheet 2). Together, these numbers resulted in an average elongation speed of 2 kb/min in living cells (Dataset S1, sheet 3), which is in line with previously reported Pol II elongation speeds determined by run-on sequencing (8, 40).
Fig. 3.
MC modeling of live cell Pol II kinetics. (A) Schematic representation of possible binding and release steps of Pol II to DNA. Modeled fraction sizes are depicted as percentages and written in italics. Modeled residence times of Pol II in different stages are written in bold. Chances for Pol II to pass through or exit a stage were calculated from kon and koff rate constants (see Materials and Methods) and are given as percentages. An asterisk (*) marks points of abortive Pol II release, ∆ marks Pol II release after transcription termination. (B) Fraction sizes and (C) residence times of short-, medium-, and long-bound Pol II fractions obtained from MC-based modeling of GFP-RPB1 FRAP data (Fig. 2B) of NT GFP-RPB1 KI cells or after 1 h of treatment with 1 µM THZ1, or 1 µM Flavopiridol, or 100 µM Cordycepin. Mean ± SD of the 10 best-fitting simulations.
Promoter-Bound Pol II Are Rapidly Turned Over.
MC modeling of RPB1 kinetics further allowed us to extract relative fraction sizes and revealed that ∼10% of Pol II are involved in initiation (short-bound fraction) and ∼23% are promoter-bound during promoter pausing (medium-bound fraction). This implied that most Pol II (93%) are chromatin-bound and that only 7% are freely diffusing in the nucleus (Fig. 3 B and C). In line with the fast recovery of photobleached Pol II after THZ1 (<10 s) and Flavopiridol (<100 s), MC modeling of Pol II kinetics in unperturbed cells revealed that initiating Pol II remains chromatin-bound for only 2.4 s (Fig. 3C, Left) and promoter-paused Pol II for merely 42 s. Interestingly, these big differences in the residence time of initiating (2.4 s), promoter pausing (42 s), and elongating (23 min) Pol II implied that only a small fraction of initiating and pausing Pol II can proceed to the subsequent step of the transcription cycle, and hence that most initiating and pausing Pol II must be released from chromatin at the promoter (Fig. 3). In line with this, analysis of the rate constants of the distinct Pol II fractions showed that only 1 of 8 Pol II that attempt initiation will proceed to promoter pausing, and that only 1 of 13 promoter-paused Pol II will continue to productive elongation (Fig. 3A). When combined, these findings imply that only 1 of 100 transcription initiation events finally results in mRNA production.
Of note, the average of three independently modeled FRAP experiments resulted in highly similar estimates of fraction sizes and residence times and illustrates the variation between biological experiments (Fig. S4A). Furthermore, the modeled parameters were hardly affected by adding artificial noise to FRAP curves (Fig. S4B), further validating our MC-based modeling approach.
Modeling of Pol II Kinetics After Transcription Inhibition.
One of the current models of Pol II promoter pausing suggests that the release of promoter-paused Pol II occurs mainly by transition to productive elongation (5). Our results, however, indicated a very rapid turnover of promoter-paused Pol II, which most likely cannot not be explained solely by release of promoter-paused Pol II into productive elongation. Therefore, we tested whether Cdk9-mediated pause release influences the half-life of promoter-paused Pol II by determining the residence time of paused Pol II after blocking pause release with Flavopiridol. Interestingly, MC-based modeling indicated that Flavopiridol did not increase the residence-time of promoter-paused Pol II, (Fig. 3C, Center), illustrating that Pol II turnover at the promoter is hardly affected by Pol II release into productive elongation, and suggesting that the half-life of paused Pol II is mainly determined by promoter-proximal termination. Although Flavopiridol is known to accumulate paused Pol II at promoters by inhibiting the transition to productive elongation (8, 16, 41), it only resulted in a 35% increase of the fraction size of paused Pol II (Fig. 3B), which is comparable to observed increases in Pol II promoter occupancy (8) (Fig. S2E). The increase of paused Pol II was concomitant with a decrease of initiating Pol II and is in line with the recent finding that paused Pol II inhibits Pol II initiation (11).
