Significance
Urban, Westernized populations suffer extensively from noncommunicable diseases such as allergies. However, the overlapping effects of living environment and lifestyle are difficult to separate. Intriguingly, also our fellow animals, dogs, suffer from analogous diseases. Therefore, we suggest that pet dogs, sharing their environment and lifestyle with humans but having a comparatively simple life, provide a valuable model for understanding origins of noncommunicable diseases. We show that living environment and lifestyle concurrently, but still independently, shape both the skin microbiota and the risk of allergic disease in dogs. Urbanized lifestyle, featuring restricted animal contacts and small family size, is allergy promoting both in rural and urban dogs. Hence, both environment and lifestyle seem to influence the microbiota and, probably consequently, immune tolerance.
Keywords: microbiome, biodiversity hypothesis, canine model, allergy, veterinary
Abstract
A rural environment and farming lifestyle are known to provide protection against allergic diseases. This protective effect is expected to be mediated via exposure to environmental microbes that are needed to support a normal immune tolerance. However, the triangle of interactions between environmental microbes, host microbiota, and immune system remains poorly understood. Here, we have studied these interactions using a canine model (two breeds, n = 169), providing an intermediate approach between complex human studies and artificial mouse model studies. We show that the skin microbiota reflects both the living environment and the lifestyle of a dog. Remarkably, the prevalence of spontaneous allergies is also associated with residential environment and lifestyle, such that allergies are most common among urban dogs living in single-person families without other animal contacts, and least common among rural dogs having opposite lifestyle features. Thus, we show that living environment and lifestyle concurrently associate with skin microbiota and allergies, suggesting that these factors might be causally related. Moreover, microbes commonly found on human skin tend to dominate the urban canine skin microbiota, while environmental microbes are rich in the rural canine skin microbiota. This in turn suggests that skin microbiota is a feasible indicator of exposure to environmental microbes. As short-term exposure to environmental microbes via exercise is not associated with allergies, we conclude that prominent and sustained exposure to environmental microbiotas should be promoted by urban planning and lifestyle changes to support health of urban populations.
Our health tends to be affected by our living environment, green space being the promoter of our well-being (1). Supporting evidence is provided by studies showing that allergies are increasing due to urbanization, especially in Western countries, while allergies remain rarer in developing countries as well as in rural areas (2, 3). As the urban and rural environmental microbiota, i.e., the diverse microbial community in a defined ecosystem, differ (4), it is suggested that increased exposure to microbes from natural environments, directly or indirectly through animals or family members, is protective against the development of allergic diseases (5). This idea is further strengthened by findings that the microbiota differs between healthy and allergic people (reviewed in ref. 6). While there is good evidence for associations between the living environment and allergic diseases (7, 8) as well as between host microbiota and allergies (e.g., refs. 6 and 9), it is unclear whether all these factors are related, and if so, how.
Unfortunately, a multitude of confounding factors associated with human life, such as a constant change both in lifestyle patterns and in living areas due to moving and traveling, makes it difficult to establish a causal relationship between living environment and lifestyle and health (8). To overcome this problem, several studies linking allergies to microbiota have been carried out in the mouse model (e.g., refs. 10 and 11). However, controlled laboratory experiments are not ideal for studying the effect of complex living environments, as they are practically impossible to replicate in artificial conditions. In an attempt to establish a feasible compromise between problematic human studies and overly simplistic laboratory models, we adopted a canine model (pet dogs). We consider dogs to be relevant model animals, as they suffer increasingly from diseases analogous to inflammatory disorders in humans, such as allergies (e.g., in ref. 12). Importantly, dogs develop these diseases spontaneously unlike in a mouse model. Also, dogs share the living environment with and are influenced by the lifestyle of their owners. However, dogs still live simpler and shorter lives than humans, excluding several confounding factors common in studies involving human subjects.
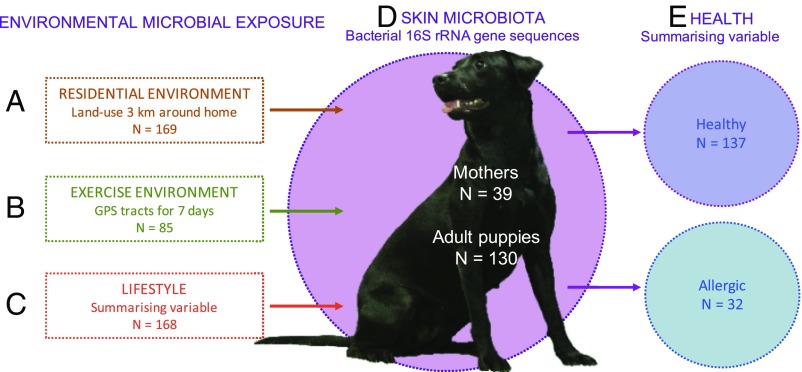
Our aim was to describe associations between the exposure to environmental microbes, skin microbiota, and allergic symptoms, utilizing the canine model. Skin microbiota was selected instead of widely studied gut microbiota, because it has more immediate contact with the environment. Recent findings also suggest that the skin microbiota can have a systemic influence on immune tolerance (10, 13, 14). We defined three factors concurrently influencing the exposure of the skin to environmental microbes: (i) land use in the residential environment, (ii) contact with different land-use types during exercise, and (iii) lifestyle of the dog owner (Fig. 1). We tested how these factors relate to the skin microbiota, characterized by bacterial 16S rRNA gene sequences, and allergies, quantified by a large questionnaire developed by canine dermatologists. We also tested the relation of residential environment at the time of birth with skin microbiota and allergies in adult dogs.
