Significance
Arabidopsis RopGEF1 acts as a negative regulator of signal transduction by the plant hormone abscisic acid (ABA). In turn, ABA treatment causes subcellular translocation and degradation of RopGEF1 protein. Interestingly, PP2C protein phosphatases, the core negative regulators of ABA signal transduction, protect RopGEF1 from degradation. This suggests that protein kinases may be involved in RopGEF1 protein removal. We find that calcium-dependent protein kinases (CPKs) including CPK4 phosphorylate RopGEF1. CPK4 promotes RopGEF1 degradation in Arabidopsis. CPK4 also negatively regulates RopGEF1 activities in root hair development. Furthermore, phosphorylation of serine residues at the N terminus of RopGEF1 is important for RopGEF1 degradation. We further discuss possible abiotic stress-triggered repression of plant growth via CPK-mediated removal of RopGEF.
Keywords: ABA, CPK, RopGEF, abiotic stress, protein phosphorylation and degradation
Abstract
Abscisic acid (ABA) plays essential roles in plant development and responses to environmental stress. ABA induces subcellular translocation and degradation of the guanine nucleotide exchange factor RopGEF1, thus facilitating ABA core signal transduction. However, the underlying mechanisms for ABA-triggered RopGEF1 trafficking/degradation remain unknown. Studies have revealed that RopGEFs associate with receptor-like kinases to convey developmental signals to small ROP GTPases. However, how the activities of RopGEFs are modulated is not well understood. Type 2C protein phosphatases stabilize the RopGEF1 protein, indicating that phosphorylation may trigger RopGEF1 trafficking and degradation. We have screened inhibitors followed by several protein kinase mutants and find that quadruple-mutant plants in the Arabidopsis calcium-dependent protein kinases (CPKs) cpk3/4/6/11 disrupt ABA-induced trafficking and degradation of RopGEF1. Moreover, cpk3/4/6/11 partially impairs ABA inhibition of cotyledon emergence. Several CPKs interact with RopGEF1. CPK4 binds to and phosphorylates RopGEF1 and promotes the degradation of RopGEF1. CPK-mediated phosphorylation of RopGEF1 at specific N-terminal serine residues causes the degradation of RopGEF1 and mutation of these sites also compromises the RopGEF1 overexpression phenotype in root hair development in Arabidopsis. Our findings establish the physiological and molecular functions and relevance of CPKs in regulation of RopGEF1 and illuminate physiological roles of a CPK-GEF-ROP module in ABA signaling and plant development. We further discuss that CPK-dependent RopGEF degradation during abiotic stress could provide a mechanism for down-regulation of RopGEF-dependent growth responses.
Environmental stressors such as drought and high salinity affect plant growth and productivity. Abscisic acid (ABA) is a vital phytohormone that regulates plant responses to environmental stresses. PYR/RCAR ABA receptors, PP2C phosphatases, and SnRK2 kinases are core components of the ABA signal transduction pathway that sense ABA and initiate a signaling cascade to regulate downstream transcriptional and ion channel activities (1–5).
In addition to SnRK2 protein kinases, calcium-dependent protein kinases (CPKs) are involved in ABA signal transduction (6–9). CPKs are serine/threonine protein kinases that are composed of four characterized domains including a variable N-terminal domain, a catalytic kinase domain, an autoinhibitory junction domain, and a calmodulin-like domain with Ca2+-binding EF-hand motifs (10). Integration of kinase activity and Ca2+ sensing motifs enables CPKs to transduce Ca2+ signals generated by environmental and developmental stimuli via a Ca2+-induced kinase activation (10–16). Genetic and biochemical analyses of CPKs have revealed biological functions of CPKs in plant development and in abiotic and biotic stress signaling (6, 14, 17–23). Specific substrates mediate CPK functions in plants. Identification of the substrates of CPKs is of present interest and a prerequisite for a full understanding of the physiological functions of CPKs.
Guanine nucleotide exchange factors (RopGEFs) are activators of small GTPases named ROPs in plants (24, 25). RopGEF activation of ROPs in turn regulates diverse cellular processes ranging from polarized cell growth, cell division, and reproduction to plant responses to environmental stress (26–34). RopGEFs can act as a bridge to link receptor-like kinases and ROP GTPases (35). However, the molecular mechanisms by which the activities of RopGEFs are modulated remain largely unknown.
ROP10 and ROP11 negatively regulate ABA signal transduction by stabilizing PP2C activity (36–39). Via activation of ROP10 and ROP11, RopGEF1 can contribute to shutting off ABA signal transduction in the absence of ABA (37, 40). Moreover, ABA causes rapid formation of intracellular RopGEF1 particles, trafficking of RopGEF1 to the prevacuolar compartment, and subsequent RopGEF1 degradation (41). ABA-induced degradation of RopGEF1 can facilitate ABA signaling (34, 41). RopGEF1 undergoes constitutive degradation in the ABA hypersensitive abi1/abi2/hab1/pp2ca PP2C protein phosphatase quadruple-mutant background (41). These findings led us to speculate that protein kinases may be potentially responsible for RopGEF1 degradation. After screening inhibitors and analyzing several higher-order protein kinase mutants, we show here that a cpk quadruple mutant, cpk3/4/6/11, disrupts ABA-triggered trafficking and degradation of RopGEF1. Additionally, cpk3/4/6/11 shows a reduced ABA response in germination. CPK4 directly interacts with and phosphorylates RopGEF1. CPK4-mediated phosphorylation facilitates RopGEF1 degradation. Moreover, CPKs affect RopGEF1 action in root hair growth. Taken together, our work reveals that RopGEF1 is a direct phosphorylation substrate of CPKs that mediate ABA-induced trafficking and degradation of RopGEF1.
Results
RopGEF1 Is Phosphorylated in Vivo.
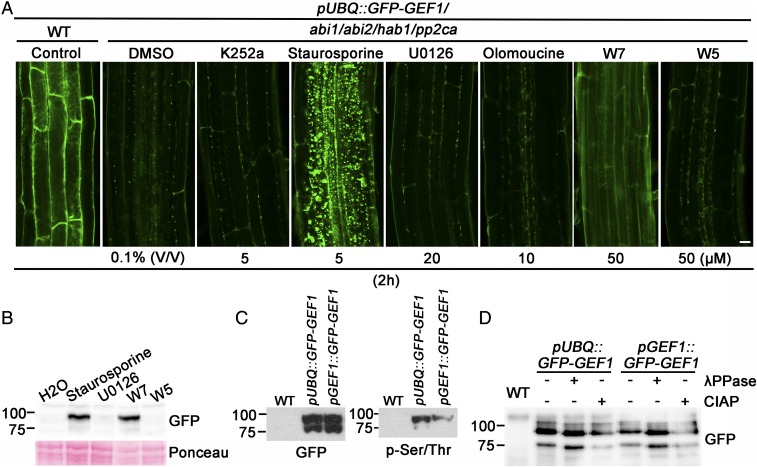
Previously, we found that RopGEF1 directly interacts with several ABA-regulated PP2C protein phosphatases and RopGEF1 is constitutively trafficked to the prevacuolar compartment and degraded in abi1/abi2/hab1/pp2ca pp2c quadruple-mutant plants (41). We speculated that an unknown protein kinase may phosphorylate GEF1 to promote its trafficking and degradation. To initially test this hypothesis, we took into account that GFP-GEF1 fluorescence is extremely low in pp2c quadruple-mutant plants due to GEF1 degradation (Fig. 1A, abi1/abi2/hab1/pp2ca DMSO). We investigated whether inhibition of potential protein kinases causes an increase of GFP-GEF1 fluorescence in pp2c quadruple-mutant plants. We treated Arabidopsis thaliana Columbia ecotype pUBQ::GFP-GEF1/abi1abi2hab1pp2ca seedlings with diverse protein kinase inhibitors and found that the Ser/Thr kinase inhibitor staurosporine and the calmodulin antagonist W7 could substantially increase GFP-GEF1 fluorescence (Fig. 1A and SI Appendix, Fig. S1A). In comparison, W5, an ineffective analog of W7, showed no such effect (Fig. 1A and SI Appendix, Fig. S1A). As reported previously, GFP-GEF1 could not be detected in immunoblots from abi1/abi2/hab1/pp2ca plants (Fig. 1B, H2O control). Immunoblot analyses showed that staurosporine and W7 treatments increased GFP-GEF1 protein abundance in 10-d-old abi1/abi2/hab1/pp2ca mutant plants (Fig. 1B). In addition, we found that immunoprecipitated GFP-GEF1 from total extracts of pUBQ::GFP-GEF1/col and pGEF1::GFP-GEF1/col plants in the WT (Col-0) background exhibited two distinct bands detected with GFP antibodies (Fig. 1C, Left). The more slowly migrating band was preferentially recognized by a phospho-Ser/Thr antibody (Fig. 1C, Right). Both bands showed slight electromobility shifts in SDS/PAGE gels when immunoprecipitated GFP-GEF1 was subjected to lamda phosphatase or calf intestinal alkaline phosphatase treatments (Fig. 1D). Mass spectrometry data indicated that both bands included GEF1 peptides and that both bands represent phosphorylated forms of GFP-GEF1. Taken together, these data suggested that GEF1 is phosphorylated in vivo in Arabidopsis plants.
