Significance
A given protease typically degrades multiple substrates that serve different functions and may be required in variable amounts under specific cellular conditions. Some substrates require adaptor proteins to be delivered to the protease. We find that changes in the abundance of an adaptor can lead to differential degradation of substrates that have different affinities for the adaptor. Thus, adaptor amounts can have a broad impact, as illustrated by the increase in antibiotic persister bacteria when the abundance of the ClpS adaptor is decreased. Specific growth conditions decrease ClpS abundance decreasing proteolysis of a subset of ClpSAP substrates.
Keywords: ClpS, ClpAP, cytoplasmic Mg2+, PhoP, proteolysis
Abstract
ATP-dependent proteases control critical cellular processes, including development, physiology, and virulence. A given protease may recognize a substrate directly via an unfoldase domain or subunit or indirectly via an adaptor that delivers the substrate to the unfoldase. We now report that cells achieve differential stability among substrates of a given protease by modulating adaptor amounts. We establish that the regulatory protein PhoP represses transcription of the gene specifying the ClpAP protease adaptor ClpS when the bacteria Salmonella enterica and Escherichia coli experience low cytoplasmic Mg2+. The resulting decrease in ClpS amounts diminishes proteolysis of several ClpSAP-dependent substrates, including the putrescine aminotransferase Oat, which heightens the formation of antibiotic persisters, and the transcriptional regulators UvrY and PhoP, which alter the expression of genes controlled by these proteins. By contrast, the decrease in ClpS amounts did not impact the abundance of the ClpSAP substrate FtsA, reflecting that FtsA binds to ClpS more tightly than to UvrY and PhoP. Our findings show how physiological conditions that reduce adaptor amounts modify the abundance of selected substrates of a given protease.
ATP-dependent proteases are present in all domains of life. They are necessary for regulated proteolysis and for the elimination of nonfunctional and potentially toxic proteins (1). ATP-dependent proteases consist of two subunits or domains: one that uses the energy derived from ATP hydrolysis to unfold and shuttle substrates into the other subunit or domain where proteolysis actually takes place (1). Degradation of particular substrates of a given protease often requires adaptor proteins to recognize such substrates and deliver them to the unfoldase (2, 3). We now report that enteric bacteria achieve differential degradation of substrates of the ATP-dependent protease ClpAP by altering the amounts of the adaptor protein ClpS under specific stress conditions.
The protease ClpAP is composed of the serine protease ClpP and the unfoldase ClpA (4). ClpA recognizes certain substrates directly but requires the adaptor ClpS to recognize others, most conspicuously those bearing leucine, tyrosine, tryptophan, or phenylalanine as the N-terminal residue. This makes ClpS the enforcer of the N-end rule degradation pathway in bacteria, chloroplasts, and mitochondria (2, 5, 6). ClpS is also required for proteolysis of the Salmonella PhoP protein even though PhoP’s N-terminal amino acid is a methionine that is neither removed nor modified (7). In addition, ClpS inhibits the degradation of a ClpAP substrate by hindering substrate access to ClpA (8). Given that some ClpAP substrates have regulatory roles, a change in the number of ClpS molecules has the potential of altering the abundance of multiple proteins.
The transcriptional regulator PhoP and the sensor PhoQ form a two-component system that governs Mg2+ homeostasis (9, 10) and virulence (11–14) in several Gram-negative species. PhoQ responds to low Mg2+ (15) and to certain antimicrobial peptides (16) in the periplasm and to a mildly acidic pH (17) in the cytoplasm (18) by activating the DNA-binding protein PhoP. In Salmonella enterica serovar Typhimurium, PhoP regulates transcription of ∼9% of the genome (19). PhoP-activated genes include mgtC, which specifies an inhibitor of the F1F0 ATPase (20), and mgtA and mgtB, which specify sequelogous (21) Mg2+ transporters (22, 23).
The mRNAs corresponding to the mgtA gene and mgtCB operon include unusually long leader sequences that prevent transcription elongation into the associated coding regions unless specific conditions are met, such as a drop in cytoplasmic Mg2+ below a certain threshold (24, 25). By importing Mg2+ into the cytoplasm and by reducing the amount of Mg2+-chelating ATP, the MgtA, MgtB, and MgtC proteins enable Salmonella to carry out protein synthesis (albeit at a lower rate) when experiencing a low cytoplasmic Mg2+ concentration that otherwise compromises ribosome assembly (26). The changes brought about by MgtA, MgtB, and MgtC, and potentially by other proteins expressed in low cytoplasmic Mg2+, dramatically reduce the bacterial growth rate from logarithmic to linear (26).
In addition to aiding translation, the MgtA and MgtC proteins further transcription of PhoP-activated genes by increasing the amount of active PhoP protein (7, 27). MgtA accomplishes this task by removing Mg2+ from the periplasm, where it acts as a negative signal for PhoQ (27). MgtC sequesters PhoP away from ClpS, thereby preventing PhoP degradation by ClpSAP (7).
We now report a mechanism that enables Salmonella and Escherichia coli to alter proteolysis of selective ClpS-dependent ClpAP substrates. This mechanism operates when bacteria face low cytoplasmic Mg2+, a condition that decreases the rate of protein synthesis (26) and hyperactivates the PhoP protein (7, 27). The uncovered mechanism has broad physiological consequences because it impacts gene transcription and the formation of antibiotic persister cells.
Results
The PhoP Protein Directly Represses Transcription of the Adaptor Gene clpS.
The PhoP-activated genes mgtA and mgtC specify products that increase the amount of active PhoP protein (7, 27), thereby positively feeding back on their transcriptional activator. Given that the ClpSAP protease degrades the PhoP protein (7), we wondered whether PhoP reduces the abundance of the ClpS and/or ClpA proteins, which are specific to the ClpSAP protease. [ClpP also associates with ClpX to form the ClpXP protease (28, 29).]
ClpS amounts were lower in wild-type Salmonella grown in low than in high Mg2+ medium (Fig. 1A), which are PhoP-inducing and -noninducing conditions, respectively (15). The reduction in ClpS amounts taking place in low Mg2+ is PhoP dependent because it was not observed in a phoP mutant (Fig. 1A). By contrast, similar amounts of the control protein OmpA were present across strains and conditions (Fig. 1A).
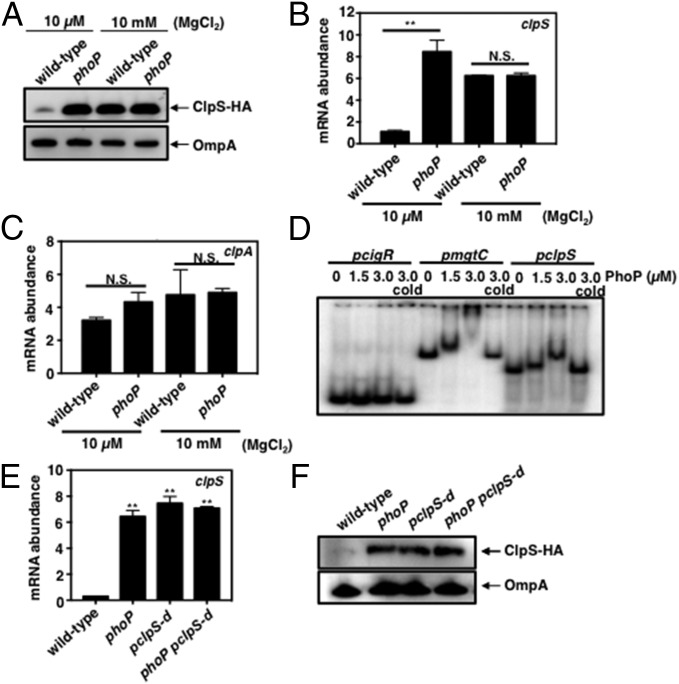
Fig. 1.
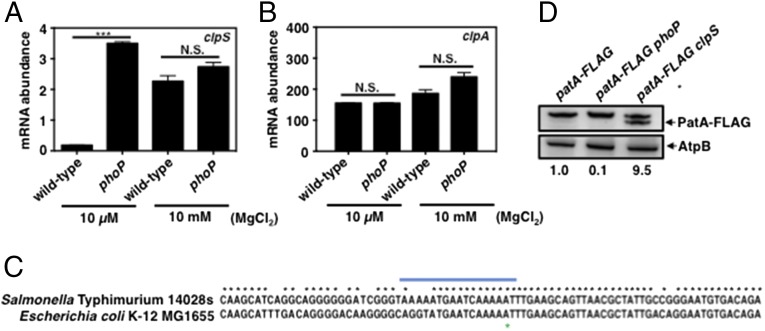
PhoP directly represses transcription of the clpS gene. (A) Western blot analysis of crude extracts prepared from clpS-HA (JY691) and clpS-HA phoP (JY854) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM or 10 mM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the HA tag or the OmpA protein. Data are representative of three independent experiments, which gave similar results. (B and C) mRNA abundance of the clpS (B) and clpA (C) genes produced by wild-type (14028s) and phoP (MS7953s) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM or 10 mM MgCl2 at 37 °C for 5.5 h. mRNA abundance was normalized to that of the ompA gene. (D) EMSA of DNA fragments carrying the Salmonella cigR, mgtC, and clpS promoter regions with the indicated amounts of purified PhoP protein. The gel is representative of three independent experiments. Samples were incubated at room temperature for 20 min and then were electrophoresed on 6% Tris/borate/EDTA gels at 100 V for 90 min. (E) mRNA abundance of the clpS gene produced by wild-type (14028s), phoP (MS7953s), pclpS-d (JY665), and phoP pclpS-d double mutant (JY939) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 5.5 h. mRNA abundance was normalized to that of the ompA gene. (F) Western blot analysis of crude extracts prepared from clpS-HA (JY691), clpS-HA phoP (JY854), clpS-HA pclpS-d (JY937), and clpS-HA phoP pclpS-d (JY980) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the HA tag or the OmpA protein. Data are representative of three independent experiments, which gave similar results. For B, C, and E, the primers used in qRT-PCR are listed in Table S2. Data shown are the mean and SD from three independent experiments. N.S., not significant. *P < 0.05, two-tailed t test with each sample vs. wild-type.
