Significance
Lacking a canonical signal peptide for translocation into the endoplasmic reticulum, galectin-3 (Gal3) is exported by unconventional secretion to play key roles in various cellular processes of fundamental importance. However, molecular details about the sorting components, the signals involved, and the underlying mechanism have remained elusive. Here, we identify a highly conserved tetrapeptide motif P(S/T)AP in the amino-terminal domain of Gal3 that directly interacts with the endosomal sorting complex required for transport (ESCRT) component Tsg101, resulting in exosomal release. This study thus defines a unique molecular mechanism based on a late domain-like motif known from many viruses by which an endogenous non-ESCRT protein is secreted in exosomes.
Keywords: unconventional secretion, galectins, exosomes, ESCRT, P(S/T)AP motif
Abstract
The beta-galactoside binding lectin galectin-3 (Gal3) is found intracellularly and in the extracellular space. Secretion of this lectin is mediated independently of the secretory pathway by a not yet defined nonclassical mechanism. Here, we found Gal3 in the lumen of exosomes. Superresolution and electron microscopy studies visualized Gal3 recruitment and sorting into intraluminal vesicles. Exosomal Gal3 release depends on the endosomal sorting complex required for transport I (ESCRT-I) component Tsg101 and functional Vps4a. Either Tsg101 knockdown or expression of dominant-negative Vps4aE228Q causes an intracellular Gal3 accumulation at multivesicular body formation sites. In addition, we identified a highly conserved tetrapeptide P(S/T)AP motif in the amino terminus of Gal3 that mediates a direct interaction with Tsg101. Mutation of the P(S/T)AP motif results in a loss of interaction and a dramatic decrease in exosomal Gal3 secretion. We conclude that Gal3 is a member of endogenous non-ESCRT proteins which are P(S/T)AP tagged for exosomal release.
The majority of newly synthesized secretory proteins are transferred via the secretory endoplasmic reticulum (ER)/Golgi pathway to the extracellular milieu. Only a minor fraction of proteins are exported by alternative mechanisms, which are summarized as unconventional secretion (1). How these proteins are tagged for unconventional secretion and how they are sorted have been challenging questions in exocytosis.
Galectins comprise 15 vertebrate members associated with a variety of different cellular functions in development, cell polarity, immunity, inflammation, apoptosis, and cancer (2). Galectin-3 (Gal3) has been detected in the cytoplasm, the nucleus, within endosomal compartments, or in the extracellular space (3). Once secreted by the cell, extracellular Gal3 plays a key role in processes of fundamental importance such as immunomodulation (4) and tumor progression, especially concerning angiogenesis (5, 6), cell adhesion (7), cell motility (8), and cell invasion (9). By manipulating cells in the microenvironment of a tumor, extracellular Gal3 affects these mechanisms converging finally to metastasis (10).
Gal3 lacks a signal peptide and is most likely secreted via an unconventional pathway as shown by its insensitivity to drugs interfering with the classical export pathway (11). However, the precise export mechanism of Gal3 has not been described yet.
Dependent on the cell type, secreted Gal3 was identified either in exosomes (12–14) or microvesicles (15). Exosomes and microvesicles can be distinguished by their origin and their physical and morphological properties. Biogenesis of exosomes starts with the formation of intraluminal vesicles (ILVs) in special late endosomes termed multivesicular bodies (MVBs). After fusion of MVBs with the plasma membrane, the ILVs are released into the extracellular space as exosomes.
The endosomal sorting complex required for transport (ESCRT) machinery is critical for budding and scission of ILVs into MVBs. In particular, the ESCRT-I component Tsg101 recruits cargo proteins through binding of the tetrapeptide motif P(S/T)AP (16, 17). Analogous to cargo selection and MVB formation, many viruses, including Ebola and HIV, use P(S/T)AP motifs interacting with Tsg101 to usurp the ESCRT machinery for virus particle budding at the plasma membrane (18).
Here, we found that in epithelial cells, Gal3 is recruited into ILVs through direct interaction with Tsg101 for apical exosome-mediated release. This direct interaction depends on a highly conserved P(S/T)AP motif in the amino-terminal region of Gal3. Specific knockdown of Tsg101 or mutation of the Gal3-P(S/T)AP motif prevented exosomal Gal3 release. To our knowledge, this is a unique expression of an endogenous non-ESCRT protein employing a P(S/T)AP motif for exosomal secretion.
Results
Gal3 Is Found in Apical Exosomes Derived from Madin–Darby Canine Kidney Cells.
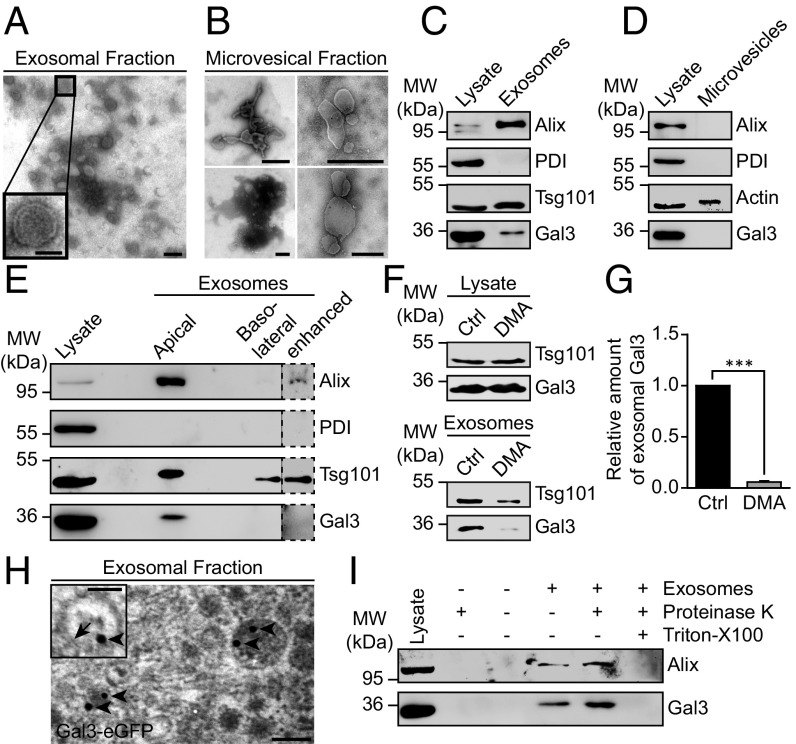
To assess whether Gal3 is secreted by Madin–Darby canine kidney (MDCK) cells in vesicular structures, the culture supernatant of these cells was subjected to sequential centrifugation steps. The resulting microvesicular fraction (10,000 × g pellet) and the exosomal fraction (100,000 × g pellet) were then analyzed by electron microscopy (Fig. 1 A and B). The exosomal fraction contained a homogenous population of small, membrane surrounded structures with a diameter of 50–90 nm (Fig. 1A), similar to those described in the literature for exosomes (19). In contrast, vesicles found in the 10,000 × g pellet were of much higher disparity regarding size and shape (Fig. 1B). Their diameter ranged from 100 to 1,000 nm. Both size and heterogeneity were in agreement with previous descriptions for microvesicles (19). Both pellets were further analyzed by immunoblot for the presence of Gal3 (Fig. 1 C and D). The exosomal pellet was enriched in the ESCRT-associated protein Alix (20) and ESCRT-I protein Tsg101, while being devoid of ER-resident protein disulfide isomerase (PDI) as negative control (Fig. 1C). The microvesicles contained neither Alix nor PDI (Fig. 1D). Gal3, endogenously expressed in MDCK cells, was detected in the exosomal, but not in the microvesicular fraction, suggesting that exosomes serve as vehicles for Gal3 secretion.
Fig. 1.
Gal3 is contained in exosomes. (A and B) The 100,000 × g pellets (exosomes) (A) or the 10,000 × g pellets (microvesicles) (B) were subjected to negative staining for electron microscopy. Inset shows the typical appearance of an exosome. (C) Western blotting analysis of the exosomal fraction. Representative results, n = 3 independent experiments. (D) Western blotting analysis of the microvesicular fraction. Actin served as loading control for microvesicles. Representative results, n = 3 independent experiments. (E) Immunoblot analysis of the exosomal fractions from filter-grown MDCK cells. Representative results, n = 3 independent experiments. (F) Western blotting analysis of DMA-treated cells. (G) Quantification of experiments as in F. Normalized to the respective cell lysate. Means ± SEM, n = 5 independent experiments. (H) Electron microscopy analysis of Gal3-eGFP localization in exosomes. A total of 15 nm gold-labeled GFP nanobodies (arrowheads) was detected in exosomes, often in close proximity to the exosomal membrane (arrow). (I) Proteinase protection assay. Representative results, n = 3 independent experiments. Statistical analysis: Student’s unpaired t test, ***P < 0.001. [Scale bars: A and H, 100 nm; A and H, (Inset), 50 nm; B, 500 nm.]
We next examined whether exosomal Gal3 is released in a polarized fashion since Gal3 is predominantly secreted from the apical side of MDCK cells (11). Consistently, the exosomal fraction isolated from the apical medium contained high amounts of Gal3 as well as the exosomal marker Alix and Tsg101 (Fig. 1E), whereas Gal3 was not detectable in the basolateral fraction. This indicates that Gal3 is secreted in exosomes from the apical membrane domain.
To confirm the notion that Gal3 is conveyed by exosomes, we selectively inhibited exosomal release by application of dimethyl amiloride (DMA) (21). DMA dramatically reduced the amount of Gal3 and the exosome marker Tsg101 in the exosome fraction (Fig. 1 F and G).
The presence of eGFP-tagged Gal3 in exosomes was then verified by immunoelectron microscopy. Here, 15 nm gold-labeled GFP nanobodies were detected in exosomes (Fig. 1H). To clarify whether Gal3 is enriched inside or associates on the surface of exosomes, purified exosomes were treated with proteinase K in the presence or absence of Triton X-100 (Fig. 1I). Gal3 as well as Alix were insensitive to proteinase K digestion, but were degraded following detergent solubilization. These findings confirmed that Gal3 is localized in the lumen of exosomes. In essence, our data point to exosomes as vehicles for Gal3 secretion.
