Significance
Advanced prostate cancer is a deadly disease made up of multiple cancer subtypes that evolve during its natural history. Unfortunately, antibody- and cell-based therapies in development that target single tumor antigens found in conventional prostate cancer do not account for this heterogeneity. Here, we show that two major subtypes of advanced prostate cancer, prostate adenocarcinoma (PrAd) and neuroendocrine prostate cancer (NEPC), exhibit distinct cell-surface expression profiles. Integrated analysis of gene expression and cell-surface protein expression of prostate cancer nominated multiple subtype-specific cell-surface antigens. We specifically characterize FXYD3 and CEACAM5 as targets for immune-based therapies in PrAd and NEPC and provide preliminary evidence of the antigen-specific cytotoxic activity of CEACAM5-directed chimeric antigen receptor T cells in NEPC.
Keywords: prostate cancer, cell surface antigens, immunotherapy
Abstract
Prostate cancer is a heterogeneous disease composed of divergent molecular and histologic subtypes, including prostate adenocarcinoma (PrAd) and neuroendocrine prostate cancer (NEPC). While PrAd is the major histology in prostate cancer, NEPC can evolve from PrAd as a mechanism of treatment resistance that involves a transition from an epithelial to a neurosecretory cancer phenotype. Cell surface markers are often associated with specific cell lineages and differentiation states in normal development and cancer. Here, we show that PrAd and NEPC can be broadly discriminated by cell-surface profiles based on the analysis of prostate cancer gene expression datasets. To overcome a dependence on predictions of human cell-surface genes and an assumed correlation between mRNA levels and protein expression, we integrated transcriptomic and cell-surface proteomic data generated from a panel of prostate cancer cell lines to nominate cell-surface markers associated with these cancer subtypes. FXYD3 and CEACAM5 were validated as cell-surface antigens enriched in PrAd and NEPC, respectively. Given the lack of effective treatments for NEPC, CEACAM5 appeared to be a promising target for cell-based immunotherapy. As a proof of concept, engineered chimeric antigen receptor T cells targeting CEACAM5 induced antigen-specific cytotoxicity in NEPC cell lines. Our findings demonstrate that the surfaceomes of PrAd and NEPC reflect unique cancer differentiation states and broadly represent vulnerabilities amenable to therapeutic targeting.
Prostate cancer is the most common non-skin cancer diagnosed in men and the second leading cause of cancer death in men (1). More than 95% of prostate cancers are diagnosed as prostate adenocarcinoma (PrAd), which is characterized by glandular epithelial architecture, expression of luminal cytokeratins (CK8 and CK18), and active androgen receptor (AR) signaling. In advanced disease, blockade of AR signaling has been the mainstay of treatment for decades, but inevitably leads to resistance in the form of castration-resistant prostate cancer (CRPC). Recent data indicate that CRPC can retain the PrAd histology or recur as a distinct subtype called neuroendocrine prostate cancer (NEPC). Recent work also indicates that a subset of CRPC assumes a double-negative (AR- and neuroendocrine-negative) phenotype that is maintained by enhanced FGF and MAPK pathway signaling (2). NEPC describes a group of neuroendocrine tumors that includes aggressive variants such as large- and small-cell carcinoma of the prostate (3). Aggressive, treatment-related NEPC evolves from PrAd in up to 20% of CRPC cases through neuroendocrine transdifferentiation, which involves epigenetic reprogramming mediated by Polycomb proteins (4, 5) and often the loss of the tumor suppressors RB1 and TP53 (6). NEPC often exhibits an anaplastic morphology, expression of neuroendocrine markers including chromogranins and synaptophysin, loss of AR signaling, overexpression and amplification of MYCN and AURKA (7–9), and a particularly poor prognosis due to rapid and progressive metastatic dissemination.
Treatments for CRPC have expanded in recent years to include second-generation antiandrogen therapies, vaccine immunotherapy, an alpha particle-emitting radioactive agent, and additional cytotoxic chemotherapy (10). Notably, the characterization of prostate cancer cell-surface antigens like prostate-specific membrane antigen (PSMA) has spurred the development of novel diagnostic imaging and targeted therapeutic strategies. For example, PSMA-based positron emission tomography has demonstrated high sensitivity and specificity in localizing recurrent prostate cancer (11). PSMA-targeted radioligand and chimeric antigen receptor T cell (CAR T) therapies are rapidly advancing to the clinic and represent a new generation of precision medicine for advanced prostate cancer. However, NEPC does not demonstrate uptake of PSMA radiotracers and is not expected to respond to PSMA-targeted therapies due to low-to-absent PSMA expression (12, 13). Similarly, prostate stem-cell antigen (PSCA) is another cell-surface antigen that is the focus of diagnostic and therapeutic development whose expression is heightened in the majority of PrAd, but down-regulated in NEPC (7).
The expression patterns of PSMA and PSCA in PrAd and NEPC represent a microcosm of the transcriptomic and epigenetic differences between these distinct states of prostate cancer differentiation (14). The differentiation-linked expression of cell-surface proteins has been extensively studied in lineage specification of normal tissues and in cancer (15, 16). The foremost example of this is the cluster of differentiation or classification determinant (CD) used to define stem/progenitor cells and the developmental hierarchy in normal hematopoiesis. The lineage-specific immunophenotyping of hematologic malignancies with CD molecules subsequently prompted the development of immune-based, targeted cancer therapies, including anti-CD20 antibodies (17) and recently CD19-directed CAR T (18, 19), which are transforming the treatment of B-cell malignancies. The identification of suitable target antigens in solid tumors has been limited by the lack of bona fide tissue-specific surface antigens and nonuniform antigen expression (20). New strategies to uncover target antigens and the combinatorial targeting of antigens will be needed to overcome the heterogeneity and plasticity inherent to solid tumors.
Here, we present a generalizable approach to discover cancer subtype-specific target antigens as vulnerabilities amenable to therapeutic exploitation. Our data demonstrate that divergent cancer differentiation states arising during prostate cancer progression are associated with large changes in the repertoire of expressed cell surface proteins (surfaceome). From these differences, multiple candidate antigens were nominated, and the expression of FXYD3, a FXYD domain-containing ion transport regulator, in PrAd and CEACAM5, a member of the carcinoembryonic antigen family, in NEPC were systematically validated. Lastly, we provide preliminary evidence demonstrating the feasibility of targeting CEACAM5 in NEPC with CAR T immunotherapy.
Results
Expression of Genes Encoding Cell-Surface Proteins Distinguishes PrAd and NEPC.
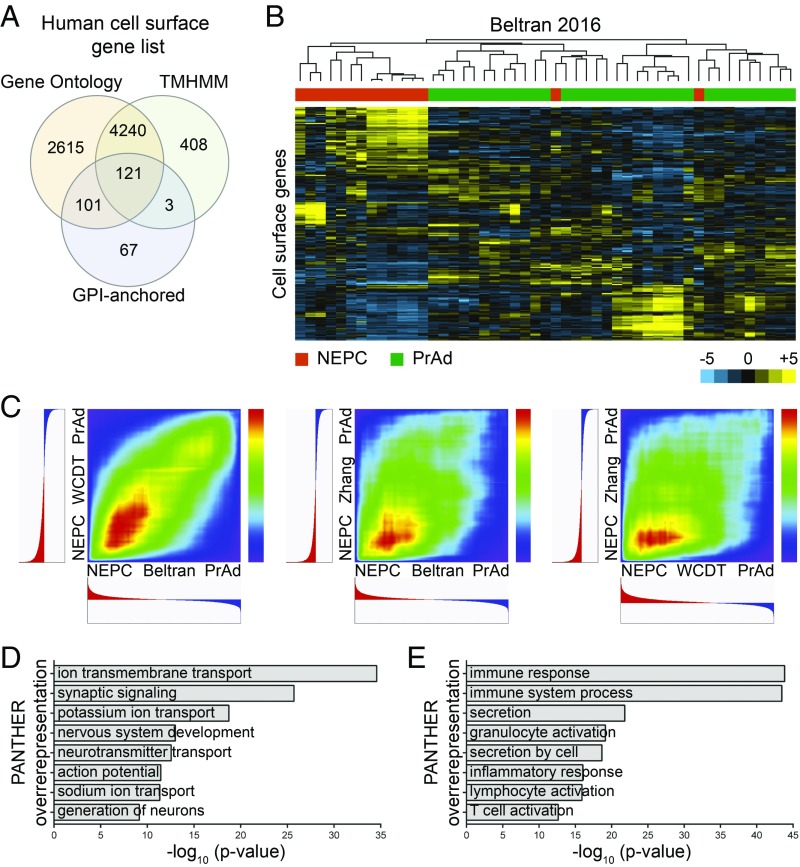
As PrAd and NEPC are distinct states of prostate cancer differentiation, we hypothesized that the composition of the cell surfaceome could discriminate these prostate cancer subtypes. To evaluate this possibility, a set of 7,555 putative human cell-surface proteins was bioinformatically derived (Dataset S1) from publicly available databases by using an adaptation of a published cell-surfaceome construction strategy (21). Human cell-surface proteins were predicted based on Gene Ontology (22), the TransMembrane prediction using hidden Markov models (TMHMM) (23), and glycosylphosphatidylinositol (GPI)-anchored protein annotations (Fig. 1A). Unsupervised hierarchical clustering of human prostate cancer RNA sequencing (RNA-seq) datasets (7, 14) and a cDNA microarray dataset of patient-derived xenografts (PDXs) (24) based on expression of the cell-surface gene set consistently differentiated PrAd and NEPC samples (Fig. 1B and SI Appendix, Fig. S1 A and B).