Fig. 1.
RopGEF1 protein is phosphorylated in vivo in the A. thaliana Columbia ecotype. (A) GFP fluorescence in root epidermal cells of 5-d-old plants expressing GFP-GEF1 in abi1/abi2/hab1/pp2ca quadruple-mutant background, treated with the indicated protein kinase and calmodulin inhibitors for 2 h. (DMSO treatment as a control; confocal images were acquired using identical confocal parameters in GFP-GEF1/abi1abi2hab1pp2ca seedlings.) (Scale bar, 10 μm.) (B) Immunoblot analysis of GFP-GEF1 protein in 10-d-old Arabidopsis seedlings expressing GFP-GEF1 in the abi1/abi2/hab1/pp2ca background after 3 h of kinase inhibitor treatments. Kinase inhibitor concentrations: staurosporine, 5 μM; U0126, 20 μM; W7 and W5, 50 μM. Total protein extracts of seedlings were probed with an anti-GFP antibody. Ponceau staining was used as a loading control. (n = 2). (C) Detection of immunoprecipitated GFP-GEF1 with GFP (Left) and phospho-Ser/Thr antibody (Right). (n = 2). (D) Phosphatase treatments of immunoprecipitated GFP-GEF1 caused migration shifts in SDS/PAGE gels. Immunoprecipitated GFP-GEF1 protein was treated with lambda (λ) phosphatase and calf intestinal alkaline phosphatase (CIAP) for 1 h. GFP antibody was used to detect GFP-GEF1 (n = 5).
CPK Mutant Plants Disrupt RopGEF1 Removal.
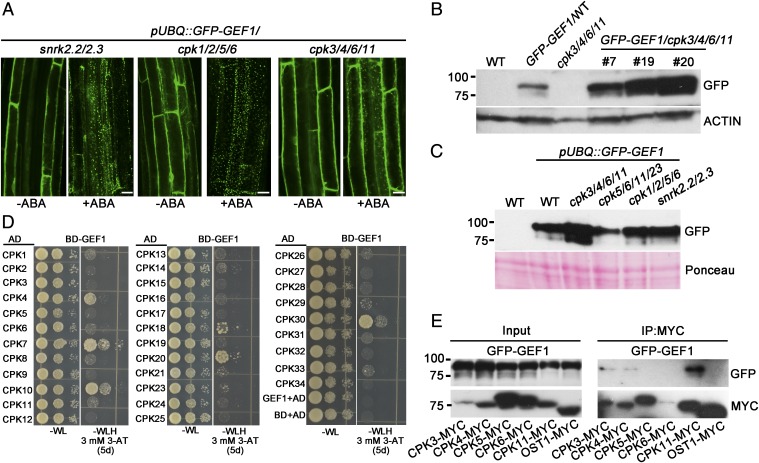
In Arabidopsis, the calmodulin antagonist W7 may impair protein kinase activities of CPKs, calcineurin B-like–linked protein kinases, and calcium and calmodulin-dependent protein kinases CCaMKs (note that CCaMK isoforms have not been found in the Arabidopsis genome). A pUBQ::GFP-GEF1 construct was transformed into several protein kinase mutants and homozygous single insertion lines were isolated to examine ABA-mediated GFP-GEF1 intracellular particle formation (41) in these mutant backgrounds. As ABA causes trafficking and degradation of GEF1, we included the protein kinase snrk2.2/2.3 double mutant that impairs ABA signal transduction in seedlings and roots (42, 43). We found that the subcellular localization of GFP-GEF1 is similar in these mutant backgrounds in the absence of exogenous ABA (SI Appendix, Fig. S1C). Interestingly, ABA-mediated GFP-GEF1 particle formation in roots is compromised in cpk3/4/6/11 quadruple-mutant plants (Fig. 2A). In contrast, other tested mutants did not disrupt this ABA response including the cpk1/2/5/6 and cpk5/6/11/23 quadruple cpk mutants and snrk2.2/2.3 double mutants (Fig. 2A and SI Appendix, Fig. S1D). Furthermore, in immunoblot analyses we found overaccumulation of GFP-GEF1 protein in cpk3/4/6/11 quadruple-mutant plants, but not in the cpk5/6/11/23, cpk1/2/5/6 and snrk2.2/2.3 backgrounds (Fig. 2 B and C). This GFP-GEF1 protein overaccumulation could not be attributed to differential transcription as shown in quantitative real-time PCR assays (SI Appendix, Fig. S2A). These data suggest that GFP-GEF1 protein abundance is regulated by members of the group of the Ca2+-dependent protein kinases CPK3, 4, 6, and 11.
Fig. 2.
CPKs directly interact with RopGEF1. (A) ABA-mediated GFP-GEF1 particle formation in root epidermal cells of 5-d-old homozygous Arabidopsis plants expressing GFP-GEF1 at a single locus in the snrk2.2/2.3, cpk1/2/5/6, and cpk3/4/6/11 backgrounds, in the absence or presence of 50 μM ABA for 1 h. Confocal images with identical imaging parameters are shown [confocal parameters: Zeiss LSM 710 (objective: 20×; laser: 488; pinhole: 90 μm; digital gain: 1; channel: 8 bit; average: line 4; zoom: 1; master gain: 800)]. (Scale bars, 10 μm.) (B and C) Immunoblot analysis of GFP-GEF1 protein in 10-d-old Arabidopsis seedlings expressing GFP-GEF1 in the indicated genotype backgrounds (n = 3). (D) GAL4-based yeast two-hybrid assays show that GEF1 interacts with members of the CPK family in yeast. Yeast colonies were grown on −L−W−H (lacking leucine, tryptophan, and histidine) selective plates with 3 mM 3-amino-1,2,4-triazole (3-AT) for 5 d. AD, activation domain; BD, binding domain. (E) Co-immunoprecipitation of GEF1 and CPKs expressed in N. benthamiana leaves. Total protein (Input) from 6-wk-old N. benthamiana leaves coexpressing GFP-GEF1 and CPK-MYC was extracted and subjected to immunoprecipitation using anti-MYC magnetic beads followed by immunoblot analysis with anti-GFP and anti-MYC antibodies (n = 2).