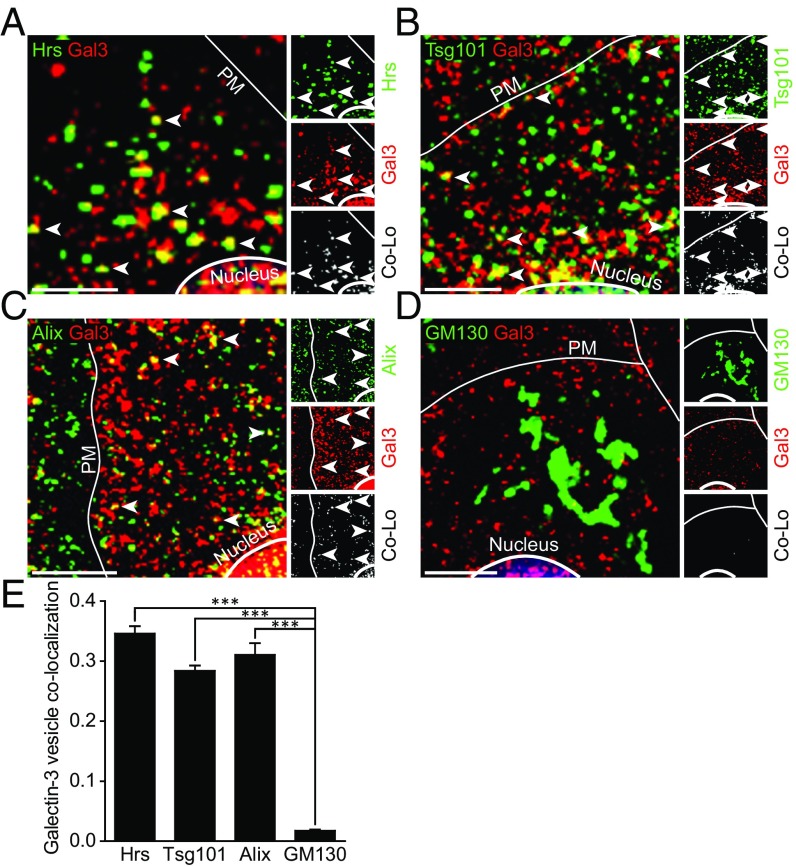
Gal3 Colocalizes with the MVB-Associated Proteins Hrs, Tsg101, and Alix.
Applying confocal immunofluorescence microscopy, we analyzed whether Gal3 was associated with the ESCRT components Hrs and Tsg101, and the ESCRT-associated protein Alix (Fig. 2 A–C). Quantification using Manders’ correlation coefficients revealed that 34.6 ± 1.2% of cytoplasmic Gal3 structures localized to Hrs-positive vesicles representing the MVB formation site (Fig. 2E). Comparable quantities of Gal3 vesicles showed substantial labeling for Tsg101 (28.4 ± 0.9%) or Alix (31 ± 1.9%). In contrast, costaining of Golgi-resident GM130 resulted in a negligible overlap (1.8 ± 0.2%). Antibody specificity of Gal3, Hrs, and Tsg101 was assessed either by CRISPR/Cas9-mediated knockout (22, 23) (MDCKΔGal3) or by siRNA-mediated knockdown (Hrs and Tsg101) of the respective protein followed by identical immunostaining and confocal imaging (SI Appendix, Fig. S1).
Fig. 2.
Gal3 colocalizes with ESCRT-associated proteins in MDCK cells. (A–D) Confocal codistribution analysis of immunostained Gal3 with Hrs (A), Tsg101 (B), Alix (C), and GM130 (D). Colocalized areas are depicted by an additional channel (white) and highlighted by arrowheads. (E) Manders’ correlation coefficient was used for quantification of experiments as in A–G. Means ± SEM, 15–20 cells per experiment, n = 3 independent experiments. Nuclei were excluded from quantification. Statistical analysis: Student’s unpaired t test, ***P < 0.001. (Scale bars: A–G, 10 µm.) PM, plasma membrane.
Our findings were further confirmed by triple staining for Gal3, Hrs, and Tsg101 (SI Appendix, Fig. S2 A and B). As expected, Hrs and Tsg101 colocalized extensively, indicated by a calculated overlap of 55.6 ± 1.7%. Thus, we conclude that either Hrs or Tsg101 staining can be used to tag MVB formation sites. Again, a comparable fraction of Gal3 vesicles were positive for Hrs (37.1 ± 1.3%) or Tsg101 (33.8 ± 1.8%). Strikingly, the majority of these colocalized structures were, in addition, triple positive for the respective MVB marker (SI Appendix, Fig. S2 A and B, arrowheads). Moreover, lactose treatment and thus elimination of internalized Gal3 (24) significantly enhanced Gal3-Hrs overlap to 59.6 ± 0.6%, thereby corroborating the notion of a substantial presence of Gal3 within MVBs that is independent of endocytic uptake (SI Appendix, Fig. S2 C and D).
These observations document that considerable quantities of Gal3 are closely related to MVBs, which are the source compartment for exosomal release.
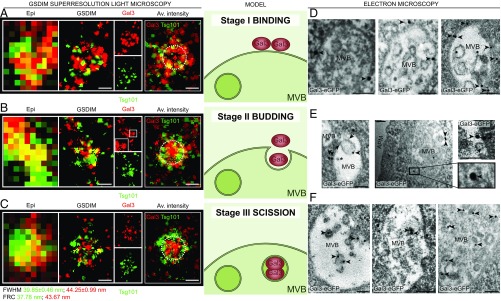
Superresolution Microscopy Depicts Precise Mechanism of Gal3 Sorting into MVBs.
To precisely determine the localization of Gal3 at MVB formation sites, we employed ground state depletion followed by individual molecule return (GSDIM) superresolution localization microscopy (25). The GSDIM technique enables the ultrastructural analysis of Gal3 by using metastable dark states of the fluorophore for temporal separation of single molecules. Therefore, GSDIM can be compared with other single molecule-based superresolution techniques such as dSTORM and PALM. The colocalizing areas of Gal3 and the MVB marker Tsg101 or Hrs were conglomerates of 511.9 ± 6.4 nm and 483.6 ± 5.8 nm size (means ± SEM, n = 3 independent experiments, five to seven cells each), corresponding to the reported size (400–500 nm) of MVBs (26). Interestingly, we were able to visualize profiles of three distinct stages of Gal3 sorting into MVBs, reproducing the ESCRT-dependent model for ILV biogenesis and budding (27) (Fig. 3 A–C and SI Appendix, Fig. S2E).
Fig. 3.
Gal3 sorting mechanism into MVBs. (A–C) Localization of immunostained Gal3 at MVB formation sites revealed by GSDIM superresolution microscopy. Tsg101 was used as marker for MVBs. Correlated areas in conventional epifluorescence (Epi) are matched with corresponding superresolved GSDIM data. (A) We identified circular arrangements of Tsg101, where Gal3 built clusters with Tsg101, documenting a first initiation stage of Gal3 recruitment. (B) In the budding stage, Gal3 staining exhibited finger-like projections toward the center of the MVB structures. The constricted conjunctions originated from protein clusters with Tsg101. The Inset (Gal3) shows finger-like projections of Gal3. (C) The final third stage was characterized by central accumulation of Gal3 vesicles generated by scission and orbital distribution of the MVB marker. An overlay of four discrete GSDIM images was used to calculate average intensity plots (Av. intensity). Dashed circles with a diameter of 450 nm approximating the MVB limiting membrane were included for orientation. Gal3 accrued at the boundary of the marker protein accumulation in the binding stage, whereas subsequent stages show progressive recruitment of Gal3 to the center of the MVB. Structural resolution for Tsg101 and Gal3 was estimated by Fourier ring correlation (FRC) (Tsg101 = 37.78 nm; Gal3 = 43.67 nm) and by average full width at half maximum (FWHM) (21 single spots; Tsg101 = 39.85 ± 0.46 nm; Gal3 = 44.25 ± 0.99 nm). Representative results, n = 3 independent experiments. The Middle exemplifies a schematic model of Gal3 sorting. (D–F) Electron microscopy analysis of Gal3-eGFP arrangement at MVB formation sites. Gal3-eGFP was detected by direct immunolabeling with gold-conjugated (15 nm) GFP nanobodies. (D) Gal3-eGFP was detected at the MVB membrane facing the cytosol (arrowheads). (E) Gal3-eGFP signals (arrowheads) were associated with ILV budding. Note also protein complexes which may correspond to ESCRT-III rings (arrows; see also SI Appendix, Fig. S4B). (Left) Gal3-eGFP clustering was also observed at the cytosolic constriction area of a budding ILV which is situated outside the focal plane (asterisk). (Middle and Right) Gal3-eGFP clustering at the cytosolic membrane constriction and in the lumen of budding ILVs. (F) Gal3-eGFP (arrowheads) was also found in the lumen of postscission ILVs. (Scale bars: GSDIM panel, 250 nm; electron microscopy panel, 100 nm.) APM, apical plasma membrane; Av. intensity, average intensity plot.
To validate the GSDIM superresolution experiments, we complemented these findings by mimicking exactly the experimental conditions with computer simulations. Intriguingly, dual-color images of the reconstructed simulation data showed a characteristic distinction between binding, budding, and scission stages, reproducing the experimentally visualized process of Gal3 recruitment into ILVs (SI Appendix, Fig. S3). Direct comparison between the distributions of fluorophores in the average intensity plots of GSDIM images with the simulated ones revealed a high concordance, thus supporting the classification of GSDIM images into three distinct stages.
We then validated the observations made with GSDIM by immunoelectron microscopy in MDCK-Gal3-eGFP cells (SI Appendix, Fig. S4A) with gold-labeled anti-GFP nanobodies. Indeed, we found stages that can be regarded as Gal3 binding (Fig. 3D), budding (Fig. 3E), and scission (Fig. 3F) into ILVs. In contrast to the GSDIM data, 100-nm thin sections allowed us to categorize each Gal3-eGFP labeling gold particle into one of the three different stages (exemplified by roman numerals in SI Appendix, Fig. S4B). Statistical evaluation of the particle staging either classified visually (SI Appendix, Fig. S4 C and D) or by distance measurement (SI Appendix, Fig. S4 E–H) revealed that the proportion between binding and scission stage particles was balanced, whereas budding stage particles were observed less frequently.