Modeling of Pol II kinetics after THZ1 or Flavopiridol revealed a substantial increase in freely diffusing Pol II, whereas the fraction of initiating Pol II was slightly reduced (Fig. 3B), suggesting that Pol II initiation frequency does not directly depend on the number of freely diffusing Pol II. The very small fraction of elongating Pol II that remained after THZ1 and Flavopiridol resided longer on the gene body (Fig. 3C, Right) and might represent a subfraction of Pol II that did not run off within the 90 min of inhibitor treatment, likely because they transcribe very long genes or genes with a low elongation speed. Interestingly, Cordycepin increased the fraction size of elongating Pol II (Fig. 2B), and reduced the number of free Pol II (Fig. 2B). In addition to the shift of fraction sizes, Cordycepin increased the residence time of elongating Pol II threefold (Fig. 3C, Right), arguing for a strongly decreased elongation speed after treatment with this elongation inhibitor, without affecting transcription initiation. MC-modeled fraction sizes of Pol II were well in line with Pol II quantities detected in the chromatin-bound or nucleoplasmic fraction following cell fractionation, in both nontreated conditions or after treatment with different transcription inhibitors (Fig. S4C).
Kinetics of Pol II After Treatment with Triptolide or α-Amanitin.
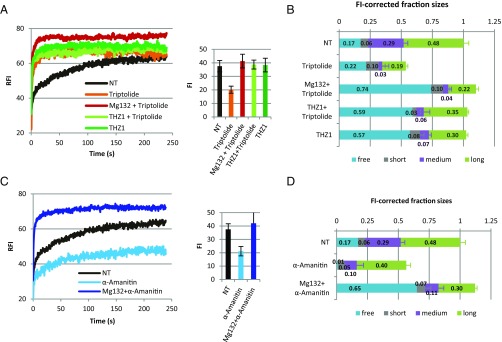
We demonstrated that GFP-RPB1 KI cells are a suitable tool to quantitatively assess real-time Pol II transcription kinetics and Pol II concentration. Next, we applied this tool to study the effects of Triptolide and α-Amanitin, two widely used compounds that inhibit Pol II transcription and induce Pol II degradation. Triptolide inhibits the ATPase activity of the TFIIH subunit XPB, thereby preventing the opening of the transcription bubble, which consequently blocks transcription and triggers Pol II degradation (18, 42, 43) (Fig. 4A, Right and Fig. S5A). Cells that were treated with 0.5 μM Triptolide for 90 min showed a sharp increase of the short-bound Pol II fraction and a loss of both the medium- and long-bound Pol II fractions (Fig. 4 A and B), similar to what we observed after THZ1 (Fig. 2C). The residence time of short-bound Pol II after Triptolide or THZ1 was similar to NT conditions (Fig. S5D), indicating that the turnover of initiating Pol II binding is barely affected by its transition to promoter pausing. In contrast to THZ1, 90 min of Triptolide resulted in a loss of ∼50% of Pol II compared with NT cells, as determined by live-cell imaging (Fig. 4A, Right). Triptolide-induced Pol II degradation could be rescued by proteasome inhibition (Mg132, 50 µM for 1 h before Triptolide), which further increased the short-bound Pol II fraction (Fig. 4A), most likely because of accumulation of free Pol II (Fig. 4B). This indicates that the fraction of Pol II that is targeted for degradation after Triptolide is highly mobile. Of note, Mg132 alone did not affect Pol II mobility or protein levels (Fig. S5B). Interestingly, rescue of Triptolide-induced Pol II degradation by Mg132 accumulated P-Ser5 Pol II, suggesting that CDK7 can phosphorylate CTD Ser5 independently of transcription bubble opening and that this Ser5-phosphorylation might be a prerequisite for Pol II degradation after Triptolide. To test this hypothesis, we inhibited the Triptolide-induced Ser5-phosphorylation with THZ1 (Fig. S5C). The addition of THZ1 completely rescued the Triptolide-induced Pol II degradation (Fig. 3A, Right), in line with a model in which in the absence of an open transcription bubble due to Triptolide, P-Ser5 Pol II fails to escape the PIC and is subsequently degraded.
Fig. 4.
Live-cell Pol II kinetics after Triptolide and α-Amanitin. FRAP of GFP-RPB1 in NT cells and cells treated with 0.5 µM Triptolide for 1 h (A) or 100 µg/mL α-Amanitin for 2 h (C). Mg132, proteasome inhibitor. Mean of n = 20 cells of two independent experiments, FI chart shows mean ± SD. Modeled, FI-corrected Pol II fraction sizes in NT KI cells or after treatment with Triptolide (B) or α-Amanitin (D). Mean ± SD of the 10 best-fitting simulations.
