Significance
Double-strand breaks (DSBs) are deleterious DNA lesions, and impairment of the DSB repair machinery can lead to devastating diseases, such as Nijmegen Breakage Syndrome (NBS). During meiosis, DSBs represent a “necessary evil”: they are required to promote formation of crossovers between homologous chromosomes. Crossovers, in turn, ensure correct chromosome inheritance during gamete formation, which is essential for viability and normal development of embryos. During meiosis, numerous DSBs are actively created, so meiotic cells must ensure that all breaks are properly repaired to ensure crossover formation and restore genomic integrity. Here, we identify Caenorhabditis elegans NBS-1 as essential to properly process meiotic DSBs to both promote crossover formation and antagonize an error-prone DSB repair pathway, thereby ensuring faithful chromosome inheritance.
Keywords: meiosis, DNA repair, double-strand breaks, non-homologous end joining, homologous recombination
Abstract
Faithful inheritance of genetic information through sexual reproduction relies on the formation of crossovers between homologous chromosomes during meiosis, which, in turn, relies on the formation and repair of numerous double-strand breaks (DSBs). As DSBs pose a potential threat to the genome, mechanisms that ensure timely and error-free DSB repair are crucial for successful meiosis. Here, we identify NBS-1, the Caenorhabditis elegans ortholog of the NBS1 (mutated in Nijmegen Breakage Syndrome) subunit of the conserved MRE11-RAD50-NBS1/Xrs2 (MRN) complex, as a key mediator of DSB repair via homologous recombination (HR) during meiosis. Loss of nbs-1 leads to severely reduced loading of recombinase RAD-51, ssDNA binding protein RPA, and pro-crossover factor COSA-1 during meiotic prophase progression; aggregated and fragmented chromosomes at the end of meiotic prophase; and 100% progeny lethality. These phenotypes reflect a role for NBS-1 in processing of meiotic DSBs for HR that is shared with its interacting partners MRE-11-RAD-50 and COM-1 (ortholog of Com1/Sae2/CtIP). Unexpectedly, in contrast to MRE-11 and RAD-50, NBS-1 is not required for meiotic DSB formation. Meiotic defects of the nbs-1 mutant are partially suppressed by abrogation of the nonhomologous end-joining (NHEJ) pathway, indicating a role for NBS-1 in antagonizing NHEJ during meiosis. Our data further reveal that NBS-1 and COM-1 play distinct roles in promoting HR and antagonizing NHEJ. We propose a model in which different components of the MRN-C complex work together to couple meiotic DSB formation with efficient and timely engagement of HR, thereby ensuring crossover formation and restoration of genome integrity before the meiotic divisions.
Maintenance of genome integrity throughout cell divisions and generations is of paramount importance for organismal survival and faithful inheritance of genetic information, and multiple mechanisms have evolved to detect and repair DNA damage. Double-strand breaks (DSBs), where both DNA strands are severed, are among the most dangerous DNA lesions, as inaccurate repair of DSBs can result in genomic rearrangements, cell death, and/or carcinogenesis. DSBs can be provoked by environmental sources, such as radiation or chemical exposure, or can result from intrinsic cellular sources, such as DNA replication errors (1).
While DSBs constitute a dangerous form of DNA damage in most cellular contexts, DSBs are deliberately induced during meiosis to promote formation of crossovers (COs) (2). Meiotic COs are critical for the balanced segregation of homologous chromosomes at meiosis I, as CO recombination events between the DNA molecules of homologous chromosomes, together with sister chromatid cohesion, establish physical connections between homologs (chiasmata), which in turn ensure their correct orientation toward opposite poles of the meiosis I spindle. Thus, the requirement for COs to ensure homolog segregation poses a challenge for sexually reproducing organisms, as meiotic recombination is initiated by the formation of DNA lesions that constitute a danger to genomic integrity. By the end of meiosis, all DSBs must be accurately repaired to (i) ensure CO formation and proper chromosome segregation and (ii) guarantee that genome integrity is restored before cell division.
Meiotic DSBs are specifically induced by the conserved topoisomerase VI-like protein SPO11 (3–5). The SPO11 protein remains covalently bound to both broken DNA ends after the break occurs and has to be removed through a process called resection for the DSB to be repaired. Resection is initiated by an endonucleolytic cleavage that leads to the release of SPO11 attached to a small oligonucleotide (6) and results in a short 3′ ssDNA tail. Additional resection of the 5′ end produces a longer ssDNA tail (7), which recruits DNA strand exchange proteins DMC1 and/or RAD51 to stimulate invasion of a homologous DNA duplex and repair of the DSB by homologous recombination (HR) (8). The first DNA cleavage event is dependent on endonuclease activity of the conserved MRE11-RAD50-NBS1/Xrs2 (MRN/X) complex as well as on the COM1/Sae2/CtIP/Ctp1 protein, which associates with MRN/X (9). Analysis of budding yeast meiosis shows that, in the second step, the Exo1 exonuclease joins in to extend the resected tracts and produce the long 3′ ssDNA-tailed intermediates (10).
An alternative mechanism for double-strand break repair (DSBR) is the nonhomologous end-joining (NHEJ) pathway, which involves protection of the broken ends by the Ku70/Ku80 heterodimer ring (11). Binding of Ku prepares DSBs for direct ligation between broken DNA ends with little or no homology, an inherently error-prone process (12). In cases where multiple DSBs on different chromosomes are present in the same cell, as occurs during meiosis, end joining can result in chromosome translocations. In contrast to NHEJ, HR is generally considered an error-free pathway of DSBR, as it uses a homologous DNA template to repair the broken molecule. A strong body of evidence indicates that there is competition between the HR and NHEJ pathways for repair of DSBs, raising the question as to how pathway choice is regulated (12). Initiation of resection by the MRN/X complex and Com1/Sae2/CtIP/Ctp1 seems to be critical for this decision, as it commits cells to homology-dependent repair (7). Interestingly, evidence from Caenorhabditis elegans indicates that such competition occurs even during meiosis, where it is absolutely critical for DSBR to occur exclusively by HR (13, 14). Thus, efficient coupling of DSB formation and DSB resection is of paramount importance for ensuring a successful outcome of meiosis.
MRE11 and RAD50 are highly conserved in eukaryotes. MRE11 is the nuclease subunit of the complex, while RAD50, which belongs to the Structural Maintenance of Chromosomes family of proteins, is required for regulating MRE11 nuclease activity in an ATP-dependent manner and may also be important for tethering of DNA ends (7). Nbs1/Xrs2 is the least conserved member of the MRN/X complex, and the high sequence divergence between mammalian NBS1 and yeast Xrs2 had precluded the identification of orthologs in many species (15), including C. elegans.
Here, we report the identification of the previously elusive C. elegans NBS1 ortholog based on a role in meiotic recombination revealed by a mutant screen. Unexpectedly, we found that the requirements for NBS-1 during meiosis are distinct from those of its complex partners. In contrast to MRE-11 and RAD-50, which are required both for formation and for resection of meiotic DSBs (14, 16, 17), NBS-1 is required for DSB resection but is dispensable for DSB formation. We further found that NBS-1 [like MRE-11 (14)] is not only important for promoting resection and HR but also, for antagonizing NHEJ during meiosis. This latter characteristic is shared with COM-1 (13, 18), a partner of the MRN complex, but our data reveal distinct roles for NBS-1 and COM-1 in promoting HR and antagonizing NHEJ. Our results support a model in which different components of the MRN-C complex work together during meiosis to couple formation and repair of meiotic DSBs to both (i) promote efficient and timely DSB resection to promote HR and (ii) antagonize NHEJ to ensure genome stability.
Results
Identification of the C. elegans NBS-1 Ortholog.