We next investigated the association of stage III gold particles with ILVs (SI Appendix, Fig. S4I). The majority of these particles (67.2 ± 6.2%) were localized in the lumen or at the membrane of ILVs, whereas a significantly lower number were found to be unrelated to ILVs. These findings correspond well with the proteinase K studies and suggest that major quantities of Gal3 are recruited into ILVs.
Moreover, it was imperative to analyze the 3D Gal3 distribution in MVBs by electron tomography (SI Appendix, Fig. S5 A and B). Morphologic analysis revealed that Gal3 localized to the limiting membrane of MVBs (SI Appendix, Fig. S5A). Notably, we found Gal3 confined to the lumen of a budding ILV (SI Appendix, Fig. S5B).
In conclusion, these data faithfully reproduce the observations made with superresolution light microscopy and documented on a molecular level that Gal3 is recruited, sorted, and packed into ILVs, possibly through interaction with ESCRT components. Finally, Gal3-positive ILVs are released as exosomes into the apical medium as illustrated by immunoelectron microscopy (SI Appendix, Fig. S5C).
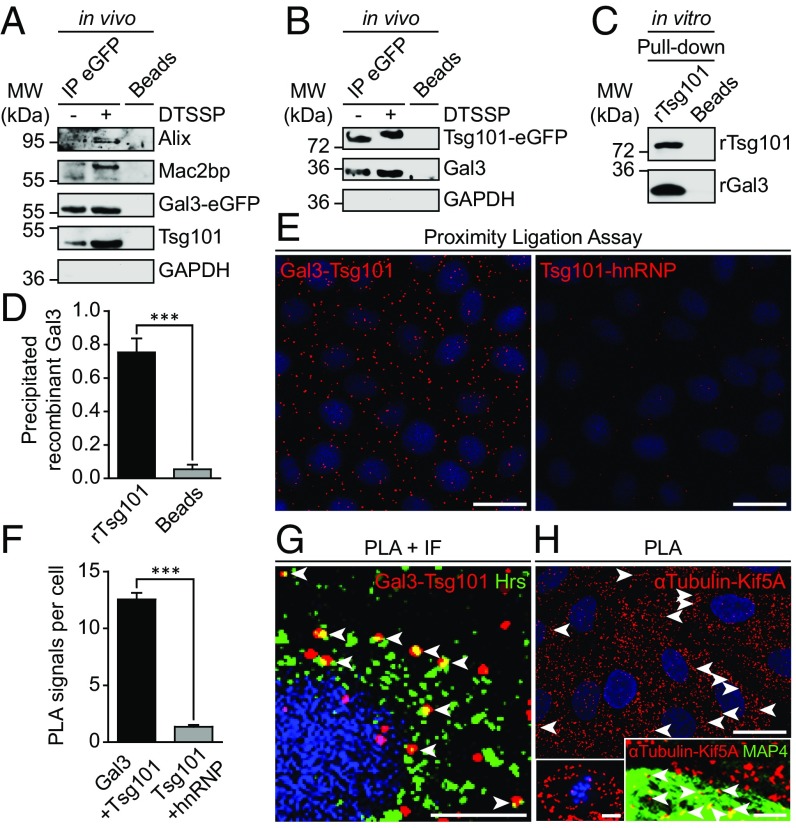
Gal3 Directly Interacts with Tsg101.
We next sought to identify the initiatory events leading to the packaging of Gal3 into exosomes. Therefore, GFP-Trap beads were used for precipitation of Gal3-eGFP from MDCK-Gal3-eGFP lysates to identify putative binding partners (Fig. 4A). Indeed, the ESCRT-I component Tsg101 was identified as a unique interaction partner of Gal3. Administration of the unspecific protein cross-linker 3,3′-dithiobis(sulfosuccinimidyl propionate) (DTSSP) resulted in a stabilization of the interaction. The known binding partners of Gal3, Alix (4), and Mac2bp (28), served as a positive control for successful Gal3 immunoprecipitation. To verify these results, we employed a complementary approach with MDCK cells expressing Tsg101-eGFP (29) (Fig. 4B). Again, Gal3 was coimmunoprecipitated, confirming our previous conclusion that Tsg101 interacts with Gal3. The selectivity of these experiments was verified by expression of noncoupled eGFP protein (SI Appendix, Fig. S5D). Furthermore, these data also revealed an increased coimmunoprecipitation of endogenous wild-type Gal3, indicating that Gal3-eGFP was still able to oligomerize properly. In this regard we sought to confirm that Gal3-eGFP similarly retains the lectin characteristics of wild-type Gal3. Therefore, we employed a lactose pull-down assay in these Gal3-eGFP expressing MDCK cells (SI Appendix, Fig. S5 E and F). Indeed, Gal3-eGFP was clearly immobilized by lactose beads, demonstrating that the carbohydrate recognition domain (CRD) was functional. Hence, these results provide, together with studies on the intracellular trafficking of Gal3-eGFP (30) and other small fusion proteins (31, 32), confirmatory evidence that regular Gal3 function was not impaired by fusion with eGFP. We also analyzed whether Gal3 interacts with syntenin, which has been shown to drive exosomal secretion of a cytoplasmic lysyl-tRNA synthetase (33). However, we did not find evidence for an interaction with Gal3.
Fig. 4.
Gal3 interacts with the ESCRT-component Tsg101. (A) Coimmunoprecipitation of Gal3-eGFP in MDCK cells. Gal3-eGFP fusion protein and its binding partners were immunoprecipitated with GFP-nanobody beads and detected by immunoblotting. Positive control: Alix, Mac2bp; negative control: GAPDH. Representative results, n = 3 independent experiments. (B) Complementary approach with Tsg101-eGFP. Negative control: GAPDH. Representative results, n = 3 independent experiments. (C) Direct rGal3 and rTsg101-GST interaction as determined by in vitro pull-down assay (rGal3: recombinant Gal3; rTsg101: recombinant Tsg101). Representative results, n = 4 independent experiments. (D) Quantification of experiments as in C. Means ± SEM, n = 4 independent experiments. (E) Physical closeness of the two proteins was evaluated by an in situ proximity ligation assay (PLA). Negative control: hnRNP. (F) Quantification of experiments as in E. Means ± SEM, 10–15 cells per experiment, n = 3 independent experiments. (G) PLA and immunofluorescence (IF) were combined to ensure that the interaction between Gal3 and Tsg101 was localized to Hrs-positive vesicles (arrowheads). (H) PLA positive control with α-tubulin and Kif5A. Single microtubules were decorated PLA signals (arrowheads). (Left Inset) Mitotic cell in metaphase with PLA signals visualizing microtubule-Kif5A proximity on the mitotic spindle apparatus. (Right Inset) Combination of PLA with MAP4 staining (arrowheads; see also SI Appendix, Fig. S6A). Statistical analysis: Student’s unpaired t test, ***P < 0.001. [Scale bars: E and H, 25 µm; G, 10 µm; H (Insets), 5 µm.]
To further investigate the interaction between Gal3 and Tsg101, we used recombinant Gal3 and Tsg101 for in vitro pull-down experiments. Remarkably, Gal3 was captured out of solution with recombinant Tsg101-GST (Fig. 4 C and D), which strongly suggests that Gal3 and Tsg101 interact through direct binding. This relevant finding was verified by a negative control with uncoupled recombinant GST (rGST) (SI Appendix, Fig. S5G) and by a proximity ligation assay (PLA). PLA signals were significantly increased if primary antibodies against the two binding partners Gal3 and Tsg101 were used (Fig. 4 E and F), indicating physical Gal3-Tsg101 closeness. Subcellular localization of this interaction was determined by combination of PLA with an immunostaining against Hrs. PLA signals colocalized extensively with the MVB marker Hrs (Fig. 4G, arrowheads), demonstrating physical proximity of Gal3 and Tsg101 at MVB formation sites. To ensure the selectivity of the PLA method, we implemented a positive control with α-tubulin and Kif5A (Fig. 4H). Single microtubules were decorated with pearl chain-like PLA signals (arrowheads), including a spindle apparatus of a mitotic cell in metaphase (Left Inset). To better visualize this observation, MAP4-eGFP was expressed as counterstain for PLA by baculovirus-mediated gene transduction (Fig. 4H, Right Inset; see also SI Appendix, Fig. S6A).
According to the results obtained by PLA and immunoprecipitation, two-color fluorescence recovery after photobleaching (FRAP) analysis showed similar recovery dynamics of Gal3-DsRed and Tsg101-eGFP at MVBs (SI Appendix, Fig. S6 B and C and Movies S1–S3). Cumulatively, our in vivo and in vitro data reveal that Gal3 and Tsg101 are dynamically recruited to MVBs for direct interaction.