Fig. 1.
Expression of genes encoding human cell-surface proteins distinguishes prostate cancer subtypes. (A) Venn diagram of a putative human cell-surface gene set bioinformatically constructed from the analysis of Gene Ontology, TMHMM, and GPI-anchored protein databases. (B) Heatmap demonstrating unsupervised hierarchical clustering of CRPC samples from the Beltran 2016 RNA-seq dataset based on the expression of cell-surface genes. Color bar represents a log2 scale. NEPC samples are labeled in orange and PrAd samples in green. (C) RRHO heatmaps showing rank overlap of differentially expressed cell-surface genes across NEPC and PrAd samples in pairwise comparisons of the Beltran 2016, SU2C/AACR/PCF West Coast Dream Team (WCDT), and Zhang 2015 gene expression datasets. (D and E) Gene enrichment analysis from PANTHER overrepresentation testing of cell-surface genes differentially expressed more than fourfold in NEPC relative to PrAd (D) and PrAd relative to NEPC (E) in the Beltran 2016 dataset.
To determine whether the cell-surface phenotype was more highly conserved in PrAd or NEPC, we used rank–rank hypergeometric overlap (RRHO) analysis (25) to compare the ranked differential expression of genes encoding cell-surface proteins (heretofore called “cell surface genes”) between prostate cancer subtypes in multiple datasets [WCDT from the Stand Up To Cancer/American Association for Cancer Research/Prostate Cancer Foundation West Coast Dream Team (26), Beltran 2016 (14), and Zhang 2015 (24)]. This rank-based methodology circumvents the complications of normalization for specific sample preparations and analysis platforms, enabling facile comparisons of gene expression across defined classes of prostate cancer in published datasets. Pairwise evaluation of the datasets revealed higher rank correlation of cell-surface genes enriched in NEPC (Fig. 1C), supporting stronger homogeneity of cell-surface phenotypes in NEPC than in PrAd.
Differential expression analysis of the Beltran 2016 dataset showed that the expression of 330 cell-surface genes was enriched fourfold or more in the PrAd samples, while expression of 438 cell-surface genes was similarly enriched in the NEPC samples. PANTHER (Protein ANnotation THrough Evolutionary Relationship) analysis was performed to compare differentially expressed genes to a reference gene list to identify enriched molecular functions and biological processes (27). PANTHER overrepresentation testing of the cell-surface genes enriched in NEPC in the Beltran 2016 dataset identified gene ontologies related to neural functions, including synaptic signaling, nervous system development, and neurotransmitter transport (Fig. 1D). In contrast, analysis of cell-surface genes enriched in PrAd from the same dataset revealed biological processes involving secretion, immune response, and inflammatory response (Fig. 1E). The results highlight substantial differences in cell-surface antigen expression linked to the cancer differentiation states of NEPC and PrAd.
Identification of Candidate Prostate Cancer Cell-Surface Antigens by Transcriptomic Analysis.
We next assembled a diverse panel of human prostate cancer cell lines to further characterize cell-surface antigens in the PrAd and NEPC subtypes. The panel included established lines including CWR22Rv1, LNCaP, NE1.3 (28), DU145, NCI-H660, and LASCPC-01 (8), as well as two developed cell lines named NB120914 and MSKCC EF1. NB120914 was initiated from an intraoperative biopsy of a metastatic castration-resistant PrAd involving the femur. While the original tumor showed a luminal phenotype with CK8 and AR expression, the resultant PDX tumor and subsequent cell-line xenograft tumors lacked expression of both luminal and neuroendocrine markers, indicative of the development of double-negative (AR- and neuroendocrine-negative) prostate cancer (SI Appendix, Fig. S2 A, B, and D) (2). MSKCC EF1 was adapted from 3D organoid culture (29) to suspension culture and formed xenograft tumors marked by NEPC histology, absence of AR, and expression of the neuroendocrine marker synaptophysin and p63 (SI Appendix, Fig. S2 C and E).
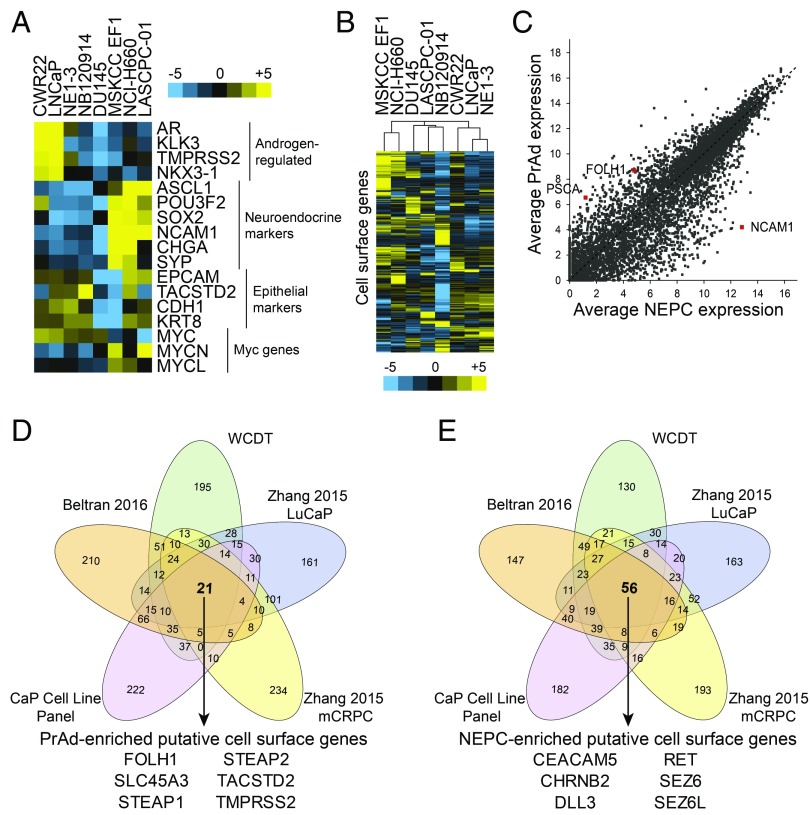
RNA-seq gene-expression analysis revealed significant heterogeneity in expression of androgen-regulated genes, neuroendocrine markers, and epithelial markers in the cell line panel (Fig. 2A), demonstrating a diverse range of molecular phenotypes. Unsupervised hierarchical clustering analysis of the prostate cancer cell lines based on the expression of cell-surface genes yielded clusters of NEPC lines (MSKCC EF1 and NCI-H660), AR-negative PrAd lines (DU145 and LASCPC-01 marked by mixed NEPC and PrAd phenotypes and NB120914), and AR-positive PrAd lines (CWR22Rv1, LNCaP, and the LNCaP-derivative NE1.3) (Fig. 2B). Differential cell-surface gene expression was evaluated in the PrAd and NEPC cell lines and identified the established PrAd markers PSCA and FOLH1 (PSMA), as well as the NEPC marker neural cell adhesion molecule 1 (NCAM1) (Fig. 2C).
Fig. 2.
