Significance
The World Health Organization recommended pilot deployment of Wolbachia-infected mosquitoes to curb viral transmission to humans. Releases of mosquitoes are underway worldwide because Wolbachia can block replication of these pathogenic viruses and deterministically spread by a drive system termed cytoplasmic incompatibility (CI). Despite extensive research, the underlying genetic basis of CI remains only half-solved. We recently reported that two prophage WO genes recapitulate the modification component of CI in a released strain for vector control. Here we show that one of these genes underpins rescue of CI. Together, our results reveal the complete genetic basis of this selfish trait and pave the way for future studies exploring WO prophage genes as adjuncts or alternatives to current control efforts.
Keywords: cytoplasmic incompatibility, rescue, Wolbachia, prophage WO, Drosophila melanogaster
Abstract
Wolbachia are maternally inherited, intracellular bacteria at the forefront of vector control efforts to curb arbovirus transmission. In international field trials, the cytoplasmic incompatibility (CI) drive system of wMel Wolbachia is deployed to replace target vector populations, whereby a Wolbachia-induced modification of the sperm genome kills embryos. However, Wolbachia in the embryo rescue the sperm genome impairment, and therefore CI results in a strong fitness advantage for infected females that transmit the bacteria to offspring. The two genes responsible for the wMel-induced sperm modification of CI, cifA and cifB, were recently identified in the eukaryotic association module of prophage WO, but the genetic basis of rescue is unresolved. Here we use transgenic and cytological approaches to demonstrate that maternal cifA expression independently rescues CI and nullifies embryonic death caused by wMel Wolbachia in Drosophila melanogaster. Discovery of cifA as the rescue gene and previously one of two CI induction genes establishes a “Two-by-One” model that underpins the genetic basis of CI. Results highlight the central role of prophage WO in shaping Wolbachia phenotypes that are significant to arthropod evolution and vector control.
The bacteria Wolbachia occur in an estimated 40–52% of arthropod species (1, 2) and 47% of the Onchocercidae family of filarial nematodes (3), making them the most widespread bacterial symbiont in the animal kingdom (2). In arthropods, Wolbachia mainly reside in the cells of the reproductive tissues, transmit transovarially (4), and often commandeer host fertility, sex ratios, and sex determination to enhance their maternal transmission via male killing, feminization, parthenogenesis, or cytoplasmic incompatibility (CI) (5, 6).
Discovered nearly half a century ago (7), Wolbachia-induced CI is the most common reproductive modification and results in embryonic lethality when an infected male mates with an uninfected female, but this lethality is rescued when the female is likewise infected (8). As such, rescue can provide a strong fitness advantage to infected females, the transmitting sex of Wolbachia (9–11). Alone, CI-induced lethality is deployed in vector control studies to crash the resident uninfected mosquito population through release of Wolbachia-infected males (12–17). Together, CI-induced lethality and rescue constitute a microbial drive system that is used in field studies worldwide to stably replace an uninfected mosquito population with an infected one via release of males and females harboring wMel Wolbachia (18), which confer resistance against dengue and Zika viruses (19, 20). The efficacy of this drive system for spreading Wolbachia in target populations critically depends on Wolbachia’s ability to rescue its own lethal sperm modifications.
While CI is gaining momentum as a natural, sustainable, and inexpensive tool for vector control, the genes that underpin this microbial adaptation are not fully known. Our previous screen of Wolbachia genomes and transcriptomes from infected ovaries identified two adjacent genes, cifA and cifB, from the wMel strain in Drosophila melanogaster as the only genes strictly associated with CI (21). These two genes occur in the eukaryotic association module of prophage WO (22) and recapitulate CI when dually expressed in uninfected male flies (21, 23). Each gene alone is incapable of inducing CI (21), and the rescue gene remains unknown. As cifA and cifB are the only two wMel genes strictly associated with CI, we previously hypothesized that the CI induction and rescue genes might be the same (21). Here we test the hypothesis that transgenic expression of cifA and/or cifB genes from wMel Wolbachia in ovaries can rescue CI and nullify the associated embryonic defects in D. melanogaster.