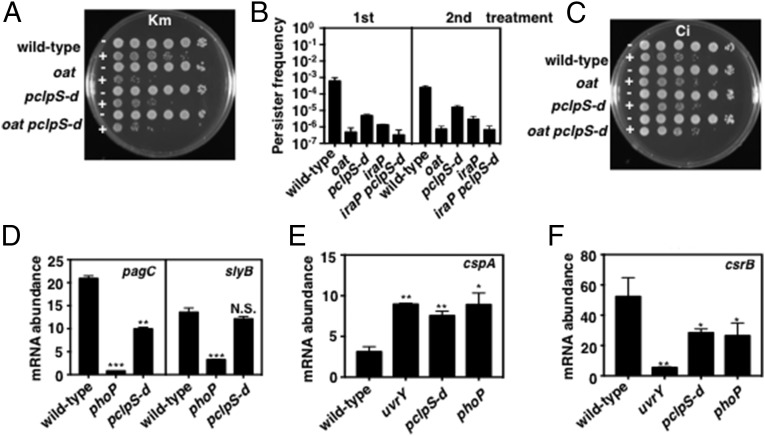
PhoP represses clpS expression at the transcriptional level because the phoP mutant had five times more clpS mRNA than the wild-type strain following growth in 10 μM Mg2+ (Fig. 1B) but similar clpS mRNA amounts when bacteria were grown in 10 mM Mg2+ (Fig. 1B). As expected, the PhoP-activated genes pagC and slyB, which were used as controls, were highly transcribed in low Mg2+ in a PhoP-dependent manner (Fig. S1 A and B). PhoP repression of the clpS gene does not appear to extend to other protease adaptor genes because the mRNA amounts of the rssB gene, which specifies a ClpXP adaptor (30), were similar in wild-type and phoP Salmonella whether grown in low or high Mg2+ (Fig. S1C).
PhoP binds to the clpS promoter, suggesting it represses clpS transcription directly. That is, the purified PhoP protein shifted a DNA fragment harboring the clpS promoter region (Fig. 1D), and excess unlabeled clpS promoter DNA outcompeted PhoP binding to the labeled clpS promoter DNA fragment (Fig. 1D). Control experiments demonstrated PhoP binding to a DNA fragment carrying the promoter of the PhoP-activated mgtC gene (Fig. 1D) but not to one corresponding to the promoter of the PhoP-independent cigR gene (Fig. 1D).
We identified a putative PhoP box (AAAAATGAATCAAAAATT) (31) upstream of the clpS coding region overlapping the predicted −10 region of the clpS promoter (Fig. S2A). Nucleotide substitutions (to AACCATGACTCAACCATT) (Fig. S2A) in this site largely abolished PhoP binding to a DNA fragment carrying the clpS promoter in vitro (Fig. S2B). Moreover, an engineered strain with these nucleotide substitutions in the chromosomal copy of the clpS promoter produced clpS mRNA amounts equivalent to those of the phoP-null mutant when bacteria were grown under PhoP-inducing conditions (Fig. 1E). [Please note that the chromosomal clpS promoter mutant has an additional 83-nt-long sequence upstream of the PhoP binding site (Fig. S3A) that does not impact clpS expression because similar clpS mRNA amounts were produced by wild-type Salmonella and by a strain carrying the 83-nt-long sequence in an otherwise wild-type genetic background (Fig. S3B).] Furthermore, a phoP clpS promoter double mutant produced the same high clpS mRNA amounts as the phoP and clpS promoter single mutants (Fig. 1E). Therefore, PhoP represses clpS transcription, likely by hindering access of RNA polymerase to the clpS promoter.
In agreement with the mRNA data discussed in the previous paragraph, ClpS protein amounts were similar in the phoP mutant, the clpS promoter mutant and in the phoP clpS promoter double mutant and were higher than those present in wild-type Salmonella (Fig. 1F). By contrast, similar amounts of the control protein OmpA were present in the four strains (Fig. 1F).
We considered the possibility that PhoP might repress the transcription of both the clpS and clpA genes because the clpA start codon is located only 31 nt downstream of the clpS stop codon (Fig. S1D). However, similar amounts of clpA mRNA were present in wild-type and phoP Salmonella following growth in 10 µM or 10 mM Mg2+ (Fig. 1C). Moreover, the clpS promoter mutation that derepressed clpS transcription did not alter clpA mRNA levels (Figs. S2C and S3C) or ClpA protein amounts (Fig. S2D). These results are consistent with the clpA gene having its own transcription start sites, as proposed in E. coli (32). Therefore, PhoP directly represses the transcription of clpS but not of clpA during growth in low Mg2+.
PhoP Represses clpS Transcription When Cytoplasmic Mg2+ Drops Below a Threshold That Triggers Synthesis of the MgtA and MgtC Proteins.
We hypothesized that PhoP represses clpS expression when the MgtA and/or MgtC proteins are produced because the number of ClpS molecules decreases as the number of MgtC molecules increases several hours following a shift from high to low Mg2+ (7) and also because transcription of the mgtA-coding region mimics that of the mgtC-coding region when bacteria experience low cytoplasmic Mg2+ (24, 25).
As hypothesized, clpS mRNA amounts were approximately eight times lower in the wild-type strain than in the phoP mutant following growth in 10 µM Mg2+ for 5.5 h (Fig. 2A) when both MgtA and MgtC are present (26) and when the mRNA for the PhoP-activated MgtA- and MgtC-dependent pagC gene is highly abundant (Fig. 2B). The decrease in clpS mRNA amounts is dependent on both the mgtC and mgtA genes (Fig. 2C), but inactivating the mgtC gene has a stronger effect than mutating the mgtA gene. That PhoP-dependent clpS repression is compromised more in the mgtC mutant than in the mgtA mutant is in contrast to the stronger phenotype that mgtA inactivation has on PhoP-dependent pagC expression (Fig. 2D).
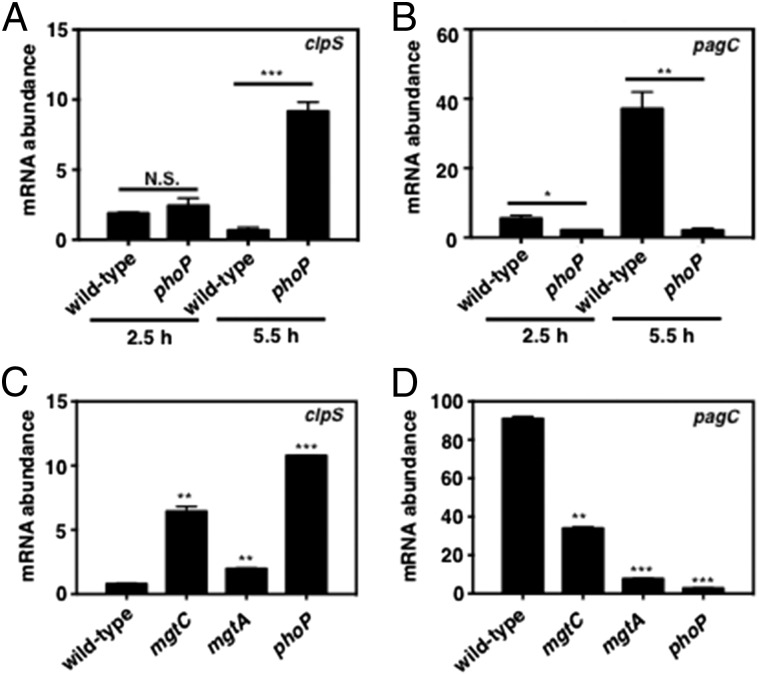
Fig. 2.
Full repression of clpS transcription by PhoP is dependent on the PhoP-activated mgtC and mgtA genes. (A and B) mRNA abundance of the clpS (A) and pagC (B) genes produced by wild-type (14028s) and phoP (MS7953s) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 2.5 or 5.5 h. mRNA abundance was normalized to that of the ompA gene. (C and D) mRNA abundance of the clpS (C) and pagC (D) genes produced by wild-type (14028s), mgtC (EL4), mgtA (EG16735), and phoP (MS7953s) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 5.5 h. mRNA abundance was normalized to that of the ompA gene. Primers used in qRT-PCR are listed in Table S2. Data shown are the mean and SD from three independent experiments. N.S., not significant. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t test with each sample vs. wild-type.
PhoP repression of clpS transcription specifically responds to low cytoplasmic Mg2+ because clpS mRNA amounts were similar in wild-type and phoP Salmonella following growth in 10 µM Mg2+ for 2.5 h (Fig. 2A), which is before the cytoplasmic Mg2+ concentration drops below the threshold that triggers production of the MgtA and MgtC proteins (26). Also, clpS mRNA abundance was not altered when bacteria were grown in the presence of 10 mM MgCl2, which is a noninducing condition for PhoQ, or when the PhoQ-inducing conditions were a mildly acidic pH or the antimicrobial peptide C18G (Fig. S1F). The latter two conditions promoted the transcription of the pagC-coding region in a PhoP-dependent manner (albeit not to the levels achieved in low Mg2+) (Fig. S1G). Thus, PhoP represses clpS transcription when Salmonella experiences a drop in cytoplasmic Mg2+ below a certain threshold.