Transcriptomic analysis identifies candidate PrAd- and NEPC-specific cell surface markers. (A) Heatmap of the gene expression of select androgen-regulated, neuroendocrine, and epithelial markers and Myc genes based on RNA-seq of a diverse panel of human prostate cancer cell lines. Color bar represents a log2 scale. (B) Heatmap showing unsupervised hierarchical clustering of human prostate cancer cell lines based on the expression of cell-surface genes. (C) Plot of average expression of genes in NEPC vs. PrAd prostate cancer cell lines with select markers (NCAM1, FOLH1, and PSCA) highlighted. Gene expression is shown in log2 scale. (D and E) Venn diagrams showing rank overlap of the top 500 differentially expressed cell-surface genes in PrAd relative to NEPC (D) and NEPC relative to PrAd (E) in each of five gene-expression datasets [prostate cancer (CaP) cell line panel, Beltran 2016, WCDT, Zhang 2015 LuCaP xenografts, and Zhang 2015 metastatic CRPC (mCRPC) samples]. Listed are the genes identified from rank overlap analysis that are enriched in all of the datasets evaluated.
We then integrated multiple prostate cancer gene expression datasets to identify differentially expressed PrAd- and NEPC-specific cell-surface markers that are conserved in prostate cancer cell lines, PDXs, and patient tumors. For each of the datasets (WCDT, Beltran 2016, Zhang 2015 LuCaP PDXs, Zhang 2015 metastatic CRPC, and prostate cancer cell line panel), we performed rank overlap analysis by ranking the top 500 differentially expressed PrAd or NEPC cell-surface genes from each dataset and evaluating the overlap of these genes across datasets. A total of 21 genes were enriched in PrAd samples in all datasets, including well-established biomarkers and target antigens for prostate cancer therapeutics such as FOLH1 (PSMA), TACSTD2 (Trop2), and STEAP1 (Fig. 2D and Dataset S2). A total of 56 genes were commonly identified in NEPC samples across the datasets (Dataset S2). Notable from this set of genes were RET, DLL3, and SEZ6 that have been identified as disease markers in neuroendocrine cancers, including medullary thyroid cancer, small- and large-cell lung cancer, and malignant pheochromocytoma (30–32).
Prioritization of High-Confidence Cell-Surface Markers by Integrated Proteomic and Transcriptomic Analysis.
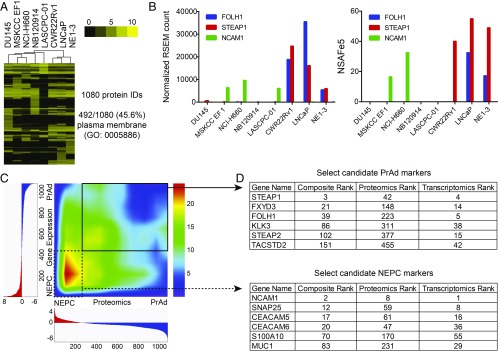
While transcriptomic analysis of the prostate cancer subsets for the identification of cell-surface antigens appeared informative, we needed to overcome (i) any inaccuracy of bioinformatic predictions of human cell-surface genes and (ii) the potential for discordance between mRNA levels and protein expression (33). We therefore directly profiled the surfaceomes of the prostate cancer cell-line panel by incubation with a membrane-impermeable biotin to label primary amines on extracellular domains of cell-surface proteins, cell lysis, streptavidin-affinity purification to enrich for biotinylated proteins, trypsin digestion, and quantitative proteomic mass spectrometry (34, 35). Then, 1,080 total proteins were identified, with 45.6% annotated for plasma membrane localization by Gene Ontology (Fig. 3A). Unsupervised clustering of the cell lines based on cell-surface proteomics showed two major clusters with NEPC and AR-negative PrAd lines segregated from AR-positive PrAd lines (Fig. 3A). To evaluate the relative concordance of the cell-surface gene expression and proteomic data from the cell lines, we compared the expression levels of known prostate cancer surface markers including FOLH1 (PSMA), STEAP1, and NCAM1. The gene- and protein-level expression of these markers in the cell-line panel were similar (Fig. 3B), with minor exceptions likely explained by protein-specific availability of primary amines for biotinylation, limitations in the sensitivity of mass spectrometry, and inherent variation in the abundance of proteins and their cognate mRNAs.
Fig. 3.
Integration of cell-surface proteomics with transcriptomics nominates high-confidence PrAd and NEPC cell surface proteins. (A) Heatmap displaying unsupervised hierarchical clustering of prostate cancer cell lines based on the expression of cell-surface proteins identified from cell-surface proteomics. Depicted are normalized protein abundance values (NSAFe5). Color bar represents a log2 scale. GO, Gene Ontology. (B) Comparison of the normalized RSEM gene expression counts and NSAFe5 values for the cell-surface markers FOLH1, STEAP1, and NCAM1 in each of the prostate cancer cell lines. (C) Rank–rank hypergeometric heatmap showing rank overlap of differentially expressed cell-surface proteins vs. cell-surface genes identified from cell-surface proteomics and RNA-seq gene-expression analysis of the prostate cancer cell line panel. (D) Select markers demonstrating concordantly enriched protein and gene expression in the PrAd or NEPC cell lines are shown with their associated composite, proteomics, and transcriptomics ranks.
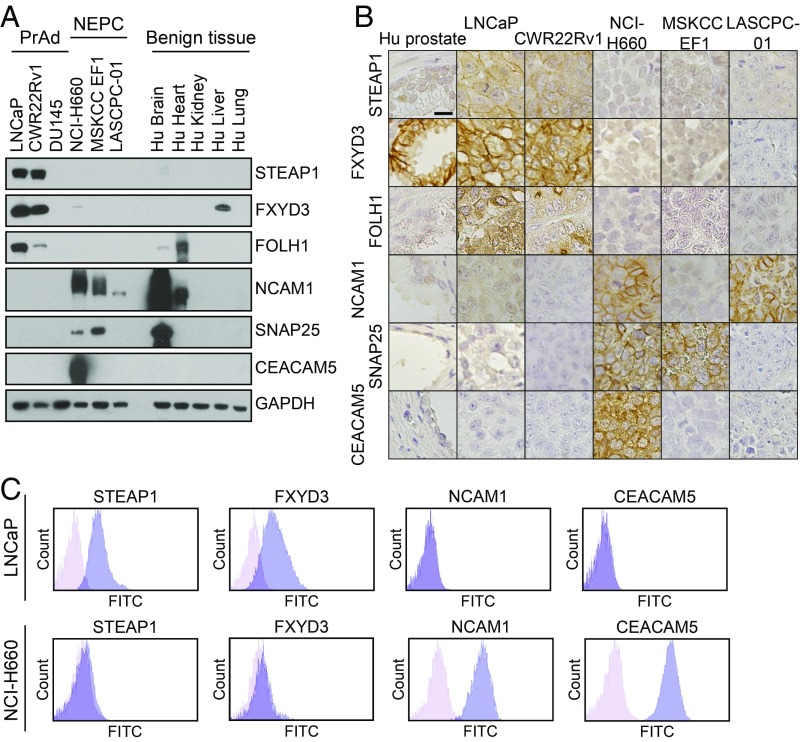
As a strategy to identify high-confidence markers in the NEPC and PrAd lines, we integrated the transcriptomic and proteomic data by RRHO analysis to prioritize surface markers that are enriched at the mRNA level and exhibit differential protein expression. Overall, the rank correlation between differentially expressed cell-surface genes and proteins was highest in the NEPC lines (Fig. 3C). We generated a composite rank by arbitrarily assigning equal weights to the proteomics and transcriptomics ranks of differentially expressed proteins or genes for each prostate cancer subset (Fig. 3D). Of the candidates with high composite ranks, the PrAd-specific expression of STEAP1, FXYD3, and FOLH1 (PSMA) and the NEPC-specific expression of NCAM1, SNAP25, and CEACAM5 were validated by immunoblot (Fig. 4A) and immunohistochemistry (IHC) (Fig. 4B) of prostate cancer cell lines and xenografts. Importantly, the expression patterns of these antigens in published human prostate cancer RNA-seq datasets mirrored the subtype-enriched expression observed in the cell-line panel (SI Appendix, Fig. S3). Flow cytometry confirmed the surface-protein expression of STEAP1 and FXYD3 on the LNCaP PrAd line, but not on the NCI-H660 NEPC line (Fig. 4C). Conversely, surface-protein expression of NCAM1 and CEACAM5 were found on NCI-H660, but not on LNCaP.
Fig. 4.
Validation of candidate prostate cancer subtype-specific cell-surface antigens. (A) Immunoblot analysis of select PrAd (LNCaP, CWR22Rv1, and DU145) and NEPC (NCI-H660, MSKCC EF1, and LASCPC-01) cell lines as well as benign human tissues (brain, heart, kidney, liver, and lung) with antibodies against STEAP1, FXYD3, FOLH1, NCAM1, SNAP25, CEACAM5, and GAPDH as a loading control. (B) Human prostate tissue (Hu prostate) or prostate cancer cell line (LNCaP, CWR22Rv1, NCI-H660, MSKCC EF1, and LASCPC-01) xenograft sections after immunohistochemical staining with antibodies for the candidate antigens from A. (Scale bar, 25 µm.) (C) Flow cytometry histogram plots of the PrAd cell line LNCaP and the NEPC cell line NCI-H660 stained with antibodies against STEAP1, FXYD3, NCAM1, and CEACAM5.
