Significance
Synthetic polymers are ubiquitous in the modern world but pose a global environmental problem. While plastics such as poly(ethylene terephthalate) (PET) are highly versatile, their resistance to natural degradation presents a serious, growing risk to fauna and flora, particularly in marine environments. Here, we have characterized the 3D structure of a newly discovered enzyme that can digest highly crystalline PET, the primary material used in the manufacture of single-use plastic beverage bottles, in some clothing, and in carpets. We engineer this enzyme for improved PET degradation capacity and further demonstrate that it can also degrade an important PET replacement, polyethylene-2,5-furandicarboxylate, providing new opportunities for biobased plastics recycling.
Keywords: biodegradation, poly(ethylene terephthalate), poly(ethylene furanoate), plastics recycling, cutinase
Abstract
Poly(ethylene terephthalate) (PET) is one of the most abundantly produced synthetic polymers and is accumulating in the environment at a staggering rate as discarded packaging and textiles. The properties that make PET so useful also endow it with an alarming resistance to biodegradation, likely lasting centuries in the environment. Our collective reliance on PET and other plastics means that this buildup will continue unless solutions are found. Recently, a newly discovered bacterium, Ideonella sakaiensis 201-F6, was shown to exhibit the rare ability to grow on PET as a major carbon and energy source. Central to its PET biodegradation capability is a secreted PETase (PET-digesting enzyme). Here, we present a 0.92 Å resolution X-ray crystal structure of PETase, which reveals features common to both cutinases and lipases. PETase retains the ancestral α/β-hydrolase fold but exhibits a more open active-site cleft than homologous cutinases. By narrowing the binding cleft via mutation of two active-site residues to conserved amino acids in cutinases, we surprisingly observe improved PET degradation, suggesting that PETase is not fully optimized for crystalline PET degradation, despite presumably evolving in a PET-rich environment. Additionally, we show that PETase degrades another semiaromatic polyester, polyethylene-2,5-furandicarboxylate (PEF), which is an emerging, bioderived PET replacement with improved barrier properties. In contrast, PETase does not degrade aliphatic polyesters, suggesting that it is generally an aromatic polyesterase. These findings suggest that additional protein engineering to increase PETase performance is realistic and highlight the need for further developments of structure/activity relationships for biodegradation of synthetic polyesters.
In less than a century of manufacturing, plastics have become essential to modern society, driven by their incredible versatility coupled to low production costs. It is, however, now widely recognized that plastics pose a dire global pollution threat, especially in marine ecosystems, because of the ultralong lifetimes of most synthetic plastics in the environment (1–9). In response to the accumulation of plastics in the biosphere, it is becoming increasingly recognized that microbes are adapting and evolving enzymes and catabolic pathways to partially degrade man-made plastics as carbon and energy sources (10–19). These evolutionary footholds offer promising starting points for industrial biotechnology and synthetic biology to help address the looming environmental threat posed by man-made synthetic plastics (19–23).
Poly(ethylene terephthalate) (PET) is the most abundant polyester plastic manufactured in the world. Most applications that employ PET, such as single-use beverage bottles, clothing, packaging, and carpeting, employ crystalline PET, which is recalcitrant to catalytic or biological depolymerization due to the limited accessibility of the ester linkages. In an industrial context, PET can be depolymerized to its constituents via chemistries able to cleave ester bonds (24, 25). However, to date, few chemical recycling solutions have been deployed, given the high processing costs relative to the purchase of inexpensive virgin PET. This, in turn, results in reclaimed PET primarily being mechanically recycled, ultimately resulting in a loss of material properties, and hence intrinsic value. Given the recalcitrance of PET, the fraction of this plastic stream that is landfilled or makes its way to the environment is projected to persist for hundreds of years (1).
In 2016, Yoshida et al. (17) reported a newly discovered bacterium, Ideonella sakaiensis 201-F6, with the unusual ability to use PET as its major carbon and energy source for growth. Especially in the past decade, there have been multiple, foundational studies reporting enzymes that can degrade PET (10, 26–31), but, to our knowledge, previous work had not connected extracellular enzymatic PET degradation to catabolism (11) in a single microbe. As illustrated in Fig. 1, Yoshida et al. (17) demonstrated that an I. sakaiensis enzyme dubbed PETase (PET-digesting enzyme) converts PET to mono(2-hydroxyethyl) terephthalic acid (MHET), with trace amounts of terephthalic acid (TPA) and bis(2-hydroxyethyl)-TPA as secondary products. A second enzyme, MHETase (MHET-digesting enzyme), further converts MHET into the two monomers, TPA and ethylene glycol (EG). Both enzymes are secreted by I. sakaiensis and likely act synergistically to depolymerize PET. Sequence analysis and recent structural studies of PETase highlight similarities to α/β-hydrolase enzymes (17, 32, 33), including the cutinase and lipase families, which catalyze hydrolysis of cutin and fatty acids, respectively. This observation provides clues to the origin of PETase, but further insights into its structural and functional evolution are needed.
Fig. 1.
PETase catalyzes the depolymerization of PET to bis(2-hydroxyethyl)-TPA (BHET), MHET, and TPA. MHETase converts MHET to TPA and EG.
Beyond PET, humankind uses a wide range of polyesters, broadly classified by aliphatic and aromatic content. PET, for example, is a semiaromatic polyester. Some aliphatic polyesters, such as polylactic acid (PLA) (34), polybutylene succinate (PBS) (35), or polyhydroxyalkanoates (36), can be produced from renewable sources and are marketed as biodegradable plastics, given their relatively low crystallinity and glass transition temperatures, in turn, providing relatively more direct enzymatic access to ester linkages. Aromatic and semiaromatic polyesters, conversely, often exhibit enhanced thermal and material properties and, accordingly, have reached substantially higher market volume but are typically not as biodegradable as their aliphatic counterparts. An emerging, biobased PET replacement is polyethylene-2,5-furandicarboxylate [or poly(ethylene furanoate); PEF], which is based on sugar-derived 2,5-furandicarboxylic acid (FDCA) (37). PEF exhibits improved gas barrier properties over PET and is being pursued industrially (38). Even though PEF is a biobased semiaromatic polyester, which is predicted to offset greenhouse gas emissions relative to PET (39), its lifetime in the environment, like that of PET, is likely to be quite long (40). Given that PETase has evolved to degrade crystalline PET, it potentially may have promiscuous activity across a range of polyesters.