PhoP-Dependent Decrease in ClpS Abundance Increases Amounts of ClpSAP Substrate Subset.
To ascertain how the decrease in ClpS abundance taking place in low cytoplasmic Mg2+ impacts Salmonella physiology, we first investigated the amounts of ClpSAP substrates in isogenic wild-type phoP and clpS strains. To date, only two native ClpSAP substrates are known in Salmonella: the putrescine aminotransferase Oat and PhoP (7). In work to be described elsewhere, we identified the UvrY and FtsA proteins as ClpSAP substrates (Fig. S4 presents the verification of this identification). [As in Caulobacter crescentus (33), the FtsA protein appears to be a substrate of both the ClpXP and ClpAP proteases in Salmonella because FtsA amounts were higher in the clpX mutant than in the wild-type strain (Fig. S4B).] A decrease in ClpS abundance revealed three distinct behaviors in C-terminally tagged versions of the Oat, UvrY, and FtsA proteins, which were expressed from their normal promoters and chromosomal locations.
Oat-FLAG was undetectable in the phoP mutant but was in great excess in the clpS mutant compared with the wild-type strain following bacterial growth for 5.5 h in low Mg2+ (Fig. 3A). The different Oat-FLAG abundance reflects decreased and increased stability in the phoP and clpS mutants, respectively (Fig. 3B), as opposed to transcription of the oat-FLAG gene because wild-type, phoP, and clpS Salmonella had similar oat-FLAG mRNA abundance (Fig. S5A). In wild-type Salmonella, Oat-FLAG amounts were much higher at 5.5 h than at 2.5 h in low Mg2+ (Fig. 3C). The low Oat-FLAG amounts present in the wild-type strain at 2.5 h were similar to those present in the phoP mutant at both 2.5 and 5.5 h (Fig. 3C).
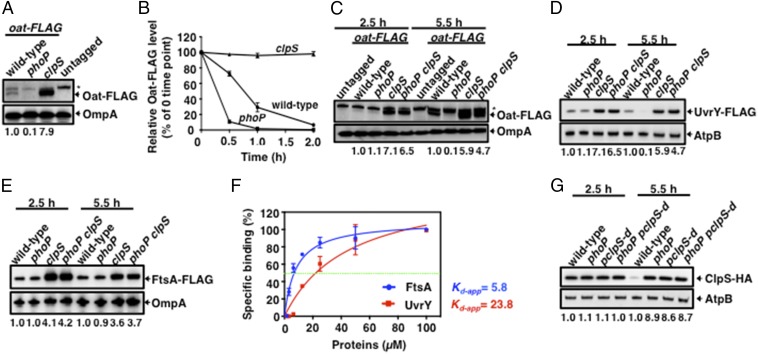
Fig. 3.
Inactivation of the phoP gene differentially affects the amounts of ClpS-dependent ClpAP substrates. (A) Western blot analysis of crude extracts prepared from wild-type (14028s), oat-FLAG (JY655), oat-FLAG phoP (JY656), and oat-FLAG clpS (JY657) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the FLAG tag or the OmpA protein. The asterisk indicates a nonspecific cross-reactive band. Data are representative of three independent experiments, which gave similar results. (B) Oat-FLAG stability was determined in oat-FLAG (JY655), oat-FLAG clpS (JY657), and oat-FLAG phoP (JY656) Salmonella following growth in 20 mL N-minimal medium containing 10 μM MgCl2 for 6 h. Protein synthesis was then inhibited with chloramphenicol (1 mg/mL), and samples were removed at the indicated times and analyzed by Western blotting using antibodies directed to the FLAG tag. Relative Oat-FLAG levels were calculated from two independent experiments. (C) Western blot analysis of crude extracts prepared from wild-type (14028s), oat-FLAG (JY655), oat-FLAG phoP (JY656), oat-FLAG clpS (JY657), and oat-FLAG clpS phoP double mutant (JY658) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 2.5 or 5.5 h. Samples were analyzed using antibodies directed to the FLAG tag or the OmpA protein. The asterisk indicates a nonspecific cross-reactive band. Data are representative of three independent experiments, which gave similar results. (D) Western blot analysis of crude extracts prepared from uvrY-FLAG (JY911), uvrY-FLAG phoP (JY912), uvrY-FLAG clpS (JY913), and uvrY-FLAG clpS phoP double mutant (JY983) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 2.5 or 5.5 h. Samples were analyzed using antibodies directed to the FLAG tag or the AtpB protein. Data are representative of three independent experiments, which gave similar results. (E) Western blot analysis of crude extracts prepared from ftsA-FLAG (JY905), ftsA-FLAG phoP (JY906), ftsA-FLAG clpS (JY907), and ftsA-FLAG clpS phoP double mutant (JY982) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 2.5 or 5.5 h. Samples were analyzed using antibodies directed to the FLAG tag or the OmpA protein. Data are representative of three independent experiments, which gave similar results. (F) In vitro protein–protein interactions determined by coimmunoprecipitation using Western blot analysis of purified ClpS-His, FtsA-FLAG, and UvrY-FLAG proteins. The Kd-app between ClpS-His and FtsA-FLAG or UvrY-FLAG was measured by Western blot analysis at different amounts of FtsA-FLAG or UvrY-FLAG and the same amount of ClpS-His. The intensity of the UvrY-FLAG, FtsA-FLAG, and ClpS-His bands in Fig. S7C was determined with purified proteins. For all binding intensity analyses, data shown are the mean and SD from three independent experiments. Please note that the purified FtsA-FLAG and UvrY-FLAG proteins are monomeric (Fig. S7 A and B). (G) Western blot analysis of crude extracts prepared from clpS-HA (JY691), clpS-HA phoP (JY854), clpS-HA pclpS-d (JY937), and clpS-HA phoP pclpS-d (JY980) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 2.5 or 5.5 h. Samples were analyzed using antibodies directed to the FLAG tag or the AtpB protein. Data are representative of three independent experiments, which gave similar results. Numbers below the blots correspond to the normalized relative amount compared with OmpA or AtpB amounts in each lane at a given time point.
In contrast to Oat-FLAG, UvrY-FLAG amounts were similar in wild-type Salmonella at 2.5 and 5.5 h in low Mg2+ (Fig. 3D) and in the phoP mutant at 2.5 h (Fig. 3D). UvrY-FLAG amounts were much reduced in the phoP mutant at 5.5 h (Fig. 3D), reflecting decreased stability (Fig. S6A) rather than decreased expression because wild-type and phoP Salmonella had similar uvrY mRNA abundance at all investigated times (Fig. S5B). That UvrY-FLAG protein amounts were similar in wild-type Salmonella at 2.5 and 5.5 h (Fig. 3D) may reflect that uvrY mRNA abundance decreased from peak values at 2 h to low values at 6 h following a shift to low Mg2+ (Fig. S6C). By contrast, mRNA abundance of the PhoP-activated pagC gene increased between 2 and 6 h in low Mg2+ in a PhoP-dependent manner (Fig. S6D). The steady-state levels of the UvrY-FLAG protein in wild-type Salmonella peaked at 6.5 h in low Mg2+ (Fig. S6B), reflecting increased stability at 5.5 h (Fig. S6A).
In contrast to both Oat-FLAG and UvrY-FLAG, FtsA-FLAG amounts were similar in wild-type and phoP Salmonella at both 2.5 and 5.5 h in low Mg2+ (Fig. 3E). Thus, the PhoP-dependent decrease in ClpS amounts taking place at 5.5 h in low Mg2+ has a strong effect on the steady-state levels of Oat, a decreased effect on UvrY (detected only in the phoP mutant), and no effect on FtsA.
Inactivation of the clpS gene dramatically increased UvrY-FLAG amounts in the phoP mutant to levels equivalent to those of the clpS single mutant (Fig. 3D). Oat-FLAG was slightly more abundant in the clpS single mutant than in the phoP clpS double mutant (Fig. 3C). FtsA-FLAG amounts were similar in clpS single and clpS phoP double mutants (Fig. 3E) and were higher than in wild-type and phoP Salmonella at both 2.5 and 5.5 h (Fig. 3E).
Differential Binding of ClpSAP Substrates to the ClpS Protein.
The degradation of ClpSAP substrates is anticipated to reflect the concentration of both ClpS and its clients as well as the affinity of ClpS for its clients. To understand the basis for the different effects that a decrease in ClpS amounts had on the UvrY-FLAG and FtsA-FLAG proteins (Fig. 3), we calculated the apparent binding affinity (Kd-app) of these proteins for ClpS (Fig. S7).
The affinity of FtsA for ClpS was four times higher than that of UvrY for ClpS (i.e., Kd-app values of 5.8 vs. 23.8 μM) (Fig. 3F), the latter being similar to that exhibited by PhoP for ClpS (Kd-app 18.8 μM) (7). These results support the notion that a reduction in ClpS levels impacts the amounts of UvrY and PhoP but not of FtsA, because FtsA displays a higher affinity for ClpS than UvrY and PhoP.
The Mutant with the clpS Promoter Refractory to Repression by PhoP Exhibits Lower Amounts of a ClpSAP Substrate Subset.
To investigate the impact of PhoP repression of clpS transcription on PhoP itself, we examined PhoP protein amounts in the clpS promoter mutant that is refractory to PhoP repression. In addition, by comparing the clpS promoter mutant with the phoP mutant, we could ascertain whether PhoP controls the amounts of other ClpSAP substrates by a mechanism independent of clpS repression.
