Significance
The germline is responsible for the reproduction of an entire organism through the recovery of totipotency after fertilization. This ability is presumably associated with a chromatin feature known as bivalent chromatin domains, which are marked with both repressive and active histone modifications. In this study, we determine the identity of bivalent domain genes in the male germline of mice and the regulatory mechanism for these genes. We demonstrate that SCML2 facilitates a repressive modification, H3K27me3, thereby establishing bivalent domains in the germline. Our study identifies a possible germline mechanism by which differentiated and unipotent germ cells give rise to a totipotent zygote following fertilization.
Keywords: germline, spermatogenesis, meiosis, Polycomb, bivalent domains
Abstract
Repressive H3K27me3 and active H3K4me2/3 together form bivalent chromatin domains, molecular hallmarks of developmental potential. In the male germline, these domains are thought to persist into sperm to establish totipotency in the next generation. However, it remains unknown how H3K27me3 is established on specific targets in the male germline. Here, we demonstrate that a germline-specific Polycomb protein, SCML2, binds to H3K4me2/3-rich hypomethylated promoters in undifferentiated spermatogonia to facilitate H3K27me3. Thus, SCML2 establishes bivalent domains in the male germline of mice. SCML2 regulates two major classes of bivalent domains: Class I domains are established on developmental regulator genes that are silent throughout spermatogenesis, while class II domains are established on somatic genes silenced during late spermatogenesis. We propose that SCML2-dependent H3K27me3 in the male germline prepares the expression of developmental regulator and somatic genes in embryonic development.
The germline is responsible for the reproduction of an entire organism through the recovery of totipotency after fertilization. This ability is directed by epigenetic mechanisms that govern heritable gene expression programs. Before fertilization, during the later stages of germ cell differentiation in the testes, germ cells go through male-specific epigenetic programming (1–5). This preprogramming is critical to license the zygote for totipotency. It remains a long-standing mystery as to the germline-based mechanism to establish unique epigenetic information that prepares totipotent zygotes.
Recent studies have revealed a prominent epigenetic signature in the germline that persists in mature sperm. Genes critical for somatic development (termed developmental genes) remain silent throughout the germline and harbor bivalent genomic domains, characterized by the concomitant enrichment in repressive H3K27me3 and active H3K4me2/3 marks (6–11). Bivalent domains were initially identified in embryonic stem (ES) cells as molecular hallmarks of developmental potential; while developmental genes are suppressed, they harbor bivalent marks that maintain their potential to differentiate into any cell lineage (12–14). Due to this potential, bivalent domains in the male germline could be vitally important for the recovery of totipotency and developmental programs in the next generation (15, 16). Although the persistence of histone modifications from sperm to embryos is debated (17–19), bivalent domains are acknowledged as chromatin features that persist into sperm without being replaced by protamines (6, 7, 9, 20), potentially serving as mediators of epigenetic inheritance across generations.
To prepare the next generation, male germ cells undergo global suppression of a somatic gene expression program in late spermatogenesis, during meiosis, and into postmeiotic stages (21, 22). During these stages, thousands of genes commonly expressed in both somatic lineages and the progenitor cells (mitotic phases) of spermatogenesis, termed somatic/progenitor genes, are largely suppressed; meanwhile, a distinct class of thousands of genes, termed late spermatogenesis genes, is activated. Of note, we recently identified a germline-specific Polycomb protein, SCML2, that suppresses somatic/progenitor genes in the later stages of spermatogenesis (21). Our recent study further demonstrated that suppression of somatic/progenitor genes is associated with the formation of bivalent domains (22).