Another widely used transcription inhibitor is α-Amanitin. α-Amanitin traps Pol II in a conformation that prevents translocation of the transcript and thereby inhibits nucleotide incorporation (42). Thus far, the effect of α-Amanitin on Pol II in living cells could not be extensively studied, as most studies using fluorescently tagged Pol II were performed using an exogenously expressed α-Amanitin–resistant Pol II mutant (24, 44, 45). FRAP of GFP-RPB1 after treatment with 100 μg/mL α-Amanitin for 90 min showed a loss of free, short-bound, and medium-bound Pol II, whereas the long-bound, elongating fraction did not change in size (Fig. 4D), but increased in residence time (Fig. S5E), illustrating the reduced productive elongation rate after α-Amanitin (46). In addition, α-Amanitin led to degradation of Pol II (Fig. 4C, Right and Fig. S5A). Interestingly, rescue of α-Amanitin-induced Pol II degradation by Mg132 tremendously increased the free fraction of Pol II (Fig. 4D), as shown by an increase of the short-bound fraction during FRAP (Fig. 4C), suggesting that stalled and presumably ubiquitylated Pol II is evicted from chromatin before proteasomal degradation.
Discussion
Detailed knowledge of the dynamics of Pol II transcription is crucial to understand the complex regulation of Pol II transcription. To study the in vivo dynamics of endogenously expressed Pol II, we generated GFP-RPB1 KI cells and used them to monitor real-time Pol II kinetics. Photobleaching experiments revealed three kinetically distinct fractions of Pol II that remained chromatin-bound for either 2.4, 42, or 1,370 s (Fig. 3C). Using dedicated inhibitors that block transcription at specific stages, we allocated these fractions to Pol II engaged in initiation, promoter pausing, or productive elongation (Fig. 2B). Allocation of these fractions allowed us to model Pol II kinetics and to construct a kinetic framework of steady-state Pol II transcription in unperturbed conditions.
FRAP Allows the Discrimination of Promoter-Paused Pol II Based on Its Distinct Kinetics.
Interestingly, a previous study on the in vivo kinetics of YFP-tagged α-Amanitin–resistant Pol II on a lacO array integrated into U2OS cells also identified three different Pol II populations with chromatin binding times of 6, 54, and 517 s, respectively (24). The authors linked the fastest Pol II fraction to Pol II transiently interacting with promoters, while the slowest fraction was allocated to elongating Pol II. However, the fraction with intermediate kinetics was linked to initiating Pol II. Although the residence time of their intermediate fraction was strikingly similar to ours (54 s vs. 42 s), which would suggest that this might represent the same population of Pol II, their fraction was insensitive to P-TEFb inhibition, while we observed an increase in the fraction sizes of initiating and promoter-paused Pol II (Fig. 3B). Hence, our study discerned the kinetic fraction of Pol II that corresponds to promoter-paused complexes. While THZ1 and Triptolide abrogate promoter pausing and productive elongation, Flavopiridol still allows Pol II initiation and pausing but inhibits the release of Pol II from pausing to productive elongation. In line with this, we observed that in Flavopiridol-treated cells, the short- and the medium-bound Pol II fractions remained, while the long-bound fraction was lost, strongly suggesting that the medium-bound fraction mainly represents promoter-paused Pol II. Of note, Flavopiridol accumulated transcription-competent but promoter-paused Pol II, as tested by performing FRAP after washing out Flavopiridol after 60 min of treatment, which resulted in Pol II kinetics similar to NT cells within minutes.
Pol II Promoter Pausing Is a Highly Dynamic Event.