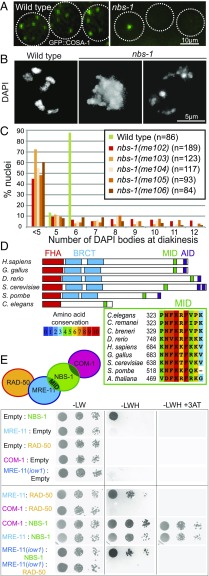
We isolated the initial nbs-1(me102) mutant allele in a genetic screen for C. elegans mutants with altered numbers of GFP::COSA-1 foci, which mark the sites of COs in C. elegans germ cells at the late pachytene stage of meiotic prophase (Fig. 1A). As each chromosome pair normally undergoes only a single CO during C. elegans meiosis, wild-type (WT) late pachytene nuclei consistently exhibit six GFP::COSA-1 foci, one for each pair of homologs (19). Furthermore, DAPI staining of WT oocytes at diakinesis, the last stage of meiotic prophase I, reveals six well-resolved DAPI bodies corresponding to the six pairs of homologs linked by chiasmata (six bivalents). The nbs-1(me102) mutant was isolated based on observation of a severe reduction in the number of GFP::COSA-1 foci by live imaging (Fig. 1A), indicating impairment of meiotic recombination. DAPI staining of diakinesis oocytes in the nbs-1(me102) mutant further revealed frayed, aggregated, and/or fragmented chromosomes (Fig. 1 B and C), indicative of defects in DNA repair. Moreover, me102 homozygous hermaphrodites produced no viable progeny (0 survivors of 1,575 eggs laid) (Table S1).
Fig. 1.
Identification of the C. elegans nbs-1 ortholog based on its requirement for meiotic DNA repair. (A) GFP::COSA-1 foci in late pachytene nuclei of live worms. Each WT nucleus has six foci (corresponding to the six CO sites), while the nbs-1 nuclei usually have one or zero foci. (B) Images of DAPI-stained chromosomes from individual diakinesis-stage oocytes. The WT oocyte displays six DAPI bodies corresponding to the six pairs of homologs connected by chiasmata, while the nbs-1(me102) mutant oocytes display chromosome aggregates (less than five DAPI bodies), indicative of defective DNA repair. (C) Graphs showing frequencies of diakinesis-stage oocytes with the indicated number of DAPI bodies in WT worms and worms homozygous for nbs-1 mutant alleles (Fig. S1). (D, Upper) Schematic depicting the C. elegans NBS-1 protein and its orthologs in other species. NBS-1 contains the conserved FHA domain and the MID but lacks the tandem BRCT domains and the ATM interacting domain (AID). (D, Lower) Alignment showing conservation of the MID among members of the NBS1/Nibrin protein family. C. breneri, Caenorhabditis breneri; C. remanei, Caenorhabditis remanei; G. gallus, Gallus gallus; H. sapiens, Homo sapiens. (E) Yeast two-hybrid assay revealing interactions between NBS-1 and its cognate partners. Interaction between prey proteins fused with the GAL4 activation domain (Left) and the baits fused with the LexA DNA binding domain (Right) assayed by growth on media lacking histidine (−LWH); growth in the presence of 3-AT, a competitive inhibitor of His3p, indicates strong interaction. Serial dilutions are spotted (1, 1:100, 1:1,000). The MRE-11(iow1) mutant protein contains an amino acid substitution (T72I) in the first phosphoesterase motif and is defective for DSBR but proficient for promoting DSB formation (14).
The causal mutation was mapped to an ∼6.8-cM region on chromosome II. Whole-genome sequencing of a 3×-backcrossed strain identified two mutations in the interval, one being a nonsense mutation in the C09H10.10 gene (SI Materials and Methods). Insertion/deletion mutant alleles were generated using CRISPR technology, creating early frame shifts and stop codons in C09H10.10 (Fig. S1). All four CRISPR-derived alleles recapitulated the diakinesis and progeny inviability phenotypes of me102, confirming that disruption of C09H10.10 is responsible for the observed phenotypes (Fig. 1C) and suggesting that all five mutant alleles (me102-6) of C09H10.10 are likely null alleles.
The predicted C09H10.10 protein contains a conserved Forkhead-associated (FHA) domain (Fig. 1D and Fig. S1A) at the N terminus, and PSI-BLAST (position-specific iterative basic local alignment search tool) searches initiated using C09H10.10 as the query sequence detected homology with the Danio rerio Nibrin protein, a predicted ortholog of mammalian NBS1. Furthermore, a small but highly conserved MRE11 interacting domain (MID) discovered in Schizosaccharomyces pombe Nbs1 (20) is clearly recognizable near the C terminus of C09H10.10 (Fig. 1D). However, Caenorhabditis NBS-1 ortholog lacks both the tandem BRCA1 C terminus (BRCT) domains found adjacent to the FHA domain in previously recognized NBS1/Xrs2 orthologs (15) and the conserved ATM interacting domain (FXF/Y motif preceded by an acid patch) from the C terminus of the protein (21). The presence of the conserved FHA domain and MID coupled with functional data presented below identify C09H10.10 as the C. elegans NBS1 ortholog, hereafter referred to as NBS-1.
Yeast two-hybrid assays revealed interactions between C. elegans NBS-1 and MRE-11 and between NBS-1 and COM-1 and confirmed the previously reported interaction between MRE-11 and RAD-50 (22), recapitulating the interaction network described in other species (Fig. 1E and Fig. S2) (23). Homozygous nbs-1 worms from heterozygous parents are fully viable and do not show any developmental phenotype in normal growth conditions, which allowed us to investigate the role of NBS-1 in DSBR during meiosis.
C. elegans NBS-1 Is Required for DSB Repair but Not for DSB Formation.
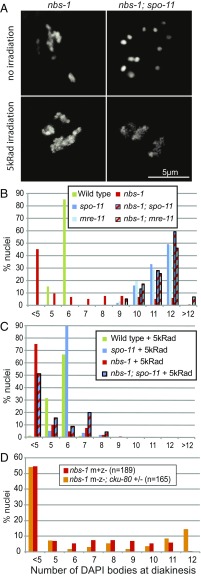
Multiple lines of evidence indicate that the presence of chromosome aggregates in nbs-1 mutants reflects a defect in repair of the SPO-11–dependent DSBs that serve as the initiating events of meiotic recombination. The spo-11 mutant lacks meiotic DSBs, resulting in lack of COs and chiasmata, which are reflected by the presence of 12 unattached chromosomes (univalents) at diakinesis (24). In contrast to the nbs-1 single mutant, the nbs-1; spo-11 double mutant displayed the canonical spo-11 phenotype, that is, it exhibited 12 DAPI bodies at diakinesis (Fig. 2 A and B) and produced a few percent viable progeny due to occasional euploid embryos arising from erratic segregation of intact chromosomes at meiosis I (Table S1). This indicates that the complete progeny lethality and the aggregated/fragmented chromosomes in diakinesis nuclei observed in nbs-1 mutants are a consequence of meiotic DSBs. Furthermore, while introduction of exogenous DSBs rescued the chiasma formation defect of the spo-11 single mutant, as shown by diakinesis nuclei displaying six DAPI bodies (24), frayed and aggregated chromosomes were observed after irradiation in nbs-1; spo-11 diakinesis nuclei, showing impaired repair of DSBs, whether SPO-11 dependent or exogenously induced, in the absence of NBS-1 (Fig. 2 A and C).
Fig. 2.
C. elegans NBS-1 is required for meiotic DSBR but dispensable for DSB formation. (A) DAPI-stained diakinesis oocytes from nbs-1 and nbs-1; spo-11 mutant worms unirradiated or exposed to 5,000-rad γ-irradiation. In contrast to the abnormal chromosome aggregates present in the nbs-1 oocytes (with or without irradiation), 12 intact chromosomes (univalents) are observed in the unirradiated nbs-1; spo-11 oocyte. Aggregation of chromosomes is, however, observed in the nbs-1; spo-11 oocyte upon introduction of exogenous DSBs by irradiation. (B) Quantification of the number of DAPI bodies in diakinesis nuclei. Fewer than 5 countable DAPI bodies reflect aggregation of chromosomes, whereas 12 DAPI bodies typically reflect intact univalents. Numbers of nuclei counted: WT, n = 92; spo-11, n = 37; nbs-1(me102), n = 42; nbs-1; spo11, n = 39; mre-11, n = 66; and nbs-1; mre-11, n = 59. (C) Quantification of DAPI bodies as in B after exposure of worms to 5,000-rad γ-irradiation, showing that irradiation-induced breaks rescue chiasma formation in the spo-11 mutant but induce chromosome aggregation in nbs-1; spo-11 mutant oocytes. Numbers of oocytes counted: WT, n = 105; spo-11, n = 57; nbs-1(me102), n = 144; and nbs-1; spo-11, n = 45. (D) Graph showing indistinguishable profiles of diakinesis DAPI body counts in nbs-1(me102) mutant worms derived from heterozygous nbs-1/+ mothers (m+z−) and nbs-1 m−z− mutant worms, which were derived from a cross using homozygous nbs-1; cku-80 double-mutant mothers (m−z−). See Fig. S3 for more details.