Fig. 1.
Exposure to environmental microbes is affected by three factors: where one lives (A), where one moves (B), and how one lives (C). We quantified these factors and studied their effects on the skin microbiota (D) and allergic symptoms (E) in dogs. The biodiversity hypothesis (42) suggests that the exposure to natural environments defines our microbial exposure (A–C), which in turn affects the composition of individual microbiota (D), and can relate to the development of inflammatory disorders (E) through immune modulation. Image courtesy of Aki Korhonen and Varpu Halonen (Foto Elukka, Lievestuore, Finland).
Results
Exposure to Environmental Microbes Shapes the Skin Microbiota.
All three factors characterizing the total exposure of a dog to environmental microbes—i.e., residential environment, exercise environment, and lifestyle (Fig. 1)—were related to skin microbiota. Interestingly, microbial communities tended to cluster according to both the residential environment and lifestyle (Fig. 2), while the effect of exercise environment was marginally significant (SI Appendix, Fig. S1). In other words, dogs living in an urban environment, exposed to urban lifestyle (i.e., living in a single-person family without other pets and having many dog-related hobbies such as agility or tracking, SI Appendix, Fig. S2), differed in their skin microbiota from dogs living in the same environment but having owners with a rural lifestyle (i.e., living in a large family with other pets, SI Appendix, Fig. S2). However, it is important to note that these three explanatory variables were somewhat correlated (SI Appendix, Fig. S3), which makes interpretation challenging.
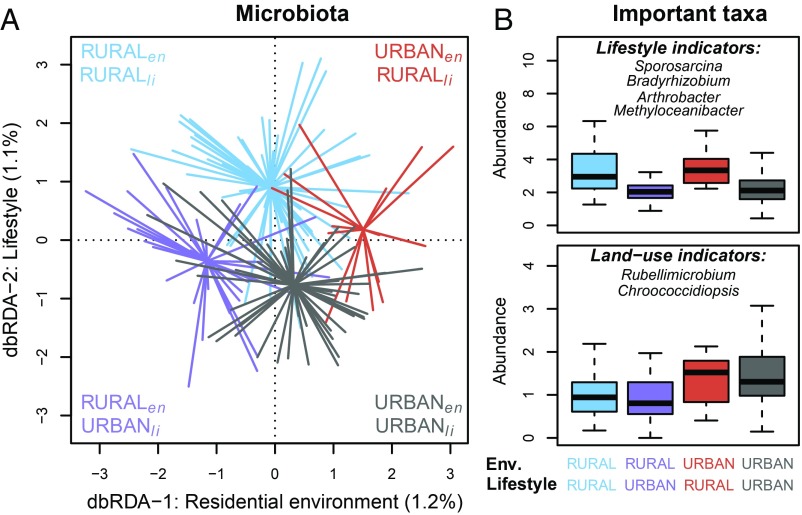
Fig. 2.
Land use in residential environment (Env) and lifestyle of dogs is associated with skin microbial communities. (A) The distance-based redundancy analysis (db-RDA) of the skin microbiota constrained by the rural–urban categories of residential environment (P = 0.0006) and lifestyle (P = 0.0024). Each line represents the distance of microbiota from a centroid of a group of an individual dog. Different colors indicate dogs as follows: blue, rural dog with rural lifestyle; purple, rural dog with urban lifestyle; red, urban dog with rural lifestyle; and gray, urban dog with urban lifestyle. Footnotes “en” and “li” mark residential environment and lifestyle, respectively. (B) Tukey boxplots of summed and square root transformed abundance (normalized counts) of taxa, which best predicted groups in A in an RF analysis. Outlier data points are not shown. Colors of the boxplots correspond to the groups in A. These groups were used to visualize the clustering of dogs’ microbiota due to the effect of constraining variables.
While the effects of residential environment and lifestyle were partly overlapping (these variables were correlated; r = 0.48, P = 3.3e–11), they also tended to be associated with independent aspects of the skin microbiota, as can be seen in Fig. 2 with separate but partly overlapping groups. More specifically, the first axis of principal component analysis (PCA1) of the land-use types correlated slightly but significantly with the second axis of principal coordinates analysis (PCoA2) of skin microbiota (Pearson’s correlation: −0.36, Padj = 0.00001). Instead, PCoA1 of lifestyle correlated with PCoA4 of skin microbiota (Pearson’s correlation: 0.25, Padj = 0.003). While these correlations were low, they indicate that lifestyle (i.e., active exposure) and the residential environment (i.e., passive exposure) independently influenced the composition of the skin microbial community.