The average of the 10 best-fitting, simulated chromatin binding times of promoter-paused Pol II was 42 s in unperturbed conditions. Although we cannot exclude that Pol II is stably paused with residence times longer than 100 s on some specific loci, as for example shown by tracking photoactivatable Pol II at the uninduced Hsp70 transgene of Drosophila polytene chromosomes (9), we found that on a genome-wide average 23% of nuclear Pol II is paused for merely 42 s in human cells (Fig. 2 B and C, Center). While this result differs strikingly from previous studies that found Pol II to be stably paused at promoters with a half-life of 5–15 min (8–12), it is in line with the very fast (<100 s) fluorescence recovery of Pol II in Flavopiridol-treated cells (Fig. 2C). To our best knowledge, the most likely explanation for this discrepancy may be that these previous studies were performed in the presence of Triptolide (8, 10–12), which disturbs transcriptional regulation and Pol II stability (17, 18) (Fig. S5A). Furthermore, Triptolide may not be fully functional instantly upon administration and it is therefore difficult to exactly time the Pol2 half-life at promoters (15, 16). Stable pausing of Pol II on promoters also has been challenged recently by DNA foot-printing experiments in Drosophila cells, that have revealed that most promoter-paused Pol II is lost within 2.5 min (the earliest time point assessed by the authors) after Triptolide (15). Rapid promoter-proximal termination was also found to be the most plausible explanation for the drastic increase of promoter-paused Pol II within a minute after H2O2 administration to U2OS cells (16). In agreement with these conclusions, our findings strongly support a nonprocessive model of transcriptional idling of promoter-paused Pol II, defined by iterative rounds of Pol II pause entry and promoter-proximal termination (13–16). As blocking the transition to productive elongation with Flavopiridol did not increase the residence-time of promoter-paused Pol II (Fig. 3C, Center), we further suggest that the Pol II turnover at the promoter is independent of its release into productive elongation but is mainly a result of continuous premature termination (15). Importantly, our findings additionally elucidate the timeframe and the quantitative set-up of these processes. In light of the increasing evidence supporting the dynamic turnover of promoter-bound Pol II, it is very tempting to speculate that the release of Pol II into productive elongation might be, at least partially, regulated by inhibiting promoter proximal Pol II termination.
Not Only Promoter-Paused, but also Initiating Pol II Complexes Are Rapidly Turned over.
In addition to termination of Pol II after promoter pausing, our results further suggested that Pol II is frequently released from chromatin after initiation (Fig. 3A). This abortive release after 2.4 s occurred even when the transition from initiation to pausing was blocked by THZ1 (Fig. 3C) or Triptolide (Fig. S5D), implying that the turnover of initiating Pol II does not depend on its transition to promoter pausing. The very fast recovery of fluorescence after photobleaching of initiating Pol II after THZ1 and Triptolide (Fig. 2C) additionally implied that the Pol II that is abortively released after initiation (which is bleached during FRAP) is not the same complex that reinitiates (which is fluorescent and causes the fast fluorescence recovery). Similar observations have been made for the TATA-box binding protein (47, 48), further corroborating a model in which Pol II PICs are rapidly assembled and disassembled, rendering initiation highly dynamic. However, it has also been reported that a subset of the transcription machinery remains at the promoter after transcription initiation, forming a reinitiation scaffold that facilitates high levels of transcription initiation (49).
Implications of Dynamic Pol II Turnover at Promoters.
As a consequence of the frequent abortive release after initiation and pausing, according to our model, only one mRNA transcript will be synthesized per 100 Pol II initiation events on average. Although this seeming inefficiency of transcriptional initiation, which is consistent with Pol II measurements on a lacO array (24), is intriguing, it highlights the importance of initiation and promoter pausing as key regulatory control events (2). However, the exact purpose of Pol II idling during initiation and pausing remains unknown. The continuous rounds of initiation and termination may contribute to keep promoters free of nucleosomes at active genes (6, 50), or alternatively, idling may be important to maintain promoter-enhancer contacts (51). Pol II idling at promoters would also facilitate transcriptional bursting, which refers to the episodic release of multiple Pol II during a short period of time (52, 53).
Quantitative Framework of the Pol II Transcription Cycle.
In addition to the assessment of Pol II dynamics, endogenous expression of GFP-RPB1 allowed us to estimate the number of nuclear Pol II complexes. This analysis resulted in ∼50,000 in a diploid cell (Fig. 1E). With the assumption that productive elongation represents a single kinetic step, the fraction size of elongating Pol II was modeled to 60%, approximating to ∼30,000 elongating Pol II. Combined with the expression frequency-corrected average gene length of 46 kb (Dataset S1, sheet 2) this suggests that on average ∼1.1 Pol II are elongating per gene body (Dataset S1, sheet 3). Of note, while some less-frequently expressed genes are probably not continuously transcribed in every cell and therefore do not have an engaged Pol II constantly, other highly expressed genes are likely continuously transcribed by multiple Pol II complexes (54). Modeling further indicated that 10% of nuclear Pol II are initiating, and 23% are paused proximal to the promoter, which equals ∼16,500 Pol II located at promoter regions. Compared with 30,000 Pol II dispersed over the average gene body of 46 kb, these 16,500 Pol II are bound to the relatively short promoter region, coinciding with the high density of Pol II at the transcription start site observed in ChIP-Seq experiments (11).