In this study, we aimed to gain a deeper understanding of the adaptations that contribute to the substrate specificity of PETase. To this end, we report multiple high-resolution X-ray crystal structures of PETase, which enable comparison with known cutinase structures. Based on differences in the PETase and a homologous cutinase active-site cleft (41), PETase variants were produced and tested for PET degradation, including a double mutant distal to the catalytic center that we hypothesized would alter important substrate-binding interactions. Surprisingly, this double mutant, inspired by cutinase architecture, exhibits improved PET degradation capacity relative to wild-type PETase. We subsequently employed in silico docking and molecular dynamics (MD) simulations to characterize PET binding and dynamics, which provide insights into substrate binding and suggest an explanation for the improved performance of the PETase double mutant. Additionally, incubation of wild-type and mutant PETase with several polyesters was examined using scanning electron microscopy (SEM), differential scanning calorimetry (DSC), and product release. These studies showed that the enzyme can degrade both crystalline PET (17) and PEF, but not aliphatic polyesters, suggesting a broader ability to degrade semiaromatic polyesters. Taken together, the structure/function relationships elucidated here could be used to guide further protein engineering to more effectively depolymerize PET and other synthetic polymers, thus informing a biotechnological strategy to help remediate the environmental scourge of plastic accumulation in nature (19–23).
Results
PETase Exhibits a Canonical α/β-Hydrolase Structure with an Open Active-Site Cleft.
The high-resolution X-ray crystal structure of the I. sakaiensis PETase was solved employing a newly developed synchrotron beamline capable of long-wavelength X-ray crystallography (42). Using single-wavelength anomalous dispersion, phases were obtained from the native sulfur atoms present in the protein. The low background from the in vacuo setup and large curved detector resulted in exceptional diffraction data quality extending to a resolution of 0.92 Å, with minimal radiation damage (SI Appendix, Fig. S1 and Table S1).
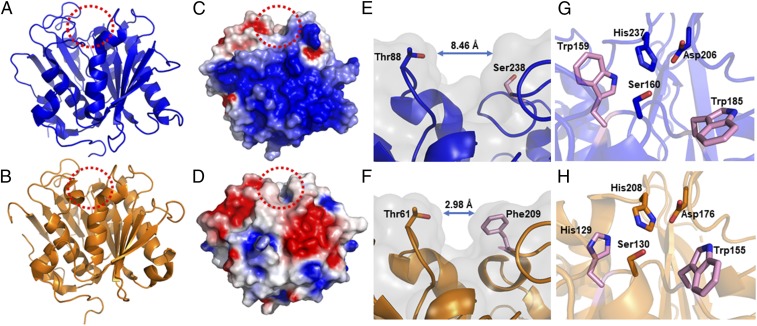
As predicted from the sequence homology to the lipase and cutinase families, PETase adopts a classical α/β-hydrolase fold, with a core consisting of eight β-strands and six α-helices (Fig. 2A). Yoshida et al. (17) noted that PETase has close sequence identity to bacterial cutinases, with Thermobifida fusca cutinase being the closest known structural representative (with 52% sequence identity; Fig. 2B and SI Appendix, Fig. S2A), which is an enzyme that also degrades PET (26, 29, 41). Despite a conserved fold, the surface profile is quite different between the two enzymes. PETase has a highly polarized surface charge (Fig. 2C), creating a dipole across the molecule and resulting in an overall isoelectric point (pI) of 9.6. In contrast, T. fusca cutinase, in common with other cutinases, has a number of small patches of both acidic and basic residues distributed over the surface, conferring a more neutral pI of 6.3 (Fig. 2D).
Fig. 2.
Structure of PETase. (A) Cartoon representation of the PETase structure at 0.92 Å resolution [Protein Data Bank (PDB) ID code 6EQE]. The active-site cleft is oriented at the top and highlighted with a dashed red circle. (B) Comparative structure of the T. fusca cutinase (PDB ID code 4CG1) (41). (C) Electrostatic potential distribution mapped to the solvent-accessible surface of PETase compared with the T. fusca cutinase as a colored gradient from red (acidic) at −7 kT/e to blue (basic) at 7 kT/e (where k is Boltzmann’s constant, T is temperature and e is the charge on an electron). (D) T. fusca cutinase in the same orientation. (E) View along the active-site cleft of PETase corresponding to the area highlighted with a red dashed circle in A and C. The width of the cleft is shown between Thr88 and Ser238. (F) Narrower cleft of the T. fusca cutinase active site is shown with the width between Thr61 and Phe209 in equivalent positions. (G) Close-up view of the PETase active site with the catalytic triad residues His237, Ser160, and Asp206 colored blue. Residues Trp159 and Trp185 are colored pink. (H) Comparative view of the T. fusca cutinase active site with equivalent catalytic triad residues colored orange. Residues His129 and Trp155 are colored pink. The residues in PETase colored pink correspond to the site-directed mutagenesis targets S238F, W159H, and W185A.
Another striking difference between PETase and the closest cutinase homologs is the broader active-site cleft, which, upon observation, we hypothesized might be necessary to accommodate crystalline semiaromatic polyesters. At its widest point, the cleft in PETase approaches threefold the width of the corresponding structure in the T. fusca cutinase. The expansion is achieved with minimal rearrangement of the adjacent loops and secondary structure (Fig. 2 E and F). A single amino acid substitution from phenylalanine to serine in the lining of the active-site cavity appears sufficient to cause this change, with the remaining cleft formed between Trp159 and Trp185 (Fig. 2G). This relative broadening of the active-site cleft is also observed in comparisons with other known cutinase structures (SI Appendix, Fig. S3 A–D).