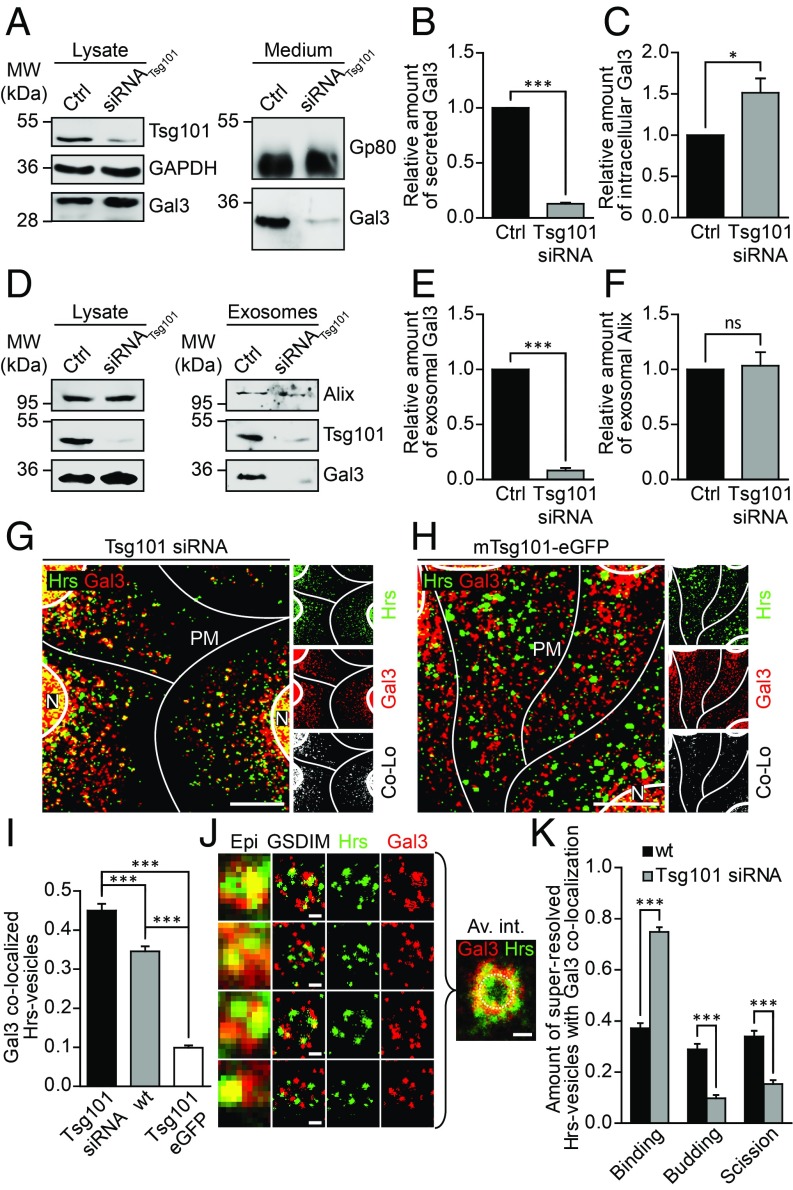
Tsg101 Knockdown Reduces Exosomal Secretion of Gal3.
To determine whether Tsg101 affects Gal3 recruitment into ILVs, Tsg101 was depleted by siRNA-mediated knockdown (Fig. 5A and SI Appendix, Fig. S7A). Culture media were collected and analyzed for the presence of Gal3 (Fig. 5B). Quantification of the data revealed that the amount of secreted Gal3 was dramatically reduced in Tsg101 knockdown cells, suggesting that exosome release of Gal3 is the predominant mechanism for secretion of this lectin in MDCK cells. In contrast, intracellular Gal3 showed a significant increase (Fig. 5C). Accordingly, Tsg101 siRNA-treated cells exhibited decreased exosomal Gal3 secretion (Fig. 5 D and E). Concomitantly, exosomal release of Alix was not affected by Tsg101 knockdown (Fig. 5F), which is surprising given the profound impact of Tsg101 knockdown on the endosomal compartment (34). However, it was shown that Tsg101 depletion rather changed the composition of exosomes (35), showing that different subpopulations of MVBs/exosomes exist (36–38). Hence, Tsg101 seems to be required for the sorting and targeting of cargos into ILVs, since Tsg101 depletion results in a modification of the overall protein content of exosomes (35). In the light of this, the differential effect of Tsg101 knockdown on exosomal Gal3 and Alix is intriguing.
Fig. 5.
Tsg101 knockdown reduces exosomal Gal3 secretion. (A) Immunoblot analysis of Tsg101 knockdown in MDCK cells and the corresponding cell culture medium. (B and C) Quantification of experiments as in A. (B) Gal3 secretion was reduced in Tsg101 knockdown cells. Normalized to Gp80. Means ± SEM, n = 3 independent experiments. (C) Intracellular Gal3 accumulated in Tsg101 siRNA-treated cells. Normalized to GAPDH. Means ± SEM, n = 3 independent experiments. (D) Immunoblot analysis of the exosomal fraction after Tsg101 knockdown in MDCK cells. (E and F) Quantification of experiments as in D. Gal3 was significantly reduced in Tsg101 siRNA-treated cells (E), whereas the exosomal marker Alix was not affected (F). Normalized to the respective cell lysate. Means ± SEM, n = 3 independent experiments. (G and H) Confocal codistribution analysis of Gal3 with Hrs in either siRNA (G) or mTsg101-eGFP (H) treated cells. Colocalized areas are shown by an additional channel (white). (I) Manders’ correlation coefficient was used for quantification of experiments as in G and H. Means ± SEM, 15–20 cells per experiment, n = 3 independent experiments. Nuclei were excluded from quantification. (J) GSDIM superresolution microscopy analysis of colocalizing structures of Gal3 and Hrs in Tsg101 siRNA-treated cells. Average intensity distribution was computed with an overlay of 35 discrete GSDIM images (Av. int.). A dashed circle with a diameter of 450 nm approximating the MVB limiting membrane was included for orientation. Representative results, n = 3 independent experiments. (K) Quantification of the three distinct sorting stages in GSDIM microscopy analysis. Means ± SEM, 5–13 GSDIM images, n = 3 independent experiments. Statistical analysis: Student’s unpaired t test, ***P < 0.001. (Scale bars: G and H, 10 µm; J, 250 nm.) Av. int., average intensity plot; Co-Lo, colocalization channel; N, Nucleus; PM, plasma membrane.
We then examined whether this reduction of exosomal Gal3 could be recovered by a rescue experiment with cells expressing a modified version of Tsg101-eGFP (mTsg101-eGFP) which does not hybridize with the utilized siRNA duplexes (SI Appendix, Fig. S7B). In fact, application of mTsg101-eGFP resulted in a compensation of exosomal Gal3 secretion in Tsg101 knockdown cells (SI Appendix, Fig. S7C). Notably, endogenous exosomal Tsg101 was similarly restored (SI Appendix, Fig. S7D). This observation documents that mTsg101-eGFP expression was functional and sufficient to restore physiological exosome release.
To further confirm the functional implication of Tsg101 in exosomal secretion of Gal3, we focused analysis on whether Gal3 was sequestered in Hrs-positive compartments after Tsg101 knockdown (Fig. 5G). We indeed observed a significant increase in Gal3/Hrs colocalization in Tsg101 siRNA-treated cells in comparison with untreated MDCK cells. On the contrary, overexpression of mTsg101-eGFP (Fig. 5H) resulted in a marked decrease in overlap (Fig. 5I). GSDIM analysis finally showed that Gal3 was retained at the boundary of Hrs-positive structures, supporting the hypothesis that Gal3 sorting into ILVs was affected by Tsg101 depletion (Fig. 5J). To verify this observation, 35 GSDIM images were used to compute an average intensity plot, illustrating that Gal3 accumulated at the boundary of the MVBs. Likewise, quantification of the superresolution data confirmed these findings (Fig. 5K). Compared with the balanced distribution in wild-type cells, budding and scission stages were strongly reduced in Tsg101 siRNA-treated cells. In agreement with the confocal data, it is also noteworthy that Gal3 was still recruited to MVBs, suggesting that alternative Gal3 binding partners, most likely Alix, take part in the initiation step.
In short, these experiments showed that Gal3 sorting into ILVs was markedly diminished in Tsg101 knockdown cells, leading to an inhibition of exosomal Gal3 secretion. Additionally, recruitment of Gal3 into exosomes is mediated by a direct interaction with Tsg101.
Expression of Dominant-Negative Vps4aE228Q-eGFP Blocks Gal3 Secretion.
To examine the importance of a functional ESCRT complex for Gal3 secretion, we established a stable cell line with inducible Vps4aE228Q-eGFP expression (SI Appendix, Fig. S7E), which is a mutated form of the AAA-type ATPase vacuolar protein-sorting-associated protein 4 (Vps4) (39). Together with ESCRT-III, Vps4 is essential in the budding process of MVB formation and topologically related processes like cytokinesis and retroviral budding (40). Particularly, it is thought that Vps4 disassembles and recycles ESCRT complexes after successful budding at the target membrane (41, 42). ATPase deficiency of the dominant-negative Vps4aE228Q-eGFP mutant (43) induces abnormal swelling of MVBs (44, 45), trapping of Tsg101 on the surface of MVBs (46) and an accumulation of nonreleased viral particles at the plasma membrane (18). By the use of this dominant-negative mutant, we indeed observed high-density accumulations of Hrs at the limiting membrane of enlarged MVBs, confirming not only ATPase deficiency but also the specificity of our Hrs immunostaining (SI Appendix, Fig. S7F). Interestingly, also Gal3 accrued, as expected, pearl chain-like at the MVB boundary, suggesting that its incorporation into MVBs was markedly impaired. Further analysis by confocal average intensity plots confirmed that large amounts of Gal3 and Hrs accumulated at the MVB surface (SI Appendix, Fig. S7G). Interestingly, compared with Hrs and Vps4a, the Gal3 distribution pattern differed slightly, with Gal3 located merely at the rim.
The correlation between Hrs and Gal3 accumulations (arrowheads) at MVBs was further confirmed by GSDIM superresolution microscopy (SI Appendix, Fig. S7H). In direct analogy to the Tsg101 knockdown data, the incorporation of Gal3 into MVBs was clearly blocked, and composite accumulations of Gal3 (asterisks) were observed that failed to be released into the MVB lumen. GSDIM microscopy also revealed that Gal3 colocalized with Hrs as budding stage patterns (arrows) likely to reflect imperfect budding from the MVB limiting membrane. The 3D GSDIM microscopy consistently showed that Hrs was exclusively localized to the MVB boundary, suggesting that the observed invaginations originated from the limiting membrane (SI Appendix, Fig. S7I and Movie S4).
To further examine the impact of ATPase deficiency and thus ESCRT functionality on exosomal Gal3 secretion, we finally analyzed the cell culture medium in mifepristone-treated Vps4aE228Q-eGFP mutants (SI Appendix, Fig. S7 J and K). Western blotting confirmed that not only exosomal Gal3 secretion upon ATPase deficiency was blocked, but also that ESCRT-mediated secretion via exosomes seemed to be the exclusive pathway for Gal3 export in MDCK cells, although we cannot fully exclude alternative routes.