Validation of FXYD3 as a Tumor Antigen in PrAd.
FXYD3 belongs to the FXYD family of regulators of Na+/K+ ATPases that contain a 35-amino acid signature sequence domain beginning with PFXYD (36). FXYD3 has previously been found to be overexpressed in a variety of cancers, including those of the breast, stomach, and pancreas (37–39). FXYD3 was strongly enriched in the integrated transcriptomic and proteomic analysis in PrAd cell lines, but not as highly ranked by the transcriptome-based rank overlap analysis of the diverse prostate cancer datasets.
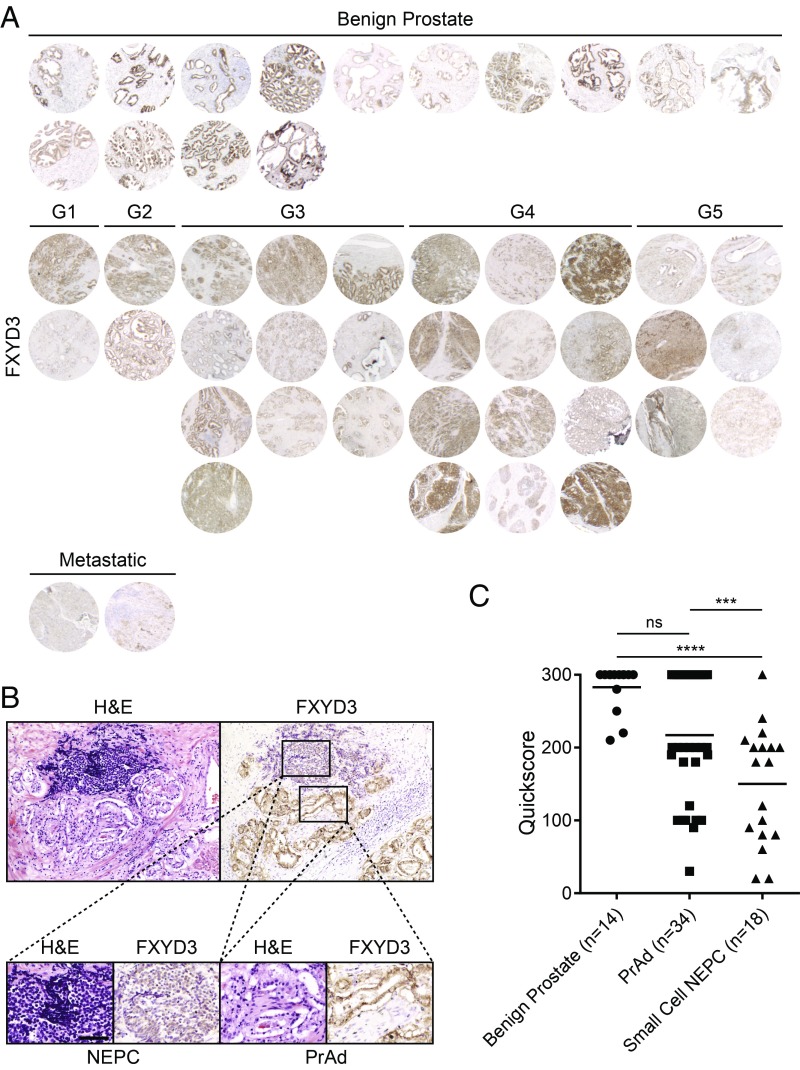
To evaluate FXYD3 expression in prostate cancer, we performed FXYD3 IHC on a tissue microarray of benign prostate samples as well as treatment-naïve primary Gleason grade 1–5 PrAd and metastatic PrAd. All 14 benign prostate tissues and 34 PrAd samples demonstrated FXYD3 expression (Fig. 5A), with the majority demonstrating moderate to strong staining specific to the normal and cancerous prostate epithelial cells (Fig. 5 A and C). FXYD3 IHC was also performed on a series of small cell NEPC tissues archived at the University of California, Los Angeles (UCLA) (SI Appendix, Fig. S4). We found that in several samples with mixed PrAd and small-cell NEPC, FXYD3 appeared to be more highly expressed in the PrAd components of the tumors (Fig. 5B). Quantitation of the FXYD3 IHC scores in all evaluated samples showed that FXYD3 protein expression was reduced on average in small-cell NEPC relative to benign prostate and PrAd (Fig. 5C).
Fig. 5.
FXYD3 is a cell-surface antigen whose expression is enriched in benign prostate epithelial cells and PrAd. (A) FXYD3 immunohistochemical stains of benign prostate tissues (n = 14), primary Gleason grade 1–5 PrAd tissues (n = 32), and metastatic PrAd samples (n = 2). (B) H&E and FXYD3 immunohistochemical stains of a section of mixed PrAd and NEPC. (Scale bar, 200 µm.) (C) Quantitation of FXYD3 IHC in benign prostate tissues (n = 14), PrAd (n = 34), and small-cell NEPC samples (n = 18) by Quickscore (intensity × percentage of positive cells; maximum score is 300). ns, nonsignificance. **P < 0.01; ****P < 0.0001 (by one-way ANOVA statistical analysis).
Evaluation of the NIH Genotype-Tissue Expression (GTEx) database showed that FXYD3 gene expression in human males is expressed in a variety of tissues including the skin, esophagus, stomach, small intestine, colon, bladder, and prostate (SI Appendix, Fig. S5A) (40). Congruent with the gene-expression data, IHC of a normal human tissue microarray demonstrated FXYD3 protein expression primarily in these organs (SI Appendix, Fig. S5B). The broad range of normal tissue expression would indicate that FXYD3 may not be suitable as a single target for highly potent cell-based immunotherapy due to the potential for on-target off-tumor toxicities. However, whether FXYD3 may be amenable to targeting by monoclonal antibodies or antibody drug conjugates (ADCs) is unknown, as dose-limiting off-tumor toxicity has not necessarily correlated with normal tissue expression (41).
Validation of CEACAM5 as a Target Antigen in NEPC.
CEACAM5 was identified as a candidate NEPC target antigen by both transcriptomic analysis of diverse prostate cancer datasets (Fig. 2E) and by integrated transcriptomic and proteomic analysis of the prostate cancer cell lines (Fig. 3C). In support of these findings, coexpression analysis of the Beltran 2016 dataset identified that the gene expression of CEACAM5 is highly correlated with the neuroendocrine marker chromogranin A (SI Appendix, Fig. S3B) (14). A similar, strong correlation was also found between CEACAM5 and the proneural pioneer transcription factor and high-grade neuroendocrine carcinoma marker ASCL1 in a metastatic prostate cancer gene-expression dataset (SI Appendix, Fig. S3C) (42, 43). CEACAM5 (CEA or carcinoembryonic antigen) is a glycophosphatidylinositol-anchored membrane protein and established tumor antigen whose expression has primarily been associated with adenocarcinomas of the colon, rectum, and pancreas. Despite case reports of detectable serum CEA in rare patients with advanced prostate cancer, a systematic study of CEACAM5 IHC in prostate tumors identified no expression in both primary and metastatic samples (44).