Rural lifestyle and environment were associated with the increasing abundance of microbes from environmental sources on skin microbiota (Fig. 2B). In contrast, both urban lifestyle and urban living environment were associated with an enrichment of taxa often found in built environments, such as Chroococcidiopsis (Fig. 2B). In addition, (human) skin-related taxa such as Propionicicella and Friedmanniella were enriched on dog skin in an urban environment, which, in turn, was associated with a homogenization of urban dog microbiota (the within-group dissimilarity of skin microbiota was smaller in urban dogs than in rural dogs; P = 0.0001). On the contrary, urban lifestyle seemed to increase within-group dissimilarity (P = 0.0026).
Allergic Symptoms Associated with Exposure to Environmental Microbes and Skin Microbiota.
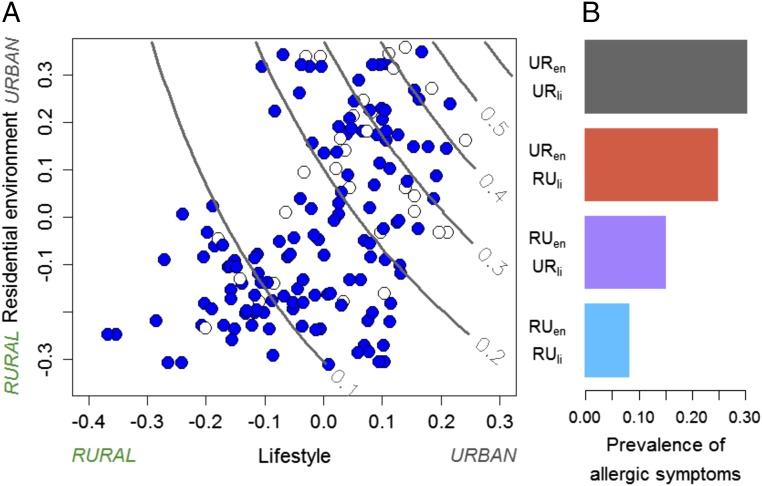
In addition to microbial composition, land use and lifestyle were also associated with the prevalence of allergic symptoms in dogs (Fig. 3), while the exercise environment was not associated with allergic symptoms. Allergies were most common in dogs living in urban environments and having an urban lifestyle, whereas the lowest prevalence was associated with rural environments and rural lifestyle (Fig. 3A). Urban-type lifestyle seemed almost to double the prevalence of allergies in dogs living in rural environments compared with dogs with rural-type lifestyle in rural environments (Fig. 3B). Furthermore, not just the prevalence of allergic symptoms (P = 0.006) but also the severity of the symptoms was positively associated with urban lifestyle (P = 0.0001). We also tested classical biomarkers of allergy and inflammation, Immunoglobulin E (IgE) and C-reactive protein (CRP) (SI Appendix, Table S1), against environment and lifestyle, but we did not discover any robust associations. Moreover, the severity and presence of allergic symptoms were not associated with IgE or CRP levels.
Fig. 3.
Residential environment and lifestyle together shaped the prevalence of allergic symptoms in dogs. (A) The prevalence of allergy (lines), predicted from data. Red and light blue symbols indicate allergic and healthy dogs, respectively. (B) The prevalence of allergic symptoms in dogs having dissimilar combinations of living environment and lifestyle (rural environment, RUen; rural lifestyle, RUli; urban environment, URen; and urban lifestyle, URli).
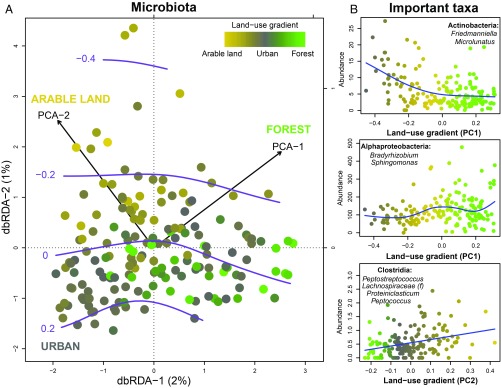
Interestingly, the predicted prevalence of allergic symptoms and the skin microbiota changed concurrently along the land use of the residential environment (Fig. 4A). An increasing area of forest and arable land correlated with a greater amount of different Proteobacteria and other soil-related taxa on skin. In urban areas, in turn, general skin-related bacteria such as Actinobacteria, especially of the genera Friedmanniella and Microlunatus (Propionibacterales), were enriched on skin (Fig. 4B). In other words, these results suggest that the living environment, skin microbiota, and allergic diseases were interrelated. This was also supported by both differing skin microbiota and prevalence of allergies in groups based on residential environment and lifestyle (Figs. 2A and 3B).