We first determined that isogenic wild type, phoP mutant, clpS promoter mutant, and phoP clpS promoter double mutant Salmonella harbored similar ClpS-HA amounts at 2.5 h in low Mg2+ medium (Fig. 3G). By contrast, ClpS-HA amounts were approximately nine times lower in the wild-type strain than in the phoP mutant, the clpS promoter mutant, and the phoP clpS promoter double mutant at 5.5 h in low Mg2+ medium (Fig. 3G). Thus, the abundance of ClpS-dependent ClpAP substrates was examined at 5.5–6 h of bacterial growth in low Mg2+ medium.
The clpS promoter mutant had lower amounts of PhoP-HA protein than wild-type Salmonella (Fig. 4A), indicating that PhoP repression of clpS transcription increases PhoP steady-state levels. PhoP-HA amounts were higher in the clpS promoter mutant than in a mutant lacking the mgtC gene (Fig. 4B), which, as discussed above, specifies a protein that protects PhoP from degradation by ClpSAP (7). Therefore, MgtC sequestration of PhoP has a stronger effect on PhoP steady-state levels than PhoP repression of clpS transcription.
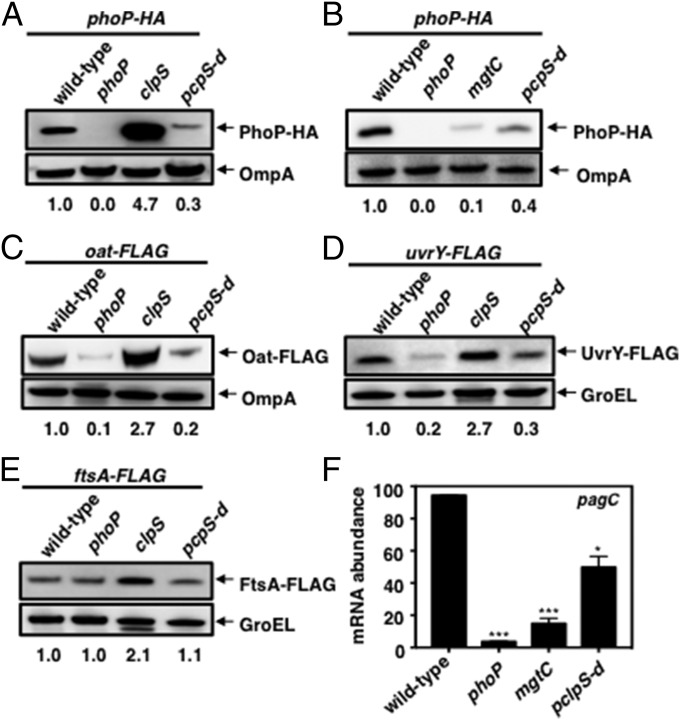
Fig. 4.
PhoP-dependent reduction in ClpS amounts alters steady-state levels of a subset of ClpAP substrates. (A) Western blot analysis of crude extracts prepared from phoP-HA (EG13918), phoP (MS7953s), phoP-HA clpS (JY881), and phoP-HA pclpS-d (JY933) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the HA tag or the OmpA protein. Numbers below the blots correspond to the normalized relative amounts compared with the OmpA or GroEL amounts in each lane. (B) Western blot analysis of crude extracts prepared from phoP-HA (EG13918), phoP (MS7953s), phoP-HA mgtC (JY879), and phoP-HA pclpS-d (JY933) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 5.5 h. Samples were analyzed using antibodies directed to the HA tag or the OmpA protein. Data are representative of three independent experiments, which gave similar results. (C) Western blot analysis of crude extracts prepared from oat-FLAG (JY655), oat-FLAG phoP (JY656), oat-FLAG clpS (JY657), and oat-FLAG pclpS-d (JY684) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the FLAG tag or the OmpA protein. (D) Western blot analysis of crude extracts prepared from uvrY-FLAG (JY911), uvrY-FLAG phoP (JY912), uvrY-FLAG clpS (JY913), and uvrY-FLAG pclpS-d (JY914) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the FLAG tag or the GroEL protein. (E) Western blot analysis of crude extracts prepared from ftsA-FLAG (JY905), ftsA-FLAG phoP (JY906), ftsA-FLAG clpS (JY907), and ftsA-FLAG pclpS-d (JY908) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the FLAG tag or the GroEL protein. (F) pagC mRNA abundance produced by the strains and under the growth conditions described in E. Primers used in qRT-PCR are listed in Table S2. Data shown are the mean and SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t test with each sample vs. wild-type.
The amounts of Oat-FLAG (Fig. 4C and Fig. S3E) and UvrY-FLAG (Fig. 4D) were lower in the clpS promoter mutant than in wild-type Salmonella but were not as low as in the phoP mutant (Fig. 4 C and D). These results argue that, in addition to repressing clpS transcription, PhoP utilizes another mechanism to increase the steady-state levels of Oat-FLAG and UvrY-FLAG by a mechanism(s) in addition to repression of clpS transcription.
In contrast to the behavior of the PhoP-HA, Oat-FLAG, and UvrY-FLAG proteins, similar amounts of FtsA-FLAG were present in wild-type, phoP mutant, and clpS promoter mutant Salmonella (Fig. 4E), likely reflecting that FtsA exhibits higher affinity toward ClpS than does UvrY (Fig. 3F) and PhoP (7). Control experiments showed that the clpS mutant had larger amounts of PhoP-HA, Oat-FLAG, UvrY-FLAG, and FtsA-FLAG than the wild-type strain (Fig. 4 A and C–E).
PhoP Promotes Antibiotic Persistence by Stabilizing the ClpS Client Oat and the ClpXP Substrate RpoS.
Putrescine is required for persistence toward aminoglycoside antibiotics in E. coli (34, 35). In Pseudomonas aeruginosa, inactivation of the putrescine aminotransferase gene spuC (a homolog of the Salmonella oat gene) reduces persister formation (36). Given that Oat is a ClpSAP substrate (Figs. 3 A–C and 4C), we reasoned that PhoP repression of clpS transcription might heighten antibiotic persistence.
We determined that the clpS promoter mutant refractory to PhoP repression exhibits less persistence to the aminoglycoside kanamycin than wild-type Salmonella following growth under PhoP-inducing conditions (i.e., 10 μM Mg2+ for 6 h) (Fig. 5A). The persistence phenotype of the clpS promoter mutant was intermediate between those exhibited by the wild-type strain and by an oat-null mutant (Fig. 5A). The lower persistence of the clpS promoter mutant appears to be due to ClpSAP’s effect on Oat, because persistence formation in an oat clpS promoter double mutant was similar to that of the oat single mutant (Fig. 5A). The bacteria growing in the presence of kanamycin appear to be true persisters (as opposed to kanamycin-resistant mutants) because they retained the persister phenotype upon subsequent exposure to kanamycin (Fig. 5B). Control experiments showed no differences among the three strains when exposed to ciprofloxacin (Fig. 5C), in agreement with previous findings (34, 35).
Fig. 5.
PhoP-dependent stabilization of ClpS clients increases antibiotic persistence and alters gene expression. (A) Kanamycin persister bacteria by wild-type (14028s), oat (JY903), pclpS-d (JY665), and oat pclpS-d (JY981) Salmonella following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h and incubation with kanamycin (Km) (100 μg/mL) for 2 h. Shown are 10-fold serial dilutions plated on LB agar plates. (B) Frequency of kanamycin persister bacterial exhibited by wild-type (14028s), oat (JY903), pclpS-d (JY665), iraP (EG17133), and iraP pclpS-d (JY936) Salmonella was determined by the number of colonies on LB agar plates after treatment with kanamycin (100 μg/mL) for 2 h. The second treatment shows persister formation by the colonies recovered from the LB agar plates following the first treatment and subjected to the same treatment. Data shown are the mean and SD from two independent experiments. (C) As in A, but cells were treated with ciprofloxacin (Ci) (2 μg/mL). (D) mRNA abundance of the pagC and slyB genes produced by wild-type (14028s), phoP (MS7953s), and pclpS-d (JY665) Salmonella following growth in in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 5.5 h. (E and F) mRNA abundance of the cspA (E) and csrB (F) genes produced by wild-type (14028s), uvrY (JY917), pclpS-d (JY665), and phoP (MS7953s) Salmonella following growth in in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 5.5 h. For D–F, mRNA abundance was normalized to that of the ompA gene. Primers used in qRT-PCR are listed in Table S2. Data shown are the mean and SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t test with each sample vs. wild-type.
The alternative sigma factor RpoS promotes persister formation toward aminoglycoside antibiotics in E. coli (34, 35). In Salmonella, PhoP increases RpoS stability by activating transcription of the iraP gene (37), which specifies an antiadaptor for RssB, the adaptor that delivers RpoS to the protease ClpXP (30). In agreement with the reduced RpoS amounts present in an iraP mutant experiencing low Mg2+ (37), an iraP mutant exhibited less persistence than the wild-type strain (Fig. 5B), and an iraP clpS promoter double mutant displayed less persistence than either iraP or clpS promoter single mutants (Fig. 5B). Therefore, PhoP promotes persister formation toward kanamycin by increasing the stability of both RpoS and Oat.
Protection of ClpS Clients PhoP and UvrY Modifies Gene Transcription.
We investigated the effects of increased PhoP amounts resulting from PhoP repression of clpS transcription by investigating the mRNA abundance of two genes directly activated by PhoP: pagC and slyB, the in vivo expression of which requires large and small amounts of active PhoP protein, respectively (27). pagC mRNA abundance was lower in the clpS promoter mutant than in wild-type Salmonella, albeit higher than that in the phoP (Fig. 5D and Fig. S3D) and mgtC (Fig. 4F) mutants. By contrast, slyB mRNA abundance was similar in wild-type and clpS promoter mutant strains (Fig. 5D). Thus, PhoP repression of clpS expression is necessary for full transcription of a subset of PhoP-activated genes.