Critical to the regulation of bivalent domains is Polycomb repressive complex 2 (PRC2), which mediates the deposition of H3K27me3. Polycomb proteins regulate heritable gene silencing, define cell type-specific gene expression (23–25), and are required for gene expression programs during spermatogenesis (21, 26, 27). PRC2 is essential in spermatogenesis for the maintenance of spermatogonial stem cells, the progression of meiosis, and the suppression of soma-specific genes (26). However, since PRC2 has broad functions in different biological contexts, an outstanding question remains as to how H3K27me3 is established at specific chromatin regions in the male germline. Here, we identify two major classes of bivalent domains: Class I is established on developmental regulators that are silent throughout spermatogenesis, while class II is established on somatic genes silenced during late spermatogenesis. We demonstrate that SCML2 is recruited to H3K4me2/3-rich hypomethylated promoters to facilitate H3K27me3 at these bivalent domains. Together, our study identifies SCML2 as a key regulator of H3K27me3 in the male germline.
Results
Establishment of Two Major Classes of Bivalent Domains in the Male Germline.
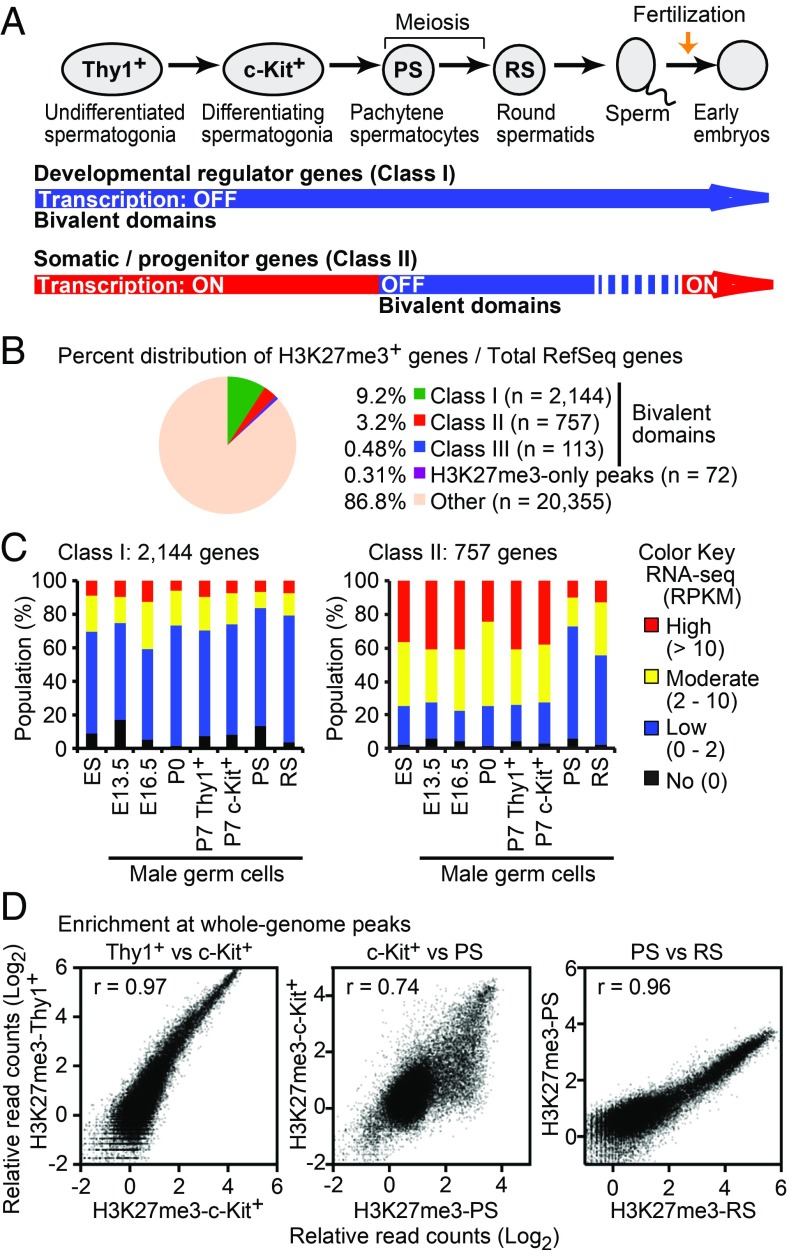
In the male germline, bivalent domains are observed on developmental regulator genes (8) and on somatic/progenitor genes (22) in late spermatogenesis. However, due to the lack of ChIP-sequencing (ChIP-seq) data of in vivo-purified spermatogonia, which comprise the mitotic stages of male germ cells, it remains unknown when these bivalent domains are established in the male germline. To define the sets of genes with bivalent domains during in vivo spermatogenesis, we performed ChIP-seq of H3K27me3, H3K4me2, and H3K4me3 using purified cells from mouse testes at representative stages of spermatogenesis. We analyzed Thy1+ undifferentiated spermatogonia from postnatal day (P) 7 testes, which contain spermatogonial stem cells and progenitor cells; c-Kit+ differentiating spermatogonia from P7 testes; purified pachytene spermatocyte(s) (PS) undergoing meiosis; and postmeiotic round spermatid(s) (RS) from adult testes (Fig. 1A). We carried out ChIP-seq for two independent biological replicates and confirmed the reproducibility between biological replicates (Fig. S1 and Table S1).