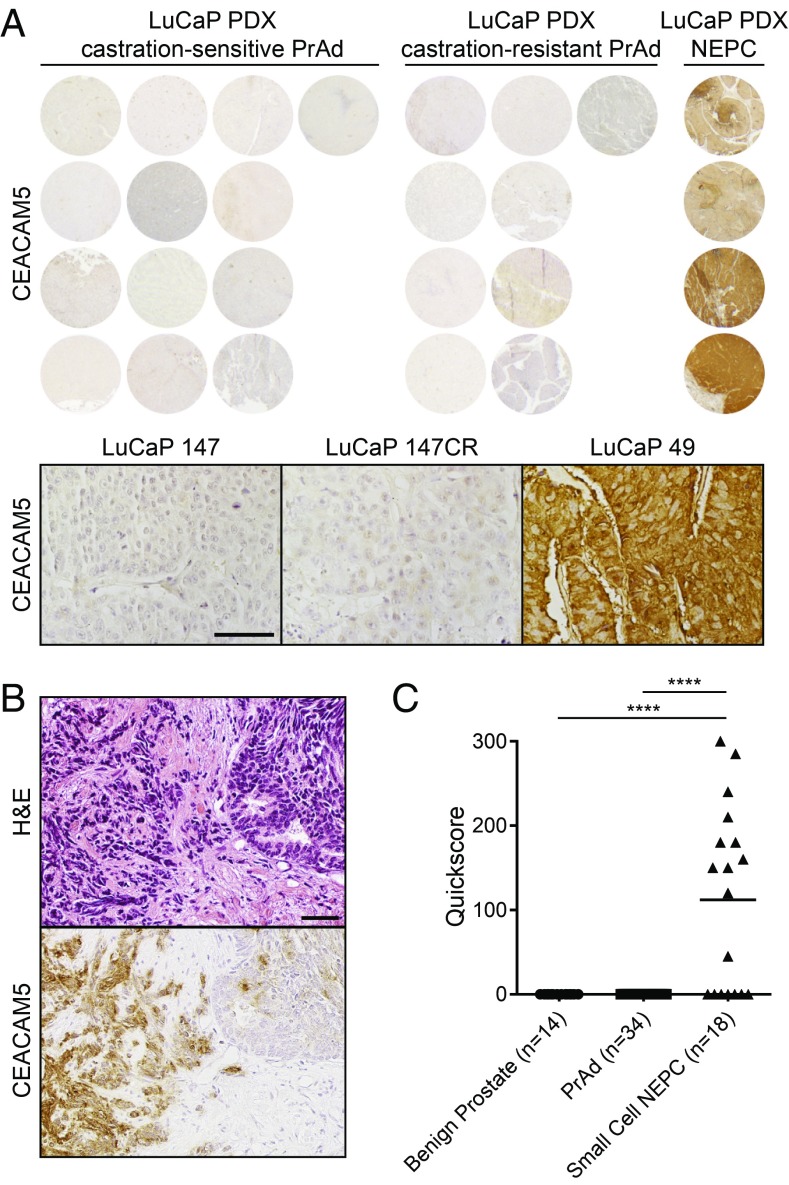
To verify protein expression of CEACAM5 in NEPC, we performed IHC on a prostate cancer tissue microarray of the LuCaP series of PDX models (45). While the 13 androgen-sensitive PrAd PDXs and 9 castration-resistant PrAd PDXs evaluated did not demonstrate CEACAM5 expression, all 4 NEPC PDXs exhibited moderate or strong CEACAM5 staining localized to the plasma membrane (Fig. 6A). We extended the IHC evaluation of CEACAM5 expression to a series of small-cell NEPC tissues archived at UCLA and a tissue microarray of benign prostate samples, as well as treatment-naïve primary Gleason grade 1–5 PrAd and metastatic PrAd. Eleven of 18 (61.1%) of the small-cell NEPC tissues stained for CEACAM5 in the plasma membrane (Fig. 6 B and C and SI Appendix, Fig. S6). In contrast, all 14 benign prostate tissues and 34 PrAd samples encompassing primary and metastatic tissues were devoid of CEACAM5 immunoreactivity (SI Appendix, Fig. S7). These IHC validation studies indicate that CEACAM5 expression appears to be prevalent in and specific to the NEPC subtype of prostate cancer.
Fig. 6.
CEACAM5 is a prostate cancer cell-surface antigen specific to the NEPC subtype. (A) CEACAM5 IHC of a LuCaP PDX tissue microarray with androgen-sensitive PrAd samples (n = 13), castration-resistant PrAd samples (n = 9), and NEPC samples (n = 4). CEACAM5 immunohistochemical stains of representative androgen-sensitive PrAd (LuCaP 147), castration-resistant PrAd (LuCaP 147CR), and NEPC (LuCaP 49) sections. (Scale bar, 100 µm.) (B) H&E and CEACAM5 immunohistochemical stains of a small cell NEPC sample archived at UCLA demonstrating adjoining regions of small-cell NEPC (left) and PrAd (right). (Scale bar, 100 µm.) (C) Quantitation of CEACAM5 IHC in benign prostate tissues (n = 14), PrAd (n = 34), and small-cell NEPC samples (n = 18) by Quickscore (intensity × percentage of positive cells; maximum score is 300). ****P < 0.0001 (by one-way ANOVA statistical analysis).
Therapeutic Targeting of CEACAM5 in NEPC.
CEACAM5 is an antigen that is the active focus of therapeutic development in colorectal cancer with ADCs and CAR T cells (46, 47). Given our findings, we sought to examine the potential for CEACAM5-targeted therapy in NEPC. We first explored safety implications by examining the systemic expression of CEACAM5 in normal human tissues at the mRNA and protein levels. Evaluation of the NIH GTEx database showed that CEACAM5 gene expression in men is limited to the colon, esophagus, and small intestine (SI Appendix, Fig. S8A) (40). A previous study of adoptive cell therapy with T cells engineered to express a high-affinity murine T cell receptor (TCR) targeting CEACAM5 in patients with metastatic colorectal cancer reported tumor regression, but also severe, transient colitis (48). However, data from a phase I trial of CEA-directed CAR T cell immunotherapy in CEA-positive metastatic colorectal cancers have indicated that CEA CAR T cell therapy may be well tolerated without evidence of colitis, even at high doses (47). In concordance with gene-expression data from the GTEx database, immunoblot analysis of a range of human tissue lysates from vital organs revealed absence of CEACAM5 protein expression in the brain, heart, kidney, liver, and lung (Fig. 4A). In addition, IHC of a normal human tissue microarray demonstrated CEACAM5 expression limited to the luminal lining of the colon and rectum in men (SI Appendix, Fig. S8B).
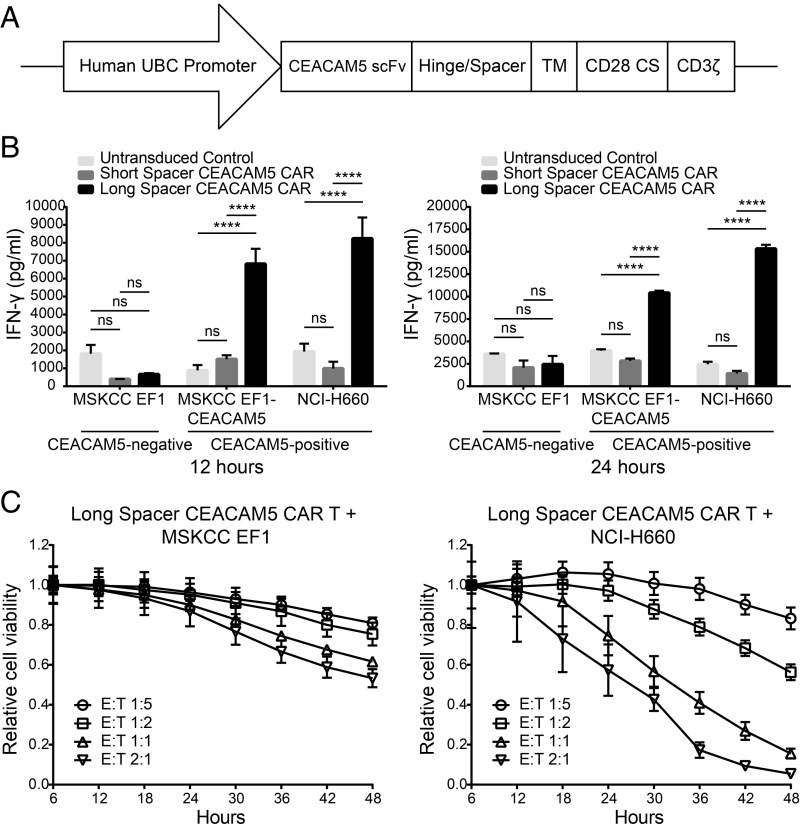
Given the relatively restricted systemic expression of CEACAM5 and the highly aggressive clinical nature of NEPC, we chose to engineer CARs targeting CEACAM5 to leverage both antigen specificity and cytotoxic potency of this technology. We generated two lentiviral CEACAM5 CAR constructs encoding a single chain variable fragment (scFv) derived from labetuzumab (49), hinge/spacer, CD28 transmembrane domain, CD28 costimulatory domain, and CD3ζ activation domain (Fig. 7A). The CEACAM5 CARs differed based on the presence of either a short spacer (IgG4 hinge) or long spacer (IgG4 hinge and CH2+CH3 spacer). We transduced T cells expanded from human peripheral blood mononuclear cells (PBMCs) with the CAR constructs and performed coculture assays with the target NEPC cell lines MSKCC EF1 (CEACAM5-negative; Fig. 4 A and B), MSKCC EF1-CEACAM5 (engineered to express CEACAM5), and NCI-H660 (CEACAM5-positive; Fig. 4) at a fixed effector-to-target ratio of 1:1. Analysis of the supernatant at 12 and 24 h by IFN-gamma (IFN-γ) ELISA revealed enhanced antigen-specific IFN-γ release associated with the long spacer CEACAM5 CAR (Fig. 7B). As the A3B3 domain of CEACAM5 recognized by the scFv is proximal to the membrane, a longer spacer may be necessary for optimal binding and T cell activation (49, 50).