Fig. 4.
Living environment is associated with skin microbiota and the prevalence of allergies. (A) Distance-based redundancy analysis (db-RDA) of canine microbiota uses land-use gradients (PC1 and PC2) as explanatory variables to constrain microbial dissimilarity. Shown are skin microbiota changes along the land-use gradient from forested to urban areas (P = 0.001 for PC1) and from arable lands to forested areas (P = 0.002 for PC2). Each dot corresponds to microbial community of an individual dog. A color gradient of dots marks the proportion of different land-use types in the residential environment of an individual dog. Purple lines mark the predicted severity of allergic symptoms showing increase along the urbanization. (B) Abundance of skin microbial taxa along the land-use gradients. The appointed taxa in are among those that best explained the corresponding land-use gradients in the RF analysis. In each panel, a dot marks the summed abundance of appointed taxa in microbiota of individual dogs. The abundance of Clostridia in the lowest panel was square root transformed. The color gradient of dots marks the proportion of different land-use types in the residential environment of an individual dog as in A. Blue lines demonstrate the fitted values.
The microbial communities of allergic dogs resembled each other more than the communities of healthy dogs (P = 0.0001), indicating commonalities in the skin microbiota in allergic individuals. However, the variation among dogs was still so high that no clear key taxa in microbiota of allergic dogs could be identified in the random forest (RF) analysis, apart from the enrichment of human-derived skin microbiota. However, the summed abundance of microbes relevant in the skin microbiota of urban individuals (defined by the RF analysis of land use against microbiota) was significantly higher in allergic dogs, but individual-level variation remained large (SI Appendix, Fig. S4).
Early-Life Environment Partly Associated with the Current Skin Microbiota and Allergic Symptoms.
Interestingly, we found that the skin microbiota differed between dogs belonging to healthy and allergic dog families (i.e., dog families with only healthy puppies vs. at least one allergic puppy, P = 0.0156; SI Appendix, Fig. S5A). This compositional difference seemed to relate to both early-life and current residential environment. Microbiota of dogs belonging to healthy dog families was significantly associated with increasing area of arable land and forest in the surroundings of the birthplace (P = 0.0007) and current home (P = 0 0.0003; SI Appendix, Fig. S5A). For example, several operational taxonomic units (OTUs) from the genus Acinetobacter were more abundant in agricultural early-life environment and were differently, although not significantly, abundant in allergic and healthy dog families (SI Appendix, Fig. S5B). Also, among allergic dog families, human skin-related Actinobacteria (genus Friedmanniella) was more abundant than in healthy dog families.
The first weeks of life also had an influence on the prevalence of allergies. Allergies seemed to be most common in dogs that had spent their entire life in an urban environment, and rarest among dogs that were born and had lived only in rural environments (SI Appendix, Fig. S5C). It is noteworthy that dogs from healthy dog families, those born and living in rural environments, had a similar skin microbial composition (i.e., “the triangle of healthy individuals” delineated by the land-use axes in SI Appendix, Fig. S5A). Mothers and their puppies tended to be more similar in their microbial composition than random pairs between adults (dog mothers) and puppies (P = 0.0001). Additionally, the dissimilarity between dogs was smaller in allergic than in healthy dog families, indicating, again, the homogenization of skin microbiota (SI Appendix, Fig. S5D, P = 0.0001). These findings suggest that puppies receive bacteria from the early-life living environment (most likely via their mother), which can affect their future health. However, we were unable to exclude the effect of genetic predisposition on the prevalence of allergies, which can be important as an allergic mother dog more likely had allergic puppies than a healthy mother (P = 7.5e–5).
Discussion
Our results suggest a vital role of residential environment and lifestyle in dog health. Exposure to environmental microbes, both passively (via the living environment i.e., where one is) and actively (through lifestyle i.e., what one does), related to the composition of skin bacterial communities. Remarkably, both passive and active exposure were also associated with the prevalence of allergies, as shown previously (15). Finally, the composition of skin microbiota differed in healthy and allergic dogs. Therefore, our results suggest a triangle of associations between skin microbiota, allergies, and exposure to environmental microbes. Recent studies trying to relate the living environment and allergic disease have produced somewhat contrasting results, most likely due to several confounding factors associated with complexities of human lifestyles (8). While previous studies on the relationships between the living environment, microbiota, and health have failed to find a robust link between environment-associated patterns in microbiota and allergic sensitization (3, 16), our simplified canine model has provided explicit empirical evidence of such interaction, as predicted by the biodiversity hypothesis of allergic diseases (5).