In terms of the active site, the well-studied catalytic triad is conserved across the lipases and cutinase families (43). In PETase, the catalytic triad comprises Ser160, Asp206, and His237, suggesting a charge-relay system similar to that found in other α/β-fold hydrolases (44). The specific location and geometry between the active site found in cutinases is also conserved in PETase (Fig. 2 G and H and SI Appendix, Fig. S4). In common with most lipases, the catalytic residues reside on loops, with the nucleophilic serine occupying a highly conserved position known as the nucleophilic elbow (45). The nucleophilic serine sits in the consensus sequence (Gly-X1-Ser-X2-Gly), and while this “lipase box” is common to most lipases (SI Appendix, Fig. S4A) and cutinases (SI Appendix, Fig. S4B), the X1 position, usually occupied by a histidine or phenylalanine in cutinases and lipases, contains a tryptophan residue, Trp159, in PETase (46) (Fig. 2G). This residue has the effect of extending the hydrophobic surface adjacent to the active site (SI Appendix, Fig. S3 E and F). In common with the Fusarium solani cutinase, PETase has two disulfide bonds, one adjacent to the active site and one near the C terminus of the protein. MD simulations have predicted that the active-site disulfide in F. solani cutinase is important for active-site stability, and it may play a similar role in PETase (47).
To explore the potential effects of crystallization conditions and packing effects, three additional crystallography datasets ranging in resolution from 1.58 to 1.80 Å provided a total of seven independent PETase chains (SI Appendix, Table S1). All domains adopt the same fold (relative rmsd values are ∼0.28 Å), and all of the residues of the catalytic triad exhibit the same conformation (SI Appendix, Fig. S5 A and B), including Trp159. In one crystal form, however, Trp185 was present in three distinct conformations, all with higher B-factors than other residues in the putative binding cleft; these results were corroborated by MD simulations of the wild-type PETase (SI Appendix, Fig. S5 C–F). In all crystal forms, the packing of PETase involves extensive packing interactions in and around the hydrophobic cleft, resulting in little space for interaction with putative ligands.
Converting PETase to a Cutinase-Like Active-Site Cleft Enables Improved Crystalline PET Degradation.
From the PETase structure, we originally hypothesized that changes in the active site relative to the T. fusca cutinase resulted from the evolution of I. sakaiensis in a PET-containing environment, thus enabling more efficient PET depolymerization. To test this hypothesis, we mutated the PETase-active site to make it more cutinase-like. Specifically, a double mutant was produced, S238F/W159H, which, based on homology modeling, was predicted to narrow the PETase active site, similar to the T. fusca cutinase. Additionally, we produced the W185A mutant to examine the role of this highly conserved dynamic residue.
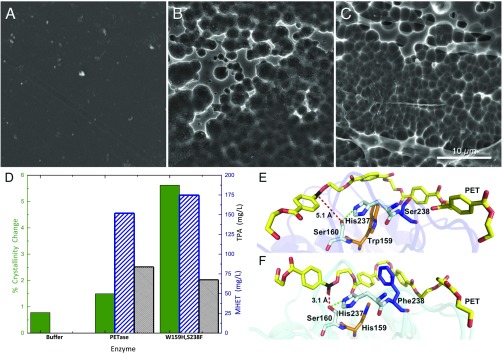
In the original report describing the discovery of PETase, Yoshida et al. (17) examined PETase digestions of amorphous PET films with a crystallinity of 1.9%, which is lower than that of most PET samples that would be encountered either in the environment or in an industrial recycling context (48). To examine the performance in the wild-type PETase relative to the two mutants, we examined PET digestion with coupons of higher crystallinity. Specifically, PET coupons with an initial crystallinity of 14.8 ± 0.2% (for reference, a commercial soft drink bottle examined via the same methods exhibits a crystallinity of 15.7% as measured by DSC) were synthesized and characterized by NMR spectroscopy to confirm their structure and by DSC to determine their crystallinity (SI Appendix, Fig. S6A). Digestions were conducted at pH 7.2 and monitored with DSC, NMR spectroscopy, and SEM, and reaction products were quantified by HPLC and NMR spectroscopy. Fig. 3 A–D shows the results of PET degradation, including a buffer-only control, the wild-type PETase, and the double mutant. It is clear that PETase induces surface erosion and pitting of a PET film with a crystallinity of 13.3 ± 0.2%, resulting in a 10.1% relative crystallinity reduction (absolute reduction of 1.5%; SI Appendix, Table S2). Surprisingly, the PETase double mutant outperforms the wild-type PETase by both crystallinity reduction and product release. The absolute crystallinity loss is 4.13% higher, and the corresponding SEM images appear to show that slightly more surface ablation occurs (Fig. 3C). After incubation, the digested PET samples for both the wild-type PETase and the double mutant exhibit a lower melting temperature over a wider temperature range (SI Appendix, Fig. S6F), indicating that the crystalline domain regions are reduced in size.
Fig. 3.
Comparison of PETase and the engineered enzyme S238F/W159H with PET. (A) Buffer-only control of PET coupon. (B) PET coupon after incubation with wild-type PETase. (C) PET coupon after incubation with the PETase double mutant, S238F/W159H. All SEM images were taken after 96 h of incubation at a PETase loading of 50 nM (pH 7.2) in phosphate buffer or a buffer-only control. (Scale bar: A–C, 10 μm.) (D) Percent crystallinity change (green, solid bar) and reaction product concentration (MHET, blue diagonal lines; TPA, black hatching) after incubation with buffer, wild-type PETase, and the S238F/W159H-engineered enzymes. (E) Predicted binding conformations of wild-type PETase from docking simulations demonstrate that PET is accommodated in an optimum position for the interaction of the carbon (black) with the nucleophilic hydroxyl group of Ser160, at a distance of 5.1 Å (red dash). His237 is positioned within 3.9 Å of the Ser160 hydroxyl (green dash). Residues Trp159 and Ser238 line the active-site channel (orange and blue, respectively). (F) Double mutant S238F/W159H adopts a more productive interaction with PET. The S238 mutation provides new π-stacking and hydrophobic interactions to adjacent terephthalate moieties, while the conversion to His159 from the bulkier Trp allows the PET polymer to sit deeper within the active-site channel. Two aromatic interactions of interest between PET and Phe238 are at optimal distance (each at 5.4 Å).