Fig. 1.
Two major classes of bivalent domains in the male germline. (A) Schematic of spermatogenesis and summary of the two major classes of bivalent domains. (B) Percent distribution of H3K27me3+ genes among total RefSeq genes. (C) Distribution of RNA-sequencing data for each class of bivalent domains in the male germline. E, embryonic day. (D) Correlation of H3K27me3 ChIP-seq signals at individual peaks between the indicated stages. Each peak was identified using model-based analysis of ChIP-Seq (MACS; P < 1 × 10−5). Enrichment levels of H3K27me3 are shown in log2 RPKM values. The Pearson correlation coefficient values (r) indicate the similarity between the stages indicated.
We identified two major classes of bivalent domains around transcription start sites (TSSs), termed class I and class II (criteria are described in SI Materials and Methods, and gene lists are shown in Dataset S1). Class I is established on developmental regulators that are silent through spermatogenesis. The class I static bivalent domains consist of 2,144 genes (9.2% of all RefSeq genes; Fig. 1B) for which H3K27me3 persists from Thy1+ spermatogonia into postmeiotic spermatids (Fig. S2 A and B). The Ebf3 locus represents class I bivalent domains that persist on TSSs throughout spermatogenesis, from Thy1+ spermatogonia through meiosis into postmeiotic spermatids (Fig. S2A). Class I genes are largely repressed throughout the male germline (Fig. 1C) and enriched for genes associated with somatic developmental contexts, such as multicellular organism development, that are not associated with spermatogenesis (Fig. S1C). By contrast, H3K27me3 is established later on class II genes, acquiring bivalent status in meiosis that persists into postmeiotic spermatids. The class II bivalent domains consist of 757 genes (3.2% of all RefSeq genes; Fig. 1B). The Sgpl1 locus represents class II bivalent domains in which H3K4me2/3 is present without H3K27me3 in undifferentiated spermatogonia (Fig. S2A). Consistent with the de novo establishment of H3K27me3 at class II genes, the genome-wide acquisition of H3K27me3 takes place during the mitosis-to-meiosis transition between c-Kit+ spermatogonia and PS (Fig. 1D). This is a unique feature of the mitosis-to-meiosis transition since H3K27me3 is highly correlated between Thy1+ and c-Kit+ spermatogonia, and between PS and RS (Fig. 1D). In association with the genome-wide acquisition of H3K27me3, class II genes are largely active before meiosis but are repressed in PS and RS (Fig. 1C). Class II genes are enriched for genes associated with the mitotic phases of germ cells, including those that regulate metabolic and signaling functions (Fig. S2C). In addition to these two major classes, we found a minor class of bivalent domain genes in which bivalent domains are present in spermatogonia that are dissolved in PS (termed class III: 113 genes, 0.48% of all RefSeq genes; Fig. 1B and Fig. S2C).