The combined effect of passive (living environment) and active (lifestyle) contact with environmental microbes on allergies in humans or dogs has previously received little attention. However, the contrasting allergy prevalence in farm and nonfarm children in rural areas (17), and in children predisposed to traditional and modern farming (18) suggest that active contact to the environment is important. Here we show that dogs living in urban environments and exposed to urban-type lifestyle—characterized by various hobbies and by living in apartments and in a single-person family without other pets—are most susceptible to allergic diseases. In opposite circumstances, i.e., when dogs are living in rural environments with a large family and frequent animal contacts, their allergies are rare. Interestingly, an urban-type lifestyle almost doubled the prevalence of allergies in dogs living in rural environments compared with dogs with rural-type lifestyle in rural environments. Furthermore, the skin microbiota of dogs differed between all combinations of rural and urban residential environment and lifestyle, suggesting that microbiota can have a role in the differing allergy prevalence in each group. This assumed interplay between microbial exposure, host microbiota, and allergies, which rises from findings here but also from previous work (3, 16–18), highlights the importance of natural environments for health. It also suggests that the exposure to a diverse microbial world can be a protective factor both in humans and dogs.
The effect of lifestyle on the skin microbiota has been suspected previously, but “lifestyle” is difficult to define. A recent study found that the living environment clearly differentiated the skin microbiota of children, but the effect disappeared in teenagers, which was likely due to shared lifestyle patterns regardless of living environment in this age group (16). Our findings highlight that where a dog lives and what a dog does can jointly influence microbiota and health. Surprisingly, the exercise environment (where a dog moves) was only marginally related to skin microbiota and did not associate with allergies. Obviously, living environment and lifestyle are more sustained factors than exercise environment, which may change on a day-to-day basis and the exposure lasts only for short periods of time. Hence, we suggest that prominent, sustained exposure is needed for health-promoting changes in skin microbiota to arise or last.
We observed that certain common members of the human skin microbiota, for example Actinobacteria, were enriched in urban canine skin microbiota. On the contrary, soil-related and other microbes from environmental sources, such as Bradyrhizobium (Alphaproteobacteria) (19), were abundant in the rural skin microbiota, indicating that skin microbiota is a relevant indicator of exposure to environmental microbes. Moreover, the urban dogs were more alike than the rural ones, which indicates homogenization of skin microbiota in urban environments. Our observations resonate with previous research, showing that urban environmental microbiota is more homogenous than rural environmental microbiota (4), and that inside a city, the built areas are homogenous compared with parks (20). Moreover, microbiota in closed indoor spaces, such as apartments and subways, are dominated by human skin-origin microbes (21, 22). Interestingly, human-derived microbes were enriched in allergic dogs compared with healthy dogs and that the skin microbiota in allergic dogs was more alike than in healthy dogs, pointing again to microbial homogenization. These findings indicate that either certain rural-dwelling microbes or a constant exposure to heterogeneous environmental microbial communities, or both, support normal immune function.
Previous studies have demonstrated the importance of early-life exposures in the development of allergic diseases in humans. The composition of gut microbiota during the first weeks of life tends to differ between those who do and do not develop allergies later on refs. 23 and 24. Importantly, there seems to be a limited window of opportunity, during which immune education by microbiota is most decisive (24). When we studied microbiota of individuals from healthy and allergic dog families, we found that in addition to the current living environment, early-life environment was associated with later composition of skin microbiota. Excitingly, we observed that the genus Acinetobacter was more prevalent in healthy dog families. These bacteria have previously been associated with protection against allergies in humans (25–27) and in a mouse model (10, 28, 29). In allergic dog families, human skin-related Friedmanniella was also more prevalent. Both genera associated with land use in early-life environment, demonstrating that early-life environment can influence the microbiota. Nevertheless, a dog’s current environment seems to be more important in development of allergy even though early environment also leaves a mark. The skin microbiota was more homogenous in allergic than in healthy dog families, which can be a primary driver in the development of allergy. Based on these observations, we propose that lack of certain early-life exposure may create a susceptibility to develop allergy, but that later lifestyle and exposure can change the final outcome of allergy in dogs.
Because dogs move from their breeder to the owner at a young age (7–8 wk old), the window of opportunity regarding immune development may still be open at that time. This complicates the interpretation of importance of early versus later life exposure. Previous studies have reported associations between the environment and allergies in dogs (12, 30), but early-life environment was not important (15). Another limitation is that canine allergy is not a well-defined disease. Allergies in dogs are commonly clustered to food allergies (FAs), canine atopic dermatitis (CAD), while allergic respiratory diseases are absent (31); however, the symptoms in FAs and CAD are overlapping (32). Neither can the diagnosis be based on immunological biomarkers, such as IgE, as these do not clearly associate with symptoms, and thus their role in canine allergies is widely debated (33, 34). Our results echoed this as we found no association between health status or severity of clinical signs and these biomarkers. Due to this lack of clarity, we have used a single measure of allergy, indicating the severity of any kind of allergic symptoms common in dogs, regardless of their origin. We were unable to exclude the effect of genetic predisposition to allergic diseases, which was concluded to be important in the development of CAD (together with environmental factors) in recent reviews (35, 36). Also, in our data the Labrador retrievers were more allergic than the Finnish lapphunds, indicating the importance of genetics (12). However, the patterns we found in microbiota and the prevalence of allergies were similar in both breeds, indicating that the environmental component operates similarly regardless of the genetic background.