Results and Discussion
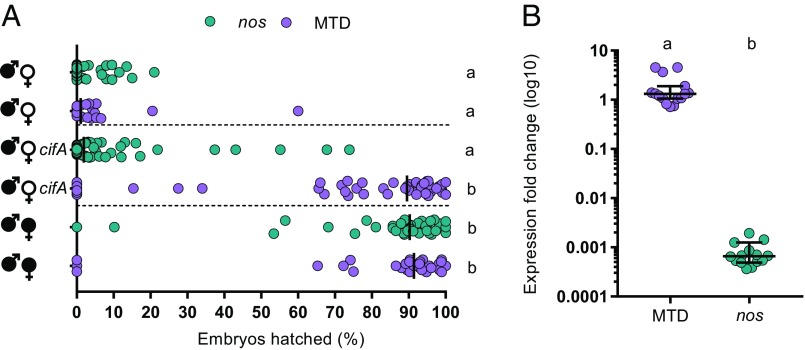
Since Wolbachia cannot be genetically transformed, we tested the ability of cifA to transgenically rescue wild-type CI using a GAL4-UAS system for tissue-specific expression in uninfected D. melanogaster females. In transcriptomes of wMel-infected D. melanogaster, cifA is a highly expressed prophage WO gene (24). As such, we conducted the transgenic experiments under the control of either nos-GAL4-tubulin in uninfected germline stem cells or the maternal triple driver, MTD-GAL4, to drive higher transgene expression throughout oogenesis. MTD-GAL4 utilizes two nos-GAL4 driver variants (including nos-GAL4-tubulin) and an ovarian tumor driver (25). Control CI and rescue crosses with either driver yielded the expected hatching rates. Crosses between infected males and uninfected females expressing cifA under the control of MTD-GAL4 showed a markedly significant increase in embryonic hatching relative to cifA expression under nos-GAL4-tubulin and at levels similar to that in control rescue crosses (Fig. 1A). These results are consistent with complete rescue of CI by cifA, in association with increased expression throughout the developing egg chambers. Similar results with nos-GAL4-tubulin expression in uninfected ovaries resulted in a small increase in hatch rate that was inconsistently significant among replicates (Fig. S1). An analysis of cifA gene expression reveals MTD-GAL4 associates with a three-order-of-magnitude increase over nos-GAL4-tubulin, supporting strength of expression as a factor for rescue (Fig. 1B).
Fig. 1.
cifA rescues cytoplasmic incompatibility when it is highly expressed throughout oogenesis. (A) Hatch rate assays were conducted with transgenic expression of cifA under the control of nos-GAL4-tubulin or MTD-GAL4 drivers. Each dot represents a replicate. Rescue occurred only under MTD-GAL4 expression. Horizontal dotted lines from top to bottom separate cross-types with CI, cifA expression, and rescue. Wolbachia infections are represented by filled sex symbols, and expressed genes are noted to the right of the corresponding sex. n = 27–59 for each experimental cross across two experiments (both shown). Vertical bars represent medians, and letters to the right indicate significant differences based on α = 0.05 calculated by Kruskal–Wallis and Dunn’s test for multiple comparisons. (B) Gene expression fold change of cifA relative to the Drosophila housekeeping gene rp49 was determined on a subset of abdomens from females expressing cifA via MTD-GAL4 or nos-GAL4-tubulin with 2−∆∆Ct. Horizontal bars represent medians with 95% confidence intervals, and letters above indicate significance based on a Mann–Whitney U test. In both cases, statistical comparisons are between all groups. Exact P values are provided in Table S2. Hatch rate experiments testing expression of cifA under MTD-GAL4 or nos-GAL4-tubulin have been repeated four and five times, respectively.
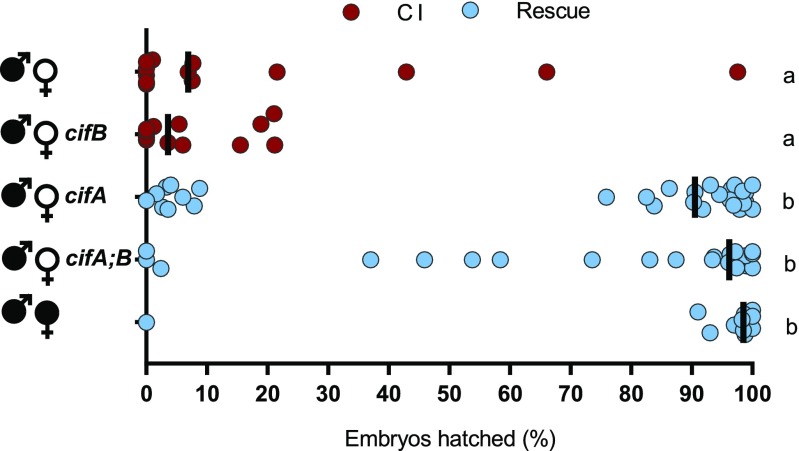
We expanded our evaluation of cif gene expression under the control of MTD-GAL4 in uninfected females to test if cifB alone or in combination with cifA impacts CI penetrance. As expected, infected males crossed to either uninfected females or females transgenically expressing cifB under MTD-GAL4 yielded similar CI penetrance (Fig. 2). These results suggest that cifB does not rescue CI when transgenically expressed in the ovaries, and its CI-related function is specific to testes. In contrast, MTD-GAL4 expression of cifA, by itself or in combination with cifB, significantly rescued CI to levels comparable to rescue by infected females (Fig. 2). These results are consistent with cifA independently functioning as the rescue factor and suggest that cifB does not inhibit cifA’s ability to rescue CI. As Wolbachia can induce phenotypes known to bias sex ratios, we collected the surviving offspring from the transgenic and control rescue crosses and sexed them to demonstrate normal sex ratios, indicating that rescue was not sex-specific (Fig. S2).