Our finding that meiotic DSBs are still formed in nbs-1 null mutant worms was unexpected, as previous studies had shown that the two other partners of the MRN complex, MRE-11 and RAD-50, are required for both DSB formation and DSBR during C. elegans meiosis (14, 16, 17). Furthermore, our own data showing that the nbs-1; mre-11 double mutant displays 12 intact univalents (and no aggregates) at diakinesis indicate that MRE-11 is still required for meiotic DSB formation in an nbs-1 mutant background (Fig. 2B). Since our analyses were conducted using nbs-1/nbs-1 worms derived from nbs-1/+ mothers (m+z− animals), we considered the possibility that DSB formation in the germ lines of nbs-1 m+z− animals could be the consequence of residual maternal NBS-1 protein. Although nbs-1/nbs-1 mutant progeny from nbs-1/nbs-1 mothers (m−z−) are normally completely inviable, we devised a crossing strategy that enabled us to generate some viable nbs-1/nbs-1 m−z− worms (Fig. S3) (see below); these m−z− nbs-1 worms displayed the same phenotype of aggregated chromosomes at diakinesis as their m+z− counterparts, indicating proficiency for DSB formation but deficiency in DSBR (Fig. 2D). These results show that C. elegans NBS-1, unlike MRE-11 and RAD-50, is dispensable for DSB formation and that MRE-11 and RAD-50 can act independently of NBS-1 to promote meiotic DSB formation.
These results are reminiscent of a previously described separation-of-function mutant mre-11(iow1) that is proficient for DSB formation but not DSBR (14). We thus tested whether the iow1 mutation might perturb the interaction between MRE-11 and NBS-1, impairing DSBR, while leaving the interaction between MRE-11 and RAD-50 intact to enable DSB formation. However, yeast two-hybrid assays showed that the mre-11(iow1) mutation weakened but did not eliminate the interaction between MRE-11 and NBS-1 and disrupted the interaction between MRE-11 and RAD-50 (Fig. 1E). This result suggests that the interaction between MRE-11 and RAD-50 might not be as crucial for DSB formation as it is for DSBR (Discussion).
NBS-1 Is Essential for DSB Resection and Loading of RAD-51 and RPA-1.
Normal repair of meiotic DSBs requires ends to be processed so that they can engage in HR-mediated repair both to form COs and to restore genome integrity. More specifically, SPO-11 protein–DNA adducts must be removed from 5′ ends through an endonucleolytic process. Furthermore, DSB ends must be further resected to yield 3′ ssDNA tails that can recruit DNA strand exchange proteins, such as RAD-51, to mediate invasion of a homologous DNA template.
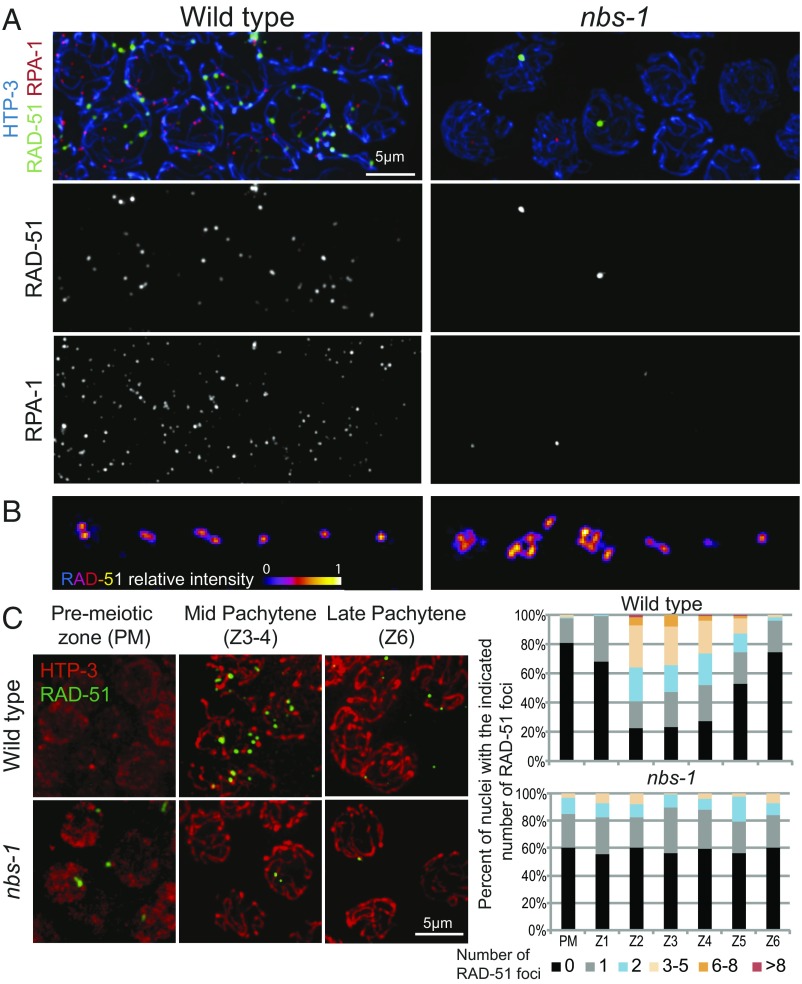
As NBS-1 is a member of the MRN complex involved in DSB resection in other species, we assessed the ability of nbs-1 mutants to process SPO-11–dependent DSBs by (i) simultaneous visualization of RAD-51 and a tagged version of RPA-1 [RPA-1::YFP (25)], a component of the eukaryotic ssDNA binding protein RPA, after nuclear spreading (Fig. 3A and Fig. S4) and (ii) quantification of RAD-51 foci in whole-mount gonads representing a time course of nuclei entering and progressing through meiosis (Fig. 3C). In WT C. elegans meiosis, RAD-51 foci appear during zygotene and early pachytene after DSB resection and become numerous by midpachytene before disappearing by late pachytene, indicative of efficient DSBR (26, 27). When observed using structured illumination microscopy (SIM), RAD-51 foci often appear as doublets, reflecting resection of both DSB ends (Fig. 3B) (28). In addition, RPA-1 foci, most of which represent post-strand-exchange recombination intermediates, rise in abundance and accumulate to higher levels than RAD-51 foci before decreasing and disappearing during late pachytene (28) (Fig. S4A).
Fig. 3.
Abundance of RAD-51 and RPA foci is greatly reduced in the absence of NBS-1. (A) Images of nuclei at the mid-pachytene stage from spread gonads from WT and nbs-1(me103) mutant worms immunostained for chromosome axis protein HTP-3, recombinase RAD-51, and a YFP-tagged version of RPA-1, a subunit of the ssDNA binding protein RPA. RAD-51 and RPA-1 foci are abundant in WT nuclei at this stage, but in the nbs-1 mutant, only a subset of nuclei has one or a few bright foci. (B) SIM images of RAD-51 foci in spread nuclei. For each genotype, examples of individually cropped RAD-51 signals are shown, illustrating the singlet or doublet organization characteristic of RAD-51 foci detected at meiotic DSB sites during WT meiosis (Left) and the more complex organization of RAD-51 foci detected at many sites in nbs-1 nuclei (Right), which are thought to reflect abnormalities arising during replication that persist into meiotic prophase. (C, Left) Representative images of germ cell nuclei in whole-mount preparations immunostained for HTP-3 and RAD-51 illustrating both the higher numbers of foci detected in mid-pachytene nuclei in the WT and the abnormal foci detected in a subset premeiotic nuclei in the nbs-1 mutant. (C, Right) Quantification of the numbers of RAD-51 foci in nuclei (from the whole-mount preparations) in seven consecutive zones along the distal–proximal axis of the gonad from the premeiotic zone (PM) through the end of pachytene (Z6) (Fig. S5A).
