Significance
Allosteric regulation, produced by the binding of a ligand at an allosteric site topographically distinct from the orthosteric site, represents a direct and efficient means for modulation of biological macromolecule function. Because allosteric modulators have advantages over classic orthosteric ligands as therapeutic agents, understanding the mechanism underlying allosteric modulation may open new therapeutic avenues. Here, we focused on allosteric regulation of P2X receptors, which are implicated in diverse pathophysiological processes, such as blood clotting, pain sensation, inflammation, and rheumatoid arthritis. Combining structural determination, molecular modeling, and mutagenesis, we identified a druggable allosteric site on P2X3. Our findings will facilitate the development of novel therapeutics targeting these receptors.
Keywords: P2X3 receptors, allosteric inhibition, X-ray crystallography, AF-219, AF-353
Abstract
Allosteric modulation provides exciting opportunities for drug discovery of enzymes, ion channels, and G protein-coupled receptors. As cation channels gated by extracellular ATP, P2X receptors have attracted wide attention as new drug targets. Although small molecules targeting P2X receptors have entered into clinical trials for rheumatoid arthritis, cough, and pain, negative allosteric modulation of these receptors remains largely unexplored. Here, combining X-ray crystallography, computational modeling, and functional studies of channel mutants, we identified a negative allosteric site on P2X3 receptors, fostered by the left flipper (LF), lower body (LB), and dorsal fin (DF) domains. Using two structurally analogous subtype-specific allosteric inhibitors of P2X3, AF-353 and AF-219, the latter being a drug candidate under phase II clinical trials for refractory chronic cough and idiopathic pulmonary fibrosis, we defined the molecular interactions between the drugs and receptors and the mechanism by which allosteric changes in the LF, DF, and LB domains modulate ATP activation of P2X3. Our detailed characterization of this druggable allosteric site should inspire new strategies to develop P2X3-specific allosteric modulators for clinical use.
Allosteric modulation of a receptor refers to changes in the action of orthosteric ligands due to the binding of allosteric modulators at a site distinct from where the orthosteric ligand binds (1). Allosteric modulators usually possess higher receptor selectivity, lower target-based toxicity, and excellent physicochemical properties, and thus exhibit smaller drug side effects and better market prospects (2, 3). In recent years, developing allosteric modulators as therapeutic agents has emerged as a promising new approach for the treatment of various disorders associated with the dysfunction of G protein-coupled receptors, ion channels, or enzymes (4, 5). The identification of allosteric sites on receptors and ion channels not only contributed to better understanding of the biophysical properties of these receptors but also offered new opportunities for rational drug designs (2, 3). Indeed, many drugs on the market or under clinical trials are allosteric modulators of certain receptors/ion channels (3).
P2X receptors are extracellular ATP-gated cation channels implicated in many physiological and pathological processes, including synaptic transmissions, hearing, thrombosis, pain perception, hypertension, immune regulation, etc. (6–8). Besides the orthosteric binding site for ATP (9–11), these receptors have additional allosteric sites where the agonist action can be modulated by other agents, such as ivermectin, lipids, and some trace metals (8, 12, 13). Due to their pathological roles in disease, great efforts have been made to search for P2X drugs for therapeutic use, leading to discovery of many P2X receptor inhibitors, and quite a few of them show promising effects in preclinical studies (13–15). However, only four P2X small molecule inhibitors, AF-219, AF-130, AZD9056, and GSK1482160, are still in the main phases of clinical studies, according to the ClinicalTrials.gov (https://clinicaltrials.gov). Among them, AF-219 is an allosteric modulator of P2X3 receptors that produced positive results in phase II clinical trial for the treatment of refractory chronic cough (16). The limited success in drug development targeting P2X receptors may be because of two reasons. First, the orthosteric site of P2X receptors is highly polarized, making it unsuitable for drug binding (9–11). Second, little is known about the mechanism of allosteric regulation at these receptors (8, 17). Thus, exploring allosteric regulation of P2X receptors may provide new insights into P2X drug design. Very recently, small-molecule probes of P2X7 have been identified to bind to an allosteric site in a groove formed between two adjacent subunits, distinct from the ATP binding pocket (18). This is believed to facilitate the development of P2X7-specific drugs (18). Here, we report the identification and characterization of an allosteric site at the interface of the left flipper (LF), lower body (LB), and dorsal fin (DF) domains of P2X3 (Fig. 1), which is completely different from the recently described site in P2X7. We demonstrate that small molecules cause negative allosteric modulation of the channel by interrupting the allosteric changes of the LF, LB, and DF domains of P2X3 receptors.
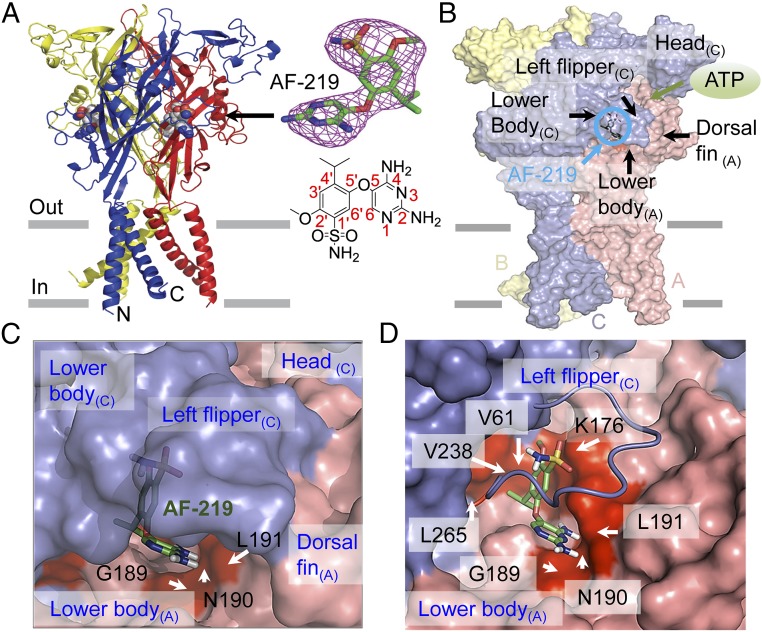
Fig. 1.