Understanding how PET binds in the PETase catalytic site is key to understanding the improved performance of the PETase double mutant. We attempted multiple trials to obtain a ligand-bound structure of PETase, to no avail. While this paper was in revision, Han et al. (32) published a study with an R132G/S160A mutant (R132 and S160 in the numbering here) that was able to accommodate monomeric ligands, alongside the publication of another PETase structure soon after (33). Here, we sought to predict PET-PETase binding modes by conducting induced fit docking (IFD) (SI Appendix, Fig. S7A). Multiple PET orientations were predicted by IFD in and around the active site of both the wild-type and double-mutant enzymes. The orientation shown in Fig. 3E and SI Appendix, Fig. S7A is one of several used to illustrate a productive PET-binding event in the wild-type enzyme: A PET carbonyl carbon is at a chemically relevant distance (5.1 Å) for nucleophilic attack from the Ser160 hydroxyl group (49, 50), His237 is at an ideal distance (3.9 Å) to activate Ser160, and Asp206 provides hydrogen bonding support to His237 (2.8 Å). This binding mode is predicted to have binding affinity (estimated by the docking score with descriptors in SI Appendix, Table S3) of −8.23 kcal/mol. Thus, our IFD-predicted binding modes are consistent with a productive Michaelis complex for PET chain cleavage. Additionally, with this low-energy, catalytically competent pose generated from flexible docking (i.e., IFD), we observe a marked difference in the position of Trp185 compared with the crystal structure (SI Appendix, Fig. S7I). The N-Cα-Cβ-C dihedral in the crystal PETase structure is −177.5°, whereas our predicted catalytically competent binding mode of PET indicates W185 rotates to accommodate aromatic interactions with PET, and thus adopts a dihedral angle of 98.4°. This dihedral rotation was observed to various extents in all docking results and in apo MD simulations (SI Appendix, Fig. S5F), and thus illustrates the necessity for flexible protein treatment during ligand binding mode prediction, especially if binding and/or catalytic hypotheses are to be posited.
IFD results also suggest potential reasons for the improved performance of the PETase double mutant over wild-type PETase, as the substrate may interact with Phe238 through several aromatic interactions, as shown in Fig. 3F. In this predicted pose (docking score of −11.25 kcal/mol, with descriptors in SI Appendix, Table S3), a PET carbonyl is at an appropriate attack distance from Ser160 (3.1 Å), Ser160 is in the range for deprotonation by His237 (2.9 Å; SI Appendix, Fig. S7C), and Asp206 is ready to accept a proton in the shuttle (2.9 Å). PET aromatic rings are within ideal π-stacking distances (51) to binding site residues (W185 and Y87), and, in particular, two aromatic interactions are formed to Phe238 (point-to-face interaction at 5.4 Å and parallel displaced interaction at 5.4 Å). The marked difference in predicted binding affinities between wild-type and double-mutant enzymes for PET is consistent with the increased activity of the PETase double mutant on PET, as observed experimentally, and we can identify aromatic interactions supported by the S238F mutation as being integral to this enhancement. All aromatic ring-ring distances for described binding modes are illustrated in SI Appendix, Fig. S7 A and C.
In contrast to the double mutant, the W185A mutant exhibits highly impaired performance relative to the wild-type PETase, as described in SI Appendix, Fig. S5 G–J and Table S2. These data confirm a critical role for this residue. From the IFD, Trp185 is predicted to play an important role by contributing π-stacking interactions to PET aromatic groups. Additionally, in all productive binding modes (i.e., when the carbonyl is oriented to be in the oxyanion hole and the carbonyl carbon is at a catalytic distance from Ser160), Trp185 is predicted to reorient relative to the crystal structure, suggesting its movement opens the active-site cleft, allowing PET binding (SI Appendix, Fig. S7I).
PETase Depolymerizes PEF, but Not Aliphatic Polyesters.
We were also interested in understanding the activity of wild-type PETase and the PETase double mutant on other polymeric substrates, including aliphatic and other semiaromatic polyesters. To that end, we synthesized, characterized (SI Appendix, Fig. S6 C and D), and conducted similar incubations with the aliphatic polyesters PBS and PLA. None of these samples showed visual differences between the control images and the PETase-treated samples (SI Appendix, Fig. S8), suggesting that PETase and the double mutant are not active on aliphatic polyesters.
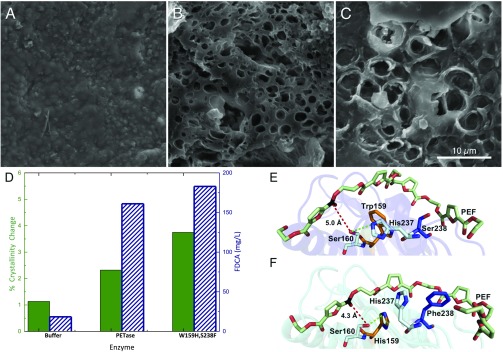
PEF is another semiaromatic polyester marketed as a biobased PET replacement (38, 39). Given the structural similarity of PET and PEF, and recent studies on PEF degradation by cutinases (52), we hypothesized that PETase may also depolymerize this substrate. Accordingly, we synthesized PEF coupons, and Fig. 4 A–D shows the results of PEF incubations with the wild-type PETase enzyme and the PETase double mutant, alongside a buffer-only control. Visually, the surface morphology of PETase-treated PEF is even more modified than PET, with SEM revealing the formation of large pits, suggesting that PETase is potentially much more active on this substrate than PET. The observation of enhanced PEF degradation by microscopy is corroborated by the DSC data for PEF, which show a reduction in relative crystallinity of 15.7% (absolute of 2.4%) compared with a relative reduction of 10.1% for PET (SI Appendix, Fig. S6E and Table S2).
Fig. 4.