Fig. 2.
Rescue of cytoplasmic incompatibility is specific to cifA. Hatch rate assays were conducted with transgenic expression of cifA, cifB, and cifA;B using the MTD-GAL4 driver for expression throughout oogenesis. Each dot represents a replicate. Wolbachia infections are represented by filled sex symbols, and expressed genes are noted to the right of the corresponding sex. n = 11–29 for each experimental cross. Vertical bars represent medians, and letters to the right indicate significant differences based on α = 0.05 calculated by Kruskal–Wallis and Dunn’s test for multiple comparisons. Statistical comparisons are between all groups. Exact P values are provided in Table S2. Hatch rate experiments testing expression of cifA under MTD-GAL4 have been repeated four times.
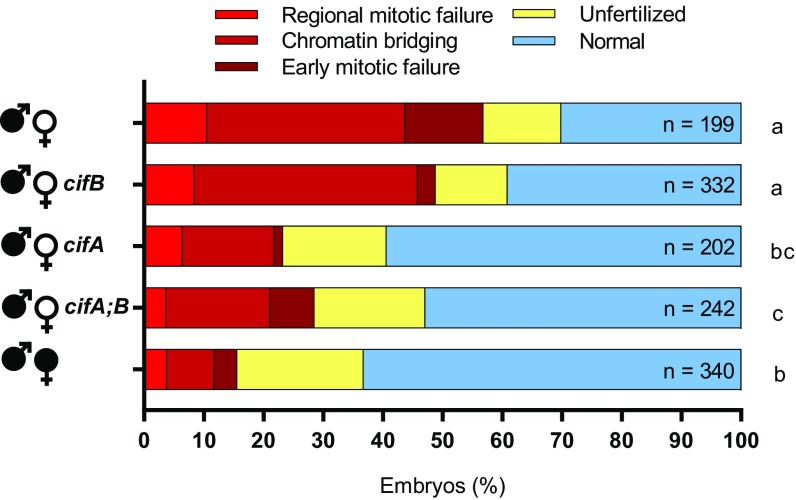
Next, we tested if the canonical cytological defects observed in early CI embryos [early mitotic failure, chromatin bridging, and regional mitotic failure (26)] were nullified under cifA-induced rescue. We examined embryos from control and transgenic crosses after 1–2 h of development and binned their cytology into one of five phenotypes as previously established for D. melanogaster CI (21). Nearly half of CI-induced lethality in embryos is the result of embryonic arrest during advanced developmental stages in Dipteran species (27–30). As expected, the control CI cross yielded high levels of all three CI-associated defects, and the embryos from the control rescue cross developed with significantly fewer abnormalities (Fig. 3). MTD-GAL4 transgenic expression of cifA in uninfected females, either alone or dually expressed with cifB, resulted in significantly fewer cytological defects (Fig. 3). These effects were not seen with transgenic cifB expression, again validating that cifA alone can recapitulate wild-type rescue by Wolbachia.
Fig. 3.
cifA rescues embryonic defects caused by cytoplasmic incompatibility. The percent of embryos with each cytological phenotype resulting from the indicated crosses are shown. All crosses were conducted in parallel and with sisters from the experiment in Fig. 2. cifA, cifB, and cifA;B transgene expression was under the control of MTD-GAL4. Wolbachia infections are represented by filled sex symbols and expressed genes are noted to the right of the corresponding sex. Letters to the right indicate significant differences based on α = 0.05 calculated by pairwise χ2 analyses comparing defects (all shades of red) against normal (blue) with Bonferroni adjusted P values. Exact P values are provided in Table S2. This experiment has been conducted once.