Fig. 7.
Targeting CEACAM5 in NEPC with CAR T cell immunotherapy. (A) Schematic of the CAR construct targeting CEACAM5. CS, costimulatory domain; TM, CD28 transmembrane domain. (B) IFN-γ quantitation in the medium at 12 and 24 h after coculture of short spacer CEACAM5 CAR-transduced, long spacer CEACAM5 CAR-transduced, or untransduced T cells with CEACAM5-negative or -positive target cell lines as shown. SE measurements for four replicate wells are displayed. Data are representative of three independent experiments with similar results. ns represents nonsignificance. ****P < 0.0001 (by two-way ANOVA statistical analysis). (C) Relative viability over time of CEACAM5-negative MSKCC EF1 target cells or CEACAM5-positive NCI-H660 target cells cocultured with long spacer CEACAM5 CAR-transduced T cells. Effector-to-target ratios varying from 1:5 to 2:1 are shown. SE measurements for three replicate wells at each timepoint are displayed. Data are representative of two independent experiments with similar results.
To quantify cytotoxicity, we performed coculture assays in an Incucyte ZOOM (51), a live-cell imaging and analysis system allowing for direct enumeration of effector and target cells based on bright-field and fluorescence imaging. Varying effector-to-target ratios of T cells transduced with the long spacer CEACAM5 CAR and either MSKCC EF1 (CEACAM5-negative) or NCI-H660 (CEACAM5-positive) target NEPC cell lines engineered to express green fluorescent protein (GFP) were cocultured. Target cell counts were calculated and plotted to show relative target cell viability over time in coculture with effector cells. Coculture of long spacer CEACAM5 CAR-transduced T cells with NCI-H660 led to >80–90% cell kill by 48 h at effector-to-target ratios of 1:1 and 2:1 (Fig. 7C). In contrast, coculture with the MSKCC EF1 caused a minor reduction in target cell viability by 48 h, which may be related to low levels of CEACAM5 expression in the MSKCC EF1 NEPC cell line. Similar coculture studies were also performed with the PrAd cell line DU145 (CEACAM5-negative) and DU145-CEACAM5 (engineered to express CEACAM5). Long spacer CEACAM5 CAR-transduced T cells had negligible effects on the DU145 cells but induced significant T cell activation and target cell death when cocultured with DU145-CEACAM5 cells (SI Appendix, Fig. S9 A and B). These findings provide preliminary support for CEACAM5 as a promising target antigen for further therapeutic development in NEPC.
Discussion
Therapeutic development for advanced prostate cancer has increased significantly over the last decade. Both antibody- and cell-based immune treatment strategies are now poised to advance to the clinic, as monoclonal antibodies, ADCs, and CAR T cells are under clinical investigation. Most of these prospective therapies are focused on PSMA and PSCA as target antigens in CRPC. However, the heterogeneity of CRPC and the potential for treatment-induced plasticity (6, 52, 53) indicate that agents targeting only PSMA and PSCA are unlikely to eradicate the disease. An additional complication is the paucity of cancer-specific antigens that are not expressed in normal tissues (54). Sequential or combinatorial treatment strategies targeting distinct antigens while optimizing safety at various stages of disease progression will likely be necessary (55). To this end, we have characterized the surfaceome of advanced prostate cancer and generated a collection of putative target antigens using a discovery pipeline based on mRNA and cell-surface protein expression data. These studies are relevant and timely with the intent of expanding the development of targeted biologic therapies for advanced prostate cancer.
We have identified significant biological differences between the PrAd and NEPC subsets based on cell-surface protein profiling. Our data indicate that PrAd and NEPC express distinct cell-surface markers that mirror their respective glandular epithelial and neuroendocrine cancer differentiation states. Global cell-surface gene-expression analysis of these subsets across multiple published prostate cancer datasets clearly indicate that the surface phenotype of NEPC is more conserved than that of PrAd. This finding is consistent with the observed heterogeneity of PrAd that demonstrates a broad spectrum of histologic features, molecular subtypes, and clinical behaviors. On the other hand, the cell-surface profile of NEPC appears relatively homogeneous, suggesting that transdifferentiation to NEPC may represent a phenotypically constraining evolutionary path.
To nominate specific target antigens in prostate cancer as potential immunotherapeutic targets for further validation, cell-surface gene-expression and proteomics data were integrated from a diverse panel of prostate cancer cell lines. However, a pitfall of this approach is that mRNA abundance does not necessarily correlate with protein abundance (56), likely due to posttranscriptional and posttranslational modifications affecting stability. In the future, technological improvements in ultrasensitive quantitative mass spectrometry in proteomics may obviate the need to consider mRNA data and enable global surfaceome analysis for target antigen discovery directly from biopsy specimens. Another limitation in the validation and therapeutic translation of candidate target antigens is the availability of specific immunoaffinity reagents against extracellular protein domains. Large-scale efforts to characterize the expression of all human proteins in both normal and cancerous cells and tissues have been fraught with issues of data reliability due to inconsistent antibody performance (57). However, advances in recombinant antibody production including the use of highly diverse phage display and antibody library technologies should help overcome this bottleneck (58, 59).
We have specifically demonstrated that FXYD3 and CEACAM5 are plasma membrane-bound antigens expressed preferentially in PrAd and NEPC, respectively, based on multilevel validation studies on prostate cancer cell lines and tissues. We evaluated the normal tissue expression of these antigens as an additional filter to determine the potential for off-tumor, on-target toxicities of antibodies or cell-based immunotherapies. Due to the clinical need for novel therapies for aggressive NEPC, combined with our characterization of CEACAM5 expression in NEPC (including small-cell prostate cancer) and normal tissues, we have engineered CEACAM5 CAR constructs and demonstrated their potent antigen-specific NEPC cytotoxicity. CEACAM5 is an attractive therapeutic target in several solid tumors, but the translation of CEACAM5-targeted therapies is overwhelmingly focused on advanced colorectal cancer. A number of immune-based strategies have shown preclinical efficacy and are under clinical investigation, including antibody–drug conjugates (46, 60), a CEACAM5 and CD3 bispecific antibody (61), adoptive TCR transfer (48), and CAR T immunotherapy (47).
Our preliminary results indicate that CEACAM5-directed CAR T immunotherapy warrants further investigation as a treatment strategy for NEPC. Future studies will need to assess the antitumor efficacy and potential for toxicity in the gastrointestinal tract (48) in relevant, immune-competent model systems. Additionally, strategies to enhance the specificity and alleviate potential off-tumor toxicity of CEACAM5 CARs in NEPC should also be explored. One such approach is to use dual-gate CARs (62), in which two CARs, one targeting CEACAM5 and the other a second tumor antigen with nonoverlapping expression with CEACAM5 in normal tissues, are coexpressed in T cells such that each individual CAR is insufficient to induce a T cell response, but when both CARs are engaged, they synergize to promote T cell activation. Lastly, CARs targeting PSCA or PSMA and CEACAM5 either together or as a single bispecific construct (63) should be evaluated for safety and efficacy as a strategy to address the heterogeneity of advanced CRPC.
Methods
Detailed descriptions of cell lines, mouse xenograft studies, prostate cancer tissue microarrays and sections, antibodies, IHC, flow cytometry, and lentiviral vectors are found in SI Appendix, SI Methods. Viable human cells and tissues were provided in a deidentified manner and were therefore exempt from Institutional Review Board approval. All animal studies were performed according to protocols approved by the Animal Research Committee at University of California, Los Angeles.
Bioinformatic Derivation of Genes Encoding the Cell Surfaceome.
Genes encoding cell-surface proteins were assembled based on Gene Ontology annotations (22) (Membrane, Plasma Membrane, Integral Components of the Membrane, and Integral Components of the Plasma Membrane), putative transmembrane proteins based on analysis of the UniProt proteome of Homo sapiens using TMHMM (Version 2.0) (23), and predictions of GPI-anchored proteins from PredGPI (64).