The Arabidopsis genome encodes 34 CPKs. The interactions of GEF1 and 33 CPKs (except CPK22) were tested in yeast two-hybrid assays. The results indicate that GEF1 may interact with some CPKs in yeast, including CPK4, CPK10, and CPK11, though to different degrees (Fig. 2D). To determine whether CPKs associate with GEF1, we immunoprecipitated CPKs expressed in Nicotiana benthamiana leaves and observed that CPK3, 4, and 11 formed association with GEF1, whereas the CPK5, CPK6, and OST1 protein kinases did not show a clear association with GFP-GEF1 (Fig. 2E).
Furthermore, we found overaccumulation of GFP-GEF1 protein in cpk4cpk11 (SI Appendix, Fig. S2B) and ABA-mediated GFP-GEF1 degradation was reduced in cpk4cpk11 double mutant compared with that in WT plants (SI Appendix, Fig. S2D). Thus far, cpk3cpk6 lines showed lower GFP-GEF1 expression levels and therefore further analyses are needed to determine whether cpk3cpk6 alone affect this ABA response. CPK4 is highly homologous with CPK11. The expression pattern and subcellular localization of CPK4 and GEF1 are similar in Arabidopsis (SI Appendix, Fig. S1 E and F). Based on these observations, we chose CPK4 to further investigate CPK effects on GEF1.
CPK4 Directly Interacts with and Phosphorylates RopGEF1 in Arabidopsis.
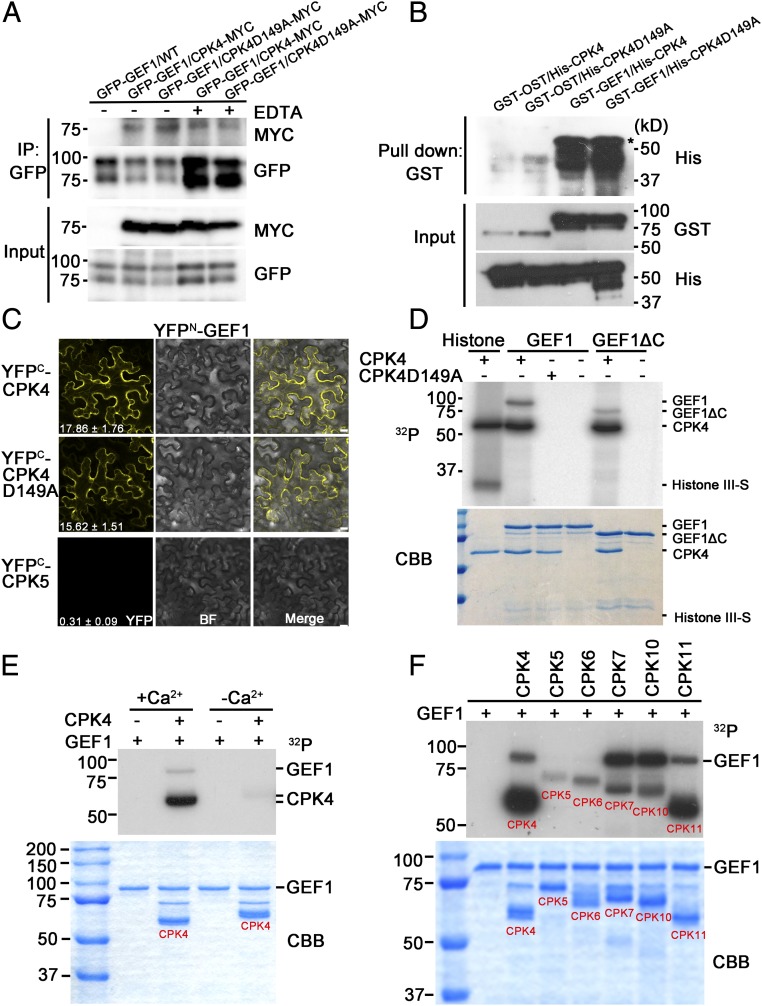
Activation analyses of CPKs have demonstrated that binding of Ca2+ to EF-hand motifs of CPKs can trigger a conformational change to release a pseudosubstrate inhibitory domain from the active site of the kinase domain (11, 12, 16, 44). To investigate whether GEF1–CPK interaction is affected by Ca2+, we examined the GEF1–CPK4 interaction in transgenic Arabidopsis plants expressing both GFP-GEF1 and mCherry-CPK4-myc generated through crossing of homozygous single insertion lines. Co-immunoprecipitation results indicate that GEF1 associates with CPK4 in Arabidopsis regardless of the Ca2+ concentration and with CPK4-D149A [a kinase-dead version of CPK4 (45)] (Fig. 3A). Furthermore, in vitro pull-down experiments indicated that GEF1 directly interacts with both CPK4 and the inactive CPK4-D149A, and that CPK4 kinase activity is not required for this interaction (Fig. 3B). In addition, CPK4–GEF1 interaction was observed at the cell periphery and cytosol in bimolecular fluorescence complementation (BiFC) assays, with CPK5 showing only a very low average fluorescence intensity as control (Fig. 3C).
Fig. 3.
CPK4 directly interacts and phosphorylates RopGEF1. (A) Co-immunoprecipitation of GEF1 and CPK4 in Arabidopsis. Total protein from 10-d-old stable transgenic homozygous Arabidopsis seedlings (F3 homozygous, single locus per transgene) generated from crosses between pUBQ::GFP-GEF1/WT and pUBQ::mCherry-CPK4/WT or pUBQ::mCherry-CPK4-D149A/WT were extracted and subjected to immunoprecipitation using anti-GFP beads followed by immunoblot analysis with anti-GFP and anti-MYC antibodies; 10 mM EDTA was added to extraction buffer to chelate calcium. CPK4-D149A is a kinase-dead version of CPK4. (n = 2). (B) GST pull-down assay shows that GEF1 interacts with CPK4 and CPK4-D149A. Asterisk shows the predicted band of His-CPK4. (C) BiFC analyses show that GEF1 and CPK4/CPK4–D149A interaction occurs in the cytosol and cell periphery. CPK5–GEF1 interaction was used as a negative control. BF, bright field. Average quantification values of YFP signals in relative units by FIJI are shown at the bottom of left panels (n = 10 images per condition). (Scale bars, 10 μm.) Confocal parameters were identical for all images. (D) CPK4 trans-phosphorylates GEF1 and C-terminal truncated version of GEF1 in vitro. (D, Lower) Coomassie blue staining (CBB) of the proteins used for the phosphorylation assay; 0.2 μg Histone III-S was used as CPK4 substrate. (n = 4). (E) Calcium-dependent phosphorylation of GEF1 by CPK4. Approximately 2 μg of CPK4 and GEF1, respectively, were mixed in kinase reaction buffer with or without 2 µM free Ca2+. After 30-min incubation, the reaction was subjected to SDS/PAGE. Note that Ca2+-bound CPK4 runs at an apparent lower molecular weight, likely due to conformational change (n = 3). (F) Phosphorylation of GEF1 by the indicated CPKs. Approximately 2 μg of CPKs and GEF1, respectively, were mixed in kinase reaction buffer with 2 µM free Ca2+. Kinase reactions were carried out for 1 h at room temperature (n = 3).