Crystal structure of hP2X3 in complex with AF-219. (A) (Upper) The structure of AF-219-bound hP2X3 (viewed in parallel to the membrane). The three subunits are colored individually in blue, red, and yellow. The omit Fo−Fc density map contoured at 3.0 σ is presented for the AF-219 molecular density. (Lower) The chemical structure of AF-219 and the atom numbering of the 4-isopropyl-2-methoxybenzenesulfonamide ring and 2,4-diaminopyrimidine ring in AF-219. (B) The van der Waals surface representation of hP2X3 bound with AF-219. Labeled domains are involved in creating the allosteric site of AF-219 binding or the ATP binding site. The deep salmon, light blue, and pale yellow colors represent the surfaces of chains A, B, and C of hP2X3, respectively. The subscript (A) in “Left flipper (A)” indicates the LF domain in chain A (similarly hereinafter). (C and D) Close-up views of surface representation of the AF-219 binding site. AF-219 is depicted by stick models. Red surface representations highlight the key residues that make contact with AF-219. AF-219 is partially buried into the allosteric site (C). To clearly view the buried part of the AF-219-binding site, the surface representation of the LF domain was replaced by a cartoon in D.
Results
The Structure of Human P2X3 in Complex with AF-219 Uncovers a Negative Allosteric Site.
To understand the mechanism underlying the negative allosteric regulation of P2X3 receptors, we crystallized human P2X3 (hP2X3) in the presence of its allosteric inhibitor for structural determination. To improve the chance of success, we tested two structurally analogous P2X3 inhibitors, AF-353 [5-(5-iodo-2-isopropyl-4-methoxyphenoxy)pyrimidine-2,4-diamine] and AF-219 [5-((2,4-diaminopyrimidin-5-yl)oxy)-4-isopropyl-2-methoxybenzenesulfonamide]. Although AF-353 is more potent and widely studied (19, 20), AF-219 has been shown to exhibit positive therapeutic effects in a phase II clinical trial for treatments of refractory chronic cough, (16) and its chemical structure (Fig. 1A and Fig. S1) was announced very recently (21, 22). We therefore synthesized AF-219 (see Methods and Fig. S1) and validated its inhibitory effect on hP2X3 [half-maximal inhibition concentration (IC50) = 0.33 ± 0.07 μM]. Although we did not obtain the structure for hP2X3 in complex with AF-353, we successfully determined the structure of hP2X3 in complex with AF-219 by vapor diffusion method (Fig. 1A). The crystals diffracted to 3.4-Å resolution (Table S1).
The crystal structure of hP2X3 in complex with AF-219 revealed the clear electron density map for AF-219, and showed that the allosteric inhibitor binds to a site fostered by the LB and DF domains in one subunit, and the LF and LB domains from the adjacent subunit (Figs. 1 B and C and 2A), which is distinct from the ATP binding site (Fig. 1B). By mapping the van der Waals surface of this crystal structure, we found that AF-219 was partially buried by the LF and DF domains (Fig. 1 B and C), where the 4-isopropyl-2-methoxybenzenesulfonamide ring (Fig. 1A and Fig. S2A) of AF-219 was trapped into the inner part while the 2,4-diaminopyrimidine ring made contacts with the outer part of the binding pocket (Fig. 1 C and D and Fig. S2A). The inner part of this allosteric site was mainly created by hydrophobic amino acids residing in the LB domain of one subunit (Fig. 1 C and D and Fig. S2A), including V61 (β1), L191 (β9) and the aliphatic side chain of K176 (β8), and the residues in the LF (L265) and LB (V238 in β11) domains from the adjacent subunit. Residues S178, G189, N190, and the main chain atoms of L191 were exposed to the solution (Figs. 1 C and D and 2A and Fig. S2A). The DF domain, which is connected with the LB domain, may stabilize this site through hydrophobic interactions with the LF domain (Fig. 1B and Fig. S2A).
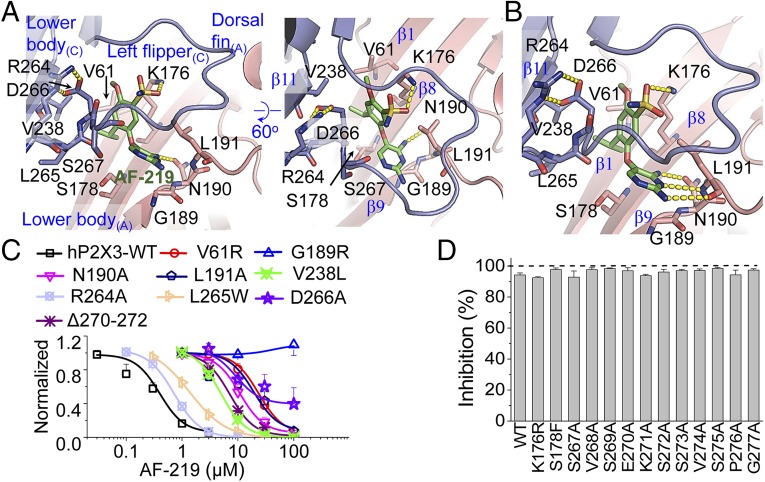
Fig. 2.