Consistent with previous reports indicating a role for MRE-11 and RAD-50 in the processing of meiotic DSBs (14, 16, 17), we found that the nbs-1 mutant is impaired for RAD-51 focus formation, exhibiting an overall reduction in the abundance of RAD-51 foci and an absence of a mid-pachytene peak in foci numbers (Fig. 3C). Furthermore, the abundance of RPA-1 foci was also severely reduced from an average of 22.7 ± 5.1 foci per nucleus (mean ± SD) at peak levels in the WT to <1 focus per nucleus in the nbs-1 mutant (Fig. 3A and Fig. S4A). Thus, nbs-1 mutant germ cells do not accumulate post-strand-exchange recombination intermediates (as occurs during WT meiosis), and they do not accumulate RPA-coated ssDNA ends [as occurs in brc-2 mutants, which are competent for DSB resection but defective in RAD-51 loading (29)]. Together, these data indicate that NBS-1 is essential for meiotic DSB resection.
Whereas numbers of RAD-51 and RPA-1 foci were reduced overall, nbs-1 mutants displayed an increased number of foci in the premeiotic zone (Fig. 3C), consistent with a role for MRN in repairing and/or preventing accumulation of DNA damage during DNA replication (17, 30). Supporting this interpretation, we found that an nbs-1; spo-11 double mutant exhibited higher levels of residual RAD-51 foci (0.44 ± 0.75 foci per nucleus in zones 1–6, n = 727) than the spo-11 single mutant (0.21 ± 0.58, n = 1,094; Mann–Whitney P < 10−4) (Fig. S5), suggesting that many of the residual RAD-51 foci detected in nbs-1 meiotic nuclei reflect DNA damage that was not of meiotic origin. Furthermore, SIM imaging revealed that RAD-51 foci in the nbs-1 mutant frequently exhibit abnormal structure (Fig. 3B and Fig. S4B). Whereas nearly all RAD-51 foci in WT germ cells are detected as doublet or singlet foci (28) (termed “simple foci”; 99.5%, n = 200), the infrequent RAD-51 foci in the nbs-1 mutant are often larger and more complex in structure (28% complex foci, n = 46). Furthermore, such complex foci are detected both in the premeiotic zone and throughout meiotic prophase, consistent with abnormalities arising during mitotic cell cycles or meiotic DNA replication and persisting after meiotic prophase entry. However, we also found that the residual level of RAD-51 foci in the nbs-1 single mutant (0.6 ± 0.9, n = 618 nuclei) was higher than in the nbs-1; spo-11 double mutant (Mann–Whitney P < 0.0001) (Fig. 3C and Fig. S5); this suggests that, although meiotic DSB resection is strongly impaired, some SPO-11–generated breaks may nevertheless load RAD-51 in the absence of NBS-1.
NBS-1 Functions both to Counteract NHEJ and to Promote Efficient HR.
DNA repair pathway choice is crucial for cellular and organismal survival: NHEJ and HR have been shown to occur cooperatively, competitively, or as backup mechanisms for DSBR in various contexts (12). As previous reports had implicated MRE-11 and COM-1 in antagonizing NHEJ (13, 14), we tested the hypothesis that the meiotic defects observed in the nbs-1 mutant might reflect inappropriate use of NHEJ for the repair of meiotic DSBs.
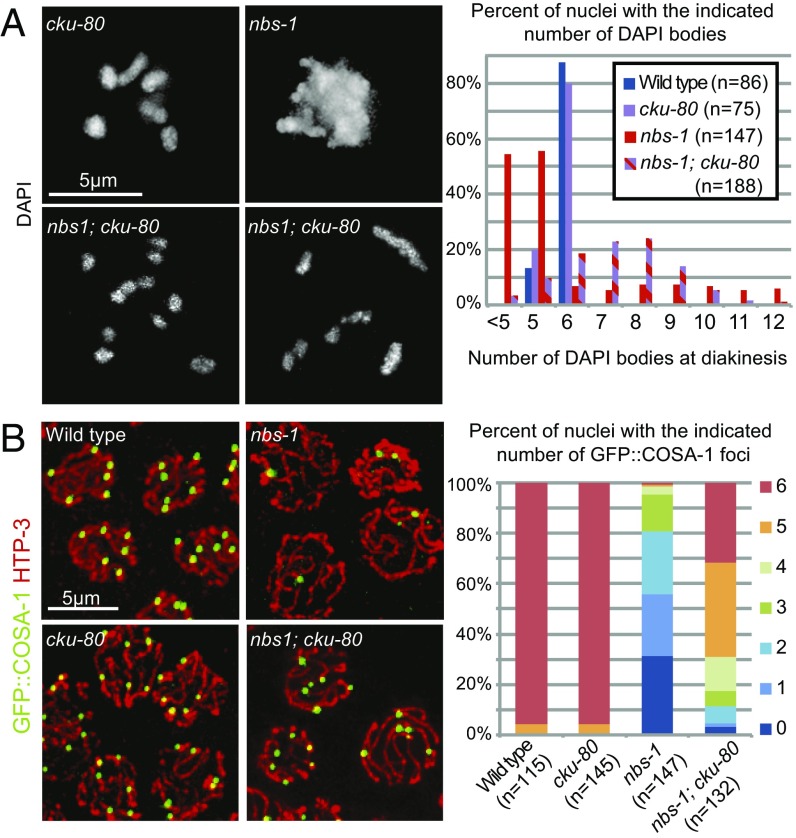
We found that mutation of cku-80, which encodes the worm ortholog of KU80 essential for NHEJ, partially alleviated multiple nbs-1 defects (Fig. 4). In contrast to the aggregated chromosomes present in the nbs-1 single mutant, diakinesis chromosomes more frequently appeared as individual univalents or bivalents in the nbs-1; cku-80 double mutant (Fig. 4A). This partial restoration of chromosome integrity was accompanied by a partial restoration of GFP::COSA-1 foci in late pachytene (Fig. 4B). While the nbs-1 mutant displayed an average of 1.4 ± 1.3 (n = 150) foci per nucleus, the nbs-1; cku-80 double mutant averaged 4.7 ± 1.5 GFP::COSA-1 foci per nucleus (n = 133, Mann–Whitney P < 0.0001). We also observed a partial rescue of progeny viability, with an average of 3.1% progeny survivorship from nbs-1; cku-80 animals (93 of 3,007 eggs laid) compared with 0% from nbs-1 animals (0 of 1,575) (Table S1). The substantial rescue of progeny viability, chromosome integrity, and GFP::COSA-1 focus formation together indicate a role for NBS-1 in preventing inappropriate utilization of NHEJ during meiosis.
Fig. 4.
NBS-1 antagonizes NHEJ and promotes efficient HR. (A, Left) Images of individual DAPI-stained diakinesis oocyte nuclei showing that the chromosome aggregation phenotype of the nbs-1 single mutant is suppressed in nbs-1; cku-80 double mutant, which instead displays a mixture of bivalents and univalents. (A, Right) Quantification of DAPI bodies in diakinesis nuclei. (B, Left) Immunolocalization of HTP-3 and GFP::COSA-1 in late pachytene nuclei in whole-mount preparations. (B, Right) Stacked bar graph showing the percentages of nuclei with the indicated numbers of COSA-1 foci, showing that COSA-1 focus formation is partially restored in the nbs-1; cku-80 double mutant compared with the nbs-1 single mutant.
Although inactivation of cku-80 attenuated the meiotic defects of the nbs-1 mutant, the rescue was not complete. This result could reflect either (i) an additional role for NBS-1 in promoting efficient HR beyond antagonizing NHEJ or (ii) a deficit of DSBs compared with the WT, which could yield a deficit in CO number. We ruled out the latter hypothesis by exposing nbs-1; cku-80 worms to 5,000-rad γ-irradiation to introduce an excess of DSBs. While this dose is more than sufficient to restore chiasmata in the spo-11 mutant background (Fig. 2C) (19, 24), it did not improve chiasma formation in the nbs-1; cku-80 mutant (Fig. S6). This result indicates that DSBs are not limiting for CO formation in the nbs-1; cku-80 mutant and instead implies that recombination intermediates cannot be efficiently processed into COs in absence of NBS-1, even when CKU-80 is absent.
NBS-1 Is Required for a Timely Resection of DSBs to Engage HR.