We next performed in vitro kinase assays to test a possible regulatory effect of CPK4 on GEF1. These experiments showed that CPK4 is autophosphorylated and trans-phosphorylates the artificial substrate histone (Fig. 3D). Furthermore, CPK4 had a trans-phosphorylation activity in vitro toward GEF1 and also toward a mutant GEF1 protein in which the C terminus was truncated (Fig. 3D). In contrast, the kinase-dead version of CPK4 (Fig. 3D) and intact OST1 (SI Appendix, Fig. S2F), a SnRK2 protein kinase in the ABA signal transduction pathway, were unable to trans-phosphorylate GEF1 in vitro. The CPK4-mediated trans-phosphorylation of GEF1 is calcium-dependent (Fig. 3E). Taken together, these findings suggest that GEF1 interacts with and could be a direct phosphorylation substrate of CPK4. Furthermore, several CPKs which showed interactions with GEF1 in yeast two-hybrid assays (Fig. 2D) also phosphorylated GEF1 (Fig. 3F).
CPKs Promote the Degradation of RopGEF1 in Arabidopsis.
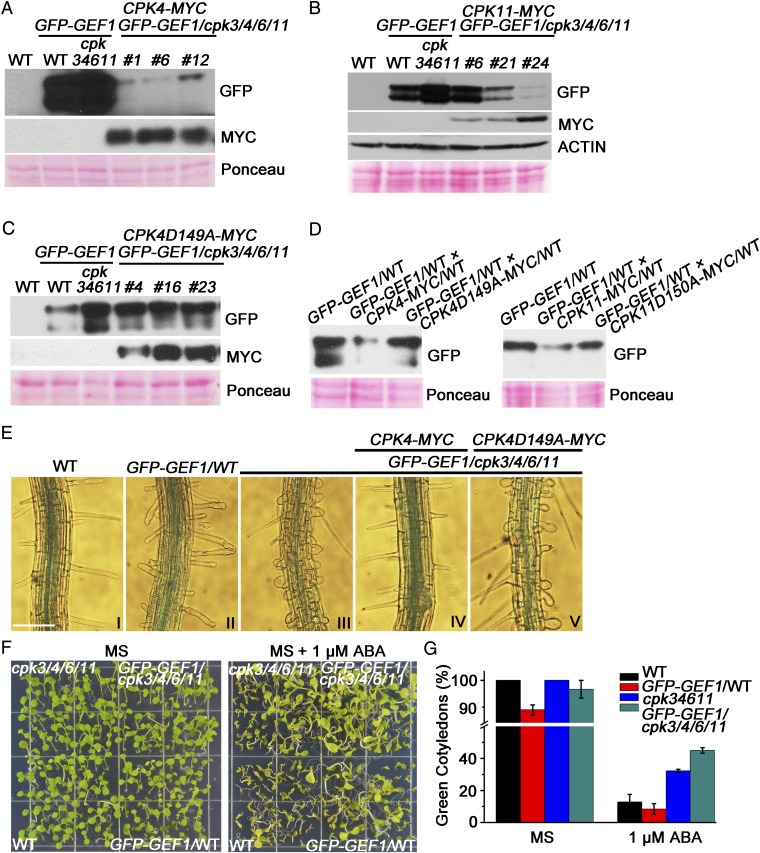
Next, we investigated the biological relevance of GEF1 phosphorylation mediated by CPKs. Considering the overaccumulation of GFP-GEF1 protein in cpk3/4/6/11 quadruple-mutant plants (Fig. 2 B and C), we next examined complementation via CPK overexpression on the protein levels of GFP-GEF1 in GFP-GEF1/cpk3/4/6/11 quadruple-mutant plants. Immunoblot analyses of homozygous transgenic plants showed that overexpression of CPK4 and CPK11, a close homolog of CPK4, but not the CPK4 kinase-dead version dramatically decreased GFP-GEF1 protein abundance in GFP-GEF1/cpk3/4/6/11 plants (Fig. 4 A–C). In controls, GEF1 transcripts were not greatly different among these lines (SI Appendix, Fig. S4, right group).
Fig. 4.
CPK4/11 overexpression promotes the degradation of RopGEF1 in Arabidopsis. (A–C) Immunoblot analyses of GFP-GEF1 protein in 10-d-old Arabidopsis seedlings expressing pUBQ::mCherry-CPK4-MYC (A, n = 3), pUBQ::mCherry-CPK11-MYC (B, n = 2), and pUBQ::mCherry-CPK4-D149A-MYC (C, n = 3) in pUBQ-GFPGEF1/cpk3/4/6/11 plants. Anti-GFP and anti-MYC antibodies were used for detecting GEF1 and CPKs, respectively. Ponceau staining was using as loading control. (D) Immunoblot analyses of GFP-GEF1 protein in Arabidopsis seedlings expressing both pUBQ::GFP-GEF1/WT and pUBQ::mCherry-CPK4/WT or pUBQ::mCherry-CPK4-D149A/WT generated through crossing (Left) pUBQ::GFP-GEF1/WT and pUBQ::mCherry-CPK11/WT or pUBQ::mCherry-CPK11-D150A/WT (Right). (E) Root hair morphology of the indicated genotypes (I–V) grown on one-fourth MS medium plates for 7 d. (Scale bar, 100 μm.) (F and G) ABA-mediated inhibition of seedling growth in the indicated genotypes grown on half MS plates lacking ABA for 6 d or half MS medium supplemented with 1 μM ABA for 14 d. Sixty seeds of each genotype were sown on each plate and counted for the appearance of green expanded cotyledons; data are average ±SD from n = 3 replicates.
We introduced overexpression of CPK4 or CPK11 into GFP-GEF1/WT lines through crossing of homozygous single-locus insertion lines. GFP-GEF1 protein levels in these lines were decreased in contrast to inactive CPK4-D149A and CPK11-D150A mutant isoforms (Fig. 4D). These results support a model in which CPKs phosphorylate GEF1 and down-regulate GEF1 protein levels in Arabidopsis.
Next we explored the importance of the CPK-mediated reduction in GEF1 protein levels in Arabidopsis. GEF1 activates the small GTPase ROP11 (40). Overexpression of GEF1 induces isotropic growth of root hair cells and produces swollen root hairs (Fig. 4 E, II), similarly but to a lesser extent than overexpression of a constitutively active ROP11 isoform (41, 46). We therefore investigated the root hair morphology in different genotypes and found that overexpression of GEF1 in the cpk3/4/6/11 quadruple-mutant background produced swollen root hairs (Fig. 4 E, III) more severely than that in the WT background (Fig. 4 E, II). In complementation tests, overexpression of CPK4 but not the kinase-dead CPK4 rescues this defective root hair phenotype in GFP-GEF1/cpk3/4/6/11 plants (Fig. 4 E, IV and V). In WT (Col-0) plants, overexpression of CPK4 exhibits normal root hair morphology (SI Appendix, Fig. S3A). Interestingly, we observed abnormally shaped root hairs in cpk3/4/6/11 quadruple-mutant plants (SI Appendix, Fig. S3B).
We investigated whether the cpk3/4/6/11quadruple mutant shows an ABA-dependent phenotype in seed germination and whether GEF1 affects this response. In the absence of ABA no clear germination phenotype was observed in 6-d-old seedlings (Fig. 4F). However, cpk3/4/6/11 seedlings displayed a decreased sensitivity to ABA inhibition of cotyledon emergence compared with WT plants (Fig. 4 F and G). Moreover, GFP-GEF1/cpk3/4/6/11 seedlings exhibited a reduced ABA inhibition of cotyledon emergence (Fig. 4 F and G) compared with WT plants and cpk3/4/6/11 plants, indicating that GEF1 overabundance in the cpk3/4/6/11 background (Figs. 2 B and C and 4 A and B) further reduces ABA sensitivity in seedling growth.