Like PhoP, UvrY is a DNA-binding transcriptional regulator (38, 39) and a ClpSAP substrate (Figs. 3D and 4D). The mRNA abundance of the UvrY-repressed gene cspA (38) was similar in the phoP mutant, the clpS promoter mutant, and the uvrY-null mutant (Fig. 5E). By contrast, the phoP and clpS promoter single mutants had mRNA amounts of the UvrY-activated gene csrB (38) intermediate between those found in wild-type Salmonella and the uvrY mutant (Fig. 5F). Thus, by hindering degradation of UvrY, PhoP impacts gene expression even at promoters not bound by the PhoP protein.
PhoP Represses clpS Transcription in E. coli, Increasing the Amount of the ClpSAP Substrate PatA.
We wondered whether the PhoP protein also represses clpS transcription in E. coli, given that E. coli lacks MgtC (40), which is required for full clpS repression in Salmonella (Fig. 2C) and also given that PhoP-regulated targets often differ across species (10). We determined that E. coli reduces clpS mRNA amounts during growth in low Mg2+ in a phoP-dependent manner (Fig. 6A). By contrast, clpA mRNA levels were similar in wild-type and phoP strains grown in high or low Mg2+ (Fig. 6B), as in Salmonella (Fig. 1C).
Fig. 6.
PhoP represses expression of the clpS gene and protects ClpS client in E. coli. (A and B) mRNA abundance of the clpS (A) and clpA (B) genes in wild-type (MG1655) and phoP (EG12976) E. coli following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 5.5 h. mRNA abundance was normalized to that of the ompA gene. Primers used in qRT-PCR are listed in Table S2. For all qRT-PCR analyses, data shown are the mean and SD from three independent experiments. (C) Alignment of the nucleotide sequences corresponding to the clpS promoter regions of E. coli MG1655 and S. enterica serovar Typhimurium 14028s. The putative PhoP box is indicated by the blue bar. The green asterisk indicates the transcription start site near the putative PhoP binding box according to the Salmonella gene-expression database (19). (D) Western blot analysis of crude extracts prepared from patA-FLAG (JY670), patA-FLAG phoP (JY671), and patA-FLAG clpS (JY681) E. coli following growth in N-minimal medium (pH 7.7) containing 10 μM MgCl2 at 37 °C for 6 h. Samples were analyzed using antibodies directed to the FLAG tag or the AtpB protein. The asterisk indicates a nonspecific cross-reactive band. Data are representative of three independent experiments, which gave similar results. Numbers below the blots correspond to the normalized relative amount compared with AtpB amounts in each lane. N.S., not significant. ***P < 0.001, two-tailed t test with each sample vs. wild-type.
PhoP appears to repress clpS transcription directly, because the E. coli clpS promoter harbors a putative PhoP-binding site like the one identified in the clpS promoter region of Salmonella (Fig. 6C). Additionally, the abundance of the ClpSAP substrate and Oat homolog PatA was lower in the phoP mutant than in wild-type E. coli but was higher in the clpS mutant (Fig. 6D). Cumulatively, these results establish that PhoP repression of clpS transcription and the resulting impact on abundance of a ClpSAP substrate are conserved between E. coli and Salmonella.
Discussion
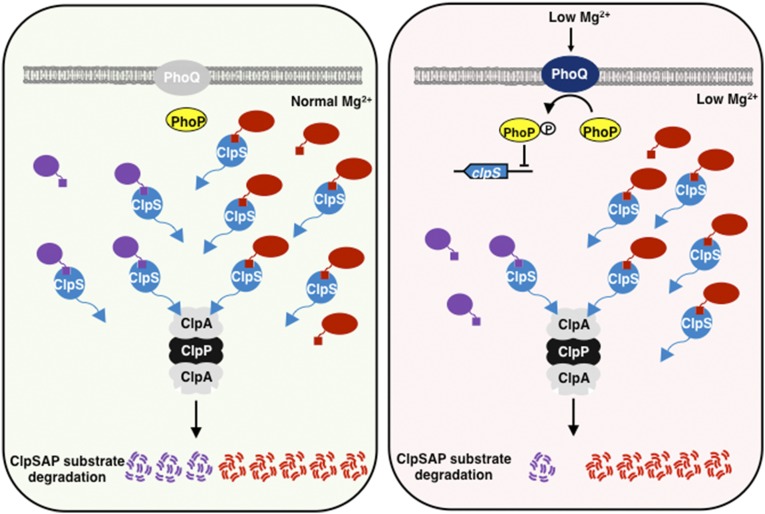
We report a mechanism that enables differential stability of ClpSAP substrates in enteric bacteria (Fig. 7). That is, the regulatory protein PhoP directly represses transcription of the adaptor-specifying clpS gene when bacteria experience low cytoplasmic Mg2+ (Fig. 1), resulting in increased steady-state levels of a subset of ClpS-dependent ClpAP substrates (Figs. 3 and 4). The increased substrate abundance furthers antibiotic persistence and alters the expression of genes controlled by different regulatory proteins (Fig. 5). Thus, by reducing adaptor amounts, organisms create a hierarchy among substrates of given protease.
Fig. 7.
A reduction in adaptor amounts creates a hierarchy among substrates delivered to a protease by an adaptor. The ClpAP protease degrades certain substrates directly but requires the adaptor ClpS to degrade others. (Left) When bacteria experience normal cytoplasmic Mg2+, there is little active (i.e., phosphorylated) PhoP protein. Thus, PhoP does not repress clpS transcription. ClpS amounts are sufficiently high to recognize substrates (purple and red) that may differ in their affinity for ClpS. Both protein synthesis and proteolysis are robust. (Right) When bacteria experience low cytoplasmic Mg2+, the amounts of active PhoP protein increase to levels that enable repression of clpS transcription. This reduces the amount of ClpS protein, which, in turn, increases the stability of some ClpS-dependent ClpAP substrates (purple) but not others (red), reflecting substrate affinity for ClpS (red substrates have higher affinity than purple substrates). The reduction in ClpS amounts increases the abundance of specific ClpSAP substrates (purple). The increase in abundance of specific proteins is in contrast to the genome-wide decrease in the rate of protein synthesis taking place in low cytoplasmic Mg2+ (26).
Changes in Adaptor Amounts Impact the Abundance of Its Clients.
Adaptor-mediated proteolysis is determined by the amounts of free adaptor and its clients, as well as by the binding affinity between adaptor and client, which often differs among clients (Fig. 3F). The affinity of an adaptor for its clients is also impacted by posttranslational modification of adaptor proteins (30) and by adaptor sequestration by antiadaptor proteins (41, 42).
Substrate amounts are determined by transcriptional, translational, and posttranslational mechanisms, usually in response to environmental and/or cellular signals. An increase in substrate amounts enhances their destruction, whereas exposure of high-affinity binding sites in a substrate does not necessarily increase protein degradation (2, 43). When a substrate is present at a low concentration, adaptors are critical for substrate degradation because they increase the local substrate concentration (2, 43). Therefore, altering adaptor amounts can be an effective way to regulate protein destruction.
We have now established that the regulatory protein PhoP represses transcription of the clpS gene both in Salmonella (Fig. 1 B and E and Fig. S1F) and E. coli (Fig. 6A) when these bacteria experience low cytoplasmic Mg2+. [Please note that others reported constant ClpS amounts in E. coli grown in different media and conditions (44).] By turning down (but not off) clpS transcription, PhoP leaves sufficient ClpS amounts to create a hierarchy among substrates of the ClpAP protease. That is, the reduction in ClpS levels is accompanied by increases in the amounts of Oat (Figs. 3 A and C and 4C), UvrY (Figs. 3D and 4D), PhoP (Fig. 4 A and B), and potentially other proteins. However, it does not alter FtsA amounts (Figs. 3E and 4E), possibly because ClpS binds more tightly to FtsA than to the other investigated proteins (Fig. 3F) (7).
By repressing clpS transcription, PhoP has the potential to alter the amounts of ClpS-independent ClpAP substrates. This is because ClpS stabilizes the ClpS-independent ClpAP substrate DnaA in Caulobacter crescentus (8) and SsrA-tagged proteins in E. coli (45) due to competition between ClpS and ClpS-independent substrates for interaction with the unfoldase ClpA.
The Phenotypic Consequences of clpS Repression in Low Cytoplasmic Mg2+.
The PhoP-dependent reduction in ClpS amounts is highly dependent on the PhoP-activated mgtA and mgtC genes (Fig. 2C). These genes are expressed when Salmonella experiences a drop in cytoplasmic Mg2+ levels below a threshold that triggers physiological changes, resulting in a dramatic slowdown in growth rate (26). These changes include a decrease in ATP levels and an increase in Mg2+ uptake, reducing the number of ribosomes and thus the rate of protein synthesis (26). What, then, are the physiological consequences of lowering ClpS amounts in low cytoplasmic Mg2+?
By lowering ClpS amounts (Fig. 1A), Salmonella increases the stability of the regulatory proteins PhoP and UvrY (Fig. 4 A and D and Fig. S4C) (7), thereby impacting the transcription of multiple genes (Fig. 5 D–F). PhoP is a major regulator of virulence and Mg2+ homeostasis in enteric bacteria (9, 10). UvrY promotes biofilm formation and cell viability in stationary phase in E. coli (46, 47) and controls several functions, including virulence, in Salmonella (48, 49).
The PhoP-mediated repression of clpS transcription resulted in increased steady-state levels of Oat (Figs. 3 A and C and 4C) but not FtsA (Figs. 3E and 4E). We determined that the increase in Oat amounts heightens bacterial persistence to the aminoglycoside kanamycin (Fig. 5 A and B), thereby providing a signaling input to Oat-mediated persistence described in other species (34, 35). That FtsA abundance was not altered by lowering ClpS amounts reflects that FtsA binds to ClpS more tightly than UvrY (Fig. 3F) and PhoP (7). These results suggest that, under the investigated conditions, FtsA levels are limited for optimal cell division, given the role of its homolog in regulating this process in C. crescentus (50), and that FtsA overexpression disrupts Z ring formation in E. coli (51).