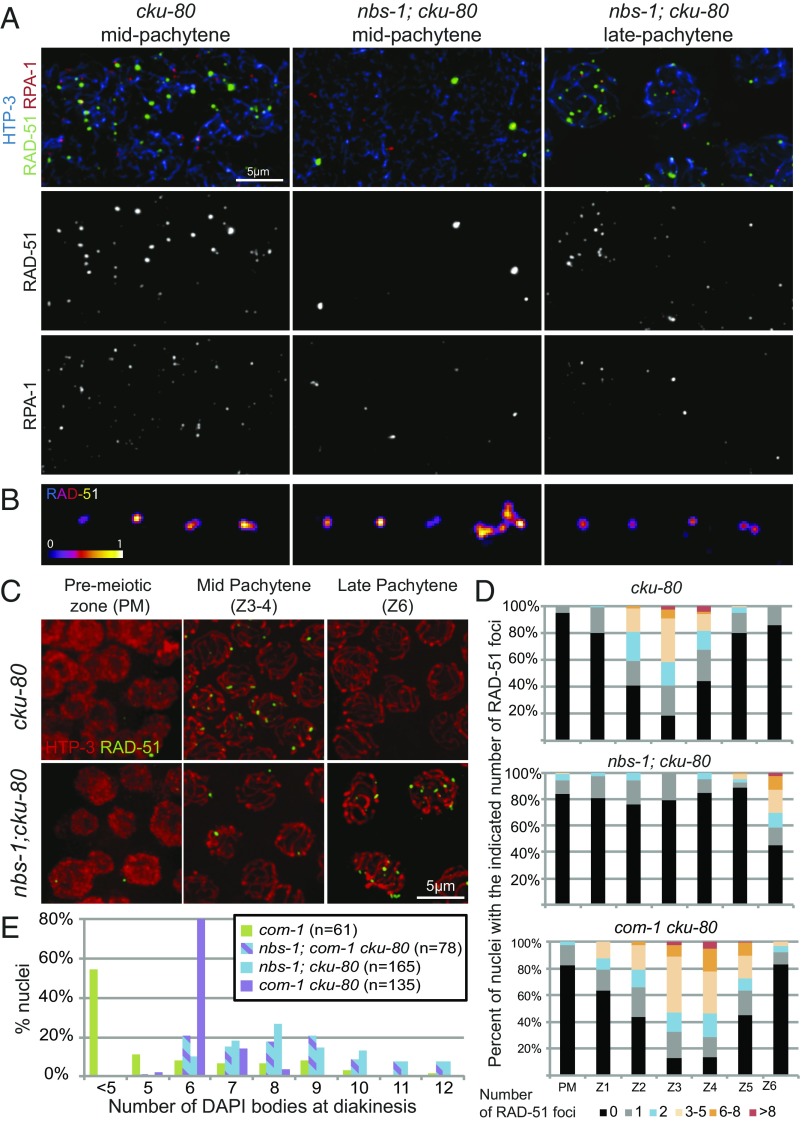
Examination of the timing of appearance of RAD-51 and RPA-1 foci in nbs-1; cku-80 double mutants indicated a role for NBS-1 in promoting timely resection of DSBs, even in absence of NHEJ (Fig. 5). The nbs-1; cku-80 double mutant differed from both the cku-80 single mutant, which exhibits WT dynamics of RAD-51 and RPA-1 foci with an enrichment in mid-pachytene (Fig. 5 A, C, and D and Fig. S4), and from the nbs-1 single mutant, which displays low levels of both types of foci throughout meiosis I. Instead, numbers of RAD-51 and RPA-1 foci in nbs-1; cku-80 remained low throughout most of meiotic prophase and then rose in abundance during late pachytene (zone 6) (Fig. 5 A, C, and D and Fig. S4A), similar to what was reported for RAD-51 foci in the mre-11(iow1); cku-80 double mutant (14). Furthermore, SIM imaging revealed that nearly all of these late pachytene RAD-51 foci were simple (doublet or singlet) rather than complex in structure (98.3% simple foci, n = 231) (Fig. 5B and Fig. S4B). Together, these results suggest that some resection of meiotic DSBs can occur independently of NBS-1 when CKU-80 is absent, but this NBS-1–independent mode of resection seems largely restricted to late pachytene and early diplotene.
Fig. 5.
NBS-1 is required for timely loading of RAD-51 and RPA-1 during pachytene. (A) Images of nuclei from spread gonads from the cku-80 mutant in mid-pachytene and from the nbs-1(me103); cku-80 mutant in mid- and late pachytene immunostained for HTP-3, RAD-51, and RPA-1. Levels of RAD-51 and RPA-1 foci in the cku-80 gonad are comparable with the WT (Fig. S4). In the nbs-1; cku-80 double mutant, few RAD-51 and RPA-1 foci are detected in mid-pachytene, but foci increase in abundance by late pachytene. (B) SIM images of RAD-51 foci from spread nuclei as in Fig. 3B. (C) Immunolocalization of HTP-3 and RAD-51 on whole-mount gonads. (D) Quantification of the numbers of RAD-51 foci in whole-mount gonads, illustrating the contrast between the nbs-1; cku-80 double mutant, in which increased abundance of RAD-51 foci is restricted to late pachytene (zone 6), and the com-1 cku-80 double mutant, which exhibits normal RAD-51 foci dynamics. (E) Quantification of DAPI bodies in diakinesis nuclei, showing that successful bivalent formation occurs much more frequently in the com-1 cku-80 double mutant than in either nbs-1; cku-80 or nbs-1; com-1 cku-80.
This late timing of appearance of RAD-51 foci may help to explain why restoration of CO formation is incomplete in the nbs-1; cku-80 double mutant. Initial loading of pro-CO factors must occur before the transition to late pachytene for DSBR intermediates to become competent to mature into COs (19). Thus, when resection is delayed, it may sometimes occur too late to engage the homolog as a repair partner and/or to enable recruitment of factors needed to generate COs.
We note that number of RAD-51 foci detected in premeiotic germ cells was significantly lower in the nbs-1; cku-80 double mutant than in the nbs-1 single mutant (Mann–Whitney P < 0.0001), suggesting that, in addition to antagonizing RAD-51 loading at meiotic DSBs, Ku may also contribute to replication-associated problems caused by lack of NBS-1.
NBS-1 and COM-1 Play Distinct Roles in Promoting HR.
Both COM-1 and NBS-1 are required for meiotic DSBR but dispensable for DSB formation, and mutation of either leads to aggregated chromosomes at the end of meiotic prophase (13, 18) (Figs. 2 and 5E). However, our data indicate that their respective roles in resection and promotion of HR are quite different. Whereas elimination of cku-80 resulted in a modest partial rescue of bivalent formation in the nbs-1 background, with 10% of diakinesis nuclei showing six bivalents (Fig. 4A), loss of cku-80 in the com-1 background resulted in much more substantial restoration of bivalent formation, with 80% of diakinesis nuclei showing six bivalents (Fig. 5E), recapitulating previous observations (13). Moreover, analysis of diakinesis nuclei in the nbs-1; com-1 cku-80 triple mutant indicated that NBS-1 is required for the efficient bivalent formation observed in the com-1 cku-80 mutant (Fig. 5E). Together, these results suggest that, while COM-1 is required to antagonize CKU-80 and prevent NHEJ-mediated repair of DSBs, it is not essential for MRN-dependent resection to yield interhomolog COs.
This conclusion is further supported by comparison of RAD-51 dynamics in the com-1 cku-80 and nbs-1; cku-80 double mutants. In contrast to nbs-1; cku-80, where RAD-51 foci did not increase in abundance until late pachytene, the com-1 cku-80 double mutant exhibited RAD-51 foci dynamics similar to the WT, with a strong peak in foci numbers in mid-pachytene and a decline in foci numbers by late pachytene (Fig. 5D) as previously described (13). This indicates that COM-1 function is essential for resection in the presence of CKU-80 but becomes dispensable in the absence of CKU-80. This result implies that COM-1 is primarily required during meiosis to antagonize CKU-80 and NHEJ but is not essential for timely MRN-dependent resection when NHEJ is abrogated (Discussion). In contrast, NBS-1 is required both for antagonizing CKU-80 and for promoting resection.
EXO-1 Is Required to Promote CO Formation and Chromosome Integrity in the nbs-1; cku-80 Double Mutant.