Interactions between AF-219 and hP2X3. (A and B) Zoom-in views of the contacts between AF-219 and hP2X3 in the crystal structure in two angles (A), and the optimized conformation after short-time MD simulations (B). The key residues and AF-219 are displayed in stick models for emphasis. Yellow dotted lines indicate H bonds between AF-219 and hP2X3. (C) Concentration−response curves for AF-219 of hP2X3-WT and its mutants (n = 3 to 6). (D) Effects of AF-219 (3 μM) on ATP (10 µM)-induced activation of the indicated hP2X3 mutants (n = 3 to 4).
The Identified Allosteric Site Is Essential for the Action of AF-219.
The crystal structure of hP2X3 in complex with AF-219 reveals that this allosteric inhibitor makes direct hydrogen bond (H-bond) contacts with the main chain atom of N190 and the side chain of K176 while keeping hydrophobic contacts with V61, V238, and L265 (Fig. 2A and Fig. S2A). This is consistent with the results that mutations, V61R (IC50 = 23.6 ± 1.4 μM), N190A (IC50 = 10.33 ± 0.12 μM), V238L (IC50 = 4.92 ± 0.15 μM), and L265W (IC50 = 1.37 ± 0.05 μM), rendered significantly decreases in the sensitivity to AF-219 compared with wild-type hP2X3 (hP2X3-WT) (IC50 = 0.33 ± 0.07 μM; Fig. 2C). However, K176R, S178F, and S267A mutations did not significantly affect the inhibition by 3 µM AF-219 (Fig. 2D), although there are an H-bond contact between AF-219 and K176, a short distance between the hydroxyl group of S267 and the N1 atom of the 2,4-diaminopyrimidine ring of AF-219 (3.7 Å; Fig. S2B), and a close localization of S178 to AF-219 (Fig. 2A).
Interestingly, although the crystal structure did not resolve a direct contact between AF-219 and the main chain atoms of L191 (Fig. 2A), a short-time molecular dynamics (MD) simulation optimization (∼10 ps to 20 ps) of this structure easily established the contacts (Fig. 2B). As revealed by the ligand torsion plot summaries (Fig. S3A) and interactions-occurring analysis in the simulation time (Fig. S3B), these contacts were very stable during 300-ns MD simulations. Consistent with this observation, L191A strongly decreased the inhibition by AF-219 (IC50 = 16.0 ± 1.1 μM; Fig. 2C), confirming the functional importance of these contacts. Additionally, although G189 did not display a contact with AF-219 in either the crystal structure (Fig. 2A) or the optimized structure after MD simulations (Fig. 2B), G189R fully abolished the inhibition by AF-219 even at concentrations up to 100 µM (Fig. 2C). This might be attributed to the bulkiness of the Arg side chain that prevented the access of AF-219 to the binding pocket, or the conformational change caused by the substitution of glycine to arginine altered orientations of adjacent residues, e.g., N190 and L191, such that they could no longer effectively interact with AF-219. Together, these data suggest that the direct contacts of AF-219 with N190 and L191 are essential for the inhibition of AF-219, with G189 also being critical for the action of the allosteric inhibitor.
We then screened the residues in the LF domain (S267 to G277) of hP2X3 using alanine substitutions. These replacements had no obvious effect on the inhibition by 3 µM AF-219 (Fig. 2D), suggesting that, most likely, the compound does not make direct contacts with these residues. However, the LF domain may serve to maintain the overall shape of the allosteric site, because shortening its length by the deletion of three residues, Δ270 to Δ272, markedly decreased the IC50 of AF-219 to 4.3 ± 1.7 μM (Fig. 2C). Furthermore, the salt bridge between R264 and D266 (Fig. 2 A and B), a conserved structure at the beginning of LF domains of all P2X receptor subtypes (10, 23), is crucial for holding the LF domain in place as the R264A (IC50 = 0.72 ± 0.01 μM; Fig. 2C) and D266A (IC50 > 30 μM; Fig. 2C) mutations right-shifted the concentration−response curve to AF-219 (Fig. 2C). Therefore, the LF domain provides the structural constraint for the shape of this allosteric site.
AF-353 and Other Structurally Analogous Allosteric Inhibitors Modulate hP2X3 Through the Same Allosteric Site.