PhoP reduces the transcription of clpS but not of the downstream clpA gene (Fig. 1 B and C and Fig. S1E). The selective repression of clpS allows proteolysis of ClpS-independent ClpAP substrates to proceed unimpeded and, as discussed above, this proteolysis might even be enhanced given that ClpS competes with ClpS-independent ClpAP substrates for ClpA (8). In addition, PhoP represses clpS expression when Salmonella changes from logarithmic to linear growth (26), thus freeing up ClpAP, which appears to play a critical role during slow growth, as ClpA and ClpP amounts increase threefold when E. coli enters the stationary phase (44). ClpAP degrades ssrA-tagged proteins in the stationary phase more than in the exponential phase (44) and the Dps and DnaA proteins at the stationary phase (8, 52) [note that the Dps and DnaA proteins are degraded by ClpXP and Lon, respectively, during exponential growth (8, 52)].
Protein Stabilization Contributes to Global Gene Transcription in Low Cytoplasmic Mg2+.
The PhoP protein controls the transcription of ∼9% of the Salmonella genome (19). However, only a subset of PhoP-dependent genes harbors PhoP-binding sites in their promoter regions. PhoP impacts global gene transcription both by stimulating the synthesis and by hindering the degradation of regulatory proteins.
The PhoP protein appears to utilize different mechanisms to increase the amounts of active regulatory proteins depending on the stress condition. For example, when Salmonella experiences low extracytoplasmic Mg2+ detected by the sensor PhoQ (15), PhoP furthers the levels of regulatory proteins transcriptionally (e.g., RstA) (53) and posttranslationally (e.g., PmrA) (54, 55). These changes take place while Salmonella is growing logarithmically and shortly after it faces low extracytoplasmic Mg2+ (26). By contrast, the linear growth triggered when cytoplasmic Mg2+ decreases below a threshold results in PhoP hindering the degradation of premade proteins. That is, PhoP stabilizes UvrY (Figs. 3D and 4D and Fig. S6 A and B) and RpoS (37) proteins by reducing the amount of the adaptor ClpS (Fig. 1A) and by promoting the synthesis of the antiadaptor IraP (37), respectively.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Bacterial strains and plasmids used in this study are presented in Table S1. All S. enterica serovar Typhimurium strains are derived from strain 14028s (56) and were constructed by phage P22-mediated transductions as described (57). All E. coli K-12 strains are derived from strain MG1655 and were constructed by phage P1-mediated transductions as described (57). DNA oligonucleotides used in this study are presented in Table S2. Bacteria were grown at 37 °C in Luria-Bertani broth (LB) and N-minimal medium (pH 7.7) (23) supplemented with 0.1% casamino acids, 38 mM glycerol, and the indicated concentrations of MgCl2. E. coli DH5α was used as the host for preparation of plasmid DNA. Ampicillin was used at 50 μg/mL, kanamycin at 50 μg/mL, chloramphenicol at 25 μg/mL except for the protein stability assays when it was used at 1 mg/mL, and tetracycline at 12.5 μg/mL except for the protein stability assays when it was used at 50 μg/mL.
Construction of Chromosomal Mutants and Plasmids.
Chromosomal mutants were constructed using the one-step disruption method (58) with minor modifications. To construct the oat::cat mutant (JY903), a cat cassette was introduced in the oat gene as follows: A cat gene fragment was amplified from plasmid pKD3 using primer pair 16494/16495 and then was introduced into wild-type Salmonella (14028s) carrying plasmid pKD46. Strain JY981 was made by transducing the oat::cat insertion into strain JY665 using a P22 lysate generated in strain JY903.
To construct the aat mutant (JY602), a cat cassette was introduced into the aat gene as follows: A cat gene fragment was amplified from pKD3 using primers 15945/15946 and then was introduced into wild-type Salmonella (14028s) carrying plasmid pKD46.
To construct the uvrY mutant (JY917), a cat cassette was introduced into the uvrY gene as follows: A cat gene fragment was amplified from pKD3 using primers 16500/16501and then was introduced into wild-type Salmonella (14028s) carrying plasmid pKD46.
To construct the PhoP-binding site clpS promoter mutant (JY664), a cat cassette was introduced into the PhoP-binding site in the clpS promoter as follows: A cat gene fragment was amplified from pKD3 using primers 15962/15963 and then was introduced into wild-type Salmonella (14028s) carrying plasmid pKD46. The resulting strain, JY664, was kept at 30 °C and then was transformed with plasmid pCP20 to remove the cat cassette, thus generating strain JY665.
To introduce the 83-nt-long FRT insertion at the clpS promoter present in strains JY937 and JY938, a cat cassette was introduced into clpS promoter region as follows: A cat gene fragment was amplified from pKD3 using primers 16627/16628 and then was introduced into wild-type Salmonella (14028s) and oat-FLAG (JY655) strains carrying plasmid pKD46. The resulting strains were transformed with pCP20 to remove the cat cassette.
To construct a strain specifying a C-terminally HA-tagged ClpS protein (JY691), a kan cassette was introduced into the clpS gene as follows: A kan gene fragment was amplified from plasmid pKD4 using primer pair 16048/16049 and then was introduced into wild-type Salmonella 14028s carrying plasmid pKD46. The resulting strain was kept at 30 °C and then was transformed with pCP20 to remove the kan cassette. Strain JY854 was made by transducing the phoP::Tn10 (59) insertion into strain JY691 using a P22 lysate generated in strain MS7953s (59). Strains JY934 and JY980 were made using a P22 lysate prepared in strain JY664 to transduce the pclpS-d::cat insertion into strains JY691 and JY854, respectively.
To construct a strain specifying a C-terminally HA-tagged ClpA protein (JY694), a kan cassette was introduced in the clpA gene as follows: A kan gene fragment was amplified from plasmid pKD4 using primer pair 16050/16051 and then was introduced into wild-type Salmonella 14028s carrying plasmid pKD46. The resulting strain was kept at 30 °C and then was transformed with pCP20 to remove the kan cassette. Strain JY935 was made using a P22 lysate generated in strain JY664 to transduce the pclpS-d::cat insertion into strain JY694.
To construct a strain specifying a C-terminally FLAG-tagged FtsA protein (JY905), a kan cassette was introduced into the ftsA gene as follows: A kan gene fragment was amplified from plasmid pKD4 using primer pair 16502/16503 and then was introduced into wild-type Salmonella (14028s) carrying plasmid pKD46. Strains JY906, JY907, JY908, JY909, JY910, and JY918 were made by transducing the phoP::Tn10 (59), clpS::cat (7), pclpS-d::cat, clpX::cat (37), clpA::cat (7), and aat::cat insertions into strain JY905 using P22 lysates generated in strains MS7953s, JY570, JY664, EG18499, JY199, and JY602, respectively. Strain JY982 was made by transducing the pclpS-d::cat insertion into strain JY906 using a P22 lysate generated in strain JY664.
To construct a strain specifying a C-terminally FLAG-tagged UvrY protein (JY911), a kan cassette was introduced in the uvrY gene as follows: A kan gene fragment was amplified from plasmid pKD4 using primer pair 16502/16503 and then was introduced into wild-type Salmonella (14028s) carrying plasmid pKD46. Strains JY912, JY913, JY914, JY915, JY916, and JY919 were made by transducing the phoP::Tn10 (59), clpS::cat (7), pclpS-d::cat, clpX::cat (37), clpA::cat (7), and aat::cat insertions into strain JY911 using P22 lysates generated in strains MS7953s, JY570, JY664, EG18499, JY199, and JY602, respectively. Strain JY983 was made by transducing the pclpS-d::cat insertion into strain JY912 using a P22 lysate generated in strain JY664.
Strains JY879, JY881, and JY933 were made by transducing the mgtC::km (60), clpS::cat (7), and pclpS-d::cat insertions into strain EG13918 using P22 lysates generated in strains EL1, JY570, and JY664, respectively. The resulting strains were transformed with pCP20 to remove the cat or kan cassettes.
Strains JY656 and JY658 were made by transducing the phoP::Tn10 (59) insertion into strains JY655 and JY657 using a P22 lysate generated in strain MS7953s.
Strain JY936 was made by transducing the pclpS-d::cat insertion into strain EG17133 using a P22 lysate generated in strain JY664.
Strain JY920 was made by transducing the aat::cat insertion into strain JY655 using a P22 lysate generated in strain JY602.
Strains JY684 and JY939 were made by transducing the pclpS-d::cat insertion into strains JY655 and MS7953, respectively, using a P22 lysate generated in strain JY664. The resulting strain was transformed with pCP20 to remove the cat cassette.
To construct the clpS mutant E. coli (JY680), a kan cassette was introduced into the clpS gene as follows: A kan gene fragment was amplified from pKD4 using primers 16009/16010 and then was introduced into wild-type E. coli carrying plasmid pKD46. The resulting strain was kept at 30 °C and transformed with pCP20 to remove the kan cassette.
To construct strains specifying a C-terminally FLAG-tagged PatA protein (JY670) mutant strain, a cat cassette was introduced in the oat gene as follows: A cat gene fragment was amplified from plasmid pKD3 using primer pair 15964/15965 and then was introduced into wild-type E. coli (MG1655) carrying plasmid pKD46. The resulting strain was kept at 30 °C and was transformed with pCP20 to remove the cat cassette. Strains JY681 and JY671 were made by transducing the patA-FLAG::cat insertion into strains EG12976 and JY680, respectively, using a P1 lysate generated in strain JY670.