RNA-Seq.
RNA was isolated from human prostate cancer cell lines by using an miRNeasy Mini Kit (Qiagen). Libraries for RNA-seq were prepared by using a TruSeq RNA Library Prep Kit (Version 2; Illumina). Sequencing was performed on an Illumina HiSeq 3000 with 2 × 150-bp reads. Demultiplexing of reads was performed by using CASAVA software (Version 1.8.2; Illumina). The Toil RNA-Seq Pipeline developed by the Computational Genomics Laboratory at the Genomics Institute of the University of California, Santa Cruz, was run locally to obtain gene- and transcript-level RSEM quantification of expression (65).
Transcriptome Analysis.
FASTQ files from the Beltran 2016 RNA-Seq dataset were downloaded from dbGaP (study accession no. phs000909.v1.p1) and analyzed with the Toil RNA-Seq Pipeline. The TCGA and NIH GTEx Toil RNAseq Recompute datasets were downloaded from the University of California, Santa Cruz, Xena Public Data Hub (65). In each prostate cancer gene expression dataset analyzed, differentially expressed cell-surface genes between NEPC and PrAd samples [false discovery rate (FDR) < 0.05] were ranked based on the magnitude of fold change. RRHO analysis was performed in pairwise comparisons of gene-expression datasets as described (25). For PANTHER analysis, cell-surface genes enriched more than eightfold in either NEPC or PrAd samples in the Beltran 2016 dataset were submitted for overrepresentation testing as described (27). Rank overlap analysis was performed by taking the 500 most differentially enriched cell-surface genes in NEPC and PrAd samples from each dataset (FDR < 0.05) and identifying genes similarly enriched across all datasets.
Proteomic Analysis.
A total of 4 × 107 cells from each cell line were subjected to cell-surface biotinylation and quenching per the Pierce Cell Surface Protein Isolation Kit (Thermo Fisher Scientific). Cells were lysed in urea lysis buffer (8 M urea, 2% SDS, and 100 mM Tris, pH 8) and DNA digested with 250 U of Benzonase endonuclease (Sigma). Biotin-labeled proteins were affinity-purified on streptavidin agarose beads (Thermo Fisher Scientific), sequentially treated with 5 mM Tris(2-carboxyethyl) phosphine and 10 mM iodoacetamide, and digested on-bead with Lys-C and trypsin proteases as described (66). Peptides were fractionated by multidimensional chromatography followed by tandem mass spectrometric analysis on a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). RAW-Xtract (Version 1.8) was used to extract peak list information from Xcalibur-generated RAW files. Database searching of the MS/MS spectra was performed by using the ProLuCID algorithm (Version 1.0). Other database search parameters included (i) precursor ion mass tolerance of ±20 ppm; (ii) fragment ion mass tolerance of ±400 ppm; (iii) only peptides with fully tryptic ends were considered candidate peptides in the search with no consideration for missed cleavages; and (iv) static modification of +57.02156 on cysteine residues. Peptide identifications were organized and filtered by using the DTASelect algorithm, which uses a linear discriminant analysis to identify peptide-scoring thresholds that yield a peptide-level FDR of <5% as estimated by using a decoy database approach. Proteins were considered present in the analysis if they were identified by two or more peptides using the 5% peptide-level FDR.
CAR T cell Engineering and Coculture Assays.
Deidentified human PBMCs were obtained from the UCLA Virology Core Laboratory and grown in T cell medium (TCM) base medium composed of AIM V medium (Thermo Fisher Scientific), 5% heat-inactivated human AB serum, 2 mM glutamine, and 55 μM 2-mercaptoethanol (Sigma). For coculture experiments involving IFN-γ release assays measured by ELISA, human PBMCs were activated in a 24-well plate coated with 1 µg/mL anti-CD3 (eBioscience OKT-3), 1 µg/mL anti-CD28 (eBioscience CD28.2), and 300 U/mL IL-2 in TCM base medium. After 48 h, cells were spin-infected daily for 2 d with CAR lentivirus at a multiplicity of infection (MOI) of ∼6–11 in TCM base medium, 300 U/mL IL-2, and 8 µg/mL polybrene. After each infection, the cells were washed and grown in TCM base medium with 300 U/mL IL-2. Ninety-six hours after final spin infection, T cell transduction efficiency was measure by flow cytometry, and T cells were cocultured with target cells at a target:effector ratio of 1:1. Supernatant was harvested at 12 and 24 h after coculture. IFN-γ was quantitated with the BD OptEIA Human IFN-γ ELISA Set (BD Biosciences) according to the manufacturer’s protocol. For coculture experiments with direct visualization of cytotoxicity by live cell imaging, human PBMCs were activated with Gibco Dynabeads Human T-Activator CD3/CD28 (Thermo Fisher Scientific) in TCM base medium with 50 U/mL IL-2 at a cell:bead ratio of 1. After 96 h, T cells were infected with CAR lentivirus by spin infection in TCM base medium with 50 U/mL IL-2 at an MOI of 0.5–50. Cells were washed 24 h after infection and cultured in TCM base medium with 50 U/mL IL-2. Dynabeads were removed 48 h after infection. Ninety-six hours after spin infection, T cell transduction efficiency was measured by flow cytometry, and T cells were cocultured with target cells at a range of target:effector ratios. Cytotoxicity was measured by Incucyte ZOOM through quantification of GFP-positive target cell counts.
Supplementary Material
Acknowledgments
We thank the Technology Center for Genomics and Bioinformatics at UCLA for RNA-seq services, the UCLA Proteome Research Center for mass spectrometry services, and the Translational Pathology Core Laboratory at UCLA for histology services. This work was supported by the Prostate Cancer Biorepository Network supported by the Department of Defense Prostate Cancer Research Program Grants W81XWH-14-2-0182, W81XWH-14-2-0183, W81XWH-14-2-0185, W81XWH-14-2-0186, and W81XWH-15-2-0062. J.K.L. is supported by a Lori Milken Prostate Cancer Foundation Young Investigator Award, a Prostate Cancer Foundation Challenge Award, Department of Defense Prostate Cancer Research Program Physician Research Award PC160018, UCLA Specialized Program of Research Excellence (SPORE) in Prostate Cancer Career Enhancement Award NIH P50 CA092131, and a STOP CANCER Foundation Seed Grant. J.W.P. is supported by NIH/National Cancer Institute (NCI) Grant K99 CA218731. T.G.G. is supported by NIH/NCI Grant P01 CA168585, UCLA SPORE in Prostate Cancer Grant NIH P50 CA092131, and American Cancer Society Research Scholar Award RSG-12-257-01-TBE. O.N.W. is supported by UCLA SPORE in Prostate Cancer Grant P50 CA092131, NCI/NIH Grant R01 CA220238, a Prostate Cancer Foundation Challenge Award, the Gaba Foundation, and the UCLA Broad Stem Cell Research Center. Portions of this research were also supported by funds from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research and the Parker Institute for Cancer Immunotherapy Grant PICI 20163828.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802354115/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Bluemn EG, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32:474–489.e6. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epstein JI, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol. 2014;38:756–767. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clermont PL, et al. Identification of the epigenetic reader CBX2 as a potential drug target in advanced prostate cancer. Clin Epigenetics. 2016;8:16. doi: 10.1186/s13148-016-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleb B, et al. Differentially methylated genes and androgen receptor re-expression in small cell prostate carcinomas. Epigenetics. 2016;11:184–193. doi: 10.1080/15592294.2016.1146851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ku SY, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltran H, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JK, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29:536–547. doi: 10.1016/j.ccell.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dardenne E, et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30:563–577. doi: 10.1016/j.ccell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komura K, et al. Current treatment strategies for advanced prostate cancer. Int J Urol. 2018;25:220–231. doi: 10.1111/iju.13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afshar-Oromieh A, et al. Comparison of PET imaging with a (68)Ga-labelled PSMA ligand and (18)F-choline-based PET/CT for the diagnosis of recurrent prostate cancer. Eur J Nucl Med Mol Imaging. 2014;41:11–20. doi: 10.1007/s00259-013-2525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty PS, et al. Metastatic poorly differentiated prostatic carcinoma with neuroendocrine differentiation: Negative on 68Ga-PSMA PET/CT. Clin Nucl Med. 2015;40:e163–e166. doi: 10.1097/RLU.0000000000000594. [DOI] [PubMed] [Google Scholar]