The presence of COSA-1 foci as well as late RAD-51 and RPA-1 foci in the nbs-1; cku-80 double mutant made us wonder what factors might be mediating resection in this context. One candidate is the exonuclease Exo1, which has been shown to be involved alongside the MRN complex in promoting extended resection in various contexts (9). C. elegans EXO-1 is dispensable for meiotic recombination in an otherwise WT background (13), but EXO-1 is required for partial restoration of RAD-51 loading, CO formation, and chromosome integrity in the mre-11(iow1); cku-80 double mutant (14), indicating that delayed DSB resection and repair via HR are dependent on EXO-1 in this context. We, therefore, investigated the potential involvement of EXO-1 in promoting resection and/or CO formation during late pachytene in the absence of NBS-1 and CKU-80 (Fig. 6).
Fig. 6.
EXO-1 is required for DSBR in the absence of CKU-80 and NBS-1. (A) Images of individual DAPI-stained diakinesis nuclei showing the presence of chromosome aggregates in the nbs-1; cku-80 exo-1 triple mutant. Asterisks in cku-80 exo-1 indicate two chromosomes on top of each other. (B) Quantification of DAPI bodies in diakinesis nuclei. (C) Immunolocalization of HTP-3 and COSA-1 in late pachytene nuclei from whole-mount gonads. (D) Stacked bar graph representing the percentage of nuclei with the indicated numbers of COSA-1 foci in the different genotypes, showing that COSA-1 focus formation is eliminated in the nbs-1(me102); cku-80 exo-1 triple mutant. (E) Quantification of RAD-51 foci from whole-mount preparations for the indicated genotypes as in Fig. 3C.
While the nbs-1; cku-80 double mutant displayed mostly univalents and bivalents in diakinesis oocytes, we frequently observed chromosome aggregates at diakinesis in the nbs-1; cku-80 exo-1 triple mutant (Fig. 6 A and B), suggesting partial redundancy of NBS-1 and EXO-1 function in maintaining genome integrity. Moreover, the partial rescue of GFP::COSA-1 focus formation observed in nbs-1; cku-80 was also dependent on EXO-1, as the nbs-1; cku-80 exo-1 triple mutant failed to form GFP::COSA-1 foci (Fig. 6 C and D). These results indicate a strict requirement for EXO-1 to form COs in the absence of both NBS-1 and CKU-80. Despite loss of CO site markers, however, many late pachytene nuclei (zone 6, Z6) with numerous RAD-51 foci were detected in the nbs-1; cku-80 exo-1 triple mutant (Fig. 6E and Fig. S7), reminiscent of phenotypes observed in the com-1 cku-80 exo-1 triple mutant (13). As our cytological analysis of RAD-51 foci does not provide information regarding the length of resection tracts, one possible explanation for this finding is that the residual resection occurring in the absence of EXO-1 in these contexts may be of insufficient length and/or occur too late to engage the homolog or recruit CO factors. Alternatively, these results may reflect an additional late function of C. elegans EXO-1 in promoting CO formation, separable from its nuclease activity, as has been observed in mouse and yeast (31, 32).
Discussion
Identification of C. elegans NBS-1 as a Compact Ortholog of NBS1/Xrs2.
The MRN complex has long been recognized as a central player in mediating HR-based repair of DSBs across species, but there are substantial differences in the degree of conservation among its subunits (33). MRE11 and RAD50 are ancient in origin, are highly conserved among eukaryotes, and have clearly identifiable orthologs in both eubacteria (SbcC and SbcD) and archae. In contrast, NBS1 orthologs are detected only in eukarya and are notoriously poorly conserved. Primary sequence conservation among orthologs from different kingdoms is mainly restricted to the N-terminal FHA domain, and conservation outside this domain is marginal even within kingdoms (e.g., Saccharomyces cerevisiae Xrs2 and S. pombe Nbs1 share only 10% identity in the 250 amino acids after the FHA domain), and the presence of tandem BRCT domains within this region had remained unrecognized in many orthologs until introduction of an algorithm specifically designed to detect such motifs (15). Indeed, when human NBS1 was first discovered, its protein size and association with MRE11 and RAD50 were crucial for recognizing NBS1 and Xrs2 as functional homologs (34).
Although C. elegans MRE-11 and RAD-50 and their roles in meiotic recombination and DNA repair have been known for some time (14, 16, 17), the nematode counterpart of NBS1/Xrs2 had remained elusive. Our identification of C09H10.10 as C. elegans NBS-1 makes the reason that it had escaped detection apparent: while it contains both the N-terminal FHA domain and the conserved MRE11 interaction domain (MID) near its C terminus, Ce NBS-1 is only about half the size of most other NBS1 orthologs and lacks the tandem BRCT domains and the ATM interaction domain. This stripped down version of NBS1 present in C. elegans is nonetheless sufficient to support the functions of MRE-11 and RAD-50 in promoting efficient and timely meiotic DSBR and in repairing/preventing accumulation of replication-associated DNA damage. The fact that such a compact version of NBS-1 can support the essential functions of MRE-11 and RAD-50 in DSBR parallels the recent finding that a 108-aa fragment of mammalian Nbs1 (which contains the MID but lacks both the tandem BRCT motifs and the N-terminal FHA domain) can substantially support essential functions of Mre11 and Rad50 in mouse cells in vitro and in vivo (35).
NBS-1–Independent Functions of MRE-11 and RAD-50 During C. elegans Meiosis.
In all species where it has been studied, the MRN complex has been shown to be crucial for repair of meiotic DSBs (36). However, involvement of MRN in the formation of such breaks varies from species to species. Whereas Mre11, Rad50, and Nbs1 are not required for meiotic DSB formation in S. pombe or Arabidopsis thaliana (37–41), all three core members of MRX are required for DSB formation during S. cerevisiae meiosis (42, 43). Interestingly, our analysis here revealed that these two meiotic functions of MRN complex components can be uncoupled. While C. elegans NBS-1 is integral to the functions of the MRN complex in promoting timely resection and repair of meiotic DSBs, we found that the previously reported roles of MRE-11 and RAD-50 in promoting DSB formation (16, 17) are independent of NBS-1 (Fig. S8).
How MRN complex components function to promote DSB formation remains unknown. However, separation-of-function mutations that uncouple DSB formation and repair activities may be informative. Missense mutations in C. elegans [mre-11(iow1)] and S. cerevisiae (mre11-D16A) that impair DSB resection but not DSB formation affect adjacent highly conserved residues in the first phosphoesterase domain, and although this domain is distant from the MRE11/RAD50 interface, both mutations destabilize the interaction between MRE11 and RAD50 (refs. 14, 23, and 44 and this study). This suggests that stable association between MRE11 and RAD50 may be less important for their DSB-promoting activity than for resection and repair, raising the possibility that MRE11 and RAD50 may function in a different conformation (35, 45) or stoichiometry (46) or even as separate proteins to influence DSB formation. Furthermore, in S. cerevisiae, Xrs2 may be required for DSB formation partially based on its role in promoting nuclear localization of Mre11 (47); conversely, the fact that C. elegans NBS-1 is dispensable for DSB formation indicates that (at least some) MRE-11 and RAD-50 must get into the nucleus without NBS-1.
Additional evidence suggests that C. elegans MRE-11 may also be able to function independently of NBS-1 in another context. Specifically, we found that late RAD-51 foci reflecting delayed end resection were present in late pachytene nuclei in nbs-1; cku-80 exo-1 mutant germ lines, whereas such foci were absent in mre-11(iow1); cku-80 exo-1 (14). This result suggests that MRE-11 may be capable of promoting some degree of end resection in late pachytene nuclei in the absence of NBS-1 and EXO-1.
Distinct Roles for MRN and COM-1 in Promoting DSB Resection and Antagonizing NHEJ.
DSBs pose a threat to genome integrity, and DNA repair machineries have evolved to prevent or limit their damaging consequences. Moreover, evidence for competition between different DSBR pathways is present in all studied species. For example, elimination of Ku in mammalian cells increases the frequency of DSB-induced HR between direct repeats (48), and conversely, mutation of the Mre11 nuclease domain results in higher incidence of NHEJ in yeast cells (49–51). The extent to which DSBs are repaired using mutagenic repair mechanisms, such as NHEJ, vs. high-fidelity mechanisms, such as HR, depends on cellular context. During meiosis, it is crucial that DSBs be repaired strictly by HR both (i) to promote the formation of interhomolog COs needed to segregate chromosomes and (ii) to restore genome integrity while minimizing introduction of new mutations. However, even during meiosis, where the outcome of DSBR is so heavily biased toward HR, abrogation of HR in C. elegans germ cells has revealed that NHEJ factors are nevertheless still present and can promote illegitimate repair (refs. 13 and 14 and this study). The MRN complex and COM-1 are crucial during meiosis to tip the balance irrevocably toward the HR outcome.