Plasmids carrying genes specifying FtsA-FLAG or UvrY-FLAG were constructed as follows: The ftsA-FLAG and uvrY-FLAG genes were amplified using primer pairs 16618/16619 and 16620/16621, respectively. The PCR products were digested with BamHI and HindIII and then were introduced between the BamHI and HindIII sites of pUHE21-2lacIq (61). Plasmids carrying genes specifying ClpS-His6, ClpA-His6, and ClpP-His6 were constructed as follows: The clpS, clpA, and clpP genes were amplified using primer pairs 16437/16438, 16441/16442, and 16439/16440, respectively. The PCR products were digested with NcoI and XhoI and then were introduced between the NcoI and XhoI sites of pET-28a(+).
Western Blot Assay.
Bacteria were grown in N-minimal medium with 10 mM MgCl2 overnight. Bacteria were harvested, washed twice with N-minimal medium, and used to inoculate N-minimal medium containing 10 μM MgCl2 (1:50 dilution). Bacteria were grown at 37 °C with 250 rpm and aeration for the indicated times. Crude extracts were prepared in B-PER reagent (Pierce) with 100 μg/mL lysozyme and EDTA-free protease inhibitor (Roche). Samples were loaded on 4–12% NuPAGE gels (Life Technologies) and transferred to nitrocellulose membrane using an iBot machine (Life Technologies). Membranes were blocked with 3% skim milk solution (50 mM Tris⋅Cl, 138 mM NaCl, 2.7 mM KCl, and 3% skim milk) at room temperature for 2 h. Then, samples were analyzed using antibodies recognizing the HA and FLAG tags, or the AtpB, GroEL, or OmpA proteins. Rabbit anti-HA and anti-FLAG antibodies were used at 1:2,000 dilution. Mouse anti-AtpB and anti-GroEL were used as control at 1:5,000 dilution. Rabbit anti-OmpA was used as control at 1:5,000 dilution. Secondary HRP-conjugated anti-rabbit or anti-mouse antiserum (GE Healthcare) was used at 1:5,000 dilution. The blots were developed with the SuperSignal West Femto Chemiluminescent system (Pierce).
Purification of the FtsA, UvrY, ClpS, ClpA, and ClpP Proteins.
For purification of the FtsA-FLAG and UvrY-FLAG proteins, 5-mL saturated cultures of E. coli BL21 (DE3) carrying plasmids pUHE-FtsA-FLAG or pUHE-UvrY-FLAG were used to inoculate 500 mL of LB medium. Cells were grown at 37 °C to logarithmic phase (OD600 ∼0.3), and expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) (0.2 mM) followed by growth at 30 °C for an additional 16 h. Cells were collected, washed twice with 1× Tris-buffered saline (TBS) [50 mM Tris⋅HCl (pH 8.0), 138 mM NaCl, and 2.7 mM KCl]. Then cells were resuspended with 1× TBS and were subjected to a French press at 18 kpsi. Cell debris was removed by centrifugation 12,800 × g at 4 °C for 60 min, and the supernatant was applied to a 2-mL anti-FLAG affinity resin (GenScript). The anti-FLAG agarose resin was washed twice with 20 mL of 1× TBS. Protein was recovered using the elution buffer (1× TBS buffer and 1 mg/mL 3×FLAG peptide), exchanged with 1× TBS buffer, followed by TBS buffer containing 10% glycerol, and concentrated using an Amicon Ultra-3 (MW 3,000; Millipore) filter.
To estimate the molecular weights of the FtsA-FLAG and UvrY-FLAG proteins, the corresponding purified proteins were analyzed using a size-exclusion column (Superdex 200 10/300 GL; GE Healthcare) at a flow rate of 0.5 mL/min in buffer containing 50 mM Tris⋅HCl (pH 8.0), 138 mM NaCl, 2.7 mM KCl, and 5% glycerol. To calculate the molecular weight, purified proteins of known molecular weight were run on the same column and used as standards. Preparation of standard proteins was conducted using the Gel Filtration Molecular Weight Markers kit (Sigma) following the manufacturer’s protocol.
For purification of the ClpS, ClpA, and ClpP proteins, 12-mL saturated cultures of E. coli BL21 (DE3) carrying plasmid pET-28a(+)-ClpS, pET-28a(+)-ClpA, or pET-28a(+)-ClpP were used to inoculate 1 L of LB medium containing 50 µg/L kanamycin. Bacteria were grown at 37 °C. Protein expression was induced at OD600 ∼0.6 with IPTG (0.5 mM). After induction, bacteria were grown at 30 °C for 5 h and were collected by centrifugation. The cell pellet was resuspended in equilibration buffer containing 50 mM Tris⋅HCl (pH 7.5), 200 mM KCl, 10% glycerol, and 2 mM DTT. Cells were broken by a French press at 25 kpsi. Clarified lysates were loaded onto a nickel- nitrilotriacetic acid (Ni-NTA) gravity column equilibrated in equilibration buffer containing 10 mM imidazole, washed with a 20-column volume of the same buffer, and eluted in 200 mM imidazole in the same buffer. ClpS-His6 was further purified through a size-exclusion column (Superdex 75 10/300 GL; GE Healthcare) in buffer containing 25 mM Hepes (pH 7.5), 150 mM KCl, 2 mM DTT, and 10% glycerol.
In Vitro Pull-Down Assays for Determination of Binding Affinity.
To measure the concentrations of FtsA-FLAG, UvrY-FLAG, and ClpS, known amounts of purified FtsA-FLAG, UvrY-FLAG, and ClpS proteins were run on the same gel and used as standards. Standard curves calculated from purified FtsA-FLAG, UvrY-FLAG, and ClpS proteins in the same blots were used for calculation of the concentrations and binding affinities of the FtsA-FLAG, UvrY-FLAG, and ClpS proteins. All proteins were incubated in 200 μL TBS at room temperature for 3 h. Then samples were pulled down with Ni-NTA magnetic agarose beads (GE Healthcare) at room temperature for 3 h. The magnetic agarose beads were washed three times with 0.2 mL TBS. Protein was recovered using elution buffer (1× TBS buffer and 500 mM imidazole). Proteins then were electrotransferred onto nitrocellulose membrane (iBlot; Life Technologies) following the manufacturer’s protocol and were detected by immunoblotting using monoclonal antibodies against FLAG or His and the secondary antibody HRP-conjugated anti-rabbit or anti-mouse IgG fragment (GE Healthcare). All proteins were visualized using the SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) and LAS-4000 (Fuji Film). The densities of protein bands were determined by quantification using ImageJ version 1.49u (NIH). The amounts of FtsA-FLAG, UvrY-FLAG, and ClpS proteins were then calculated from standard curves derived from serial dilutions of purified protein standards run on the same blot (Fig. S7C).
qRT-PCR.
To determine mRNA amounts of the genes of interest, bacteria were grown in N-minimal medium containing with 10 μM or 10 mM MgCl2 at 37 °C. To test the effect of mildly acidic pH on the expression of PhoP-dependent genes, bacteria were grown in N-minimal medium (pH 5.8) with 10 mM MgCl2. To test the effect of an antimicrobial peptide on the expression of PhoP-dependent genes, bacteria were grown in N-minimal medium (pH 7.7) with 10 mM MgCl2 and C18G (7 μg/mL). Total RNA was purified by using the RNeasy Kit (Qiagen) with on-column DNase treatment, and cDNA was synthesized using VILO Super Mix (Life Technologies). Quantification of transcripts was carried out by qRT-PCR using SYBR Green PCR Master Mix (Applied Biosystems) in a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). The mRNA amount was determined by using a standard curve obtained from PCR products generated with serially diluted genomic DNA, and results were normalized to the levels of the ompA gene. Data shown are an average from at least three independent experiments. The primers used in the qRT-PCR assay are presented in Table S2.
In Vivo Protein Degradation Assay.
To measure Oat-FLAG stability, bacteria were grown in 20 mL N-minimal medium containing 10 μM MgCl2 for 6 h. Bacteria were treated with chloramphenicol (1 mg/mL), and 1.5-mL samples were removed at the indicated times and harvested at 4 °C. Pelleted cells were kept on dry ice for 30 min. To measure stability of the FtsA and UvrY proteins, bacteria were grown in 10 mL N-minimal medium containing 10 μM MgCl2 for 5 h. Bacteria were treated with tetracycline (50 μg/mL) or chloramphenicol (1 mg/mL), and 1-mL samples were removed at the indicated times and harvested at 4 °C. Pelleted cells were kept on dry ice for 30 min. Samples were then resuspended in B-PER reagent (Pierce) with 100 μg/mL lysozyme and EDTA-free protease inhibitor (Roche). After the addition of the same volume of SDS sample buffer, samples were separated on 4–12% SDS-polyacrylamide gel and analyzed by Western blotting. For quantitative analysis of the blots, we used ImageJ version 1.49u (NIH).
In Vitro Substrate Degradation Assay.
In vitro substrate degradation assays were performed as described (44) with some modifications. The assay was performed in a solution containing 25 mM Tris⋅HCl (pH 7.5), 100 mM KCl, 12 mM MgCl2, and 2 mM DTT. Purified proteins were used at 0.2 μM ClpS, 0.2 μM ClpA, 0.2 μM ClpP, 0.2 μM FtsA-FLAG, and 0.5 μM UvrY-FLAG. Reactions were carried out at 30 °C for the indicated times in the presence of an ATP regeneration system (2 mM ATP, 20 μg/mL pyruvate kinase, and 4 mM pyruvate phosphate) started by the addition of substrates. Samples were removed from the reactions at the indicated times, and reactions were stopped by the addition of sample buffer. After separation by SDS/PAGE, proteins were detected by Coomassie blue staining (Invitrogen).