In brief, dominant-negative Vps4aE228Q-eGFP blocks exosomal Gal3 sorting, resulting in a sequestration of Gal3 on the MVB limiting membrane.
Mutation of a Conserved PSAP Motif Affects Tsg101 Interaction and Exosomal Release of Gal3.
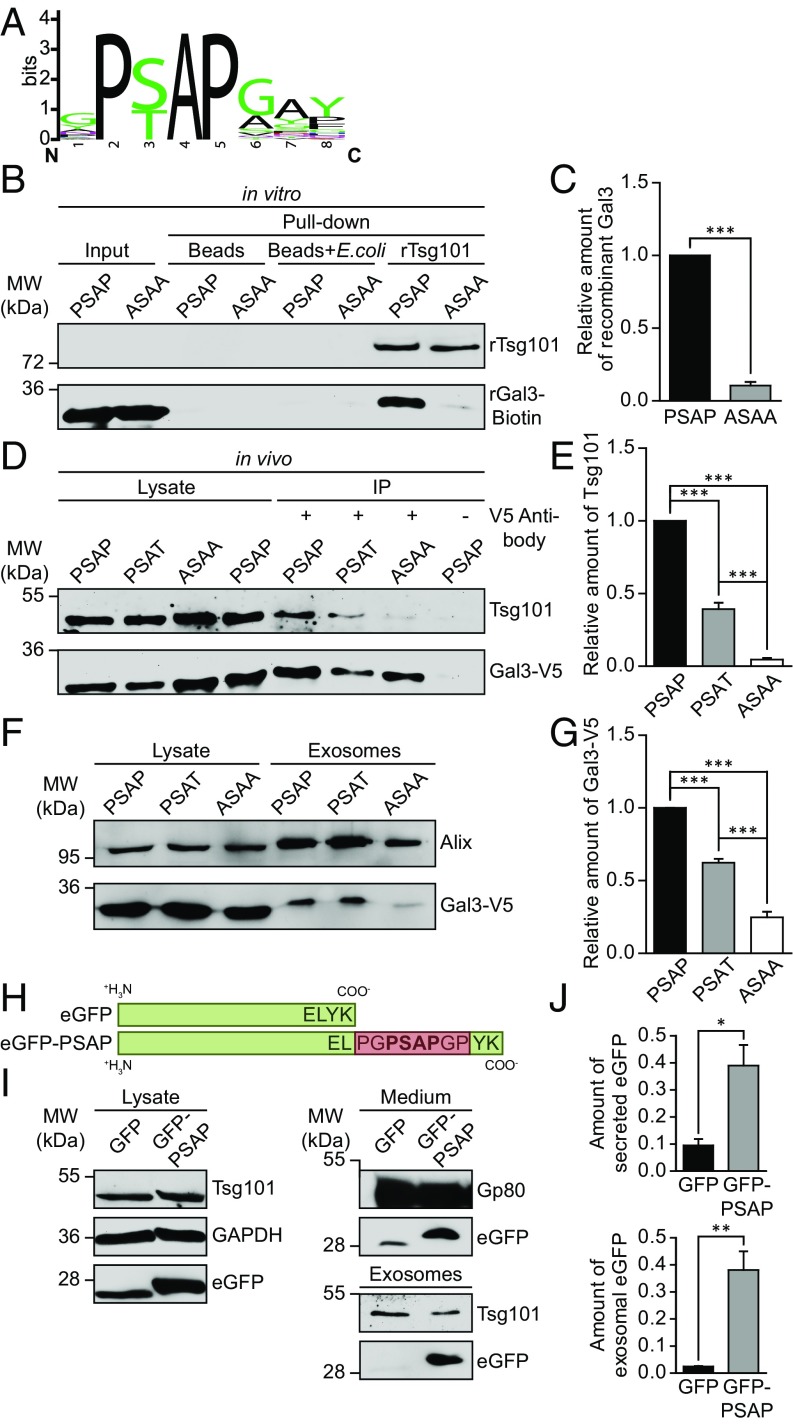
Previous studies demonstrated that a small amino-terminal octapeptide YPSAPGAY plays a pivotal role in the secretion of hamster Gal3 (47). We therefore aligned Gal3 sequences from 34 species and identified a highly conserved P(S/T)AP domain at several “hot spot” segments in the amino terminus of Gal3 (Fig. 6A and SI Appendix, Fig. S8 A–C and Supporting File). P(S/T)AP motifs were described in ESCRT-dependent proteins like Hrs and in so-called late domains of multiple viruses, inducing viral budding at the plasma membrane through direct binding to Tsg101 (48). Importantly, Gal3 seems to be the only galectin which features a late domain-like motif. The Gal3 P(S/T)AP motif showed extensive multiplications in some species, up to two PSAP and two PTAP domains in rats. Intriguingly, we observed a distinctive redundancy between PSAP and PTAP sequences, which is similar to viral late domains, indicating a convergent evolution of this motif in Gal3.
Fig. 6.
The PSAP domain of Gal3 is responsible for the interaction with Tsg101 and required for exosomal secretion. (A) An alignment of Gal3 late domain-like motifs found in 34 vertebrates was used to generate a sequence logo of the octapeptide (94)QPSAPGAY(101) (SI Appendix, Fig. S8). (B) In vitro pull-down assay with recombinant Gal3 mutants and Tsg101-GST [PSAP, human recombinant Gal3, labeled with biotin; ASAA, human recombinant Gal3 with a mutated late domain-like motif (95)PSAP(98)→(95)ASAA(98), labeled with biotin; rTsg101, recombinant human Tsg101]. Negative controls: beads only; beads which were incubated with Escherichia coli lysate. (C) Quantification of experiments as in B, normalized to input. Means ± SEM, n = 3 independent experiments. (D) Coimmunoprecipitation of Gal3-V5 mutants in MDCK cells [PSAT, human Gal3-V5 with a mutated late domain-like motif (95)PSAP(98)→(95)PSAT(98), which is a natural variant in humans]. (E) Quantification of experiments as in D. Negative control, Gal3-V5-PSAP lysate without anti-V5 antibody. Normalized to the respective cell lysate. Means ± SEM, n = 3 independent experiments. (F) Immunoblot analysis of exosomal fractions in the Gal3-V5 mutants. (G) Quantification of experiments as in F. Normalized to the respective cell lysate. Means ± SEM, n = 6 independent experiments. (H) Schematic drawing of the eGFP-PSAP polypeptide in comparison with eGFP. (I) At 48 h following transient transfection, cell culture media from MDCK cells expressing eGFP or eGFP-PSAP were collected to purify exosomes. Cell lysates, media, and isolated exosomes were analyzed by immunoblot. (J) Quantification of experiments as in I. Normalized to eGFP or eGFP-PSAP levels in the respective cell lysate. Means ± SEM, n = 3 independent experiments. Statistical analysis: Student’s unpaired t test, ***P < 0.001, **P < 0.01, *P < 0.05.
Because viruses exploit the tetrapeptide P(S/T)AP for direct interaction with Tsg101 at the plasma membrane, it was imperative to elucidate whether direct interaction and exosomal secretion of Gal3 are similarly based on the conserved P(S/T)AP domain. We therefore mutated the P(S/T)AP motif of human Gal3 into Gal3-ASAA, mimicking a mutation that is known to abrogate the release of Marburg virus-like particles (49). As a prerequisite to this study, we confirmed that the ASAA mutation of recombinant Gal3 affected neither the clustering behavior based on the amino terminus nor the functionality of the CRD (SI Appendix, Fig. S9 A and B). These findings were also supported by circular dichroism spectroscopy, showing that the ASAA mutation induced no significant alterations in the secondary structure of the protein (SI Appendix, Fig. S9C and Table S1). We also verified the binding capacity of recombinant Tsg101 (rTsg101) by a successful in vivo pull-down of Hrs (SI Appendix, Fig. S9D). After these preliminary studies, we indeed found that the direct interaction of Gal3 and rTsg101 was restrained by ASAA mutation in vitro (Fig. 6 B and C). Accordingly, in vivo coimmunoprecipitation analysis revealed that Gal3-PSAP precipitated much larger quantities of Tsg101 than the ASAA mutant (Fig. 6 D and E). The natural variant Gal3-PSAT (dbSNP:rs4652) showed intermediate precipitation capacity.
To further implicate the abrogation of direct interaction by ASAA mutation, we visualized this process by using the PLA assay. Importantly, the number of PLA signals were significantly lower in cells expressing the Gal3-ASAA variant (SI Appendix, Fig. S9 E and F). In short, these experiments clearly document that the PSAP motif of Gal3 is essential for the interaction with Tsg101.
We then explored the impact of the ASAA mutation on the localization of Gal3 at MVBs by GSDIM superresolution microscopy (SI Appendix, Fig. S9G). PSAP deletion had no apparent effect on the accumulation of Gal3-V5 at the MVB boundary, whereas neither budding nor scission stage archetypes were detectable. To clarify this, we again calculated an average intensity plot of 18 MVBs. As expected, Gal3-ASAA-V5 exhibited an orbital distribution, resembling the pattern in Vps4aE228Q-eGFP cells and particularly in Tsg101 knockdown cells. Therefore, we conclude that the interaction of Gal3 and Tsg101, based on the late domain-like motif, is central for its incorporation into MVBs, implying the involvement of other binding factors in the recruitment of Gal3 to the limiting membrane of MVBs.
To biophysically validate the above results and to characterize the binding affinities, we performed an in vitro association study based on equilibrium titrations between the recombinant Gal3-V5 mutants and recombinant Tsg101 (1–145 aa) by microscale thermophoresis (MST) (50). Strikingly, rGal3-PSAP exhibited an equilibrium dissociation constant of 8.8 ± 0.9 µM (Table 1 and SI Appendix, Fig. S10 A and C), which is in excellent agreement with previously reported affinities e.g., for HIV p6 and Tsg101 (18). Conversely, rGal3-ASAA showed a significantly reduced affinity (Kd 45.8 ± 4.7 µM; SI Appendix, Fig. S10 B and C), confirming our in vivo and in vitro studies. Lastly, we determined the binding affinity of rat Gal3, since rat Gal3 exhibits four late domain-like P(S/T)AP motifs (SI Appendix, Fig. S10 D and E). Indeed, we found that rat Gal3 displayed a greatly increased binding affinity to rTsg101 (1–145 aa; 0.3 ± 0.1 µM), which is in complete agreement with the notion that a higher number of P(S/T)AP motifs enforces protein–protein interaction between Gal3 and Tsg101, and strongly suggests cooperative binding.