This work integrated with previous findings (13, 14) shows that MRN and COM-1 make distinct contributions to promoting HR and antagonizing NHEJ during C. elegans meiosis (Fig. S8). NBS-1, MRE-11, and COM-1 are all required to prevent meiotic catastrophe resulting from inappropriate engagement of the NHEJ pathway. As com-1 meiotic defects were not suppressed by loss of LIG-4, which functions in NHEJ downstream of Ku recruitment (13), the meiotic catastrophe observed in the nbs-1, mre-11(iow1), or com-1 mutants likely reflects blocking of DNA ends by Ku in the absence of a functional MRN-C complex, thereby preventing engagement of HR. However, in the absence of Ku, differences in the roles of MRN and COM-1 are revealed. When Ku is removed in an nbs-1 or mre-11(iow1) mutant background, RAD-51 loading (indicative of end resection) is delayed, and CO formation is inefficient. However, when Ku is removed in a com-1 mutant background, RAD-51 foci levels and timing seem normal, and CO formation is much more efficient. These findings indicate that COM-1 is required primarily to antagonize Ku yet is substantially dispensable for SPO-11 removal and MRN-mediated end resection when Ku is absent (Fig. S8). Whereas MRN can promote efficient and timely end resection without COM-1 (in combination with EXO-1; see below), however, MRN cannot function without COM-1 to antagonize Ku. We interpret these findings in light of reports that the S. pombe Nbs1 FHA domain directly engages COM-1 ortholog Ctp1 and that Ctp1/CtIP is recruited to DSB sites through NBS1 in both S. pombe and human cells (52–54). Specifically, we propose that, during C. elegans meiosis, NBS-1 couples resection initiation and inhibition of NHEJ both by participating in MRN-mediated end resection and by recruiting COM-1 to DSB sites. Furthermore, based on structural analysis of S. pombe Ctp1 suggesting an ability to form bridges between MRN-C complexes on opposite sides of a DSB (55, 56), we propose that MRN-C may play a dual role in antagonizing NHEJ both by promoting endonucleolytic cleavage to initiate resection and by mediating bridging between DNA ends, thereby preventing the loading of the preformed Ku ring.
Redundancy in HR Machinery Contributes to Robustness of Repair.
Genome integrity of germ cells is paramount to perpetuation of species. As faithful chromosome inheritance during sexual reproduction depends on meiotically induced DSBs, it is crucial that DSBR in germ cells be highly robust. Synthesis of this work with prior analyses of MRN-C function in the C. elegans germ line suggests that partial redundancy among factors and activities promoting DSB resection may contribute to robustness of the system.
From the onset of meiotic prophase through the end of the early pachytene stage, DSB end resection is highly dependent on MRN (refs. 14 and 17 and this study). As DSBs must be processed and engage the homolog during early prophase to be competent for CO formation (19), the timely participation of MRN in DSB resection is thus crucial for efficient CO formation. In contrast, C. elegans EXO-1 is not essential for CO formation in otherwise WT germ cells. However, EXO-1 can mediate resection during late prophase in the absence of MRN activity, and either MRN or EXO-1 can mediate resection during late prophase in the absence of Ku. Furthermore, in a com-1 cku-80 double mutant, DSBs can undergo timely resection during early prophase, but now, both MRN and EXO-1 are required for this to occur. This indicates that, when the system is compromised by loss of COM-1, EXO-1 is available and can participate in resection during early prophase either through its own exonuclease function or by enhancing the activity of MRN. We suggest that, although EXO-1 is largely dispensable for successful meiosis in C. elegans, it likely does collaborate with MRN-C during normal meiosis to promote optimal resection, thereby helping to ensure a reliable outcome.
Materials and Methods
Strains and Genetics.
All C. elegans strains were cultivated at 20 °C under standard conditions. Strains used in this study are listed here.
AV630 meIs8 [gfp::cosa-1] II
AV727 meIs8 [gfp::cosa-1] II, ruIs32 [unc-119(+); pie-1::mcherry::histoneH2B] III; itIs38 [pAA1; pie-1::GFP::PH::unc-119(+)]
AV828 nbs-1(me102) meIs8/mIn1 [mIs14 dpy-10(e128)] II; backcrossed three times from the original balanced strain
AV845 spo-11(me44)/nT1 [unc(n754dm) let] IV
AV846 nbs-1(me102) meIs8/mIn1 [mIs14 dpy-10(e128)] II; spo-11(me44)/nT1 IV
AV860 nbs-1(me103)/mIn1 [mIs14 dpy-10(e128)] II
AV861 nbs-1(me104)/mIn1 [mIs14 dpy-10(e128)] II
AV862 nbs-1(me105)/mIn1 [mIs14 dpy-10(e128)] II
AV863 nbs-1(me106)/mIn1 [mIs14 dpy-10(e128)] II
AV865 nbs-1(me102) meIs8/mIn1 [mIs14 dpy-10(e128)] II; mre-11(ok179)/nT1 V
AV874 meIs8 II; cku-80(ok861) III
AV875 meIs8 II; exo-1(tm1842) III
AV876 meIs8 II; cku-80(ok861) exo-1(tm1842) III
AV877 nbs-1(me102) meIs8/mIn1[mIs14 dpy-10(e128)] II; cku-80(ok861) III
AV878 nbs-1(me102) meIs8/mIn1 [mIs14 dpy-10(e128)] II; exo-1(tm1842) III
AV879 nbs-1(me102) meIs8/mIn1 [mIs14 dpy-10(e128)] II; cku-80(ok861) exo-1(tm1842) III
AV904 nbs-1(me103)/mIn1 [mIs14 dpy-10(e128)] II; opIs263[rpa-1::yfp, unc-119+]
AV905 cku-80(ok861) III; opIs263[rpa-1::yfp, unc-119+]
AV947 nbs-1(me103)/mIn1 [mIs14 dpy-10(e128)] II; cku-80(ok861) III; opIs263[rpa-1::yfp, unc-119+]
WS4581 opIs263[rpa-1::yfp, unc-119+]
XF0644 com-1(t1626) unc-32(e189)/hT2 [bli-4(e937) let-?(q782) qIs48] III
XF0697 com-1(t1626) unc-32(e189 cku-80(tm1524)/hT2 cku-80(tm1524) III
Isolation and identification of the nbs-1(me102) mutation, generation of CRISPR alleles, and yeast two-hybrid experiments are described in SI Materials and Methods.
Cytological Analysis.
Numbers of DNA bodies present in diakinesis oocytes were assessed in intact adult hermaphrodites at 24 h post-L4 larval stage, fixed in ethanol, and stained with DAPI as in ref. 57. Immunostaining for GFP::COSA-1 and RAD-51 in whole-mount gonads was conducted as in ref. 58. All experiments were performed on gonads dissected at 24–26 h post-L4 at 20 °C. The following primary antibodies were used at the indicated dilutions in PBS with 0.1% Tween: chicken anti–HTP-3 [1:500 (59)], rabbit anti-GFP [1:200 (19)], and rat anti–RAD-51 [1:500 (60)]. Dual RPA-1::YFP/RAD-51 immunostaining was performed on spread gonads as in ref. 61, with YFP being detected by the rabbit anti-GFP antibody. All images were acquired using a 100× N.A. 1.40 objective on a DeltaVison OMX Blaze microscopy system, deconvolved, and corrected for registration using SoftWoRx. Gonads were subsequently assembled using the “Grid/Collection” plugin (62) in ImageJ. Wide-field images were obtained as 200-nm spaced Z stacks, while 3D SIM images were obtained as 125-nm spaced Z stacks. For display, contrast and brightness were adjusted in individual color channels using ImageJ.
For quantification of RAD-51 foci in whole-mount gonads, at least three gonads were counted per genotype. Gonads were divided into seven zones: the premeiotic zone, where HTP-3 appears diffuse, and six equal-sized zones based on physical distance from meiotic entry (where HTP-3 signal forms tracks along chromosomes) to late pachytene (end of cell rows). For the GFP::COSA-1 experiments, nuclei within the last six cell rows were counted; numbers of nuclei counted were as follow: WT (n = 115), cku-80 (n = 145), exo-1 (n = 205), cku-80 exo-1 (n = 69), nbs-1(me102) (n = 147), nbs-1; cku-80 (n = 132), nbs-1; exo-1 (n = 150), and nbs-1; cku-80 exo-1 (n = 127).