CPK4 Phosphorylates RopGEF1 at the N Terminus.
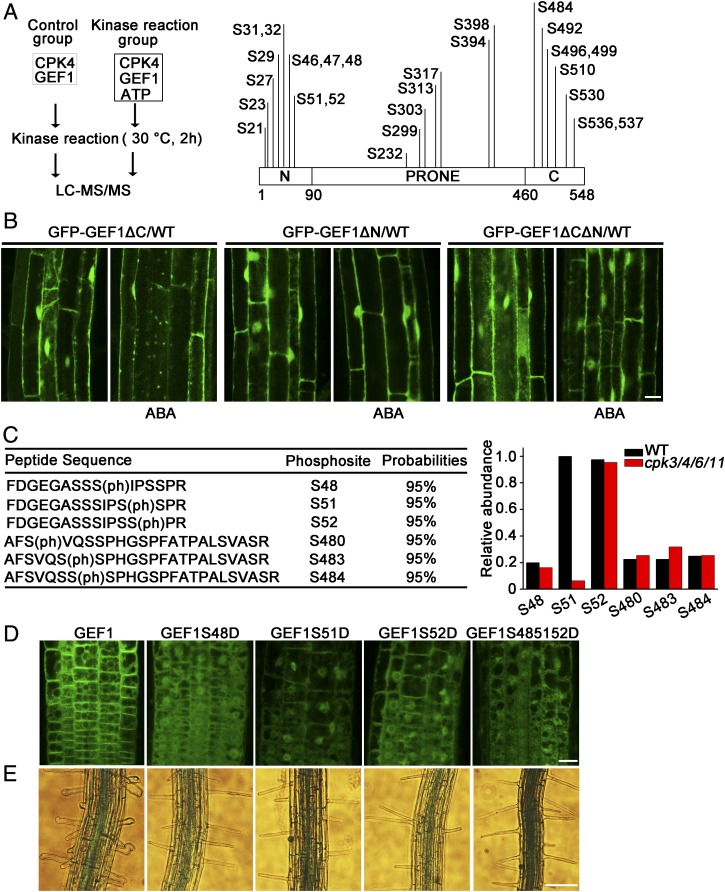
The above analyses suggest that GEF1 is a direct phosphorylation substrate of CPK4. Next, we sought to map which residues are phosphorylated by CPK4 and test whether these phosphorylated residues are responsible for GEF1 degradation. To identify the GEF1 phosphorylation site resulting from CPK4 phosphorylation, in vitro kinase assays were performed with recombinant GEF1 and CPK4. GEF1 and CPK4 mixtures without ATP were used as a control. Phosphorylation sites were detected using mass spectrometry. GEF1-dependent phosphorylation sites by CPK4 were determined by subtracting phosphorylation sites detected in the control group (Fig. 5A, Left). In these in vitro analyses, we detected 26 phosphorylated serine residues in GEF1 by CPK4 and most phosphorylated residues were distributed in the N- and C-terminal variable domains (Fig. 5A) and not in a classical ФxRxxS/T CPK-recognition motif (47, 48).
Fig. 5.
CPK-mediated N-terminal phosphorylation of RopGEF1 promotes its degradation. (A) Summary of in vitro phosphorylation sites identified in GST-GEF1 by His-CPK4 through MS/MS analyses. A GEF1 and CPK4 mixture without ATP was used as a control. GEF1 phosphorylation sites by CPK4 were determined through subtracting phosphorylation sites detected in the control group from the kinase reaction group. (B) GFP-GEF1 expression pattern and ABA-mediated GFP-GEF1 particle formation in root epidermal cells of 5-d-old WT plants expressing the indicated GEF1 fragments in the absence or presence of 50 μM ABA for 1 h. (GEF1ΔC: residues 1–460; GEF1ΔN: residues 91–548; GEF1ΔCΔN: residues 91–460). (Scale bar, 10 μm.) (C) Summary of GEF1 phosphorylation sites identified in vivo in Arabidopsis. GFP-GEF1 was immunoprecipitated from pUBQ::GFP-GEF1/WT and pUBQ::GFP-GEF1/cpk3/4/6/11 plants and subjected to MS/MS analyses. Two independent MS experiments were performed, each containing immunoprecipitated GFP-GEF1 protein samples from both WT and cpk3/4/6/11 background. Phosphorylated residues are shown as S(ph). Relative abundance (C, Right) was determined by dividing the mean number of phosphorylated spectra for each residue by the number of spectra showing phosphorylation of the most abundant phophopeptide (S51/WT) (n = 2). (D and E) Subcellular localization (D) and root hair morphology (E) of plants expressing the indicated phosphorylation-mimic GEF1 isoforms. (Scale bars, 10 μm in D and 100 μm in E.)
The N- and C-terminal variable regions of RopGEFs have regulatory functions (33, 49, 50). Both N and C termini of GEF1 harbor many serine and threonine residues. We therefore explored which termini of GEF1 are relevant for ABA-mediated GEF1 particle formation and degradation. We observed that a C-terminally truncated GFP-GEF1ΔC (GEF1, amino acids 1–460) still causes an ABA-induced reduction in fluorescence and forms cytosolic particles in response to ABA (Fig. 5B). ABA did not cause intracellular particle formation in Arabidopsis roots that were stably transformed with an N-terminally truncated GFP-GEF1ΔN (GEF1, amino acids 91–548) or with a doubly truncated GFP-GEF1ΔCΔN (GEF1, amino acids 91–460), indicating a possible role for the N terminus of GEF1 in ABA-induced trafficking of GEF1 (Fig. 5B). CPK4 did not interact with the C-terminal domain of GEF1 but the N-terminal domain of GEF1 promoted GEF1-CPK4 interaction in yeast two-hybrid assays (SI Appendix, Fig. S3C).
Next we purified GFP-GEF1 fusion proteins from 2-wk-old homozygous single locus pUBQ::GFP-GEF1/WT and pUBQ::GFP-GEF1/cpk3/4/6/11 transgenic Arabidopsis plants grown on half Murashige and Skoog (MS) medium to study GEF1 ;phosphorylation in vivo using tandem mass spectrometry. Six phosphorylated serine residues (S48, S51, S52, S480, S483, and S484) could be reproducibly detected (Fig. 5C). These phosphorylated serine residues were clustered and were detected in phosphopeptides of the N- and C-terminal domains of GEF1. Interestingly, a similar peptide containing the clustered serine residues (NDKLPRVSSSDSMEA) in the N terminus of RopGEF11 was also shown to be phosphorylated by CPK34 in a CPK34 substrate screen (51). We synthesized peptides containing S48, S51, and S52 and different mutations in these three serine residues and performed in vitro kinase assays on these synthesized peptides with CPK4 using peptide arrays. CPK4 phosphorylated these three serine residues in these peptides (SI Appendix, Fig. S3D). Furthermore, we found that the relative abundance of S51 phosphorylation was reduced significantly by ∼90% when GFP-GEF1 was isolated from cpk3/4/6/11 mutant plants compared with WT (Fig. 5C).