In summary, we show that GFP-RPB1 KI cells provide a powerful tool to study endogenous Pol II in living cells and uncover valuable insights into the mechanism and the dynamics of Pol II initiation and promoter pausing. We demonstrate that photobleaching combined with modeling is a valuable extension of the existing tools to study the dynamics of Pol II transcription.
Materials and Methods
Generation of GFP-RPB1 KI Cells and Cell Culture.
KI cells were generated from Sv40-immortalized MRC-5 fibroblasts, as described here (19). Detailed protocol can be found in SI Materials and Methods. Cells were cultured in a 1:1 mixture of Ham’s F-10 and DMEM (Gibco) supplemented with antibiotics and 10% FCS, at 37 °C; 20% O2, and 5% CO2 in a humidified incubator.
Immunofluorescence and EU Incorporation.
Click it-based EU incorporation was performed as described previously (55). A detailed protocol is in SI Materials and Methods. Immunofluorescence was performed as described in SI Materials and Methods. Transcription inihibitors, antibodies, and respective concentrations are listed in Dataset S2.
Cellular Fractionation, Western Blot Analysis.
The fractionation procedure was adapted from ref. 56 and is described in detail in SI Materials and Methods. Fractions were separated on a 6% SDS Page gel, blotted overnight at 40 V. Blots were blocked with 1.5% BSA in PBS and stained with primary antibodies listed in Dataset S2. Secondary antibodies were coupled to IRDyes (LiCor) and imaged with an Odyssey CLx infrared scanner (LiCor).
Live-Cell Confocal Microscopy and Quantification of Nuclear Pol II.
Live-cell imaging was performed on a Leica SP5 confocal laser scanning microscope with a HCX PL APO CS 63×, 1.40-NA oil-immersion lens. Images were recorded with a 488-nm Argon laser and a 500- to 600-nm bandpass filter. For the detailed RPB1 FRAP procedure, please see SI Materials and Methods. For Pol II concentration measurements eGFP (Biovision) was dissolved in PBS, serially diluted in culture medium, and the fluorescence intensity was measured alongside nuclear GFP-RPB1 fluorescence. For a detailed procedure, see SI Materials and Methods.
MC Modeling.
Experimental FRAP curves were fit by least squares, to a large set of computer simulation-generated FRAP curves that were computed based on a model that simulates diffusion of molecules (here Pol II), and binding to and releasing from immobile elements, (here chromatin) in an ellipsoidal volume (here the nucleus). For details, see SI Materials and Methods.
Detailed protocols for RNA-Seq data analysis, Chromatin immunoprecipitation, siRNA transfection, and RT-PCR can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank W. Vermeulen and F. Grosveld for critical reading of the manuscript; the Optical Imaging Center for their support with microscopes and image analysis; and A. Burnez for assistance with generating the GFP-RPB1 knockin cells. This work is part of the Oncode Institute, which is partly financed by the Dutch Cancer Society and was funded by Dutch Cancer Society Grant 10506/2016-1. This study was supported by Dutch Organization for Scientific Research ZonMW TOP Grant 912.12.132, TOP ALW Grant 854.11.002, Horizon Zenith Grant 935.11.042, VIDI ALW Grant 864.13.004, and an Erasmus Medical Center fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 4810.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717920115/-/DCSupplemental.