These data are in contrast with previous work reporting the inability to transgenically rescue CI in D. melanogaster (23); however, there are three critical differences between the studies. First, wPip’s homologs from Culex pipiens were used in the prior work instead of wMel’s cif genes from D. melanogaster here. Thus, differences in host background interactions could explain the discrepancy. Second, a T2A sequence between the wPip gene homologs was used to allow for bicistronic expression, but ribosome skipping results in a C-terminal sequence extension to the first protein and a proline addition to the second protein that generates sequence artifacts and could alter function (31). Finally, different insertion sites are capable of different levels of expression due to their local chromatin environment (32), thus the chosen sites may produce insufficient product to cause rescue.
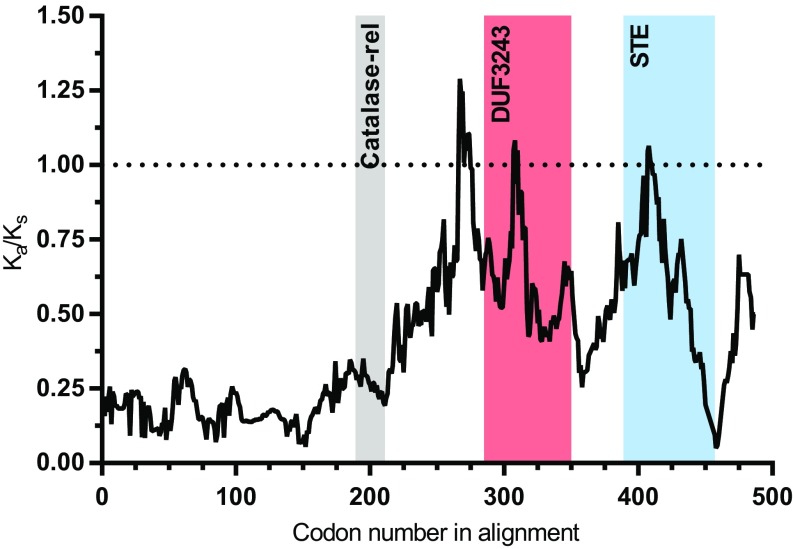
cifA encodes a putative catalase-rel domain, a sterile-like transcription factor (STE) domain, and a domain of unknown function (DUF3243) that shares homology with a putative Puf-family RNA binding domain in cifA-like homologs (33), whereas cifB has nuclease and deubiquitilase domains (23, 33). Only the deubiquitilase annotation has been functionally tested and confirmed (23). Based on subcellular localization (PSORTb) and transmembrane helix predictors (TMbase), CifA is a cytoplasmic protein without transmembrane helices (Fig. S3). Codon-based and Fisher’s exact tests of neutrality demonstrate that closely related (76.2–99.8% pairwise nucleotide identity) type I cifA homologs (21) largely evolve by purifying selection (Fig. S4 A and B), and sliding window analyses [sliding window analysis of Ka and Ks (SWAKK) and Java codon delimited alignment, JCoDA] reveal that purifying selection is strongest on the catalase-rel domain and the unannotated region at the N terminus, with considerably weaker purifying selection on the putative DUF3243 and STE domains (Fig. 4 and Fig. S4C). This is supported by prior work reporting stronger amino acid conservation within the type I CifA N terminus relative to the C terminus (33).
Fig. 4.
Ka/Ks sliding window analysis identifies cifA regions evolving under negative selection. A comparison between cifA homologs from wMel and wHa rejects the neutral expectation of Ka/Ks = 1 using a 25-amino-acid sliding window across most of cifA. Strong purifying selection is observed in several cifA regions including the sequence preceding the catalase-rel domain. Shaded regions denote previously described protein domain predictions (33).
These findings illustrate that the Wolbachia prophage WO gene cifA recapitulates rescue of wild-type CI. As cifA is one of two genes involved in induction of CI, results support the hypothesis that a gene involved in CI induction is also the rescue gene (21). In addition, transgenic expression of cifA in yeast inhibits a temperature-dependent growth defect caused by cifB expression (23). The discovery that CI is induced by cifA and cifB and rescued by cifA motivates a Two-by-One model of CI where two genes act as the CI modification factors (in the male), and one of these same genes acts as the rescue factor (in the female). This modification-rescue model posits that each strain of Wolbachia has its own set of cifA- and cifB-associated CI modifications and one cifA rescue factor. The different roles of cifA in CI and rescue are intriguing. We predict that the function of cifA is dependent on differential localization and/or modification of gene products in testes/sperm (CI) relative to ovaries/embryos (rescue). Moreover, one could speculate that the putative antioxidant catalase-rel domain of the CifA protein acts as a functional switch in response to reactive oxygen species, known to be higher in Wolbachia-infected testes (34), whereas the Puf-family RNA binding domain and STE are involved in RNA binding and transcriptional (mis)regulation of an unknown host factor.