To investigate the effect of CPK-mediated phosphorylation on GEF1 function, we generated stable homozygous transgenic Arabidopsis lines overexpressing the S48D, S51D, S52D, and S48D/S51D/S52D phosphomimic GEF1 variants. These phosphomimic GEF1 variants interacted with CPK4 in yeast two-hybrid assays similarly to WT GEF1 (SI Appendix, Fig. S3E). We observed that these phosphomimic GEF1 variants were nonfunctional, as shown by the normal root hair phenotype compared with lines that overexpress GEF1 (Fig. 5E). In addition, we also failed to detect the protein of these phosphomimic GEF1 isoforms by immunoblots in plants despite a weak GFP fluorescence that could be observed in these lines (Fig. 5E and SI Appendix, Fig. S3F). These lines expressed GEF1 mRNA in controls (SI Appendix, Fig. S4, middle group). These assays indicated that either phosphorylation or the structure of this N-terminal serine cluster is important for GEF1 removal in Arabidopsis.
Discussion
Previous findings showed that RopGEFs, including GEF1, act as negative regulators of ABA signaling and that GEF1 is translocated to the prevacuolar compartment for degradation in response to ABA (41). Core group A PP2C negative regulators of ABA signal transduction, including the ABI1 PP2C phosphatase, directly interact with GEF1 and protect GEF1 from degradation (41). However, how ABA initiates trafficking and degradation of RopGEF1 is unknown. In the present study, through pharmacological, genetic, and biochemical analyses, we identified GEF1 as a direct phosphorylation substrate of the CPK CPK4. Complementation and phosphorylation analyses show that CPK4-mediated phosphorylation of GEF1 promotes GEF1 degradation. In addition, we found that ABA-induced trafficking and degradation of GEF1 in Arabidopsis was disrupted in cpk3/4/6/11 quadruple-mutant roots but continued to occur in snrk2.2/snrk2.3 double-mutant and other tested cpk mutant roots.
Together the present study suggests that CPK-mediated phosphorylation can trigger GEF1 degradation. ABA-induced degradation of GEF1 would require inactivation of PP2C phosphatases by ABA. PP2C inhibition may cause GEF1 to lose the protection from PP2C phosphatase-mediated dephosphorylation. Besides CPK4, yeast two-hybrid experiments implicate that GEF1 interacts with multiple CPKs. These CPKs may function redundantly or additively with CPK4 or some CPKs might contribute to physiological regulation of GEFs by other Ca2+-signaling pathways.
Previous research has shown that CPK3/6 and CPK4/11 function as positive regulators in ABA signaling (6, 8, 9). The disruption of ABA-mediated GEF1 trafficking in cpk3/4/6/11 indicates a possible key regulation network in ABA signal transduction. In this model active CPKs phosphorylate GEF1, causing GEF1 trafficking and degradation. GEF removal in turn would inactivate ROP10 and ROP11 that function as negative regulators of ABA signal transduction (36–39). Thus, ABA-induced GEF removal via CPKs would facilitate ABA signal transduction via removal of a negative regulation loop consisting of PP2C-GEF-ROP10/ROP11 (36–38, 41). Consistent with this model gef1/4/10/14 quadruple mutant, gef1/4/10 triple mutant, and rop10, rop11, and rop10/11 mutants have all been shown to exhibit ABA hypersensitivity in several responses (37–39, 41) and ABA down-regulates small GTP-binding protein activity (34). Moreover, cpk3/4/6/11 mutant seedlings exhibited a reduced ABA sensitivity (Fig. 4 F and G).
Note, however, that the present study also opens another intriguing question for investigation. It is conceivable that other signaling pathways that activate stress-linked CPKs could result in RopGEF removal and thus amplification or up-regulation of ABA signal transduction. GEFs are associated with stimulation of growth (31, 32, 37, 52). As members of the CPK family play roles in several stress signaling pathways (13, 14, 17, 23, 53–56), it is tempting to speculate that GEF1 degradation may contribute to reduced plant growth in response to environmental stress conditions. Further research would be needed to test this more general hypothesis for a role of CPKs in GEF degradation.
Recent studies have shown that RopGEFs act as a bridge to link receptor-like kinases with downstream ROP small GTPase. The receptor-like kinase FERONIA interacts with GEF1/4/10 and in turn activates ROP11 to relay an auxin signal and facilitate root hair growth (31, 57); the pollen-specific receptor-like kinase PRK6 that senses the LURE1 attractant peptide interacts with pollen-expressed RopGEF12 to activate ROP1 for pollen tube reorientation (52). However, whether the receptor-like kinases directly phosphorylate RopGEFs and how the activities of RopGEFs are modulated remain to be directly determined.
The C termini of RopGEFs are important for their activity and the interaction of GEFs with receptor-like kinases (33). For ABA responses the N terminus of RopGEF1 is indispensible for CPK4–GEF1 interaction. CPK4 phosphorylates N-terminal serine residues including S48, S51, and S52 in vitro. Furthermore, phosphorylation of residue S51 in planta depends on CPKs (Fig. 5C). Phosphomimic isoforms of these residues caused RopGEF1 inactivity in root hair growth regulation, suggesting that N- or C-terminal phosphorylation of RopGEFs could play different roles in regulation of GEF activities. Our work shows the biological relevance of CPKs in RopGEF1 removal. Considering the essential roles of both CPKs and GEFs in polarized growth of pollen tubes and root hairs, pathogen defense, and abiotic stress responses, further research will be of interest to probe other CPK–GEF combinations and their functions in plant development and stress responses.
Materials and Methods
Plant Material and Growth Conditions.
The Arabidopsis thaliana accession used was Columbia (Col-0). Arabidopsis seeds were surface-sterilized in 20% bleach for 30 min followed by four washes with sterile water and sown on half MS media (pH 5.8) supplemented with 1% sucrose and 0.8% Phyto Agar. Plates with sterilized seeds were stratified in the dark for 3 d at 4 °C and then transferred to the growth room under a 16/8 h light/dark cycle, 80 μmol⋅m−2⋅s−1 light intensity, 22–24 °C, and 40% relative humidity. One-week-old seedlings were transplanted into soil (Sunshine Mix1; Planet Natural) in 2.25-inch square pots (McConkey). Seeds of cpk5/6/11 were kindly provided by Jen Sheen, Harvard Medical School, Boston (13) and the cpk5/6/11/23 quadruple mutant was further constructed as described through crossing cpk5/6/11 with cpk23-1 (6); seeds of cpk1/2/5/6 were kindly provided by Ping He, Texas A&M University, College Station, TX (53) and snrk2.2/2.3 seeds were provided by Jian-Kang Zhu, Chinese Academy of Sciences, Shanghai (43).
RNA Extraction and qPCR.
Total RNA was extracted from 2-wk-old Arabidopsis seedlings using the SpectrumTM Plant Total RNA kit (Sigma). Approximately 3-μg RNA samples were treated with 1 μL DNase I (NEB) for 30 min and converted to cDNA using a First-Strand cDNA Synthesis kit (GE Healthcare). Synthesized cDNA was diluted four times and 2 μL was used for PCR templates. qPCR analyses were performed on a plate-based BioRad CFX96 qPCR System using SYBR Select Master Mix for CFX (Applied Biosystems) with gene-specific primers (GEF1 forward: tgcttgccgaaatggagattccc; GEF1 reverse: agacattccttcccgctcttgg; GAPC forward: tcagactcgagaaagctgctac; GAPC reverse: cgaagtcagttgagacaacatcatc). Expression levels are shown relative to WT GEF1 controls.
Plant Transformation and Confocal Microscopy.