We have demonstrated here that pet dogs can be a convenient model for understanding the relation between living environment and inflammatory diseases. While free of most confounding factors associated with human life, pet dogs suffer increasingly from inflammatory diseases similar to those in humans, and they share the environment and lifestyle with their owners. This means that our results are likely to apply also to humans. However, the validation of this canine model requires comparative research efforts on dog and human microbiota and health. Finally, we want to emphasize the need for longitudinal or interventional studies, focusing on the triangle of interactions between health, microbiota, and the exposure to environmental microbes. Future studies should concentrate on better understanding the right timing, quality, and duration of exposure to environmental microbes, as well as the contribution of the microbiota in different body parts to health. Experimental studies should follow these, to confirm causality that cannot be established in our cross-sectional study. We suggest that model animal-based research is needed to speed up the definition process of optimal exposure, with pet dogs serving as a valuable real-life model with spontaneous allergy.
Materials and Methods
Data Collection.
Breeders of either Labrador retrievers or Finnish lapphunds were invited in the study. These breeds are equally common in rural and urban areas and their large size eases blood sampling. The owners of puppies from selected litters were recruited via the breeders. Our sample included 169 dogs (39 were mother dogs). Skin microbial (swab) samples were taken from the inner side of the front leg, at about carpus level (3 × 3 cm2 area; SI Appendix, SI Methods). Next, DNA was extracted and the V1–V3 region of 16S rRNA gene was sequenced as previously described (16). Blood serum was collected for the analysis of IgE and CRP. Owners filled out a questionnaire (SI Appendix, Form S1), concerning their dogs’ environment, lifestyle, and allergic symptoms. A subset of owners (n = 96) followed their dogs’ movement for a week with a passive tracker (iTrail, SleuthGear). Details about participants and data collection are provided in SI Appendix, SI Methods.
Bioinformatic Analysis.
The processing of 16S rRNA gene sequences, formation of OTUs, and taxonomic classification are described in SI Appendix, SI Methods. The processed sequence counts were normalized with cumulative-sum scaling (37) and contaminant OTUs were removed as described in SI Appendix, SI Methods. Finally, our data included 45,905 normalized OTUs that were used in further statistical analysis. The large number of OTUs is explained by the strict similarity threshold we used. All 16S rRNA gene sequences have been deposited in the National Center for Biotechnology Information (accession no. SRP 133457).
Simplification of Complex Data.
The quantification of residential (birth and current) and exercise environment land use was done as described in Lehtimäki et al. (16), using PCA. From each PCA, the first or second component, or both, were extracted for further analysis (SI Appendix, Table S2). Higher values along the first axis indicated more rural, especially forested areas, while lower values indicated a higher proportion of urban such as built environments (SI Appendix, Table S2). Higher values along the second axis indicated a transition from forested areas to agricultural areas.
As no single question regarding allergic symptoms was considered a reliable measurement of canine allergy, these questions (questions 5.1–6.14, SI Appendix, Form S1), containing several mutually correlating variables, were summarized to a single continuous variable using PCoA (based on Gower’s distance, accepting a mixture of numeric and categorical variables). The first PCoA axis captured the variation in the severity of allergic symptoms, with higher values indicating more severe symptoms. Similarly, we selected a set of family- and lifestyle-related questions (marked with * in SI Appendix, Form S1), which, according our knowledge, influence how dogs are exposed to environmental microbes, to create a lifestyle variable. The first PCoA axis allocated dogs according to the lifestyle of the owner, which showed a gradient from rural to urban lifestyle. Briefly, rural lifestyle correlated with living in bigger (human) families, having more animal contacts, and living in town houses, while urban lifestyle correlated with having many hobbies, traveling a lot, and living in an apartment (SI Appendix, Fig. S2).
The resulting continuous variables were also categorized. If the location of the dog’s current home or exercise environment was loaded positively (or negatively) on the PCA1 axis of land use, the dog was classified as rural (or urban). Similarly, if the score of the PCoA1 component of lifestyle was positive (or negative), the corresponding dog was classified as having an urban-type (or rural-type) lifestyle (SI Appendix, Fig. S2). Finally, a dog was defined allergic if its corresponding value along the PCoA1 axis was higher than 0.5 (a seemingly natural cutoff point based on the distribution of scores). A dog family (i.e., mother and its puppies) was defined allergic if at least one of the puppies in the litter was allergic (families with only one puppy were excluded, n = 6).
The Analysis of Relationships.