Table 1.
Deduced binding affinities of the Gal3-V5 mutants and rat Gal3 obtained by MST
| Tsg101+ | Kd [µM] | SD | Ka [µM−1] | SD |
| Gal3 (PSAP) | 8.848 | 0.851 | 0.11302 | 0.01087 |
| Gal3 (ASAA) | 45.792 | 4.658 | 0.021838 | 0.002221 |
| Gal3 (rat) | 0.307 | 0.057 | 3.257329 | 0.604781 |
We finally addressed the critical importance of the PSAP-based direct interaction by investigation of the exosomal Gal3 release in MDCK cells expressing Gal3-PSAP, -PSAT and -ASAA (Fig. 6F). Corresponding to the binding efficiencies and kinetics, Gal3-PSAP showed the highest degree of exosomal secretion, whereas Gal3-ASAA secretion was drastically reduced (Fig. 6G). Thus, the PSAP motif of Gal3 is essential for the direct interaction with Tsg101, which is required for efficient exosomal release.
To further confirm the functionality of the PSAP motif in ILV recruitment, we designed an eGFP variant containing this motif at the C terminus (eGFP-PSAP; Fig. 6H). Following transient transfection, cell culture media were collected from MDCK cells expressing either eGFP-PSAP or eGFP as a control and directly analyzed by immunoblot or used for the purification of exosomes (Fig. 6I). Remarkably, significant enrichment of eGFP-PSAP in media as well as in purified exosomes strongly supports the hypothesis that the PSAP motif is sufficient for Tsg101 binding, ILV recruitment, and exosomal release of a polypeptide (Fig. 6J).
In conclusion, our data reveal herein a precise mechanism for the biochemical sorting of Gal3 into ILVs to eventually reach the extracellular space by exosomal secretion (SI Appendix, Fig. S10F).
Discussion
Various mechanisms have been proposed to explain the unconventional secretion of Gal3. However, the involved compartments and biochemical components required for this process have not been defined yet. Our results place the focus onto MVBs and the ESCRT-dependent biogenesis of exosomes.
Here, we have uncovered a functional role of the ESCRT-I component Tsg101 in exosome recruitment of Gal3. Tsg101 contains an amino-terminal ubiquitin E2 variant (UEV) domain that specifically recognizes P(S/T)AP sequences (16). Our data suggest that the Gal3-PSAP motif directly interacts with this UEV domain, coupling Gal3 delivery into ILVs by ESCRT-mediated reverse-topology membrane fission. This mechanism resembles then the direct interaction of syntenin and Alix in exosomal syndecan sorting which depends on a LYPX(n)L late domain-like motif in syntenin (20), suggesting that multiple mechanisms coexist/concur to import cargo proteins into ILVs.
Once merged with the plasma membrane, MVBs release Gal3-laden ILVs into the extracellular space to become exosomes. This seems to be the major pathway for apical Gal3 secretion in MDCK cells. Little is known about the regulation of MVB fusion with the plasma membrane. It was shown that the release of exosomes can be stimulated by a low extracellular pH microenvironment (51) or increasing intracellular calcium levels (21). Likewise, we found that a DMA-mediated reduction in the intracellular calcium concentration blocks the release of exosomal Gal3. Very recently, Verweij et al. (52) demonstrated that histamine-mediated G protein-coupled receptor activation triggers MVB fusion with the plasma membrane. In these studies, a basal and a regulated MVB fusion activity have been described. It therefore remains to be discovered which functions a triggered MVB fusion might fulfill in the apical release of exosomes.
The experiments reported here suggest the possibility that Gal3-containing exosomes could be conveyed to neighboring cells in the vicinity or even other tissues and the bloodstream, from where they are taken up by recipient target cells by plasma membrane fusion. Several studies demonstrate the importance of communication through the transfer of exosomal proteins from one cell to the other (51, 53), which becomes particularly evident in tumor progression and metastasis. The now cytoplasmic Gal3 can modulate a plethora of functions, including apoptosis, a number of signal transduction pathways, and gene expression (54). Corresponding to the notion that Gal3 is delivered by exosomes, recent evidence suggests that artificial exosomes exhibited calculated half-lives of about 2 min in body fluids (55, 56). Although the decisive mechanism of exosomal uptake and disruption is currently under great debate in the literature, several mechanisms are reported to be responsible for the uptake of exosomes, including Clathrin-mediated endocytosis (57), phagocytosis (58), micropinocytosis (59), and plasma or endosomal membrane fusion (60). These mechanisms support the theory that exosomal cargo is delivered by internalization.
It is still not clear whether and how Gal3 can be liberated from exosomes and exist in the extracellular space in free form. Extracellular free Gal3 is involved in a variety of physiological and pathological processes in the extracellular space, including interactions with hensin (61), laminin (62), β1-integrin (63), and fibronectin (64), as well as signal transduction of EGF, TGF-β (65), and VEGF (66). Remarkably, the Gal3 plasma level correlates with the quantities of exosomal Gal3, and both are similarly elevated in patients with arteriosclerosis (67). Nevertheless, the stability of exosomes seems to be very high in human plasma (68). Although the release of proteins possibly emanating from exosomes such as hsp60 (69) has been examined and discussed, the underlying mechanism is still unknown and further work will be necessary to elucidate how exosomal proteins can be liberated.
We propose three different mechanisms by which luminal Gal3 could be liberated from exosomes: (i) direct translocation of Gal3 across the exosomal membrane. In this context, Gal3 was reported to directly traverse the lipid bilayer of liposomes by a process that seemed to be energy independent and devoid of exogenous factors (70). A related mechanism was discussed for Gal3 translocation across the membrane of microvesicles, possibly associated with chaperones such as hsp70 (15). (ii) Internalization of Gal3-laden exosomes followed by fusion with the lysosomal compartment. Disruption of the exosomal membrane by lysosomal lipases could then release Gal3, which is in turn recycled back to the endosomal compartment. Furthermore, several studies showed that Gal3 interacts with lysosome-associated membrane proteins LAMP-1 and LAMP-2 (28, 71) which are translocated to the plasma membrane in lysosomal exocytosis and membrane repair/resealing (72). The interaction with highly glycosylated proteins like LAMPs could also explain how Gal3 may be protected against degradation through lysosomal proteases. In support of this liberation mechanism, Gal3 was found in secretory lysosomes and melanosomes (73, 74). (iii) Disruption of exosomes in the extracellular space potentially by secreted lipases. This mechanism has been discussed for IL-1β (75) and could explain how free Gal3 is delivered directly to the site of action. Clearly, additional evidence is needed to clarify the relevance of these exosomal release mechanisms in the liberation of Gal3.
Inward budding of ILVs in exosome formation and viral budding at the plasma membrane are topologically and mechanistically related processes that involve identical cellular machinery. Viral proteins exploit the Tsg101 UEV domain to recruit components of the ESCRT vesicle fission machinery for viral budding at the plasma membrane (48). Particularly, Tsg101 binding to the PSAP motif of arrestin domain-containing protein 1 induces direct budding of microvesicles from the plasma membrane (76), offering a blueprint for the evolutionary recruitment of the ESCRT complex. However, a PSAP-dependent sorting of endogenous non-ESCRT cargo molecules at MVBs has not been described before.
Furthermore, Gal3 seems to promote HIV budding at the plasma membrane through interaction with Alix (77). Remarkably, Alix overexpression rescues the PSAP-Tsg101 interaction in PSAP-defective virus release (78). Deletion of a carboxyl-terminal region in the proline-rich domain of Alix blocked this rescue, suggesting that a yet unidentified binding partner is required for Alix-mediated viral budding. Indeed, it is this interaction motif that accounts for Gal3 binding to Alix (77). Interaction between Gal3 and Alix has also been described in other cellular systems (4, 79). However, coimmunoprecipitation studies of Wang et al. (77) with flag-tagged Gal3 in HEK293T cells did not reveal Tsg101 precipitation. This negative result could be explained by apparently more efficient anti-Alix compared with anti-Tsg101 antibodies and consequently faint Tsg101 labeling or by the prominent bands of the heavy chain, possibly obscuring positive Tsg101 signals. To receive a comprehensive impression, it would be interesting to investigate in renal epithelial cells whether an interaction between Gal3 and Alix can recruit Tsg101 to viral budding sites through direct binding to the Gal3-PSAP domain. Moreover, it remains to be demonstrated whether Alix is stabilizing the Tsg101-Gal3 interaction for inward budding of ILVs in exosome formation.
In summary, this study identifies a functionally relevant PSAP-mediated direct Tsg101 interaction with an endogenous cargo molecule and provides a unique model of how proteins destined for exosomal secretion can be recruited and opens unique avenues for the polarized unconventional secretion of cytoplasmic proteins.
Materials and Methods
Antibodies.
Antibodies are detailed in SI Appendix, SI Materials and Methods. Gal3 was detected with rabbit polyclonal antibodies as described before (30).
Plasmids, Recombinant Expression, Purification, and Labeling.
A detailed description of the DNA constructs and the expression, purification, and labeling of recombinant proteins can be found in SI Appendix, SI Materials and Methods.
Cell Culture, Transfection, and Treatments.
A detailed description of cell culture and transfection can be found in SI Appendix, SI Materials and Methods. MAP4-eGFP expression in MDCK cells was induced through transduction with BacMam (Invitrogen) particles according to the manufacturer’s instructions. Exosomal release in MDCK cells was inhibited by 15 nM DMA (Sigma-Aldrich) treatment overnight. Vps4aE228Q-eGFP expression was induced with 10 nM mifepristone (ChemPur) for the indicated time intervals.
Preparation of Exosomes.