Comparison of PETase and the engineered enzyme S238F/W159H with PEF. (A) Buffer-only control of PEF coupon. (B) PEF coupon after incubation with wild-type PETase. (C) PEF coupon after incubation with the PETase double mutant, S238F/W159H. All SEM images were taken after 96 h of incubation at a PETase loading of 50 nM (pH 7.2) in phosphate buffer or a buffer-only control. (Scale bar: A–C, 10 μm.) (D) Percent crystallinity change (green, solid bar) and reaction product concentration (FDCA, blue diagonal lines) after incubation with buffer, wild-type PETase, and the S238F/W159H double mutant. (E) Predicted binding conformations of wild-type PETase from docking simulations demonstrate that PEF is accommodated in an optimum position for the interaction of the carbon (black) with the nucleophilic hydroxyl group of Ser160, at a distance of 5.0 Å (red dash). His237 is positioned within 3.7 Å of the Ser160 hydroxyl (green dash). Residues Trp159 (orange) and Ser238 (blue) line the active-site channel. (F) In contrast, the double mutant S238F/W159H significantly alters the architecture of the catalytic site for PEF binding. Residue His237 rotates away from Ser160, and instead forms an aromatic interaction with PEF chain at 5.1 Å. Surprisingly, the mutated His159 becomes an alternative productive H-bond partner at 3.2 Å. Similar to interactions with PET, Phe238 also provides additional hydrophobic interactions to an adjacent furan ring of the extended PEF polymer, creating a more intimate binding mode with the cleft, with a parallel displaced aromatic interaction at 5.2 Å.
To predict how a PEF oligomer interacts with the wild-type and double-mutant PETase-active sites, IFD was again performed. The expected PETase activity was again captured from a structural standpoint, with the PEF ester oriented within nucleophilic attack distance of Ser160 (Fig. 4 E and F and SI Appendix, Fig. S7B). As with PET IFD results, we were able to identify interactions to support increased activity of the PETase double-mutant enzyme. In the PEF wild-type binding mode (docking score of −9.07 kcal/mol), two aromatic interactions are formed to Trp185 and Trp159 (SI Appendix, Fig. S7B). However, in the PEF double-mutant binding mode (docking score of −10.07 kcal/mol), three aromatic interactions were observed: parallel displaced to Trp185 (5.7 Å), point to face to His237 (5.1 Å), and parallel displaced to Phe238 (5.2 Å). Additionally, Tyr87 is within range for a potential aromatic interaction at 6.2 Å. One interesting interaction was observed in the PEF double-mutant binding mode: His237 flipped “up,” out of the catalytic triad, to play an aromatic stabilization role (replacing the wild-type Trp159 stabilization), and, instead, His159 supported Ser160 via hydrogen bonding at 3.2 Å. This interaction between Ser160 and His159 is also observed in apo MD simulations of the double-mutant structure. It could thus be postulated that His159 serves as an additional means for shuttling protons in the PETase double mutant, which will be examined in a future study. As seen with PET, docking scores predict increased binding affinity of PEF to the double-mutant PETase (SI Appendix, Table S3), and, structurally, we can relate this to aromatic interactions supported by F238 and a potential alternative pathway for proton shuttling during catalysis.
Discussion
The high-resolution structure described in the present study reveals the binding site architecture of the I. sakaiensis 201-F6 PETase, while the IFD results provide a mechanistic basis for both the wild type and PETase double mutant toward the crystalline semiaromatic polyesters PET and PEF. Changes around the active site result in a widening of the cleft compared with structural representatives of three thermophilic cutinases (SI Appendix, Fig. S3), without other major changes in the underlying secondary or tertiary structure. Furthermore, we demonstrated that PETase is active on PET of ∼15% crystallinity; while this observation is encouraging, it is envisaged that its performance would need to be enhanced substantially, perhaps via further active-site cleft engineering similar to ongoing work on thermophilic cutinases and lipases (26, 30, 53, 54). Enzyme scaffolds capable of PET breakdown above the glass transition temperature (≥∼70 °C for PET) (20) will also be pursued in future studies. Coupling with other processes such as milling or grinding, which can increase the available surface area of the plastic, also merits investigation toward enzymatic solutions for PET and PEF recycling. Furthermore, in light of recent studies that demonstrate the impressive synergistic effect of combining multiple PET-active lipases (26, 30, 53, 54), we expect that incorporation of I. sakaiensis MHETase will further increase the performance (55), and this will be pursued in future work. The highly basic surface charge of PETase requires further investigation since it is not observed in other close structural homologs, but it is noteworthy that the MHETase partner is predicted to be a fairly acidic protein, with a pI in the region of 5.2.
Both the IFD results and MD simulations independently indicate the PETase binding site is characterized by highly flexible, large aromatic side chains, such as Trp185, Tyr87, and Trp159, and Phe238 in the PETase double mutant. Binding of PET and PEF induces conformational changes in these residues relative to the crystal structure; thus, modeling protein flexibility in response to PET/PEF is critical to predict catalytically relevant binding modes. Additionally, results of these flexible docking studies agree with experimentally observed trends in performance in the wild type relative to the double mutant, and provide structural insight to explain this enhancement.
PETase activity on both PET and PEF, but not on aliphatic polyesters such as PBS and PLA, provides the basis for characterizing this enzyme more broadly as an aromatic polyesterase rather than solely as a PETase. It is likely that the enhanced gas barrier properties of PEF will lead to its adoption for beer bottles, and that this recalcitrant material will thus ultimately find its way to the environment. It is therefore encouraging that PETase is also natively capable of PEF degradation. It is also noteworthy that in this study, PETase was freeze-dried and shipped between continents, and that it retained similar performance profiles after freeze-drying, which is a positive feature for its potential use in applications that require enzyme production and use be distinct, as it would potentially be the case for most biobased recycling options.