A number of structural analogs of AF-219 (Fig. 3A) haven been shown to inhibit P2X3 receptors (13, 24). To learn how different P2X3 inhibitors interact with the identified site, we docked (in silico) at this site the AF-219 structural analogs, RO-51, RO-3, TC-P 262, and AF-353 (Fig. 3A), as well as two structurally distinct P2X3 inhibitors, suramin and pyridoxalphosphate-6-azophenyl-2′,4′-disulfonic acid tetrasodium salt (PPADS) (Fig. S4A), as controls. As revealed by the docking scores and the molecular mechanics/generalized born surface area (MM/GBSA) binding free-energy calculations, all tested compounds except for suramin, which is too bulky to fit into the pocket, have their interaction modes with the identified allosteric site (Fig. S4 B–D). RO-51, RO-3, TC-P 262, and AF-353 were docked into the pocket with similar interaction modes to that of AF-219; however, PPADS was only docked outside the pocket. Consistent with these predictions, G189R, which abolished the inhibition by AF-219 (Fig. 2C), also became insensitive to the structural analogs, AF-353, RO-51, RO-3, and TC-P 262 (Fig. 4A and Fig. S4E), but did not affect the inhibition of structurally distinct inhibitors, suramin and PPADS (Fig. S4E). Therefore, P2X3 antagonists structurally analogous to AF-219 also bind to the same allosteric site to modulate the channel function, although the precise binding modes can vary from one drug to another, due to differences in the compound structures.
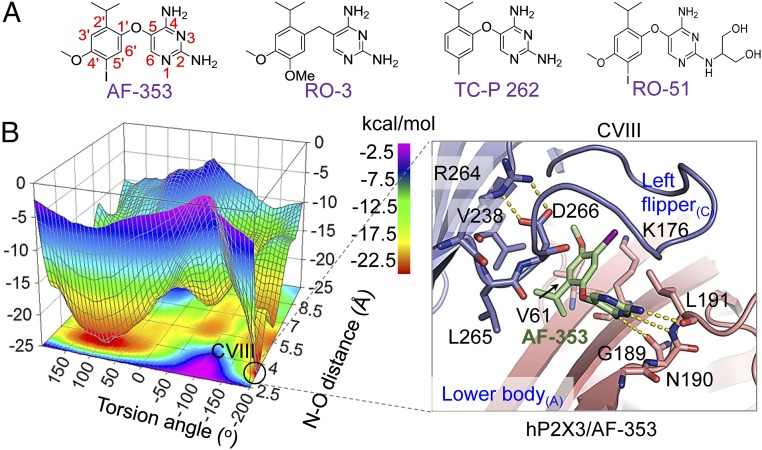
Fig. 3.
Possible interactions between hP2X3 and AF-353 and other structurally analogous allosteric inhibitors. (A) Structures of AF-219 analogs used, including AF-353, RO-3, TC-P 262, and RO-51. (B) Free-energy surface (Left) showing one of the possible binding modes of AF-353 with hP2X3 (CVIII, Right) determined by metadynamics simulations. Yellow dotted lines indicate H bonds between AF-353 and hP2X3 and the salt bridge interactions between D266 and R264.
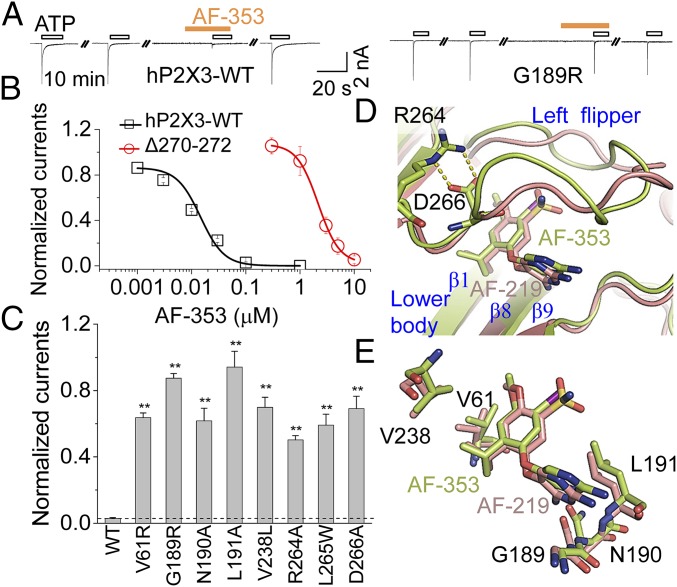
Fig. 4.
Comparison between AF-353 and AF-219 bindings to hP2X3. (A) Representative traces of AF-353 (0.1 µM) effects on ATP (10 µM)-evoked currents of hP2X3-WT and its G189R mutant. (B) Concentration−response curves for AF-353 of hP2X3-WT and Δ270 to Δ272 (n = 3 to 4). Solid lines are fits to Hill equation. (C) Effects of AF-353 (0.1 µM) on ATP (10 µM)-evoked currents of hP2X3-WT and selected mutants affecting the identified allosteric site (mean ± SEM, n = 3 to 5). **P < 0.01 vs. WT (dashed line), Student’s t test. (D) A close-up view of superimposed hP2X3 models with AF-219 (deep salmon) or AF-353 (lemon). hP2X3 and allosteric small molecules are depicted by cartoon and stick models, respectively. (E) Superimposed key residues of hP2X3 that make contacts with AF-219 (deep-salmon) and AF-353 (lemon).