To purify exosomes, cells were washed three times with PBS and incubated overnight with MEM and 10% FCS. To avoid contamination of the exosomal fraction by bovine serum exosomes, FCS was subjected to centrifugation at 100,000 × g for 2 h. Medium was collected and submitted to a series of centrifugation steps. First, detached cells were removed by a short centrifugation step at 300 × g for 6 min. Then, cellular debris and microvesicles were removed at 5,000 × g and 20,000 × g for 30 min, respectively. To allow pelleting of exosomes in a 1.5-mL reaction tube scale, the cell culture supernatant was concentrated in Amicon Ultra-15 100 K centricons (Millipore) and subsequently centrifuged at 100,000 × g for 1 h. The resulting pellet was washed in PBS++ (PBS supplemented with 1 mM CaCl2 and 1 mM MgCl2), repelleted again at 100,000 × g for 1 h, and then resuspended in either PBS++ or in SDS/PAGE sample buffer for further use. All steps were performed at 4 °C.
Preparation of Microvesicles.
Purification of microvesicles was performed as described for exosomes, whereas microvesicles were already pelleted at 10,000 × g for 30 min. After centrifugation, the pellet was washed in PBS++, repelleted at 10,000 × g for 30 min, and then resuspended in either PBS++ or SDS/PAGE sample buffer. Actin served as positive control for Western blotting (80).
Proteinase K Protection Assay.
Exosomal pellets were resuspended in PBS++, pooled, and subsequently split into three identical aliquots. Proteinase digestion was then performed with 0.5 mg/mL to 1 mg/mL proteinase K (Fermentas) in presence or absence of 1% Triton X-100 for 30 min at 37 °C. As control, one of the aliquots was incubated without proteinase K.
Immunofluorescence and Image Processing.
Cells grown on glass coverslips were washed twice with PBS++ and fixed with 4% paraformaldehyde (PFA) (20 min) or ice-cold methanol (5 min). Cells were permeabilized with 0.2% Triton X-100 (20 min). Samples were blocked with 5% goat serum or 5% skim milk powder in PBS++ for 1 h. Immunostaining was then performed with the indicated primary antibodies. As secondary antibodies, goat anti-mouse conjugated to Alexa 647 or Alexa 488 (Invitrogen) and goat anti-rabbit conjugated to Alexa 555 (Invitrogen) were used. Triple staining was performed with chicken anti-goat tagged with Alexa 647 (Invitrogen). After the Hrs chicken anti-goat Alexa 647 staining had been completed, Gal3 and Tsg101 were sequentially marked in a second staining procedure. To remove endocytosed Gal3, cells were incubated twice with cell culture medium supplemented with 150 mM lactose for 40 min. An additional washing step with 150 mM lactose in PBS++ was also implemented. Confocal images were acquired on a Leica TCS SP2 microscope with a 40× or 63× oil immersion objective. Processing of images was done with Leica LAS AF, ImageJ, Imaris (Bitplane), and Volocity 5 (PerkinElmer). We calculated colocalization between markers as Manders’ coefficient using Volocity. Structures with coefficients <0.5 were classified as “not colocalized.” A detailed description of the FRAP protocol can be found in SI Appendix, SI Materials and Methods.
Electron Microscopy.
Purified exosomes resuspended in PBS++ were absorbed onto formvar-coated metal grids and subsequently negatively stained with 2% phosphotungstate for 30 s. Cells or pellets of the exosomal fraction were fixed in a mixture of 2.5% glutaraldehyde, 2.5% paraformaldehyde, and 0.05% picric acid in 67 mM cacodylate buffer (pH 7.4). Postfixation was performed in 1% osmium tetroxide followed by an overnight incubation with 0.3% uranyl acetate dissolved in 50 mM maleate buffer (pH 5). Samples were embedded in Epon according to standard procedures. Thin sections were contrasted with lead citrate. For Gal3-eGFP labeling, thin sections were etched with 10% hydrogen peroxide for 10 min to unveil eGFP epitopes. Gold-labeled GFP nanobodies (Chromotek, Cytodiagnostics) were finally incubated for 2 h. Images were acquired with a Zeiss EM 109S electron microscope. A detailed description of the electron tomography studies can be found in SI Appendix, SI Materials and Methods.
GSDIM Superresolution Light Microscopy.
Superresolution microscopy was performed with a Leica SR GSD microscope equipped with a Sumo Stage for drift-free imaging and an oil immersion objective (HC PL Apo 160×). Collection of images was achieved with an Andor iXon DU-897 EMCCD camera (Andor Technology). Between 15,000 and 40,000 frames were recorded to reconstruct a superresolved image, with a frame rate of 100 Hz. Samples were embedded in a mixture of 20% Vectashield H-1000 (Vector Laboratories) and glycerol-Tris buffer (81) (glycerol with 50 mM Tris pH 8). Gal3 and either Tsg101 or Hrs were labeled by immunofluorescence procedure with secondary antibodies coupled to Alexa 555 and Alexa 647, respectively. Average intensity plots were computed with the ImageJ Stacks tool. Single MVBs were matched to a stack without additional alignment or processing. Finally, the average intensity of the stack was calculated. We estimated the achieved structural resolution using Fourier ring correlation (FRC) (82, 83) analysis and additionally by measuring the average full width at half maximum (FWHM) of 21 single spots. GSDIM data were acquired, processed, and quantified with the Leica LAS AF GSD Wizard. Chromatic aberration and drift were corrected. Image processing and 3D reconstructions were done with Leica LAS AF, ImageJ, and Imaris (Bitplane).
Simulation of Superresolution GSDIM Data.
The simulations were performed by using the simulation module of Picasso (84). All simulation parameters were chosen to match imaging conditions and calculated resolution of the experimental GSDIM data. Detailed description can be found in SI Appendix, SI Materials and Methods.
Coimmunoprecipitation.
MDCK cells stably expressing either eGFP fusion proteins or Gal3-V5 variants were washed with PBS++, followed by mechanical detachment of the cells in PBS++. The cells were pelleted by centrifugation at 500 × g for 3 min at 4 °C and rinsed twice with PBS++. Cell lysis was achieved by application of lysis buffer (10 mM Hepes, 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, 0.5% SDS and proteinase inhibitor mixture, pH 7.5). Tris buffers were avoided due to DTSSP cross-linker treatment. Cleared lysates (17,000 × g, 15 min, 4 °C) were incubated with GFP-nanobody agarose (GFP-Trap, Chromotek) for 1 h at 4 °C. After the fusion proteins were bound to the beads, we applied 3 mM DTSSP cross-linker (Thermo Fisher Scientific) for 2 h at 4 °C for unspecific stabilization of protein–protein interactions. Finally, beads were rinsed four times with Co-IP washing buffer (10 mM Hepes, 150 mM NaCl, 0.5 mM EDTA) and boiled in SDS/PAGE loading buffer for Western blot analysis.
GST Pull-Down Assay.
For analysis of direct protein interaction, recombinant Gal3 (1.5 µM; 10 nM for analysis of the binding capacity of Gal3-PSAP and Gal3-ASAA) was incubated for 2 h at 4 °C with Tsg101-GST bound to glutathione-Sepharose in Co-IP washing buffer. After four rinse cycles with Co-IP washing buffer, samples were boiled in SDS/PAGE loading buffer and analyzed by Western blot.
RNA Interference.
Depletion of either Tsg101 or Hrs was achieved after transfection of siRNA using Lipofectamine RNAiMAX (Invitrogen). Cells were transfected at day 1 after seeding with siRNA duplexes described in SI Appendix, SI Materials and Methods. Subsequent experiments were done at day 4 after seeding. Cell viability was assessed by Trypan blue exclusion (Invitrogen) according to the manufacturer’s instructions and automatically analyzed by a Countess cell counter (Invitrogen).
CRISPR/Cas9 Knockout of Gal3.
Gal3 CRISPR/Cas9 knockout is described in SI Appendix, SI Materials and Methods.
PLA.
PLA was used to identify the interaction between Gal3 and Tsg101. Cells were fixed, permeabilized, blocked and incubated with primary antibodies against Gal3 and Tsg101. The PLA probes anti-mouse minus and anti-rabbit plus were used (Duolink). Proximity ligation was conducted according to the manufacturer’s instructions. After hybridization, enzymatic ligation, and polymerase amplification with fluorescent oligonucleotides, fluorescent emissions were quantified by confocal microscopy. To combine PLA and immunofluorescence, goat anti-Hrs antibodies were applied in parallel to localize the interaction sites of Gal3 and Tsg101.
Microscale Thermophoresis.
MST was performed on a Monolith NT.115 (NanoTemper Technologies GmbH) at 21 °C (red LED power was set between 20% and 40% and infrared laser power to 60%). The recombinant Tsg101-UEV (1–145 aa) domain was labeled with the dye NT 647 according to the supplier (NanoTemper Technologies). A total of 9 nM rTsg101-UEV was titrated with unlabeled Gal3 in PBS supplemented with 5 mM DTT, 10 µg/mL BSA, and 0.05% Tween. Nine independent MST experiments were recorded at 680 nm and processed by NanoTemper Analysis 1.2.009 and Origin8.
Recombinant Gal3 and Tsg101 Functionality.
Preliminary studies (circular dichroism, turbidity assay, fluorescence polarization, and GST pull-down) assessing folding and functionality of recombinant Gal3 and Tsg101 are detailed in SI Appendix, SI Materials and Methods.
Alignment and Sequence Logos.
Gal3 sequences were aligned with ClustalOmega (85). Sequence logos aligned to the octapeptide (94)QPSAPGAY(101) were generated by WebLogo (86, 87). For reference accession codes (UniProt) see SI Appendix, Supporting File.
Supplementary Material
Acknowledgments
We thank B. Y. Kim (Yonsei University College of Medicine) and W. Sundquist (University of Utah) for the generous supply of the Tsg101-eGFP and Vps4aE228Q-eGFP plasmids; H. Leffler (Lund University) for providing us with fluorescently labeled A-Tetra and rat Gal3 plasmid; R. Tikkanen (Giessen University) for providing the Tsg101 expression plasmid; and W. Ackermann, M. Dienst, and U. Lehr for technical assistance. We gratefully acknowledge the technical support of the core facility Protein Spectroscopy and Protein Biochemistry (Philipps University Marburg). M.T.S. acknowledges support from the International Max Planck Research School for Molecular and Cellular Life Sciences. This work was supported by the Deutsche Forschungsgemeinschaft (Bonn, Germany; Grants JA 1033 and Graduiertenkolleg 2213).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718921115/-/DCSupplemental.