- 13.Tosoian JJ, et al. Correlation of PSMA-targeted18F-DCFPyL PET/CT findings with immunohistochemical and genomic data in a patient with metastatic neuroendocrine prostate cancer. Clin Genitourin Cancer. 2017;15:e65–e68. doi: 10.1016/j.clgc.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beltran H, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeVeale B, et al. Surfaceome profiling reveals regulators of neural stem cell function. Stem Cells. 2014;32:258–268. doi: 10.1002/stem.1550. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann A, et al. Proteomic cell surface phenotyping of differentiating acute myeloid leukemia cells. Blood. 2010;116:e26–e34. doi: 10.1182/blood-2010-02-271270. [DOI] [PubMed] [Google Scholar]
- 17.Maloney DG, et al. IDEC-C2B8 (rituximab) anti-CD20 monoclonal antibody therapy in patients with relapsed low-grade non-Hodgkin’s lymphoma. Blood. 1997;90:2188–2195. [PubMed] [Google Scholar]
- 18.Kalos M, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brentjens RJ, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5:177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadelain M, Brentjens R, Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Cunha JP, et al. Bioinformatics construction of the human cell surfaceome. Proc Natl Acad Sci USA. 2009;106:16752–16757. doi: 10.1073/pnas.0907939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 24.Zhang X, et al. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21:4698–4708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: Identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal R, et al. Targeting adaptive pathways in metastatic treatment-resistant prostate cancer: Update on the Stand Up 2 Cancer/Prostate Cancer Foundation-supported West Coast Prostate Cancer Dream Team. Eur Urol Focus. 2016;2:469–471. doi: 10.1016/j.euf.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong S-K, Kim J-H, Lin M-F, Park J-I. The Raf/MEK/extracellular signal-regulated kinase 1/2 pathway can mediate growth inhibitory and differentiation signaling via androgen receptor downregulation in prostate cancer cells. Exp Cell Res. 2011;317:2671–2682. doi: 10.1016/j.yexcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoro M, et al. The ret proto-oncogene is consistently expressed in human pheochromocytomas and thyroid medullary carcinomas. Oncogene. 1990;5:1595–1598. [PubMed] [Google Scholar]
- 31.Saunders LR, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med. 2015;7:302ra136. doi: 10.1126/scitranslmed.aac9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waldmann J, et al. Microarray analysis reveals differential expression of benign and malignant pheochromocytoma. Endocr Relat Cancer. 2010;17:743–756. doi: 10.1677/ERC-09-0118. [DOI] [PubMed] [Google Scholar]
- 33.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elia G. Biotinylation reagents for the study of cell surface proteins. Proteomics. 2008;8:4012–4024. doi: 10.1002/pmic.200800097. [DOI] [PubMed] [Google Scholar]
- 35.Nunomura K, et al. Cell surface labeling and mass spectrometry reveal diversity of cell surface markers and signaling molecules expressed in undifferentiated mouse embryonic stem cells. Mol Cell Proteomics. 2005;4:1968–1976. doi: 10.1074/mcp.M500216-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Crambert G, Geering K. FXYD proteins: New tissue-specific regulators of the ubiquitous Na,K-ATPase. Sci STKE. 2003;2003:RE1. doi: 10.1126/stke.2003.166.re1. [DOI] [PubMed] [Google Scholar]
- 37.Morrison BW, et al. Mat-8, a novel phospholemman-like protein expressed in human breast tumors, induces a chloride conductance in Xenopus oocytes. J Biol Chem. 1995;270:2176–2182. doi: 10.1074/jbc.270.5.2176. [DOI] [PubMed] [Google Scholar]
- 38.Kayed H, et al. FXYD3 is overexpressed in pancreatic ductal adenocarcinoma and influences pancreatic cancer cell growth. Int J Cancer. 2006;118:43–54. doi: 10.1002/ijc.21257. [DOI] [PubMed] [Google Scholar]
- 39.Zhu ZL, et al. Expression and significance of FXYD-3 protein in gastric adenocarcinoma. Dis Markers. 2010;28:63–69. doi: 10.3233/DMA-2010-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GTEx Consortium The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinrichs MJ, Dixit R. Antibody drug conjugates: Nonclinical safety considerations. AAPS J. 2015;17:1055–1064. doi: 10.1208/s12248-015-9790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altree-Tacha D, Tyrrell J, Li F. mASH1 is highly specific for neuroendocrine carcinomas: An immunohistochemical evaluation on normal and various neoplastic tissues. Arch Pathol Lab Med. 2017;141:288–292. doi: 10.5858/arpa.2015-0489-OA. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blumenthal RD, Leon E, Hansen HJ, Goldenberg DM. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen HM, et al. LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease and serve as models for evaluating cancer therapeutics. Prostate. 2017;77:654–671. doi: 10.1002/pros.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dotan E, et al. Phase I/II trial of labetuzumab govitecan (anti-CEACAM5/SN-38 antibody-drug conjugate) in patients with refractory or relapsing metastatic colorectal cancer. J Clin Oncol. 2017;35:3338–3346. doi: 10.1200/JCO.2017.73.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C, et al. Phase I escalating-dose trial of CAR-T therapy targeting CEA+ metastatic colorectal cancers. Mol Ther. 2017;25:1248–1258. doi: 10.1016/j.ymthe.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parkhurst MR, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19:620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein R, Goldenberg DM. A humanized monoclonal antibody to carcinoembryonic antigen, labetuzumab, inhibits tumor growth and sensitizes human medullary thyroid cancer xenografts to dacarbazine chemotherapy. Mol Cancer Ther. 2004;3:1559–1564. [PubMed] [Google Scholar]
- 50.Guest RD, et al. The role of extracellular spacer regions in the optimal design of chimeric immune receptors: Evaluation of four different scFvs and antigens. J Immunother. 2005;28:203–211. doi: 10.1097/01.cji.0000161397.96582.59. [DOI] [PubMed] [Google Scholar]
- 51.Artymovich K, Appledorn DM. A multiplexed method for kinetic measurements of apoptosis and proliferation using live-content imaging. Methods Mol Biol. 2015;1219:35–42. doi: 10.1007/978-1-4939-1661-0_4. [DOI] [PubMed] [Google Scholar]
- 52.Mu P, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miao L, et al. Disrupting androgen receptor signaling induces snail-mediated epithelial-mesenchymal plasticity in prostate cancer. Cancer Res. 2017;77:3101–3112. doi: 10.1158/0008-5472.CAN-16-2169. [DOI] [PubMed] [Google Scholar]
- 54.Priceman SJ, Forman SJ, Brown CE. Smart CARs engineered for cancer immunotherapy. Curr Opin Oncol. 2015;27:466–474. doi: 10.1097/CCO.0000000000000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadelain M, Rivière I, Riddell S. Therapeutic T cell engineering. Nature. 2017;545:423–431. doi: 10.1038/nature22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kosti I, Jain N, Aran D, Butte AJ, Sirota M. Cross-tissue analysis of gene and protein expression in normal and cancer tissues. Sci Rep. 2016;6:24799. doi: 10.1038/srep24799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berglund L, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7:2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Li K, et al. A fully human scFv phage display library for rapid antibody fragment reformatting. Protein Eng Des Sel. 2015;28:307–316. doi: 10.1093/protein/gzv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frenzel A, Schirrmann T, Hust M. Phage display-derived human antibodies in clinical development and therapy. MAbs. 2016;8:1177–1194. doi: 10.1080/19420862.2016.1212149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouillon-Pichault M, et al. Translational model-based strategy to guide the choice of clinical doses for antibody-drug conjugates. J Clin Pharmacol. 2017;57:865–875. doi: 10.1002/jcph.869. [DOI] [PubMed] [Google Scholar]
- 61.Bacac M, et al. A novel carcinoembryonic antigen T-cell bispecific antibody (CEA TCB) for the treatment of solid tumors. Clin Cancer Res. 2016;22:3286–3297. doi: 10.1158/1078-0432.CCR-15-1696. [DOI] [PubMed] [Google Scholar]
- 62.Kloss CC, Condomines M, Cartellieri M, Bachmann M, Sadelain M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol. 2013;31:71–75. doi: 10.1038/nbt.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zah E, Lin MY, Silva-Benedict A, Jensen MC, Chen YY. T cells expressing CD19/CD20 bispecific chimeric antigen receptors prevent antigen escape by malignant B cells. Cancer Immunol Res. 2016;4:498–508. doi: 10.1158/2326-6066.CIR-15-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pierleoni A, Martelli PL, Casadio R. PredGPI: A GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vivian J, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35:314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oberholzer M, et al. Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol Cell Proteomics. 2011;10:M111.010538. doi: 10.1074/mcp.M111.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.