To further illustrate the interactions of the allosteric antagonist with hP2X3 at the identified binding site, we focused on AF-353, which did not give us the crystal structure when cocrystalized with hP2X3. AF-353 is another promising P2X3 inhibitor with therapeutic potentials. It has been demonstrated to be a noncompetitive inhibitor in the affinity-labeling assay and a potent allosteric regulator of P2X3 in many functional pharmaco and in vivo studies (20). Although AF-219 and AF-353 share the same structural core (sulfonamide group changed to iodine; Figs. 1A and 3A), these two compounds exhibit distinct inhibitory efficiencies and abilities to cross the blood–brain barrier (BBB) (25). At 0.1 µM, AF-353 fully abolished ATP-induced current of hP2X3 (Fig. 4A), displaying ∼20- to 30-fold higher potency (IC50 = 12.9 ± 0.5 nM; Fig. 4B) than AF-219. Meanwhile, AF-219 is less lipophilic than AF-353. For this reason, AF-219 is preferred for use in vivo/clinically for treating peripheral disorders, with which central nervous system (CNS) side effects may be avoided because of the low probability of crossing BBB (25). However, for CNS diseases, AF-353 may be more preferred. To obtain information on AF-353 interactions with hP2X3, we applied metadynamics (Fig. 3B), an approach based on the free-energy reconstruction and accelerating rare events that have been applied to study chemical reaction courses, protein−small molecule interactions, and the optimal routes of conformational changes of macromolecules (26). To avoid the influence by the binding modes of AF-219, we used a structural model of hP2X3 at the apo state (10) with a filled-in missed loop in the LF domain. To escape the local energy minima and avoid deficiency that might result from in silico docking of hP2X3, we defined two variables based on the initial docking pose of AF-353 in hP2X3. One is the dihedral angle formed by the C6 and C5 atoms of pyrimidine, the oxygen atom, and the C1′ atom of the phenyl group (Fig. 3A). The other is the distance between the nitrogen atom of −NH2 substituted at position 2 of AF-353 and the main chain oxygen atom of L191. Using these definitions, the stretching of AF-353 from the identified allosteric site and all of the binding modes between AF-353 and hP2X3 were extensively studied, with the binding free energy measured simultaneously for each pose. This analysis revealed three poses for AF-353 interaction with hP2X3, namely CV1, CVII, and CVIII (Fig. 3B and Fig. S5), that exhibited lowest binding free energy. Among them, CVIII exhibited a nearly identical interaction mode with that shown by AF-219 as revealed by the crystal structure, in which the hydrophobic moiety of AF-353 was tightly locked by the side chains of V61, V238, and L265 (Fig. 3B, Right), and H bonds formed between AF-353 and the main chain atoms of N190 and L191 (Fig. 3B, Right; compare with Fig. 2A and Fig. S2A for AF-219). Consistent with the above structural prediction, V61R, N190A, L191A, V238L, and L265W mutations significantly attenuated or abolished the inhibition of hP2X3 induced by the saturating concentration of AF-353 (0.1 μM; Fig. 4C). R264A and D266A also strongly attenuated AF-353 inhibition of hP2X3 (Fig. 4C), indicating the essential role of the R264:D266 salt bridge in holding the LF domain in place for the inhibitory action of AF-353, just like in the case of AF-219.
The superimposition of CVIII with the AF-219-bound hP2X3 crystal structure (Fig. 4 D and E) indicated similar side-chain orientations of V61, N190, L191, and V238 and in these two conformations (Fig. 4E), even after the hP2X3/AF-353 complex had been extensively sampled using metadynamics simulations. However, the conformations of the LF domain, which is a flexible loop, are distinct (Fig. 4D), especially in the middle region, as opposed to the beginning part of this domain where the salt bridge constrains fluctuations. This observation prompted us to propose that the flexible LF domain might contribute to the difference in the potency between AF-219 and AF-353. Indeed, the triple amino acid deletion in the middle region of the LF domain, Δ270 to Δ272, caused ∼160-fold increase in the IC50 of AF-353 from 12.9 ± 0.5 nM for hP2X3-WT to 2.17 ± 0.09 μM for Δ270 to Δ272 (Fig. 4B), while the increase in IC50 of AF-219 was only ∼10-fold by the same mutation (Fig. 2B), showing that AF-219 and AF-353 can be differentially affected by structural changes in the LF domain. Thus, the flexible LF domain might enable the allosteric site to adjust its conformation to accommodate substitutions of functional groups at the C1′ atom of the 4-isopropyl-2-methoxybenzene ring (Fig. 1A), yielding different affinities for different drugs. In this case, the smaller iodine atom linked to the C5′ atom in AF-353 (Fig. 3A) seemed to be more preferred by the flexible LF domain than the bulkier and polar sulfonamide group in AF-219 (Fig. 1A) to keep the strain energy low when the drug binds hP2X3 at this allosteric site.
Covalently Linking Small Molecules to the Identified Allosteric Site Inhibits the Activation of hP2X3.