During meiosis, H3K27me3 is excluded from the sex chromosomes due to meiotic sex chromosome inactivation (22), an epigenomic event unique to the sex chromosomes. Thus, class I and II bivalent domains are located exclusively on autosomes (Dataset S1). Together, these results reveal two major classes of bivalent domains on autosomes. Class I bivalent domains are associated with genes involved in somatic developmental processes, distinct from germ cell developmental processes. Class II bivalent domains are associated with somatic/progenitor genes that are expressed in the mitotic phases of germ cells.
H3K27me3 Is Established on H3K4me2/3-Rich Promoters.
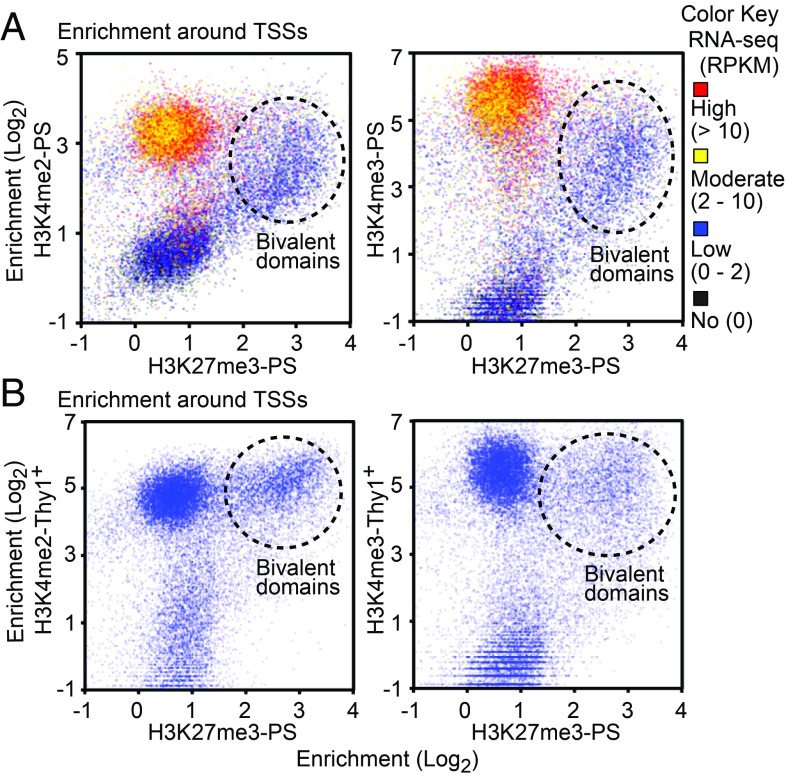
To elucidate how bivalent domains are established, we determined the distribution of the active modifications H3K4me2 and H3K4me3, which form bivalent domains together with H3K27me3. In PS, enrichment of H3K27me3 at TSSs is correlated with that of H3K4me2 and H3K4me3 and with low gene expression [less than 2 read counts per kilobase per million (RPKM); Fig. 2A], indicating that most H3K27me3 peaks form bivalent domains with H3K4me2/3 in the context of gene repression. Indeed, genes with only H3K27me3 peaks and without H3K4me2/3 are rare (72 genes, 0.31% of all RefSeq genes; Fig. 1B). Therefore, most H3K27me3-enriched TSSs are bivalent (i.e., enriched with H3K4me2/3). On the other hand, H3K4me2/3-enriched TSSs are not always enriched with H3K27me3. For example, H3K4me2/3 enrichment on moderately or highly expressed genes (i.e., those with ≥2 RPKM) is largely devoid of H3K27me3 (Fig. 2A). Notably, in PS, H3K27me3 was enriched on TSSs where H3K4me2/3 was previously enriched in Thy1+ spermatogonia (Fig. 2B), suggesting that H3K27me3 is established at a portion of H3K4me2/3-rich promoters. Taken together, these results suggest that, on class I bivalent domains, H3K27me3 is retained on H3K4me2/3-rich promoters from Thy1+ spermatogonia to PS. In comparison, class II bivalent domains are established in PS through the deposition of H3K27me3 on promoters that are preloaded with H3K4me2/3 in Thy1+ spermatogonia. Since deposition of H3K27me3 is preceded by the presence of H3K4me2/3, we next sought to determine the mechanism by which H3K27me3 is established on H3K4me2/3-rich promoters.