References
- 1.Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- 2.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Scheidegger A, Nechaev S. RNA polymerase II pausing as a context-dependent reader of the genome. Biochem Cell Biol. 2016;94:82–92. doi: 10.1139/bcb-2015-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 8.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckley MS, Kwak H, Zipfel WR, Lis JT. Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev. 2014;28:14–19. doi: 10.1101/gad.231886.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriques T, et al. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao W, Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet. 2017;49:1045–1051. doi: 10.1038/ng.3867. [DOI] [PubMed] [Google Scholar]
- 12.Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 2015;29:39–47. doi: 10.1101/gad.246173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brannan K, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagschal A, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150:1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebs AR, et al. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol Cell. 2017;67:411–422.e4. doi: 10.1016/j.molcel.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilson KA, et al. Oxidative stress rapidly stabilizes promoter-proximal paused Pol II across the human genome. Nucleic Acids Res. 2017;45:11088–11105. doi: 10.1093/nar/gkx724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Lu JJ, He L, Yu Q. Triptolide (TPL) inhibits global transcription by inducing proteasome-dependent degradation of RNA polymerase II (Pol II) PLoS One. 2011;6:e23993. doi: 10.1371/journal.pone.0023993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manzo SG, et al. Natural product triptolide mediates cancer cell death by triggering CDK7-dependent degradation of RNA polymerase II. Cancer Res. 2012;72:5363–5373. doi: 10.1158/0008-5472.CAN-12-1006. [DOI] [PubMed] [Google Scholar]
- 19.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulon S, et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell. 2010;39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson DA, Pombo A, Iborra F. The balance sheet for transcription: An analysis of nuclear RNA metabolism in mammalian cells. FASEB J. 2000;14:242–254. [PubMed] [Google Scholar]
- 22.Hieda M, Winstanley H, Maini P, Iborra FJ, Cook PR. Different populations of RNA polymerase II in living mammalian cells. Chromosome Res. 2005;13:135–144. doi: 10.1007/s10577-005-7720-1. [DOI] [PubMed] [Google Scholar]
- 23.Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 26.van Royen ME, et al. Fluorescence recovery after photobleaching (FRAP) to study nuclear protein dynamics in living cells. Methods Mol Biol. 2009;464:363–385. doi: 10.1007/978-1-60327-461-6_20. [DOI] [PubMed] [Google Scholar]
- 27.Müller WE, et al. Effect of cordycepin on nucleic acid metabolism in L5178Y cells and on nucleic acid-synthesizing enzyme systems. Cancer Res. 1977;37:3824–3833. [PubMed] [Google Scholar]
- 28.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 29.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilson KA, et al. THZ1 reveals roles for Cdk7 in co-transcriptional capping and pausing. Mol Cell. 2015;59:576–587. doi: 10.1016/j.molcel.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao SH, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 32.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 33.Sigova AA, et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci USA. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sobell HM. Actinomycin and DNA transcription. Proc Natl Acad Sci USA. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geverts B, van Royen ME, Houtsmuller AB. Analysis of biomolecular dynamics by FRAP and computer simulation. Methods Mol Biol. 2015;1251:109–133. doi: 10.1007/978-1-4939-2080-8_7. [DOI] [PubMed] [Google Scholar]
- 38.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 39.Andrade-Lima LC, Veloso A, Paulsen MT, Menck CF, Ljungman M. DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res. 2015;43:2744–2756. doi: 10.1093/nar/gkv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veloso A, et al. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu M, et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350:1383–1386. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alekseev S, et al. Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol Cell. 2017;65:504–514.e4. doi: 10.1016/j.molcel.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Becker M, et al. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lux C, et al. Transition from initiation to promoter proximal pausing requires the CTD of RNA polymerase II. Nucleic Acids Res. 2005;33:5139–5144. doi: 10.1093/nar/gki802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudd MD, Luse DS. Amanitin greatly reduces the rate of transcription by RNA polymerase II ternary complexes but fails to inhibit some transcript cleavage modes. J Biol Chem. 1996;271:21549–21558. doi: 10.1074/jbc.271.35.21549. [DOI] [PubMed] [Google Scholar]
- 47.Sprouse RO, et al. Regulation of TATA-binding protein dynamics in living yeast cells. Proc Natl Acad Sci USA. 2008;105:13304–13308. doi: 10.1073/pnas.0801901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Graaf P, et al. Chromatin interaction of TATA-binding protein is dynamically regulated in human cells. J Cell Sci. 2010;123:2663–2671. doi: 10.1242/jcs.064097. [DOI] [PubMed] [Google Scholar]
- 49.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 50.Gilchrist DA, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghavi-Helm Y, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 52.Fukaya T, Lim B, Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tantale K, et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat Commun. 2016;7:12248. doi: 10.1038/ncomms12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakazawa Y, Yamashita S, Lehmann AR, Ogi T. A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA Repair (Amst) 2010;9:506–516. doi: 10.1016/j.dnarep.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 56.Aygün O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.