The problem of plastics depolymerization by enzymes closely mirrors that of enzymes that depolymerize polysaccharides, such as cellulose and chitin (56, 57). Indeed, strategies that have been used to understand and improve glycoside hydrolases, including the development of quantitative assays for measuring enzyme (or enzyme cocktail) performance on solid substrates, likely can serve as inspiration for more quantitative metrics for comparing plastics-degrading enzymes and enzyme mixtures, which will be reported in future studies. Moreover, the method of PETase action is of keen interest for further protein and enzyme mixture engineering studies. The direct catalytic mechanism could be studied with mixed quantum mechanical/molecular mechanics MD-based approaches similar to previous work on carbohydrate-active enzymes (58). Beyond the active site, the enzyme may interact with and cleave the substrate in an endofashion by cleaving PET (or PEF) chains internal to a polymer or in an exofashion by only cleaving PET from the chain ends. Methods employed in the cellulase and chitinase research community, such as substrate labeling with easily detected reporter molecules or examination of product ratios, could potentially shed light on this question, and will be pursued in future efforts (59). Lastly, at low substrate loadings, many polysaccharide-active enzymes rely on multimodular architectures, with a carbohydrate-binding module attached to the catalytic domain (57). For polyesterase enzymes, hydrophobins, carbohydrate-binding modules, and polyhydroxyalkanoate-binding modules have been used to increase the catalytic efficiency of cutinases for PET degradation (60, 61). Certainly, further opportunities exist for engineering or evolving for higher binding affinity of accessory modules to increase the overall surface concentration of catalytic domains on the PET surface.
Given the fact that PET was only patented roughly 80 y ago and put into widespread use in the 1970s, it is likely that the enzyme system for PET degradation and catabolism in I. sakaiensis appeared only recently, demonstrating the remarkable speed at which microbes can evolve to exploit new substrates: in this case, waste from an industrial PET recycling facility. Moreover, given the results obtained for the PETase double mutant, it is likely that significant potential remains for improving its activity further. This enzyme thus provides an exciting platform for additional protein engineering and evolution to increase the efficiency and substrate range of this polyesterase, as well as to provide clues of how to further engineer thermophilic cutinases to better incorporate aromatic polyesters, toward to the persistent challenge of highly crystalline polymer degradation.
Conclusions
The discovery of a bacterium that uses PET as a major carbon and energy source has raised significant interest in how such an enzymatic mechanism functions with such a highly resistant polymeric substrate that appears to survive for centuries in the environment. This work shows that a collection of subtle variations on the surface of a lipase/cutinase-like fold has the ability to endow PETase with a platform for aromatic polyester depolymerization. These findings open up the possibility to further utilize and combine the extensive platform of cutinase and lipase research over the past decades with directed protein engineering and evolution to adapt this scaffold further and tackle environmentally relevant polymer bioaccumulation and biobased industrial polyester recycling.
Methods
Cloning and Protein Production.
Codon optimized Escherichia coli expression clones were constructed for PETase as described in SI Appendix, Fig. S2B.
Crystallization and Structure Determination.
PETase was crystallized in five conditions, and long-wavelength sulfur–single-wavelength anomalous diffraction and high-resolution X-ray data collection was performed in vacuo at beamline I23. Standard X-ray data collection was performed at beamlines I03 and I04 at the Diamond Light Source. Detailed methods and statistics are provided in SI Appendix, Table S1.
Substrate Docking.
The PETase crystal structure, PETase double mutant, and PET and PEF tetramers were modeled using tools from Schrödinger. Protein preparation and ligand preparation where conducted using tools in Schrödinger, along with IFD, to predict PET and PEF binding modes to PETase wild type and double mutant. Additional details can be found in SI Appendix (62, 63).
Polymer Synthesis.
PET and PEF were produced via the polycondensation of EG with TPA and FDCA, respectively. Following polycondensation, the polymers were dissolved in trifluoroacetic acid, precipitated in methanol, and subsequently redissolved in trifluoroacetic acid for film casting. Following casting, the coupons were annealed in a vacuum oven at 90 °C (above their glass transition temperature). Additional details can be found in SI Appendix.
PETase Digestion of Polymer Films.
Coupons sized ∼6 mm in diameter of each polymer film were placed in a 1.5-mL Eppendorf tube with 500 μL of 50 nM PETase in 50 mM phosphate buffer at pH 7.2. The digestions were carried out at 30 °C. Analysis of the films and supernatant was done after 96 h of digestion.
SEM.
Polymer coupons sized ∼6 mm in diameter were examined by SEM, both before and after PETase treatment for 96 h. PETase-treated samples were rinsed with 1% SDS, followed by dH2O and then ethanol. Samples were sputter-coated with 8 nm of iridium. Coated samples were mounted on aluminum stubs using carbon tape, and conductive silver paint was applied to the sides of the samples to reduce charging. SEM imaging was performed using an FEI Quanta 400 FEG instrument under low vacuum (0.45 torr) operating with a gaseous solid-state detector. Imaging was performed with a beam-accelerating voltage of 15 keV.
Supplementary Material
Acknowledgments
We thank the staff at the Diamond Light Source for their support and Simon Cragg for helpful comments on the manuscript. B.S.D., N.A.R., G.D., W.E.M., A.A., M.F.C., C.W.J., and G.T.B. thank the National Renewable Energy Laboratory (NREL) Directed Research and Development Program for funding. J.E.M. was supported by Grant BB/P011918/1 from the Biotechnology and Biological Sciences Research Council. H.P.A. was funded through an NREL subcontract and University of Portsmouth Faculty of Science bursary. R.L.S. was supported by a postdoctoral fellowship abroad (Grants 2013/08293-7, 2014/10448-1, and 2016/22956-7) from the Sao Paulo Research Foundation and Center for Computational Engineering & Sciences. H.L.W. was supported by Grant DE-SC0011297TDD from the US Department of Energy (DOE) and the National Science Foundation (NSF) under Grant CHE–1464946. F.L.K. acknowledges support from the NSF Graduate Research Fellowship Program (Grant 3900101301). Computer time was provided by Extreme Science and Engineering Discovery Environment (XSEDE) allocation MCB-090159 at the San Diego Supercomputing Center and the Texas Advanced Computing Center, and by the NREL Computational Sciences Center supported by the DOE Office of Energy Efficiency and Renewable Energy under Contract DE-AC36-08GO28308. The publisher, by accepting the article for publication, acknowledges that the US Government retains a nonexclusive, paid up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for US Government purposes.
Footnotes
Conflict of interest statement: H.P.A., M.D.A., B.S.D., N.A.R., C.W.J., J.E.M., and G.T.B. have filed a patent application on the PETase double mutant.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6EQD, 6EQE, 6EQF, 6EQG, and 6EQH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718804115/-/DCSupplemental.