It has been hypothesized that divergence in modification and rescue genes leads to bidirectional CI (21, 35, 36), which is a reciprocal incompatibility between males and females infected with different Wolbachia strains (7, 37–40). Comparative genomic analyses of cifA and cifB genes reveal extremely high levels of amino acid divergence (21), strong codivergence (21, 33), and recombination (36), consistent with the very rapid evolution of bidirectional CI across Wolbachia that can contribute to reproductive isolation and speciation (40, 41). Indeed, divergence of the cifA and cifB genes into several phylogenetic types correlates with bidirectional CI patterns in Drosophila and Culex (21, 36). There are at least two explanations for how simple genetic changes in these genes can contribute to bidirectional CI. First, a single mutation in the cifA gene could produce variation in the modification and rescue components that render two Wolbachia strains incompatible. For instance, given an ancestral and derived allele of cifA, males and females with Wolbachia carrying the same cifA allele are compatible; however, males with Wolbachia carrying the ancestral cifA allele cause a sperm modification that is unable to be rescued by embryos with Wolbachia carrying the derived cifA allele, and vice versa. Thus, a single mutation in cifA alone can enable the switch from being compatible to incompatible Wolbachia. Second, mutations in both cifA and cifB could be required for the evolution of bidirectional CI. For example, CifA-CifB protein binding (23) and/or differential localization in the sperm and egg may underpin bidirectional CI between Wolbachia strains. In this model, amino acid divergence in the Cif proteins may contribute to weakened binding, which in turn yields Wolbachia strains incapable of CI but capable of rescuing CI by the ancestral variant (42, 43). A compensatory substitution in the other Cif protein could in theory restore binding and yield bidirectional incompatibility with the ancestral Cif variants. Codivergence between amino acid sequences of these proteins is consistent with this model. Under both models, the presence of multiple WO prophages carrying cifA genes may also promote incompatibilities through the production of multiple CI product complexes simultaneously (21). In support of these hypotheses, complex diversification and duplication of cifA and cifB have been reported in Drosophila and C. pipiens that harbor a variety of incompatible Wolbachia strains (21, 36).
In conclusion, our findings reveal the connected genetic basis of CI and rescue and highlight the fundamental impact of prophage WO genes on the adaptive phenotypes of an obligate intracellular bacteria. In addition to genetically dissecting this widespread form of reproductive parasitism and microbial drive, we also establish a Two-by-One model to explain the modification and rescue components of CI. Finally, beneficial applications of CI and rescue genes as transgenic drive constructs may be possible as adjuncts or alternatives to pest control or vector control strategies currently deploying Wolbachia-infected mosquitoes (15–18).
Materials and Methods
Fly Rearing and Strains.
D. melanogaster stocks y1w* (BDSC 1495), nos-GAL4-tubulin (BDSC 4442), MTD-GAL4 (containing nos-GAL4-tubulin, nos-GAL4-VP16, and otu-GAL4-VP16; BDSC 31777), and UAS transgenic lines homozygous for cifA, cifB, and cifA;B (21) were maintained at 12:12 light:dark at 25 °C and 70% relative humidity (RH) on 50 mL of a standard media. GAL4 lines were found to be infected with wMel Wolbachia, and uninfected lines were produced through tetracycline treatment as previously described (21). Infection status was frequently confirmed via PCR using WolbF and WolbR3 primers (44) (Table S2). During virgin collections, flies were stored at 18 °C overnight to slow eclosion rate, and virgin flies were kept at room temperature.
Hatch Rate and Sex Ratio Assays.
Virgin MTD-GAL4 females were collected for the first 3 d of emergence and aged 9–11 d before crossing to nonvirgin homozygous UAS (cifA, cifB, or cifA;B) males. The start of collections for the maternal and paternal lineages was staggered by 7 d. Single pair matings occurred in 8-oz bottles, and a grape-juice agar plate was smeared with yeast and affixed to the opening of each bottle with tape. The flies and bottles were then stored at 25 °C and 70% RH for 24 h, at which time the plates were replaced with freshly smeared plates and again stored for 24 h. Plates were then removed and the number of embryos on each plate were counted and stored. After 30 h the remaining unhatched embryos were counted (Fig. S6). The hatch rate was calculated by dividing the number of hatched embryos by the initial embryo count and multiplying by 100. Hatch rate was plotted against clutch size for 3 MTD-GAL4 and 4 nos-GAL4-tubulin rescue crosses conducted in this study to reveal a significant correlation (Fig. S5), and a threshold clutch size for analysis was set equal to exclusion of 99% of plates with a hatch rate of 0 for each genotype (31 for nos-GAL4-tubulin and 48 for MTD-GAL4). Larvae were moved into vials of standard media and the offspring sex ratio determined after 15–18 d (Fig. S6). Hatch rates testing MTD-GAL4 or nos-GAL4-tubulin expression of cifA were conducted four and five times, respectively. Sex ratio experiments were conducted once.