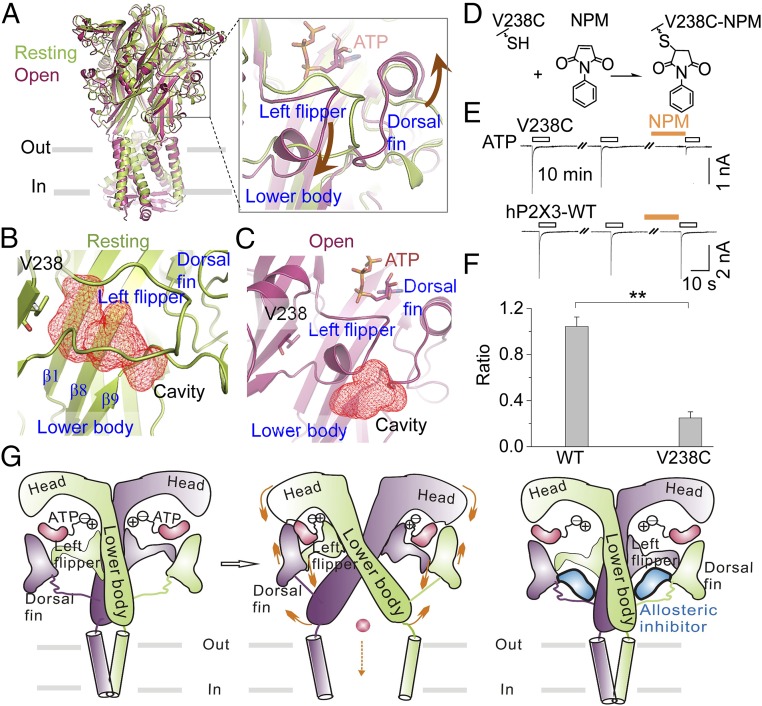
We further explored the mechanism that caused small-molecule binding to the allosteric site to inhibit P2X3 channel activation. It has been reported that relative movements between the LF and DF domains are pivotal for channel gating of P2X4 receptors (27). As revealed by the crystal structures of hP2X3 (10), the DF domain moves up, while the LF domain moves down during the gating process by ATP (Fig. 5A). A cavity outlined mainly by amino acids residing in the LB and LF domains (Fig. 5B and Fig. S6A), namely the identified allosteric site, can be observed only at the resting state of hP2X3 [note that a loop in the LF domain is missing in the published structure (PDB ID code 5SVJ), which was filled in using homology modeling]. Accompanied by the downward movement of the LF domain, this cavity undergoes structural rearrangements and collapses after ATP binding (Fig. 5C and Fig. S6B). The binding of AF-219 not only can prevent the collapsing/size reduction of the cavity but also can enlarge its volume (Fig. S6C). Thus, allosteric modulator binding at the identified site may interfere with the conformation changes of the LF, DF, and LB domains associated with gating and therefore block the coupling of ATP binding to channel activation.
Fig. 5.
Mechanism of allosteric inhibition of P2X3 receptors by the identified site. (A) Superimposition of structures at the resting (lemon, PDB ID code 5SVJ; the missing loop of the LF domain was filled in using homology modeling) and open (warm pink, PDB ID code 5SVK) states of hP2X3 receptors. Brown arrows in the enlarged box indicate the relative motions of the DF and LF domains during ATP-induced channel gating. (B and C) Zoom-in views of cavities, depicted in red mesh for emphasis, fostered by the LF and LB domains of hP2X3 at the resting (B) and open (C) states. (D) Illustration of NPM covalently linked to hP2X3V238C. (E and F) Representative current traces (E) and summary data (F) (n = 4 to 5) for NPM (1 mM) effects on ATP-evoked currents for hP2X3-WT and V238C. **P < 0.01 vs. WT, Student’s t test. (G) Illustrations of the allosteric changes during channel gating of P2X receptors and an allosteric inhibition strategy on P2X3 identified here. Only two chains are shown, for clarity. Orange arrows denote the movements of key domains crucial for channel gating by ATP.
To verify the above argument, we employed a strategy of covalently linking a small molecule to a specific amino acid inside the binding pocket. We mutated V238, which is deeply buried in the allosteric site (Fig. 5B and Fig. S6A), to cysteine (V238C) and used N-phenylmaleimide (NPM), which is reactive to cysteines, to form a covalent and irreversible bond (Fig. 5D), to examine its effect on channel function. As expected, NPM treatment significantly attenuated ATP-evoked current of hP2X3V238C, but not that of hP2X3-WT (Fig. 5 E and F), indicating that not only is the identified allosteric site accessible to allosteric small molecules but also the occupation of this site exerts a negative effect on hP2X3 gating (Fig. 5G).
Discussion
As a class of ion channels involved in diverse physiological and pathological processes, P2X receptors have drawn wide attention as novel drug targets (15). However, it is difficult to design drugs targeting the highly polar ATP binding site, and the similarity of this site among P2X subtypes also makes it unideal for selectivity. Therefore, allosteric regulation presents better opportunities for developing subtype-specific drugs of P2X receptors (13). Recently, a number of crystal structures representing different P2X receptor isoforms and active states were made available (9–11, 18, 28, 29). These structures suggest that multiple allosteric changes may be associated with ATP binding to channel gating (27, 30–35), and, presumably, each of the allosteric changes can be targeted by small molecules for functional perturbation. However, it remains challenging to define the function of a potential allosteric site, identify the drug, and determine the drug’s mechanism of action at the specific site (36, 37). In addition, some of the domains that exhibit allosteric changes, for instance the head and DF domains, are not ideal for targeting because of their partial involvement in making up the ATP binding site (30, 36, 37). Here, we focused on a potential allosteric site which forms intersubunit pockets fostered by the LF, LB, and DF domains (Figs. 1B and 2A and Fig. S2A). When bound by noncompetitive inhibitors of P2X3, the relative motion between the LF and LB−DF domains was blocked, leading to inhibition of P2X3 function (Fig. 5 A and G).