Between-sample similarity of canine microbiota was calculated using the Bray–Curtis dissimilarity index (not sensitive to shared absence of OTUs between samples). Dissimilarity was analyzed against explanatory variables with multiple regression on distance matrices (MRM) (vegan package in R, ref. 38). To study relationships between microbiota, exposure to microbes, and allergic symptoms, we used PCoA and distance-based redundancy analysis (db-RDA) in the vegan package (38). First, we determined whether the current residential environment and lifestyle were associated with microbiota. The rural–urban categorical variables of environment and lifestyle were used to constrain the microbial dissimilarity in db-RDA. These categories were also used in visualization of the groups in db-RDA ordination. Pearson’s correlations between the principal coordinates of microbiota and land use as well as lifestyle were estimated. Second, to study the effect of current residential land use on microbial composition, we used both PCA1 and PCA2 of the land use as explanatory variables in db-RDA. The severity of allergic symptoms was fitted on top of the ordination, assuming a Gaussian error distribution. Finally, we used db-RDA to assess microbial compositional difference among dog families and the effect of early-life environment on microbiota. In this analysis, PCA2 of land use in early-life environment and the categorization of healthy and allergic families were explanatory variables.
In support of each db-RDA, we used RF analysis (randomForest package in R; ref. 39) to specify those genera or OTUs that best predict environmental land use and lifestyle. If analysis was based on genera, such as in Figs. 2 and 4, the OTUs belonging to the same genus were merged prior to analysis. In RF analysis based on OTUs as in SI Appendix, Fig. S5, OTUs with greater abundance than third quartile of all OTUs were considered. All analyses were done in R version 3.3.2 (40). The significance level was set to P < 0.05. In the case of multiple comparisons, P values were adjusted using the false discovery rate (FDR) method (41) in R.
Ethics Statement.
Sample collection was ethically approved, concerning dogs, by the Animal Ethics Committee of the State Provincial Office of Southern Finland, Hämeenlinna, Finland (ESAVI/6054/04.10.03/2012), and concerning dog owners, by the Coordinating Ethics Committee, University of Helsinki Central Hospital (188/13/03/00/14). Sample collection and all subsequent experimental procedures were conducted in accordance with relevant guidelines and regulations. Before the sampling, we asked the owner of each dog to provide a signed informed consent.
Supplementary Material
Acknowledgments
We thank our technical assistants Sanna Karumaa and especially Petra Jaakonsaari; nurses Tanja Ekholm-Venäläinen and Nina Voutilainen; research assistants Tuuli Laukkanen, Emma Hakanen, Anette Lehtola, Emma Thiz, and Eini Nieminen; laboratory staff Laura Lund, Annukka Ruokolainen, Laura Häkkinen, Toscha Nyman, Tomi Issakainen, and Elina Poutanen; and especially all dogs and their owners who used their time to participate in this study. This study was funded by the Jane and Aatos Erkko Foundation, the University of Helsinki (ERCStG 260997), the Academy of Finland (268019 and 286405), Biocentrum Helsinki, and the Jenny and Antti Wihuri Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All 16S rRNA-gene sequences have been deposited in the National Center for Biotechnology Information (accession no. SRP133457).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719785115/-/DCSupplemental.
References
- 1.van den Berg M, et al. Health benefits of green spaces in the living environment. A systematic review of epidemiological studies. Urban For Urban Green. 2015;14:806–816. [Google Scholar]
- 2.Von Hertzen LC, Haahtela T. Asthma and atopy:–The price of affluence? Allergy. 2004;59:124–137. doi: 10.1046/j.1398-9995.2003.00433.x. [DOI] [PubMed] [Google Scholar]
- 3.Ruokolainen L, et al. Green areas around homes reduce atopic sensitization in children. Allergy. 2015;70:195–202. doi: 10.1111/all.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barberán A, et al. Continental-scale distributions of dust-associated bacteria and fungi. Proc Natl Acad Sci USA. 2015;112:5756–5761. doi: 10.1073/pnas.1420815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haahtela T, et al. WAO Special Committee on Climate Change and Biodiversity The biodiversity hypothesis and allergic disease: World allergy organization position statement. World Allergy Organ J. 2013;6:3. doi: 10.1186/1939-4551-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang YJ, et al. The microbiome in allergic disease: Current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J Allergy Clin Immunol. 2017;139:1099–1110. doi: 10.1016/j.jaci.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruokolainen L, Fyhrquist N, Haahtela T. The rich and the poor: Environmental biodiversity protecting from allergy. Curr Opin Allergy Clin Immunol. 2016;16:421–426. doi: 10.1097/ACI.0000000000000304. [DOI] [PubMed] [Google Scholar]
- 8.Ruokolainen L. Green living environment protects against allergy, or does it? Eur Respir J. 2017;49:1700481. doi: 10.1183/13993003.00481-2017. [DOI] [PubMed] [Google Scholar]
- 9.Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 10.Fyhrquist N, et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J Allergy Clin Immunol. 2014;134:1301–1309.e11. doi: 10.1016/j.jaci.2014.07.059. [DOI] [PubMed] [Google Scholar]