Gene Expression.
To compare the level of UAS-cifA expression between MTD-GAL4 and nos-GAL4-tubulin flies, mothers from hatch rate assays were collected after the allotted laying period, abdomens were immediately dissected, and samples were frozen in liquid nitrogen and stored at −80 °C until processing. RNA was extracted using the Direct-zol RNA MiniPrep Kit (Zymo), DNase treated with DNA-free (Ambion, Life Technologies), and cDNA was generated with SuperScript VILO (Invitrogen). Quantitative PCR was performed on a Bio-Rad CFX-96 Real-Time System using iTaq Universal SYBR Green Supermix (Bio-Rad). Forty cycles of PCR were performed against positive controls (extracted DNA), negative controls (water), no RT control (RNA), and cDNA with the following conditions: 50 °C 10 min, 95 °C 5 min, 40× (95 °C 10 s, 55 °C 30 s), 95 °C 30 s. Primers used were cifA opt and rp49 forward and reverse (Table S1). Fold expression of UAS-cifA relative to the D. melanogaster housekeeping gene rp49 was determined with 2−∆∆Ct. This experiment and corresponding hatch rate were performed once.
Embryo Cytology.
Flies were collected as described for the hatch rate assays, but with 60 females and 12 males in each bottle with a grape-juice agar plate attached. All flies used were siblings of those from the hatch rate, grape-juice plates replaced as described above, and embryos collected in parallel to egg laying by hatch rate females. Embryos were collected, dechorionated, washed, methanol fixed, stained with propidium iodide, imaged, and categorized as previously described (21) (Fig. S6). This experiment was performed once.
Putative CifA Localization.
The PSORTb v3.0.2 web server (45) was used to predict subcellular localization of the wMel CifA protein to either the cytoplasm, cytoplasmic membrane, periplasm, outer membrane, or extracellular space. A localization score is provided for each location, with scores of 7.5 or greater considered probable localizations. The TMpred web server (46) was used to predict transmembrane helices in wMel CifA. TMpred scores were generated for transmembrane helices spanning from inside-to-outside (i-o) and outside-to-inside (o-i), and scores above 500 are considered significant.
cifA Selection Analyses.
Selection analyses were conducted using four independent tests of selection: codon-based Z test of neutrality (47), Fisher’s exact test of neutrality (47), Sliding window analysis of Ka and Ks (SWAKK) (48), and Java Codon Delimited Alignment (JCoDA) (49). The first two analyses were conducted using the MEGA7 desktop app with a MUSCLE translation alignment generated in Geneious v5.5.9. The SWAKK 2.1 web server and the JCoDA v1.4 desktop app were used to analyze divergence between wMel and wHa cifA with a sliding window of 25 codons and a jump size of 1 codon for SWAKK and 5 codons for JCoDA.
Statistical Analyses.
All statistical analyses were conducted in GraphPad Prism (Prism 7 or online tools). Hatch rate and sex ratio statistical comparisons were made using Kruskal–Wallis followed by a Dunn’s multiple comparison test. Expression was compared using a Mann–Whitney U test. Correlations between hatch rate and clutch size were determined using Spearman rho. Pairwise χ2 analyses were used for cytology studies to compare defective and normal embryos followed by generation of Bonferroni adjusted P values. An unpaired t test was used for statistical comparison of RNA fold expression. All P values are reported in Table S2.
Data Availability.
All source data and replicate data are available as Supporting Information along with this publication.
Supplementary Material
Acknowledgments
We thank Daniel LePage for preliminary work, Sarah R. Bordenstein and Jessamyn Perlmutter for manuscript preparation, and Jared Nordman for kindly providing the MTD-GAL4 line. This work was supported by National Institutes of Health (NIH) Awards R01 AI132581 and R21 HD086833 (to S.R.B.), National Science Foundation Award IOS 1456778 (to S.R.B.), and a National Science Foundation Graduate Research Fellowship (to J.D.S.). Imaging was performed in part using the Vanderbilt University Medical Center Cell Imaging Shared Resources (supported by NIH Grants CA68485, DK20593, DK58404, DK59637, and EY08126). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect the views of the NIH or the National Science Foundation.