References
- 1.Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- 2.Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. Introduction to galectins. Glycoconj J. 2002;19:433–440. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- 3.Delacour D, Koch A, Jacob R. The role of galectins in protein trafficking. Traffic. 2009;10:1405–1413. doi: 10.1111/j.1600-0854.2009.00960.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen H-Y, et al. Galectin-3 negatively regulates TCR-mediated CD4+ T-cell activation at the immunological synapse. Proc Natl Acad Sci USA. 2009;106:14496–14501. doi: 10.1073/pnas.0903497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nangia-Makker P, et al. Galectin-3 induces endothelial cell morphogenesis and angiogenesis. Am J Pathol. 2000;156:899–909. doi: 10.1016/S0002-9440(10)64959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nangia-Makker P, et al. Cleavage of galectin-3 by matrix metalloproteases induces angiogenesis in breast cancer. Int J Cancer. 2010;127:2530–2541. doi: 10.1002/ijc.25254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boscher C, et al. Galectin-3 protein regulates mobility of N-cadherin and GM1 ganglioside at cell-cell junctions of mammary carcinoma cells. J Biol Chem. 2012;287:32940–32952. doi: 10.1074/jbc.M112.353334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boscher C, Nabi IR. Galectin-3- and phospho-caveolin-1-dependent outside-in integrin signaling mediates the EGF motogenic response in mammary cancer cells. Mol Biol Cell. 2013;24:2134–2145. doi: 10.1091/mbc.E13-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuboi K, et al. Galectin-3 expression in colorectal cancer: Relation to invasion and metastasis. Anticancer Res. 2007;27:2289–2296. [PubMed] [Google Scholar]
- 10.Yamamoto-Sugitani M, et al. Galectin-3 (Gal-3) induced by leukemia microenvironment promotes drug resistance and bone marrow lodgment in chronic myelogenous leukemia. Proc Natl Acad Sci USA. 2011;108:17468–17473. doi: 10.1073/pnas.1111138108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindstedt R, Apodaca G, Barondes SH, Mostov KE, Leffler H. Apical secretion of a cytosolic protein by Madin-Darby canine kidney cells. Evidence for polarized release of an endogenous lectin by a nonclassical secretory pathway. J Biol Chem. 1993;268:11750–11757. [PubMed] [Google Scholar]
- 12.Théry C, et al. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 13.Fei F, et al. B-cell precursor acute lymphoblastic leukemia and stromal cells communicate through Galectin-3. Oncotarget. 2015;6:11378–11394. doi: 10.18632/oncotarget.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welton JL, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–1338. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehul B, Hughes RC. Plasma membrane targetting, vesicular budding and release of galectin 3 from the cytoplasm of mammalian cells during secretion. J Cell Sci. 1997;110:1169–1178. doi: 10.1242/jcs.110.10.1169. [DOI] [PubMed] [Google Scholar]
- 16.Pornillos O, et al. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundquist WI, et al. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 18.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 19.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: Mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baietti MF, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 21.Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 22.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straube T, et al. pH-dependent recycling of galectin-3 at the apical membrane of epithelial cells. Traffic. 2013;14:1014–1027. doi: 10.1111/tra.12086. [DOI] [PubMed] [Google Scholar]
- 25.Fölling J, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 26.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 27.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 28.Inohara H, Raz A. Identification of human melanoma cellular and secreted ligands for galectin-3. Biochem Biophys Res Commun. 1994;201:1366–1375. doi: 10.1006/bbrc.1994.1854. [DOI] [PubMed] [Google Scholar]
- 29.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delacour D, et al. Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic. 2007;8:379–388. doi: 10.1111/j.1600-0854.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- 31.Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–1426. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- 33.Kim SB, et al. Caspase-8 controls the secretion of inflammatory lysyl-tRNA synthetase in exosomes from cancer cells. J Cell Biol. 2017;216:2201–2216. doi: 10.1083/jcb.201605118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyotte A, Russell MRG, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian “class E” compartment: A multicisternal early endosome with multiple sorting defects. J Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- 35.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 36.White IJ, Bailey LM, Aghakhani MR, Moss SE, Futter CE. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006;25:1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buschow SI, et al. MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways. Traffic. 2009;10:1528–1542. doi: 10.1111/j.1600-0854.2009.00963.x. [DOI] [PubMed] [Google Scholar]
- 38.Bobrie A, Colombo M, Krumeich S, Raposo G, Théry C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1:18397. doi: 10.3402/jev.v1i0.18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller M, Adell MA, Teis D. Membrane abscission: First glimpse at dynamic ESCRTs. Curr Biol. 2012;22:R603–R605. doi: 10.1016/j.cub.2012.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat Struct Mol Biol. 2015;22:492–498. doi: 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adell MAY, et al. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife. 2017;6:e31652. doi: 10.7554/eLife.31652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheuring S, et al. Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J Mol Biol. 2001;312:469–480. doi: 10.1006/jmbi.2001.4917. [DOI] [PubMed] [Google Scholar]
- 44.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita H, et al. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: Class E vps phenotype in mammalian cells. J Cell Sci. 2003;116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 46.Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 47.Menon RP, Hughes RC. Determinants in the N-terminal domains of galectin-3 for secretion by a novel pathway circumventing the endoplasmic reticulum-Golgi complex. Eur J Biochem. 1999;264:569–576. doi: 10.1046/j.1432-1327.1999.00671.x. [DOI] [PubMed] [Google Scholar]
- 48.Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-I in retroviral budding. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolnik O, Kolesnikova L, Stevermann L, Becker S. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol. 2010;84:7847–7856. doi: 10.1128/JVI.00476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 51.Parolini I, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verweij FJ, et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol. 2018;217:1129–1142. doi: 10.1083/jcb.201703206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funasaka T, Raz A, Nangia-Makker P. Galectin-3 in angiogenesis and metastasis. Glycobiology. 2014;24:886–891. doi: 10.1093/glycob/cwu086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi Y, et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol. 2013;165:77–84. doi: 10.1016/j.jbiotec.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Saunderson SC, Dunn AC, Crocker PR, McLellan AD. CD169 mediates the capture of exosomes in spleen and lymph node. Blood. 2014;123:208–216. doi: 10.1182/blood-2013-03-489732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng D, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 59.Fitzner D, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 60.Montecalvo A, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hikita C, et al. Induction of terminal differentiation in epithelial cells requires polymerization of hensin by galectin 3. J Cell Biol. 2000;151:1235–1246. doi: 10.1083/jcb.151.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Q, Cummings RD. The S-type lectin from calf heart tissue binds selectively to the carbohydrate chains of laminin. Arch Biochem Biophys. 1990;281:27–35. doi: 10.1016/0003-9861(90)90408-q. [DOI] [PubMed] [Google Scholar]
- 63.Fukumori T, et al. CD29 and CD7 mediate galectin-3-induced type II T-cell apoptosis. Cancer Res. 2003;63:8302–8311. [PubMed] [Google Scholar]
- 64.Sato S, Hughes RC. Binding specificity of a baby hamster kidney lectin for H type I and II chains, polylactosamine glycans, and appropriately glycosylated forms of laminin and fibronectin. J Biol Chem. 1992;267:6983–6990. [PubMed] [Google Scholar]
- 65.Partridge EA, et al. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 66.Markowska AI, Jefferies KC, Panjwani N. Galectin-3 protein modulates cell surface expression and activation of vascular endothelial growth factor receptor 2 in human endothelial cells. J Biol Chem. 2011;286:29913–29921. doi: 10.1074/jbc.M111.226423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madrigal-Matute J, et al. Galectin-3, a biomarker linking oxidative stress and inflammation with the clinical outcomes of patients with atherothrombosis. J Am Heart Assoc. 2014;3:e000785. doi: 10.1161/JAHA.114.000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalra H, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 69.Malik ZA, et al. Cardiac myocyte exosomes: Stability, HSP60, and proteomics. Am J Physiol Heart Circ Physiol. 2013;304:H954–H965. doi: 10.1152/ajpheart.00835.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lukyanov P, Furtak V, Ochieng J. Galectin-3 interacts with membrane lipids and penetrates the lipid bilayer. Biochem Biophys Res Commun. 2005;338:1031–1036. doi: 10.1016/j.bbrc.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 71.Dong S, Hughes RC. Macrophage surface glycoproteins binding to galectin-3 (Mac-2-antigen) Glycoconj J. 1997;14:267–274. doi: 10.1023/a:1018554124545. [DOI] [PubMed] [Google Scholar]
- 72.Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. doi: 10.1016/s0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 73.Schmidt H, et al. Effector granules in human T lymphocytes: The luminal proteome of secretory lysosomes from human T cells. Cell Commun Signal. 2011;9:4. doi: 10.1186/1478-811X-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Basrur V, et al. Proteomic analysis of early melanosomes: Identification of novel melanosomal proteins. J Proteome Res. 2003;2:69–79. doi: 10.1021/pr025562r. [DOI] [PubMed] [Google Scholar]
- 75.Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179:1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 76.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang SF, et al. Galectin-3 promotes HIV-1 budding via association with Alix and Gag p6. Glycobiology. 2014;24:1022–1035. doi: 10.1093/glycob/cwu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Usami Y, Popov S, Göttlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu W, et al. Galectin-3 regulates intracellular trafficking of EGFR through Alix and promotes keratinocyte migration. J Invest Dermatol. 2012;132:2828–2837. doi: 10.1038/jid.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olivier N, Keller D, Rajan VS, Gönczy P, Manley S. Simple buffers for 3D STORM microscopy. Biomed Opt Express. 2013;4:885–899. doi: 10.1364/BOE.4.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nieuwenhuizen RPJ, et al. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–562. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Banterle N, Bui KH, Lemke EA, Beck M. Fourier ring correlation as a resolution criterion for super-resolution microscopy. J Struct Biol. 2013;183:363–367. doi: 10.1016/j.jsb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 84.Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 85.McWilliam H, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.