Arabidopsis was transformed using the floral dip method. Single insertion lines were isolated by Hygromycin or Basta selection. Homozygotes were determined by lack of segregation of antibiotic resistance with the T3 generation. For transient expression in N. benthamiana leaves, overnight cultures of Agrobacteria were collected by centrifugation at 2,200 × g for 5 min. The pellets were washed twice with 1 mL buffer (10 mM MES, pH5.6, 10 mM MgCl2, and 100 μM acetosyringone) and resuspended to OD600 = 1. Equal volumes of bacterial suspensions were mixed and infiltrated into 6-wk-old N. benthamiana leaves with a syringe and needle. After infiltration, plants were kept in the dark overnight and then grown in the growth room for 48 h before harvesting for immunoprecipitation or microscopy observation. Fluorescence signals were detected using a confocal laser scanning microscope (LSM710; Carl Zeiss). Note that ABA-mediated GFP-GEF1 particle formation was not uniformly generated in primary roots under ABA treatment. Some cells responded more rapidly to ABA treatment than neighboring cells. Initially, low magnification (10× objective) was used to view the overall responses of each mutant. Contiguous epidermal cells in the mature region of primary roots that represented differences among genotypes were further analyzed. The depicted images show examples of differences that were observed between genotypes. Average fluorescence intensity of each cell was calculated through Image J software. Further preliminary analyses suggest that a 10-alanine linker between GFP and GEF1 may enhance ABA-mediated GFP-GEF1 relocation to particles.
Yeast Two-Hybrid Assays.
Yeast two-hybrid assays were performed as previously described (41); 10- and 200-fold dilutions of transformants were spotted on drop-out medium and grown for the indicated times.
In Vitro Pull-Down Assays.
Full-length CDS of CPK4, CPK4-D149A, and GEF1 were introduced into pET30a (for His-CPK4/CPK4-D149A fusion) and pGEX6P-1 (for GST-GEF1fusion) using USER enzyme. His-CPK4/CPK4-D149A and GST-GEF1 fusion proteins were produced in Escherichia coli Rosetta (DE3) pLysS (Novagen) cells with the induction conditions [0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 18 °C for His-CPK4/CPK4-D149A or 0.5 mM IPTG for 5 h at 25 °C for GST-GEF1]. Pull-down assays were performed as described previously (41).
In Vitro Phosphorylation Assays.
Recombinant CPK4/4-D149A and GEF1/GEF1ΔC proteins prepared from E. coli were incubated in phosphorylation buffer [50 mM Tris⋅HCl, 10 mM MgCl2, 2 µM free Ca2+ buffered by 1 mM EGTA,and CaCl2 (https://web.stanford.edu/∼cpatton/webmaxc/webmaxcE.htm), 0.1% Triton X-100, and 1 mM DTT at pH 7.5]. The in vitro phosphorylation reactions were started by the addition of 200 µM ATP and 0.1 µCi⋅µL−1 [γ-32P]ATP (PerkinElmer). The reactions were stopped by the addition of SDS/PAGE sample buffer after 30-min incubation at room temperature. Proteins were separated by SDS/PAGE, and the radioactivity of incorporated 32P in phosphorylated proteins was detected using an FLA-5000 Phosphor Imager (Fujifilm). The protein level was analyzed by Coomassie Brilliant Blue staining.
Immunoprecipitation.
Two grams of 2-wk-old Arabidopsis seedlings or N. benthamiana leaves were ground in liquid nitrogen and homogenized in 5 mL extraction buffer [50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM DTT, 0.1% Nonidet P-40, 1 mM phenylmenthylsulphonyl fluoride, and protease inhibitor mixture (5 mL/tablet; Roche), or supplemented with 10 mM EDTA]. Lysates were incubated on ice for 30 min and clarified by two 10-min centrifugations at 20,000 × g at 4 °C. Supernatant was further filtered through a 0.45-μm filter and incubated with 30 μL Chromotek-GFP-Trap magnetic beads (ACT-CM-GFM0050; Allele Biotechnology) or Pierce Anti-c-Myc magnetic beads (88843; Thermo Fisher Scientific) and rotated at 4 °C for 3 h. Beads were washed four times with ice-cold extraction buffer. The bound proteins were eluted with 2× SDS/PAGE sample buffer by heating at 95 °C for 7 min and analyzed by immunoblot. The primary antibodies used in this study are anti-GFP (1:4,000, SAB5300167; Sigma), anti-MYC (1:4,000, SAB1305535; Sigma), anti-His (1:1,000, SAB1306084; Sigma) anti-GST (1:1,000, RPN1236; GE Healthcare), anti-p-Ser/Thr (1:2,000, 612548; BD Transduction Laboratories), and anti-Actin (1:2,000, A0480; Sigma); second antibodies are goat anti-rabbit IgG (H + L)-HRP, (1:4,000, 1706515; Bio-Rad) and goat anti-mouse IgG (H + L)-HRP (1:4,000, 1706516; Bio-Rad). The PVDF membrane was incubated in the buffer supplied by SuperSignal West Pico PLUS Chemiluminescent substrate (34577; Pierce) and immunoblot signals were developed by ChemiDoc XRS+ (Bio-Rad).
Liquid Chromatography–Tandem Mass Spectrometry.
Approximately 9 g and 25 g of 2-wk-old seedlings from pUBQ::GFP-GEF1/cpk3/4/6/11 and pUBQ::GFP-GEF1/col were ground for total protein extraction. Immunoprecipitated proteins with GFP magnetic beads were separated by SDS/PAGE, after staining with Pierce Silver Stain for Mass Spectrometry (24600; Thermo Fisher Scientific). Trypsin-digested peptides were analyzed by ultra-high-pressure liquid chromatography (UPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nanospray ionization. The nanospray ionization experiments were performed using a Orbitrap fusion Lumos hybrid mass spectrometer (AB Sciex) interfaced with nanoscale reversed-phase UPLC (Thermo Dionex UltiMate 3000 RSLC nano system) using a 25-cm, 75-μm i.d. glass capillary packed with 1.7-µm C18 (130) BEHTM beads (Waters Corporation). Peptides were eluted from the C18 column into the mass spectrometer using a linear gradient (5–80%) of ACN (acetonitrile) at a flow rate of 375 μL/min for 1 h. The buffers used to create the ACN gradient were buffer A (98% H2O, 2% ACN, and 0.1% formic acid) and buffer B (100% ACN and 0.1% formic acid). Mass spectrometer parameters are as follows. An MS1 survey scan using the orbitrap detector [mass range (m/z): 400–1,500 (using quadrupole isolation), 120,000 resolution setting, spray voltage of 2,200 V, ion transfer tube temperature of 275 °C, AGC target of 400,000, and maximum injection time of 50 ms] was followed by data-dependent scans [top speed for most intense ions, with charge state set to only include +2–5 ions, and 5-s exclusion time, while selecting ions with minimal intensities of 50,000 at in which the collision event was carried out in the high-energy collision cell (HCD collision energy of 30%), and the fragment masses were analyzed in the ion trap mass analyzer (with ion trap scan rate of turbo, first mass m/z was 100, AGC target 5,000, and maximum injection time of 35 ms)]. Data analysis was carried out using the Byonic (Protein Metrics Inc.). Probability of a phosphorylation site was measured based on presence and intensity of site determining peaks in the MS/MS spectra. Phosphorylated peptides were accepted if their probability was over 90% based on the peptide prophet algorithm and if the neutral loss of a phosphoric acid was evident in the associated mass spectra. Datasets are deposited in the PRoteomics IDEntifications (PRIDE) database.