Quantitation of YFP::RPA-1 foci in spread gonads.
For WT and cku-80, RPA-1 foci were counted in 20 non-overlapping nuclei per gonad (at least three gonads per genotype) within the five cell rows that follow the early to mid-pachytene peak of RAD-51 foci present in these genotypes (28). For nbs-1 and nbs-1; cku-80 mutants, for each gonad, foci were counted in three cohorts of 20 nuclei each distributed across the first three-quarters of the pachytene region (defined based on HTP-3 staining). For the nbs-1; cku-80 mutant gonads, foci were counted in an additional cohort of 20 nuclei coinciding with the late pachytene RAD-51 peak.
Scoring of RAD-51 foci in SIM images.
RAD-51 foci in projected SIM images were classified into two categories, “simple” and “complex,” as illustrated in Fig. 3B. The simple category included singlet or doublet foci with either a single maximum or two adjacent ovoid maxima; five instances where three adjacent maxima were observed were also included in this category, as they most likely represented a doublet next to a singlet. Foci were scored as complex when they contained four or more adjacent maxima and/or exhibited irregular (rather than ovoid) shapes; all foci scored as complex were bigger/brighter than any singlets or doublets seen in the WT.
γ-Irradiation.
Worms were exposed to 5,000 rad (50 Gy) of γ-irradiation using a Cs-137 source at 20 h post-L4 stage. RAD-51 immunostaining was performed on gonads dissected and fixed 1 h after irradiation. Diakinesis DAPI body counts were done using worms fixed at 18–20 h post-irradiation.
Data Availability.
All strains and primary images used in this research are available on request from A.M.V. (annev@stanford.edu).
Supplementary Material
Acknowledgments
We thank N. Jain for help with sequence analysis; S. Tzouanas and S. Zimmerman for help with CRISPR methods; A. Woglar for instruction regarding chromosome spreading and SIM imaging; and C. Akerib, S. Ramakrishnan, and A. Woglar for comments on the manuscript. We thank S. Smolikove, V. Jantsch, A. Dernburg, M. Tijsterman, C. Kraft, and the Caenorhabditis Genetics Center (funded by NIH Office of Research Infrastructure Programs Grant P40 OD010440) for reagents and strains and R. Guerois for helpful discussions. We also thank the Stanford Cell Sciences Imaging Facility, which is partially funded by National Center for Research Resources Award 1S10OD01227601. This work was supported by a CJS (Contrat Jeune Scientifique) fellowship from INRA (Institut National de la Recherche Agronomique) (C.G.), an award from the Bettencourt-Schueller Foundation (to C.G.), NIH Grant R01GM67268 (to A.M.V.), and American Cancer Society Research Professor Award RP-15-209-01-DDC (to A.M.V.). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the funders, who had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719029115/-/DCSupplemental.
References
- 1.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: Setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev. 2013;23:147–155. doi: 10.1016/j.gde.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 4.Vrielynck N, et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science. 2016;351:939–943. doi: 10.1126/science.aad5196. [DOI] [PubMed] [Google Scholar]
- 5.Robert T, et al. The TopoVIB-like protein family is required for meiotic DNA double-strand break formation. Science. 2016;351:943–949. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- 6.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 8.Brown MS, Bishop DK. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol. 2014;7:a016659. doi: 10.1101/cshperspect.a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Crit Rev Biochem Mol Biol. 2016;51:195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimitou EP, Yamada S, Keeney S. A global view of meiotic double-strand break end resection. Science. 2017;355:40–45. doi: 10.1126/science.aak9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 12.Chapman JR, Taylor MRG, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 13.Lemmens BBLG, Johnson NM, Tijsterman M. COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet. 2013;9:e1003276. doi: 10.1371/journal.pgen.1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin Y, Smolikove S. Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol Cell Biol. 2013;33:2732–2747. doi: 10.1128/MCB.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker E, Meyer V, Madaoui H, Guerois R. Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics. 2006;22:1289–1292. doi: 10.1093/bioinformatics/btl075. [DOI] [PubMed] [Google Scholar]
- 16.Chin GM, Villeneuve AM. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 2001;15:522–534. doi: 10.1101/gad.864101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penkner A, et al. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 2007;26:5071–5082. doi: 10.1038/sj.emboj.7601916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoo R, et al. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiller CB, et al. Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat Struct Mol Biol. 2012;19:693–700. doi: 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulton SJ, et al. Combined functional genomic maps of the C. elegans DNA damage response. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 23.Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog Biophys Mol Biol. 2015;117:182–193. doi: 10.1016/j.pbiomolbio.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dernburg AF, et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 25.Stergiou L, Eberhard R, Doukoumetzidis K, Hengartner MO. NER and HR pathways act sequentially to promote UV-C-induced germ cell apoptosis in Caenorhabditis elegans. Cell Death Differ. 2011;18:897–906. doi: 10.1038/cdd.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- 27.Colaiácovo MP, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 28.Woglar A, Villeneuve A. Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell. 2018 doi: 10.1016/j.cell.2018.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin JS, Winkelmann N, Petalcorin MIR, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruhn C, Zhou Z-W, Ai H, Wang Z-Q. The essential function of the MRN complex in the resolution of endogenous replication intermediates. Cell Rep. 2014;6:182–195. doi: 10.1016/j.celrep.2013.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Zakharyevich K, et al. Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kan R, et al. Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol Reprod. 2008;78:462–471. doi: 10.1095/biolreprod.107.065771. [DOI] [PubMed] [Google Scholar]
- 33.Stracker TH, Petrini JHJ. The MRE11 complex: Starting from the ends. Nat Rev Mol Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carney JP, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 35.Kim JH, et al. The Mre11-Nbs1 interface is essential for viability and tumor suppression. Cell Rep. 2017;18:496–507. doi: 10.1016/j.celrep.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 37.Uanschou C, et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akamatsu Y, et al. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol. 2008;28:3639–3651. doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell. 2004;16:1968–1978. doi: 10.1105/tpc.104.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bleuyard JY, Gallego ME, White CI. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma. 2004;113:197–203. doi: 10.1007/s00412-004-0309-1. [DOI] [PubMed] [Google Scholar]
- 41.Waterworth WM, et al. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007;52:41–52. doi: 10.1111/j.1365-313X.2007.03220.x. [DOI] [PubMed] [Google Scholar]
- 42.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shima H, Suzuki M, Shinohara M. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics. 2005;170:71–85. doi: 10.1534/genetics.104.037580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furuse M, et al. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paull TT, Deshpande RA. The Mre11/Rad50/Nbs1 complex: Recent insights into catalytic activities and ATP-driven conformational changes. Exp Cell Res. 2014;329:139–147. doi: 10.1016/j.yexcr.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res. 2009;37:1580–1588. doi: 10.1093/nar/gkn1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh J, Al-Zain A, Cannavo E, Cejka P, Symington LS. Xrs2 dependent and independent functions of the Mre11-Rad50 complex. Mol Cell. 2016;64:405–415. doi: 10.1016/j.molcel.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng SK, Yin Y, Petes TD, Symington LS. Mre11-Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol Cell. 2015;60:500–508. doi: 10.1016/j.molcel.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams RS, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lloyd J, et al. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You Z, et al. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davies OR, et al. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol. 2015;22:150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andres SN, et al. Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat Struct Mol Biol. 2015;22:158–166. doi: 10.1038/nsmb.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bessler JB, Reddy KC, Hayashi M, Hodgkin J, Villeneuve AM. A role for Caenorhabditis elegans chromatin-associated protein HIM-17 in the proliferation vs. meiotic entry decision. Genetics. 2007;175:2029–2037. doi: 10.1534/genetics.107.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nabeshima K, Villeneuve AM, Hillers KJ. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–1292. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MacQueen AJ, et al. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–1050. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosu S, et al. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 2013;9:e1003674. doi: 10.1371/journal.pgen.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pattabiraman D, Roelens B, Woglar A, Villeneuve AM. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 2017;13:e1006670. doi: 10.1371/journal.pgen.1006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains and primary images used in this research are available on request from A.M.V. (annev@stanford.edu).