- 11.Schaub B, Vercelli D. Environmental protection from allergic diseases: From humans to mice and back. Curr Opin Immunol. 2015;36:88–93. doi: 10.1016/j.coi.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Nødtvedt A, Egenvall A, Bergvall K, Hedhammar A. Incidence of and risk factors for atopic dermatitis in a Swedish population of insured dogs. Vet Rec. 2006;159:241–246. doi: 10.1136/vr.159.8.241. [DOI] [PubMed] [Google Scholar]
- 13.Prescott SL, et al. The skin microbiome: Impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ J. 2017;10:29. doi: 10.1186/s40413-017-0160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues Hoffmann A. The cutaneous ecosystem: The roles of the skin microbiome in health and its association with inflammatory skin conditions in humans and animals. Vet Dermatol. 2017;28:60–e15. doi: 10.1111/vde.12408. [DOI] [PubMed] [Google Scholar]
- 15.Hakanen E, et al. Urban environment predisposes dogs and their owners to allergic symptoms. Sci Rep. 2018;8:1585. doi: 10.1038/s41598-018-19953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehtimäki J, et al. Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Sci Rep. 2017;7:45651. doi: 10.1038/srep45651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun-Fahrländer C, et al. Swiss Study on Childhood Allergy and Respiratory Symptoms with Respect to Air Pollution Prevalence of hay fever and allergic sensitization in farmer’s children and their peers living in the same rural community. SCARPOL team. Clin Exp Allergy. 1999;29:28–34. doi: 10.1046/j.1365-2222.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- 18.Stein MM, et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanInsberghe D, et al. Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J. 2015;9:2435–2441. doi: 10.1038/ismej.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mhuireach G, et al. Urban greenness influences airborne bacterial community composition. Sci Total Environ. 2016;571:680–687. doi: 10.1016/j.scitotenv.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 21.Lax S, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afshinnekoo E, et al. Geospatial resolution of human and bacterial diversity with city-scale metagenomics. Cell Syst. 2015;1:72–87. doi: 10.1016/j.cels.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrahamsson TR, et al. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434–440, 440.e1-2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 24.Arrieta M-C, et al. CHILD Study Investigators Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 25.Hanski I, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci USA. 2012;109:8334–8339. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ege MJ, et al. Environmental bacteria and childhood asthma. Allergy. 2012;67:1565–1571. doi: 10.1111/all.12028. [DOI] [PubMed] [Google Scholar]
- 27.Ruokolainen L, et al. Significant disparities in allergy prevalence and microbiota between the young people in Finnish and Russian Karelia. Clin Exp Allergy. 2017;47:665–674. doi: 10.1111/cea.12895. [DOI] [PubMed] [Google Scholar]
- 28.Brand S, et al. Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. J Allergy Clin Immunol. 2011;128:618–625.e1-7. doi: 10.1016/j.jaci.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 29.Debarry J, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119:1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 30.Meury S, et al. Role of the environment in the development of canine atopic dermatitis in Labrador and golden retrievers. Vet Dermatol. 2011;22:327–334. doi: 10.1111/j.1365-3164.2010.00950.x. [DOI] [PubMed] [Google Scholar]
- 31.Pucheu-Haston CM. Atopic dermatitis in the domestic dog. Clin Dermatol. 2016;34:299–303. doi: 10.1016/j.clindermatol.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Verlinden A, Hesta M, Millet S, Janssens GPJ. Food allergy in dogs and cats: A review. Crit Rev Food Sci Nutr. 2006;46:259–273. doi: 10.1080/10408390591001117. [DOI] [PubMed] [Google Scholar]
- 33.Griot-Wenk ME, et al. Total serum IgE and IgA antibody levels in healthy dogs of different breeds and exposed to different environments. Res Vet Sci. 1999;67:239–243. doi: 10.1053/rvsc.1999.0314. [DOI] [PubMed] [Google Scholar]
- 34.Lauber B, et al. Total IgE and allergen-specific IgE and IgG antibody levels in sera of atopic dermatitis affected and non-affected Labrador- and Golden retrievers. Vet Immunol Immunopathol. 2012;149:112–118. doi: 10.1016/j.vetimm.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Bizikova P, et al. Review: Role of genetics and the environment in the pathogenesis of canine atopic dermatitis. Vet Dermatol. 2015;26:95–e26. doi: 10.1111/vde.12198. [DOI] [PubMed] [Google Scholar]
- 36.Craig JM. Atopic dermatitis and the intestinal microbiota in humans and dogs. Vet Med Sci. 2016;2:95–105. doi: 10.1002/vms3.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oksanen J, et al. 2016 vegan: Community Ecology Package. R package Version 2.3-2. Available at https://cran.r-project.org/web/packages/vegan/index.html. Accessed September 10, 2016.
- 39.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 40.R Core Team 2016 R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna). Available at https://www.R-project.org/. Accessed September 10, 2016.
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 42.von Hertzen L, Hanski I, Haahtela T. Natural immunity. Biodiversity loss and inflammatory diseases are two global megatrends that might be related. EMBO Rep. 2011;12:1089–1093. doi: 10.1038/embor.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.