Supplementary Material
Acknowledgments
We thank Drs. Jen Sheen, Ping He, and Jian-Kang Zhu for providing the cpk5/6/11, cpk1/2/5/6, and snrk2.2/2.3 mutant seeds, respectively; Dr. Majid Ghassemian for conducting mass spectrometry experiments at the Biomolecular and Proteomics Mass Spectrometry Facility core, Department of Chemistry and Biochemistry [University of California, San Diego (UCSD)]; Jason Del-Rio in Dr. Susan Taylor’s laboratory (UCSD) for synthesizing small peptides and performing kinase assays shown in SI Appendix, Fig. S2E; and Mark Estelle (UCSD) for use of a confocal microscope. This research was supported by National Institutes of Health Grant GM060396-ES010337 and National Science Foundation Grant MCB-1616236 (to J.I.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The datasets reported in this paper have been deposited in the PRoteomics IDEntifications (PRIDE) database (accession nos. PXD009421 and PXD009422).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719659115/-/DCSupplemental.
References
- 1.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 2.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 4.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Joshi-Saha A, Valon C, Leung J. A brand new START: Abscisic acid perception and transduction in the guard cell. Sci Signal. 2011;4:re4. doi: 10.1126/scisignal.2002164. [DOI] [PubMed] [Google Scholar]
- 6.Brandt B, et al. Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife. 2015;4:e03599, and erratum (2015) 4:e10328. doi: 10.7554/eLife.03599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiger D, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu SY, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori IC, et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 2006;4:e327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper JF, et al. A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science. 1991;252:951–954. doi: 10.1126/science.1852075. [DOI] [PubMed] [Google Scholar]
- 11.Huang JF, Teyton L, Harper JF. Activation of a Ca(2+)-dependent protein kinase involves intramolecular binding of a calmodulin-like regulatory domain. Biochemistry. 1996;35:13222–13230. doi: 10.1021/bi960498a. [DOI] [PubMed] [Google Scholar]
- 12.Yoo BC, Harmon AC. Intramolecular binding contributes to the activation of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1996;35:12029–12037. doi: 10.1021/bi9606612. [DOI] [PubMed] [Google Scholar]
- 13.Boudsocq M, et al. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubiella U, et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA. 2013;110:8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers C, et al. Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009;59:528–539. doi: 10.1111/j.1365-313X.2009.03894.x. [DOI] [PubMed] [Google Scholar]
- 16.Harmon AC, Yoo BC, McCaffery C. Pseudosubstrate inhibition of CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33:7278–7287. doi: 10.1021/bi00189a032. [DOI] [PubMed] [Google Scholar]
- 17.Liu KH, et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature. 2017;545:311–316. doi: 10.1038/nature22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matschi S, Hake K, Herde M, Hause B, Romeis T. The calcium-dependent protein kinase CPK28 regulates development by inducing growth phase-specific, spatially restricted alterations in jasmonic acid levels independent of defense responses in Arabidopsis. Plant Cell. 2015;27:591–606. doi: 10.1105/tpc.15.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monaghan J, et al. The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe. 2014;16:605–615. doi: 10.1016/j.chom.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Xie K, Chen J, Wang Q, Yang Y. Direct phosphorylation and activation of a mitogen-activated protein kinase by a calcium-dependent protein kinase in rice. Plant Cell. 2014;26:3077–3089. doi: 10.1105/tpc.114.126441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu N, et al. CALCIUM-DEPENDENT PROTEIN KINASE5 associates with the truncated NLR protein TIR-NBS2 to contribute toexo70B1-mediated immunity. Plant Cell. 2017;29:746–759. doi: 10.1105/tpc.16.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao LN, et al. Ca2+-dependent protein kinase11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell. 2013;25:649–661. doi: 10.1105/tpc.112.103184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadota Y, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Berken A, Thomas C, Wittinghofer A. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature. 2005;436:1176–1180. doi: 10.1038/nature03883. [DOI] [PubMed] [Google Scholar]
- 25.Gu Y, Li S, Lord EM, Yang Z. Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell. 2006;18:366–381. doi: 10.1105/tpc.105.036434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Lavagi I. Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking. Curr Opin Plant Biol. 2012;15:601–607. doi: 10.1016/j.pbi.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schepetilnikov M, et al. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J. 2017;36:886–903. doi: 10.15252/embj.201694816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oda Y, Fukuda H. Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science. 2012;337:1333–1336. doi: 10.1126/science.1222597. [DOI] [PubMed] [Google Scholar]
- 29.Humphries JA, et al. ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell. 2011;23:2273–2284. doi: 10.1105/tpc.111.085597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoefle C, et al. A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell. 2011;23:2422–2439. doi: 10.1105/tpc.110.082131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan Q, Kita D, Li C, Cheung AY, Wu HM. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA. 2010;107:17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan Q, et al. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat Commun. 2014;5:3129. doi: 10.1038/ncomms4129. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, McCormick S. A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:18830–18835. doi: 10.1073/pnas.0705874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemichez E, et al. Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 2001;15:1808–1816. doi: 10.1101/gad.900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyawaki KN, Yang Z. Extracellular signals and receptor-like kinases regulating ROP GTPases in plants. Front Plant Sci. 2014;5:449. doi: 10.3389/fpls.2014.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, et al. ROP11 GTPase negatively regulates ABA signaling by protecting ABI1 phosphatase activity from inhibition by the ABA receptor RCAR1/PYL9 in Arabidopsis. J Integr Plant Biol. 2012;54:180–188. doi: 10.1111/j.1744-7909.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu F, et al. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proc Natl Acad Sci USA. 2012;109:14693–14698. doi: 10.1073/pnas.1212547109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng ZL, et al. Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell. 2002;14:2787–2797. doi: 10.1105/tpc.005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Kang J, Sui N, Liu D. ROP11 GTPase is a negative regulator of multiple ABA responses in Arabidopsis. J Integr Plant Biol. 2012;54:169–179. doi: 10.1111/j.1744-7909.2012.01100.x. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, Liu D. ROPGEF1 and ROPGEF4 are functional regulators of ROP11 GTPase in ABA-mediated stomatal closure in Arabidopsis. FEBS Lett. 2012;586:1253–1258. doi: 10.1016/j.febslet.2012.03.040. [DOI] [PubMed] [Google Scholar]
- 41.Li Z, Waadt R, Schroeder JI. Release of GTP exchange factor mediated down-regulation of abscisic acid signal transduction through ABA-induced rapid degradation of RopGEFs. PLoS Biol. 2016;14:e1002461. doi: 10.1371/journal.pbio.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harper JF, Breton G, Harmon A. Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol. 2004;55:263–288. doi: 10.1146/annurev.arplant.55.031903.141627. [DOI] [PubMed] [Google Scholar]
- 45.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: Kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 46.Bloch D, et al. Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol Biol Cell. 2005;16:1913–1927. doi: 10.1091/mbc.E04-07-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129:469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachmann M, et al. Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell. 1996;8:505–517. doi: 10.1105/tpc.8.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akamatsu A, et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe. 2013;13:465–476. doi: 10.1016/j.chom.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Chang F, Gu Y, Ma H, Yang Z. AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol Plant. 2013;6:1187–1201. doi: 10.1093/mp/sss103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Curran A, et al. Calcium-dependent protein kinases from Arabidopsis show substrate specificity differences in an analysis of 103 substrates. Front Plant Sci. 2011;2:36. doi: 10.3389/fpls.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeuchi H, Higashiyama T. Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature. 2016;531:245–248. doi: 10.1038/nature17413. [DOI] [PubMed] [Google Scholar]
- 53.Gao X, et al. Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 2013;9:e1003127. doi: 10.1371/journal.ppat.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheen J. Ca2+-dependent protein kinases and stress signal transduction in plants. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- 55.Boudsocq M, Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013;18:30–40. doi: 10.1016/j.tplants.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romeis T, Ludwig AA, Martin R, Jones JD. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001;20:5556–5567. doi: 10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J, et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:E5519–E5527. doi: 10.1073/pnas.1608449113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.