The crystal structures for the open states of P2X4 and P2X3 receptors clearly show a downward shift of the LF domain toward the LB−DF domain of the neighboring subunit compared with the structures at the apo state, suggesting that the downward motion of the LF domain upon ATP binding may constitute an important step of P2X channel gating by its endogenous ligand, ATP (9–11). Previously, combining computational stimulation and mutational studies, we showed that manipulations that prevented the motion between the LF and DF domains all disrupted ATP gating of P2X4 (23, 27). Not surprisingly, by allowing a disulfide bond formation between the LF and DF domains of P2X3, the activation of P2X3K201C/V274C by ATP was also largely suppressed (34). Therefore, the P2X receptors use similar gating mechanisms in which the freedom for the LF domain that sits underneath the ATP binding site to move toward the neighboring LB−DF domains appears to be essential for the transition from the closed to the open state. Preventing such a movement would exert negative allosteric modulation on P2X receptors. With this in mind, we show here that a number of allosteric inhibitors of P2X3 indeed bind to the pocket fostered by the LF, LB, and DF domains of P2X3. This finding is supported by the structural determination of hP2X3 in complex with one of these allosteric inhibitors, AF-219, by X-ray crystallography, computational simulations to optimize interactions between hP2X3 and various allosteric small molecules, and a large number of mutational studies to validate the allosteric site for inhibitor binding and critical amino acids involved in the drug−receptor interactions. In addition to further strengthening the importance of the coordinated motions of the LF, LB, and DF domains during P2X receptor gating, our finding reveals a mechanism of allosteric regulation of these receptors by small molecules and sheds lights on new strategies to develop P2X drugs for disease therapy.
The identification of the negative allosteric site provides unique opportunities for future drug design of P2X receptor small-molecule probes. First, the identified pocket is the base for combining virtual screening and patch clamp recordings to search for lead compounds with new structure cores. This will greatly improve the probability of finding more novel drug candidates. Second, the ligand−receptor interactions revealed for AF-353 and AF-219 binding to P2X3 provide key information for further structure-based rational drug design. For example, the interactions between AF-219/AF-353 and L191/N190 play important roles in compound binding to the pocket. Therefore, the strength of the H bonds or the immobilization of the torsion angles of the compound to reduce strain energy of both the ligand and the receptor may be exploited to optimize the lead compounds. Finally, the shape of the hydrophobic moiety of small molecules may be optimized according to the architecture of the pocket to create new compounds with high selectivity. This is particularly relevant since collapsing the identified allosteric site represents a crucial step of P2X receptor gating (Fig. 5 B and C). Introducing small molecules into this site to effectively block its collapse will disrupt channel gating (Fig. 5G). The architectural differences at this site among different P2X receptor subtypes (23) provide an excellent opportunity to develop subtype specific allosteric inhibitors.
Methods
Methods are fully described in SI Methods. Briefly, all constructs were expressed in cultured HEK-293 cells (27). Whole-cell recordings were performed on HEK-293 cells 24 h to 48 h after transfection (27). Homology modeling was carried out using MODELLER (23, 38). MD and metadynamics were performed using DESMOND (39). Crystallization of P2X3 in complex with AF-219 was achieved by vapor diffusion method (10).
Supplementary Material
Acknowledgments
We thank the staff from BL41XU beamline at SPring-8 (Proposals 2017A2523 and 2017B2523), from BL19U1 beamline of National Facility for Protein Science Shanghai at Shanghai Synchrotron Radiation Facility (SSRF) (Proposal 2016-NFPS-PT-001047), and from BL17U1 at SSRF (Proposals 15ssrf02687 and 2016-SSRF-PT-005911), for assistance during data collection. This study was supported by Grants 2016YFA0502800 and 2014CB910300/02 from the National Key R&D Program of China, Grants 31570832, 31570838, 31170787, 31400707, 31222018, and 31650110469 from the National Natural Science Foundation of China, Grant BX201700306 from the National Postdoctoral Program for Innovative Talents, and Grant SIMM1601KF-02 from the Opening Project of State Key Laboratory of Drug Research in Shanghai Institute of Materia Medica.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5YVE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800907115/-/DCSupplemental.
References
- 1.Guarnera E, Berezovsky IN. Allosteric sites: Remote control in regulation of protein activity. Curr Opin Struct Biol. 2016;37:1–8. doi: 10.1016/j.sbi.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Wenthur CJ, Gentry PR, Mathews TP, Lindsley CW. Drugs for allosteric sites on receptors. Annu Rev Pharmacol Toxicol. 2014;54:165–184. doi: 10.1146/annurev-pharmtox-010611-134525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Changeux JP, Christopoulos A. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell. 2016;166:1084–1102. doi: 10.1016/j.cell.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Wu P, Clausen MH, Nielsen TE. Allosteric small-molecule kinase inhibitors. Pharmacol Ther. 2015;156:59–68. doi: 10.1016/j.pharmthera.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Nickols HH, Conn PJ. Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol Dis. 2014;61:55–71. doi: 10.1016/j.nbd.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coddou C, Yan Z, Obsil T, Huidobro-Toro JP, Stojilkovic SS. Activation and regulation of purinergic P2X receptor channels. Pharmacol Rev. 2011;63:641–683. doi: 10.1124/pr.110.003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khakh BS, North RA. Neuromodulation by extracellular ATP and P2X receptors in the CNS. Neuron. 2012;76:51–69. doi: 10.1016/j.neuron.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habermacher C, Dunning K, Chataigneau T, Grutter T. Molecular structure and function of P2X receptors. Neuropharmacology. 2016;104:18–30. doi: 10.1016/j.neuropharm.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 9.Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485:207–212. doi: 10.1038/nature11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansoor SE, et al. X-ray structures define human P2X3 receptor gating cycle and antagonist action. Nature. 2016;538:66–71. doi: 10.1038/nature19367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasuya G, et al. Structural insights into divalent cation modulations of ATP-gated P2X receptor channels. Cell Rep. 2016;14:932–944. doi: 10.1016/j.celrep.2015.12.087. [DOI] [PubMed] [Google Scholar]