References
- 1.Gregory MR, Andrady AL. Plastics in the marine environment. In: Andrady AL, editor. Plastics and the Environment. Wiley; New York: 2003. pp. 379–402. [Google Scholar]
- 2.Law KL, et al. Plastic accumulation in the North Atlantic subtropical gyre. Science. 2010;329:1185–1188. doi: 10.1126/science.1192321. [DOI] [PubMed] [Google Scholar]
- 3.Cózar A, et al. Plastic debris in the open ocean. Proc Natl Acad Sci USA. 2014;111:10239–10244. doi: 10.1073/pnas.1314705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jambeck JR, et al. Marine pollution. Plastic waste inputs from land into the ocean. Science. 2015;347:768–771. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann M, Sandhop N, Schewe I, D’Hert D. Observations of floating anthropogenic litter in the Barents Sea and Fram Strait, Arctic. Polar Biol. 2016;39:553–560. [Google Scholar]
- 6.Napper IE, Thompson RC. Release of synthetic microplastic plastic fibres from domestic washing machines: Effects of fabric type and washing conditions. Mar Pollut Bull. 2016;112:39–45. doi: 10.1016/j.marpolbul.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Geyer R, Jambeck JR, Law KL. Production, use, and fate of all plastics ever made. Sci Adv. 2017;3:e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worm B, Lotze HK, Jubinville I, Wilcox C, Jambeck J. Plastic as a persistent marine pollutant. Annu Rev Environ Resour. 2017;42:1–26. [Google Scholar]
- 9.Lamb JB, et al. Plastic waste associated with disease on coral reefs. Science. 2018;359:460–462. doi: 10.1126/science.aar3320. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y, Yanful EK, Bassi AS. A review of plastic waste biodegradation. Crit Rev Biotechnol. 2005;25:243–250. doi: 10.1080/07388550500346359. [DOI] [PubMed] [Google Scholar]
- 11.Sasoh M, et al. Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl Environ Microbiol. 2006;72:1825–1832. doi: 10.1128/AEM.72.3.1825-1832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronkvist ÅM, Xie W, Lu W, Gross RA. Cutinase-catalyzed hydrolysis of poly (ethylene terephthalate) Macromolecules. 2009;42:5128–5138. [Google Scholar]
- 13.Yang J, Yang Y, Wu W-M, Zhao J, Jiang L. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms. Environ Sci Technol. 2014;48:13776–13784. doi: 10.1021/es504038a. [DOI] [PubMed] [Google Scholar]
- 14.Restrepo-Flórez J-M, Bassi A, Thompson MR. Microbial degradation and deterioration of polyethylene–A review. Int Biodeterior Biodegrad. 2014;88:83–90. [Google Scholar]
- 15.Yang Y, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: Part 1. Chemical and physical characterization and isotopic tests. Environ Sci Technol. 2015;49:12080–12086. doi: 10.1021/acs.est.5b02661. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: Part 2. Role of gut microorganisms. Environ Sci Technol. 2015;49:12087–12093. doi: 10.1021/acs.est.5b02663. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida S, et al. A bacterium that degrades and assimilates poly(ethylene terephthalate) Science. 2016;351:1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- 18.Bombelli P, Howe CJ, Bertocchini F. Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella. Curr Biol. 2017;27:R292–R293. doi: 10.1016/j.cub.2017.02.060. [DOI] [PubMed] [Google Scholar]
- 19.Dvořák P, Nikel PI, Damborský J, de Lorenzo V. Bioremediation 3.0: Engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol Adv. 2017;35:845–866. doi: 10.1016/j.biotechadv.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Wei R, Zimmermann W. Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb Biotechnol. 2017;10:1302–1307. doi: 10.1111/1751-7915.12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wierckx N, et al. Plastic waste as a novel substrate for industrial biotechnology. Microb Biotechnol. 2015;8:900–903. doi: 10.1111/1751-7915.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei R, Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics: How far are we? Microb Biotechnol. 2017;10:1308–1322. doi: 10.1111/1751-7915.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narancic T, O’Connor KE. Microbial biotechnology addressing the plastic waste disaster. Microb Biotechnol. 2017;10:1232–1235. doi: 10.1111/1751-7915.12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paszun D, Spychaj T. Chemical recycling of poly(ethylene terephthalate) Ind Eng Chem Res. 1997;36:1373–1383. [Google Scholar]
- 25.Fukushima K, et al. Organocatalytic depolymerization of poly(ethylene terephthalate) J Polym Sci A. 2011;49:1273–1281. [Google Scholar]
- 26.Müller RJ, Schrader H, Profe J, Dresler K, Deckwer WD. Enzymatic degradation of poly (ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromol Rapid Commun. 2005;26:1400–1405. [Google Scholar]
- 27.Vertommen MAME, Nierstrasz VA, Veer Mv, Warmoeskerken MM. Enzymatic surface modification of poly(ethylene terephthalate) J Biotechnol. 2005;120:376–386. doi: 10.1016/j.jbiotec.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 28.Araújo R, et al. Tailoring cutinase activity towards polyethylene terephthalate and polyamide 6,6 fibers. J Biotechnol. 2007;128:849–857. doi: 10.1016/j.jbiotec.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 29.Herrero Acero E, et al. Enzymatic surface hydrolysis of PET: Effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules. 2011;44:4632–4640. [Google Scholar]
- 30.Ribitsch D, et al. Characterization of a new cutinase from Thermobifida alba for PET-surface hydrolysis. Biocat Biotransform. 2012;30:2–9. [Google Scholar]
- 31.Sharon C, Sharon M. Studies on biodegradation of polyethylene terephthalate: A synthetic polymer. J Microbiol Biotechnol. 2017;2:248–257. [Google Scholar]
- 32.Han X, et al. Structural insight into catalytic mechanism of PET hydrolase. Nat Commun. 2017;8:2106. doi: 10.1038/s41467-017-02255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joo S, et al. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat Commun. 2018;9:382. doi: 10.1038/s41467-018-02881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drumright RE, Gruber PR, Henton DE. Polylactic acid technology. Adv Mater. 2000;12:1841–1846. [Google Scholar]
- 35.Xu J, Guo BH. Poly(butylene succinate) and its copolymers: Research, development and industrialization. Biotechnol J. 2010;5:1149–1163. doi: 10.1002/biot.201000136. [DOI] [PubMed] [Google Scholar]
- 36.Chen G-Q. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev. 2009;38:2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- 37.Sheldon RA. Green and sustainable manufacture of chemicals from biomass: State of the art. Green Chem. 2014;16:950–963. [Google Scholar]
- 38.de Jong E, Dam MA, Sipos L, Gruter GJM. 2012. Furandicarboxylic acid (FDCA), a versatile building block for a very interesting class of polyesters. Biobased Monomers, Polymers, and Materials, ACS Symposium Series (American Chemical Society, Washington, DC), Vol 1105, pp 1–13.