EMSA.
DNA fragments corresponding to the clpS, mgtC and cigR promoter regions were generated by PCR using wild-type Salmonella (14028s) or pclpS-d mutant (JY665) strains as templates and primer pairs 15939/15940, 15933/15936, and 15937/15938, respectively. PCR products were separated on a 1.0% agarose gel and purified with the QIAquick Gel Extraction Kit (Qiagen). One hundred nanograms of DNA labeled with T4 polynucleotide kinase (New England BioLabs) and [γ-32P]-ATP (PerkinElmer). Unincorporated [γ-32P]-ATP was removed by using G-50 microcolumns (GE Healthcare). Ten femtomoles of labeled DNA probes were mixed with various amounts of purified PhoP protein in binding buffer [20 mM Tris HCl (pH 8.0), 2 mM MgCl2, 10 mM KCl2, 10% (vol/vol) glycerol, 0.1 mM DTT, 60 μg/mL BSA, and 10 μg/mL of poly (dI-dC)] in a total volume of 20 μL. Following incubation at room temperature for 20 min, samples were electrophoresed on 6% Tris/borate/EDTA gels (Life Technologies) at 100 V for 90 min.
Antibiotic Persistence Assay.
Salmonella cells were grown in N-minimal medium containing 10 μM MgCl2 for 6 h at 37 °C. Then kanamycin (100 μg/mL) or ciprofloxacin (2 μg/mL) was added, and incubation continued for 2 h. Cells were washed with fresh N-minimal medium containing 10 μM MgCl2. Persistence cell formation was determined by spotting 10-fold serial dilutions onto LB agar plates. Plates then were incubated at 37 °C for 16 h.
Supplementary Material
Acknowledgments
We thank Arthur L. Horwich for useful advice on this work, Xinyu Hong for the identification of the PhoP-binding site in the clpS promoter region, Susan Gottesman, Michael R. Maurizi, and Patricia Sanchez-Vazquez for critically reading an earlier version of this manuscript, and two anonymous reviewers for their useful comments. This research was supported by NIH Grant AI49561 (to E.A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722246115/-/DCSupplemental.
References
- 1.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 2.Kirstein J, Molière N, Dougan DA, Turgay K. Adapting the machine: Adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 3.Kuhlmann NJ, Chien P. Selective adaptor dependent protein degradation in bacteria. Curr Opin Microbiol. 2017;36:118–127. doi: 10.1016/j.mib.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- 5.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Dougan DA, Truscott KN, Zeth K. The bacterial N-end rule pathway: Expect the unexpected. Mol Microbiol. 2010;76:545–558. doi: 10.1111/j.1365-2958.2010.07120.x. [DOI] [PubMed] [Google Scholar]
- 7.Yeom J, Wayne KJ, Groisman EA. Sequestration from protease adaptor confers differential stability to protease substrate. Mol Cell. 2017;66:234–246.e5. doi: 10.1016/j.molcel.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Francis LI, Jonas K, Laub MT, Chien P. ClpAP is an auxiliary protease for DnaA degradation in Caulobacter crescentus. Mol Microbiol. 2016;102:1075–1085. doi: 10.1111/mmi.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groisman EA, et al. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet. 2013;47:625–646. doi: 10.1146/annurev-genet-051313-051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez JC, et al. Evolution of a bacterial regulon controlling virulence and Mg(2+) homeostasis. PLoS Genet. 2009;5:e1000428. doi: 10.1371/journal.pgen.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derzelle S, et al. The PhoP-PhoQ two-component regulatory system of Photorhabdus luminescens is essential for virulence in insects. J Bacteriol. 2004;186:1270–1279. doi: 10.1128/JB.186.5.1270-1279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barchiesi J, Castelli ME, Di Venanzio G, Colombo MI, García Véscovi E. The PhoP/PhoQ system and its role in Serratia marcescens pathogenesis. J Bacteriol. 2012;194:2949–2961. doi: 10.1128/JB.06820-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groisman EA, Chiao E, Lipps CJ, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci USA. 1989;86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García Véscovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: Environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 16.Bader MW, et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 17.Prost LR, et al. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Choi J, Groisman EA. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol Microbiol. 2016;101:1024–1038. doi: 10.1111/mmi.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colgan AM, et al. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar typhimurium. PLoS Genet. 2016;12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee E-J, Pontes MH, Groisman EA. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell. 2013;154:146–156. doi: 10.1016/j.cell.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varshavsky A. ‘Spalog’ and ‘sequelog’: Neutral terms for spatial and sequence similarity. Curr Biol. 2004;14:R181–R183. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Snavely MD, Gravina SA, Cheung TT, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J Biol Chem. 1991;266:824–829. [PubMed] [Google Scholar]
- 23.Snavely MD, Miller CG, Maguire ME. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. 1991;266:815–823. [PubMed] [Google Scholar]
- 24.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 25.Spinelli SV, Pontel LB, García Véscovi E, Soncini FC. Regulation of magnesium homeostasis in Salmonella: Mg(2+) targets the mgtA transcript for degradation by RNase E. FEMS Microbiol Lett. 2008;280:226–234. doi: 10.1111/j.1574-6968.2008.01065.x. [DOI] [PubMed] [Google Scholar]
- 26.Pontes MH, Yeom J, Groisman EA. Reducing ribosome biosynthesis promotes translation during low Mg2+ stress. Mol Cell. 2016;64:480–492. doi: 10.1016/j.molcel.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SY, Groisman EA. Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol Microbiol. 2014;91:135–144. doi: 10.1111/mmi.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottesman S, Clark WP, de Crecy-Lagard V, Maurizi MR. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 29.Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 30.Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol. 2012;84:463–485. doi: 10.1111/j.1365-2958.2012.08036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottesman S, Clark WP, Maurizi MR. The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J Biol Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- 33.Williams B, Bhat N, Chien P, Shapiro L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol Microbiol. 2014;93:853–866. doi: 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider BL, Hernandez VJ, Reitzer L. Putrescine catabolism is a metabolic response to several stresses in Escherichia coli. Mol Microbiol. 2013;88:537–550. doi: 10.1111/mmi.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tkachenko AG, Kashevarova NM, Karavaeva EA, Shumkov MS. Putrescine controls the formation of Escherichia coli persister cells tolerant to aminoglycoside netilmicin. FEMS Microbiol Lett. 2014;361:25–33. doi: 10.1111/1574-6968.12613. [DOI] [PubMed] [Google Scholar]
- 36.De Groote VN, et al. Novel persistence genes in Pseudomonas aeruginosa identified by high-throughput screening. FEMS Microbiol Lett. 2009;297:73–79. doi: 10.1111/j.1574-6968.2009.01657.x. [DOI] [PubMed] [Google Scholar]
- 37.Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc Natl Acad Sci USA. 2006;103:13503–13508. doi: 10.1073/pnas.0606026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zere TR, et al. Genomic targets and features of BarA-UvrY (-SirA) signal transduction systems. PLoS One. 2015;10:e0145035. doi: 10.1371/journal.pone.0145035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pernestig AK, Melefors O, Georgellis D. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J Biol Chem. 2001;276:225–231. doi: 10.1074/jbc.M001550200. [DOI] [PubMed] [Google Scholar]
- 40.Blanc-Potard AB, Groisman EA. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battesti A, et al. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 2013;27:2722–2735. doi: 10.1101/gad.229617.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68:298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 43.McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 44.Farrell CM, Grossman AD, Sauer RT. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol. 2005;57:1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- 45.Dougan DA, Reid BG, Horwich AL, Bukau B. ClpS, a substrate modulator of the ClpAP machine. Mol Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 46.Mitra A, Palaniyandi S, Herren CD, Zhu X, Mukhopadhyay S. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS One. 2013;8:e55492. doi: 10.1371/journal.pone.0055492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakamoto A, et al. Enhanced biofilm formation and/or cell viability by polyamines through stimulation of response regulators UvrY and CpxR in the two-component signal transducing systems, and ribosome recycling factor. Int J Biochem Cell Biol. 2012;44:1877–1886. doi: 10.1016/j.biocel.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 49.Ahmer BM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin ME, Trimble MJ, Brun YV. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol Microbiol. 2004;54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- 51.Pichoff S, Lutkenhaus J. Identification of a region of FtsA required for interaction with FtsZ. Mol Microbiol. 2007;64:1129–1138. doi: 10.1111/j.1365-2958.2007.05735.x. [DOI] [PubMed] [Google Scholar]
- 52.Stephani K, Weichart D, Hengge R. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli. Mol Microbiol. 2003;49:1605–1614. doi: 10.1046/j.1365-2958.2003.03644.x. [DOI] [PubMed] [Google Scholar]
- 53.Choi E, Groisman EA, Shin D. Activated by different signals, the PhoP/PhoQ two-component system differentially regulates metal uptake. J Bacteriol. 2009;191:7174–7181. doi: 10.1128/JB.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kox LF, Wösten MM, Groisman EA. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000;19:1861–1872. doi: 10.1093/emboj/19.8.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato A, Groisman EA. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004;18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis RW, Botstein D, Roth JR. 1980. Advanced Bacterial Genetics (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY)
- 58.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 60.Pontes MH, Lee EJ, Choi J, Groisman EA. Salmonella promotes virulence by repressing cellulose production. Proc Natl Acad Sci USA. 2015;112:5183–5188. doi: 10.1073/pnas.1500989112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soncini FC, Véscovi EG, Groisman EA. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 63.Zwir I, et al. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 65.Shin D, Groisman EA. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem. 2005;280:4089–4094. doi: 10.1074/jbc.M412741200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.