- 12.Coddou C, Stojilkovic SS, Huidobro-Toro JP. Allosteric modulation of ATP-gated P2X receptor channels. Rev Neurosci. 2011;22:335–354. doi: 10.1515/RNS.2011.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller CE. Medicinal chemistry of P2X receptors: Allosteric modulators. Curr Med Chem. 2015;22:929–941. doi: 10.2174/0929867322666141210155610. [DOI] [PubMed] [Google Scholar]
- 14.Bartlett R, Stokes L, Sluyter R. The P2X7 receptor channel: Recent developments and the use of P2X7 antagonists in models of disease. Pharmacol Rev. 2014;66:638–675. doi: 10.1124/pr.113.008003. [DOI] [PubMed] [Google Scholar]
- 15.North RA, Jarvis MF. P2X receptors as drug targets. Mol Pharmacol. 2013;83:759–769. doi: 10.1124/mol.112.083758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdulqawi R, et al. P2X3 receptor antagonist (AF-219) in refractory chronic cough: A randomised, double-blind, placebo-controlled phase 2 study. Lancet. 2015;385:1198–1205. doi: 10.1016/S0140-6736(14)61255-1. [DOI] [PubMed] [Google Scholar]
- 17.Kawate T. P2X receptor activation. Adv Exp Med Biol. 2017;1051:55–69. doi: 10.1007/5584_2017_55. [DOI] [PubMed] [Google Scholar]
- 18.Karasawa A, Kawate T. Structural basis for subtype-specific inhibition of the P2X7 receptor. eLife. 2016;5:e22153. doi: 10.7554/eLife.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford AP, Undem BJ. The therapeutic promise of ATP antagonism at P2X3 receptors in respiratory and urological disorders. Front Cell Neurosci. 2013;7:267. doi: 10.3389/fncel.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gever JR, et al. AF-353, a novel, potent and orally bioavailable P2X3/P2X2/3 receptor antagonist. Br J Pharmacol. 2010;160:1387–1398. doi: 10.1111/j.1476-5381.2010.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ford AP, McCarthy BG. 2015. Eur Patent Appl EP3035932A.4 (WO15027212)
- 22.Szántó G, et al. New P2X3 receptor antagonists. Part 1: Discovery and optimization of tricyclic compounds. Bioorg Med Chem Lett. 2016;26:3896–3904. doi: 10.1016/j.bmcl.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, et al. Intersubunit physical couplings fostered by the left flipper domain facilitate channel opening of P2X4 receptors. J Biol Chem. 2017;292:7619–7635. doi: 10.1074/jbc.M116.771121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bölcskei H, Farkas B. P2X3 and P2X2/3 receptor antagonists. Pharm Pat Anal. 2014;3:53–64. doi: 10.4155/ppa.13.70. [DOI] [PubMed] [Google Scholar]
- 25.Pijacka W, et al. Purinergic receptors in the carotid body as a new drug target for controlling hypertension. Nat Med. 2016;22:1151–1159. doi: 10.1038/nm.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barducci A, Bussi G, Parrinello M. Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Phys Rev Lett. 2008;100:020603. doi: 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
- 27.Zhao WS, et al. Relative motions between left flipper and dorsal fin domains favour P2X4 receptor activation. Nat Commun. 2014;5:4189. doi: 10.1038/ncomms5189. [DOI] [PubMed] [Google Scholar]
- 28.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460:592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasuya G, et al. Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel. Nat Commun. 2017;8:876. doi: 10.1038/s41467-017-00887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang R, et al. Tightening of the ATP-binding sites induces the opening of P2X receptor channels. EMBO J. 2012;31:2134–2143. doi: 10.1038/emboj.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lörinczi É, et al. Involvement of the cysteine-rich head domain in activation and desensitization of the P2X1 receptor. Proc Natl Acad Sci USA. 2012;109:11396–11401. doi: 10.1073/pnas.1118759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts JA, et al. Agonist binding evokes extensive conformational changes in the extracellular domain of the ATP-gated human P2X1 receptor ion channel. Proc Natl Acad Sci USA. 2012;109:4663–4667. doi: 10.1073/pnas.1201872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Yu Y. Insights into the channel gating of P2X receptors from structures, dynamics and small molecules. Acta Pharmacol Sin. 2016;37:44–55. doi: 10.1038/aps.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowalski M, et al. Conformational flexibility of the agonist binding jaw of the human P2X3 receptor is a prerequisite for channel opening. Br J Pharmacol. 2014;171:5093–5112. doi: 10.1111/bph.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Kawate T, Silberberg SD, Swartz KJ. Pore-opening mechanism in trimeric P2X receptor channels. Nat Commun. 2010;1:44. doi: 10.1038/ncomms1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang LD, et al. Inherent dynamics of head domain correlates with ATP-recognition of P2X4 receptors: Insights gained from molecular simulations. PLoS One. 2014;9:e97528. doi: 10.1371/journal.pone.0097528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang R, Taly A, Grutter T. Moving through the gate in ATP-activated P2X receptors. Trends Biochem Sci. 2013;38:20–29. doi: 10.1016/j.tibs.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 38.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 39.Shaw DE. A fast, scalable method for the parallel evaluation of distance-limited pairwise particle interactions. J Comput Chem. 2005;26:1318–1328. doi: 10.1002/jcc.20267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.