- 39.Eerhart A, Faaij A, Patel MK. Replacing fossil based PET with biobased PEF; process analysis, energy and GHG balance. Energy Environ Sci. 2012;5:6407–6422. [Google Scholar]
- 40.Wu H, et al. Synthesis and degradability of copolyesters of 2, 5-furandicarboxylic acid, lactic acid, and ethylene glycol. Polym Degrad Stabil. 2015;121:100–104. [Google Scholar]
- 41.Roth C, et al. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl Microbiol Biotechnol. 2014;98:7815–7823. doi: 10.1007/s00253-014-5672-0. [DOI] [PubMed] [Google Scholar]
- 42.Wagner A, Duman R, Henderson K, Mykhaylyk V. In-vacuum long-wavelength macromolecular crystallography. Acta Crystallogr D Struct Biol. 2016;72:430–439. doi: 10.1107/S2059798316001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rauwerdink A, Kazlauskas RJ. How the same core catalytic machinery catalyzes 17 different reactions: The serine-histidine-aspartate catalytic triad of α/β-hydrolase fold enzymes. ACS Catal. 2015;5:6153–6176. doi: 10.1021/acscatal.5b01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, et al. Identification and characterization of bacterial cutinase. J Biol Chem. 2008;283:25854–25862. doi: 10.1074/jbc.M800848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ollis DL, et al. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 46.Jaeger K-E, et al. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 47.Matak MY, Moghaddam ME. The role of short-range Cys171-Cys178 disulfide bond in maintaining cutinase active site integrity: A molecular dynamics simulation. Biochem Biophys Res Commun. 2009;390:201–204. doi: 10.1016/j.bbrc.2009.09.073. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, Yang J, Jiang L. Comment on “A bacterium that degrades and assimilates poly(ethylene terephthalate)”. Science. 2016;353:759. doi: 10.1126/science.aaf8305. [DOI] [PubMed] [Google Scholar]
- 49.Sadiq SK, Coveney PV. Computing the role of near attack conformations in an enzyme-catalyzed nucleophilic bimolecular reaction. J Chem Theory Comput. 2015;11:316–324. doi: 10.1021/ct5008845. [DOI] [PubMed] [Google Scholar]
- 50.Burgi HB, Dunitz JD, Shefter E. Geometrical reaction coordinates. II. Nucleophilic addition to a carbonyl group. J Am Chem Soc. 1973;95:5065–5067. [Google Scholar]
- 51.Chelli R, Gervasio FL, Procacci P, Schettino V. Stacking and T-shape competition in aromatic-aromatic amino acid interactions. J Am Chem Soc. 2002;124:6133–6143. doi: 10.1021/ja0121639. [DOI] [PubMed] [Google Scholar]
- 52.Weinberger S, et al. Enzymatic surface hydrolysis of poly(ethylene furanoate) thin films of various crystallinities. Green Chem. 2017;19:5381–5384. [Google Scholar]
- 53.Gamerith C, et al. Enzymatic degradation of aromatic and aliphatic polyesters by P. pastoris expressed cutinase 1 from Thermobifida cellulosilytica. Front Microbiol. 2017;8:938. doi: 10.3389/fmicb.2017.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sulaiman S, et al. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl Environ Microbiol. 2012;78:1556–1562. doi: 10.1128/AEM.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Castro AM, Carniel A, Nicomedes Junior J, da Conceição Gomes A, Valoni É. Screening of commercial enzymes for poly(ethylene terephthalate) (PET) hydrolysis and synergy studies on different substrate sources. J Ind Microbiol Biotechnol. 2017;44:835–844. doi: 10.1007/s10295-017-1942-z. [DOI] [PubMed] [Google Scholar]
- 56.Eijsink VG, Vaaje-Kolstad G, Vårum KM, Horn SJ. Towards new enzymes for biofuels: Lessons from chitinase research. Trends Biotechnol. 2008;26:228–235. doi: 10.1016/j.tibtech.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Payne CM, et al. Fungal cellulases. Chem Rev. 2015;115:1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 58.Knott BC, et al. The mechanism of cellulose hydrolysis by a two-step, retaining cellobiohydrolase elucidated by structural and transition path sampling studies. J Am Chem Soc. 2014;136:321–329. doi: 10.1021/ja410291u. [DOI] [PubMed] [Google Scholar]
- 59.Horn SJ, Sørlie M, Vårum KM, Väljamäe P, Eijsink VGH. Measuring processivity. In: Gilbert HJ, editor. Methods in Enzymology. Vol 510. Academic; San Diego: 2012. pp. 69–95. [DOI] [PubMed] [Google Scholar]
- 60.Ribitsch D, et al. Fusion of binding domains to Thermobifida cellulosilytica cutinase to tune sorption characteristics and enhancing PET hydrolysis. Biomacromolecules. 2013;14:1769–1776. doi: 10.1021/bm400140u. [DOI] [PubMed] [Google Scholar]
- 61.Ribitsch D, et al. Enhanced cutinase-catalyzed hydrolysis of polyethylene terephthalate by covalent fusion to hydrophobins. Appl Environ Microbiol. 2015;81:3586–3592. doi: 10.1128/AEM.04111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 63.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.