Footnotes
Conflict of interest statement: J.D.S. and S.R.B. are listed as inventors on a provisional patent relevant to this work. S.R.B. is a coinventor on two other pending patents related to controlling arthropods.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800650115/-/DCSupplemental.
References
- 1.Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc Biol Sci. 2015;282:20150249. doi: 10.1098/rspb.2015.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One. 2012;7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferri E, et al. New insights into the evolution of Wolbachia infections in filarial nematodes inferred from a large range of screened species. PLoS One. 2011;6:e20843. doi: 10.1371/journal.pone.0020843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frydman HM, Li JM, Robson DN, Wieschaus E. Somatic stem cell niche tropism in Wolbachia. Nature. 2006;441:509–512. doi: 10.1038/nature04756. [DOI] [PubMed] [Google Scholar]
- 5.Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 6.LePage D, Bordenstein SR. Wolbachia: Can we save lives with a great pandemic? Trends Parasitol. 2013;29:385–393. doi: 10.1016/j.pt.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yen JH, Barr AR. The etiological agent of cytoplasmic incompatibility in Culex pipiens. J Invertebr Pathol. 1973;22:242–250. doi: 10.1016/0022-2011(73)90141-9. [DOI] [PubMed] [Google Scholar]
- 8.Serbus LR, Casper-Lindley C, Landmann F, Sullivan W. The genetics and cell biology of Wolbachia-host interactions. Annu Rev Genet. 2008;42:683–707. doi: 10.1146/annurev.genet.41.110306.130354. [DOI] [PubMed] [Google Scholar]
- 9.Turelli M, Hoffmann AA. Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol Biol. 1999;8:243–255. doi: 10.1046/j.1365-2583.1999.820243.x. [DOI] [PubMed] [Google Scholar]
- 10.Hancock PA, Sinkins SP, Godfray HC. Strategies for introducing Wolbachia to reduce transmission of mosquito-borne diseases. PLOS Negl Trop Dis. 2011;5:e1024. doi: 10.1371/journal.pntd.0001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock PA, Sinkins SP, Godfray HC. Population dynamic models of the spread of Wolbachia. Am Nat. 2011;177:323–333. doi: 10.1086/658121. [DOI] [PubMed] [Google Scholar]
- 12.Atyame CM, et al. Cytoplasmic incompatibility as a means of controlling Culex pipiens quinquefasciatus mosquito in the islands of the south-western Indian Ocean. PLoS Negl Trop Dis. 2011;5:e1440. doi: 10.1371/journal.pntd.0001440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobson SL, Bordenstein SR, Rose RI. Wolbachia mosquito control: Regulated. Science. 2016;352:526–527. doi: 10.1126/science.352.6285.526-b. [DOI] [PubMed] [Google Scholar]
- 14.O’Connor L, et al. Open release of male mosquitoes infected with a Wolbachia biopesticide: Field performance and infection containment. PLoS Negl Trop Dis. 2012;6:e1797. doi: 10.1371/journal.pntd.0001797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Zheng X, Xi Z, Bourtzis K, Gilles JR. Combining the sterile insect technique with the incompatible insect technique: I-impact of Wolbachia infection on the fitness of triple- and double-infected strains of Aedes albopictus. PLoS One. 2015;10:e0121126. doi: 10.1371/journal.pone.0121126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie SA, Townsend M, Paton CJ, Callahan AG, Hoffmann AA. Application of wMelPop Wolbachia strain to crash local populations of Aedes aegypti. PLoS Negl Trop Dis. 2015;9:e0003930. doi: 10.1371/journal.pntd.0003930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zabalou S, et al. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci USA. 2004;101:15042–15045. doi: 10.1073/pnas.0403853101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann AA, et al. Stability of the wMel Wolbachia Infection following invasion into Aedes aegypti populations. PLOS Negl Trop Dis. 2014;8:e3115. doi: 10.1371/journal.pntd.0003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker T, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 20.Dutra HL, et al. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe. 2016;19:771–774. doi: 10.1016/j.chom.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LePage DP, et al. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature. 2017;543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordenstein SR, Bordenstein SR. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun. 2016;7:13155. doi: 10.1038/ncomms13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beckmann JF, Ronau JA, Hochstrasser M. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2017;2:17007. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutzwiller F, et al. Dynamics of Wolbachia pipientis gene expression across the Drosophila melanogaster life cycle. G3 (Bethesda) 2015;5:2843–2856. doi: 10.1534/g3.115.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- 26.Landmann F, Orsi GA, Loppin B, Sullivan W. Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog. 2009;5:e1000343. doi: 10.1371/journal.ppat.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lassy CW, Karr TL. Cytological analysis of fertilization and early embryonic development in incompatible crosses of Drosophila simulans. Mech Dev. 1996;57:47–58. doi: 10.1016/0925-4773(96)00527-8. [DOI] [PubMed] [Google Scholar]
- 28.Callaini G, Riparbelli MG, Giordano R, Dallai R. Mitotic defects associated with cytoplasmic incompatibility in Drosophila simulans. J Invertebr Pathol. 1996;67:55–64. [Google Scholar]
- 29.Wright JD, Barr AR. Wolbachia and the normal and incompatible eggs of Aedes polynesiensis (Diptera: Culicidae) J Invertebr Pathol. 1981;38:409–418. [Google Scholar]
- 30.Duron O, Weill M. Wolbachia infection influences the development of Culex pipiens embryo in incompatible crosses. Heredity (Edinb) 2006;96:493–500. doi: 10.1038/sj.hdy.6800831. [DOI] [PubMed] [Google Scholar]
- 31.Donnelly ML, et al. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 32.Akhtar W, et al. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Lindsey ARI, et al. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol Evol. 2018;10:434–451. doi: 10.1093/gbe/evy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brennan LJ, Haukedal JA, Earle JC, Keddie B, Harris HL. Disruption of redox homeostasis leads to oxidative DNA damage in spermatocytes of Wolbachia-infected Drosophila simulans. Insect Mol Biol. 2012;21:510–520. doi: 10.1111/j.1365-2583.2012.01155.x. [DOI] [PubMed] [Google Scholar]
- 35.Charlat S, Calmet C, Merçot H. On the mod resc model and the evolution of Wolbachia compatibility types. Genetics. 2001;159:1415–1422. doi: 10.1093/genetics/159.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonneau M, et al. Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat Commun. 2018;9:319. doi: 10.1038/s41467-017-02749-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Neill SL, Karr TL. Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature. 1990;348:178–180. doi: 10.1038/348178a0. [DOI] [PubMed] [Google Scholar]
- 38.Bordenstein SR, Werren JH. Bidirectional incompatibility among divergent Wolbachia and incompatibility level differences among closely related Wolbachia in Nasonia. Heredity (Edinb) 2007;99:278–287. doi: 10.1038/sj.hdy.6800994. [DOI] [PubMed] [Google Scholar]
- 39.Poinsot D, Bourtzis K, Markakis G, Savakis C, Merçot H. Wolbachia transfer from Drosophila melanogaster into D. simulans: Host effect and cytoplasmic incompatibility relationships. Genetics. 1998;150:227–237. doi: 10.1093/genetics/150.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bordenstein SR, O’Hara FP, Werren JH. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature. 2001;409:707–710. doi: 10.1038/35055543. [DOI] [PubMed] [Google Scholar]
- 41.Brucker RM, Bordenstein SR. Speciation by symbiosis. Trends Ecol Evol. 2012;27:443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Zabalou S, et al. Natural Wolbachia infections in the Drosophila yakuba species complex do not induce cytoplasmic incompatibility but fully rescue the wRi modification. Genetics. 2004;167:827–834. doi: 10.1534/genetics.103.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourtzis K, Dobson SL, Braig HR, O’Neill SL. Rescuing Wolbachia have been overlooked. Nature. 1998;391:852–853. doi: 10.1038/36017. [DOI] [PubMed] [Google Scholar]
- 44.Casiraghi M, Anderson TJ, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: Comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122:93–103. doi: 10.1017/s0031182000007149. [DOI] [PubMed] [Google Scholar]
- 45.Yu NY, et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hofmann K. TMbase-A database of membrane spanning proteins segments. Biol Chem Hoppe Seyler. 1993;374:166. [Google Scholar]
- 47.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang H, Zhou W, Landweber LF. SWAKK: A web server for detecting positive selection in proteins using a sliding window substitution rate analysis. Nucleic Acids Res. 2006;34:W382–W384. doi: 10.1093/nar/gkl272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinway SN, Dannenfelser R, Laucius CD, Hayes JE, Nayak S. JCoDA: A tool for detecting evolutionary selection. BMC Bioinformatics. 2010;11:284. doi: 10.1186/1471-2105-11-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data and replicate data are available as Supporting Information along with this publication.