Fig. 2.
H3K27me3 is established on H3K4me2/3-rich promoters. (A) Enrichment of ChIP-seq signal (±2 kb around TSSs) and its comparison with RNA-sequencing (RNA-seq) data for PS (RNA-seq signal is indicated by color, as shown in the legend). (B) Enrichment of ChIP-seq signal (±2 kb around TSSs).
SCML2 Binds to TSSs Where Bivalent Domains Are Formed.
During spermatogenesis, chromatin-bound SCML2 is enriched at somatic/progenitor gene loci in undifferentiated spermatogonia; these genes are later suppressed by SCML2 after the mitosis-to-meiosis transition (21) and acquire bivalent domains (22). Since SCML2 is critical for the suppression of somatic/progenitor genes (21), we hypothesized that SCML2 regulates class II bivalent domains for the suppression of somatic/progenitor genes. To determine whether the deposition of SCML2 in undifferentiated spermatogonia leads to the establishment of bivalent domains later in spermatogenesis, we compared the distribution of SCML2 and H3K27me3 by ChIP-seq. We analyzed our SCML2 ChIP-seq data (21) in germline stem (GS) cells, an in vitro culture of spermatogonial stem cells (28), due to the technical demands of obtaining sufficient amounts of undifferentiated spermatogonia from testes for SCML2 ChIP-seq. The validity of comparing GS cells with Thy1+ spermatogonia is supported by our previous studies, which show that GS cells and Thy1+ spermatogonia have comparable gene expression profiles (21, 22). At the Sgpl1 locus, a locus representative of class II genes, SCML2 binds near TSSs where H3K4me2/3 is enriched. The region subsequently gains H3K27me3 in PS, becoming a bivalent domain (Fig. S2A). Notably, at the Ebf3 locus, a locus representative of class I genes, SCML2 also accumulates near the TSS, which correlates with the accumulation of H3K27me3 in Thy1+ spermatogonia (Fig. S2A).
To determine the link between SCML2 binding and the establishment of H3K27me3, we examined the relationship between SCML2 enrichment and H3K27me3 enrichment by ChIP-seq analyses. SCML2 enrichment was clearly correlated with H3K27me3 enrichment both in Thy1+ spermatogonia and in PS (Fig. 3A), as well as in c-Kit+ spermatogonia and in RS (Fig. S3A). Taken together, we conclude that SCML2 binds not only class II bivalent domain genes but also class I bivalent domain genes in undifferentiated spermatogonia. Further, the deposition of SCML2 predicts the establishment of H3K27me3 on bivalent domain genes during spermatogenesis.
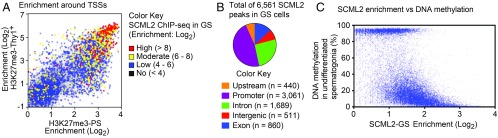
Fig. 3.
SCML2 binds to TSSs where bivalent domains are formed and is enriched at hypomethylated loci. (A) Enrichment of H3K27me3 ChIP-seq reads in Thy1+ spermatogonia (Thy1+) and PS, and their comparison with SCML2 ChIP-seq reads in GS cells (±2 kb around TSSs). (B) Proportions of SCML2 peaks in GS cells. (C) SCML2 enrichment near TSSs (±2 kb around TSSs) largely correlates with DNA hypomethylation in undifferentiated spermatogonia.
SCML2 Is Enriched at Hypomethylated Loci.
Since the deposition of SCML2 predicts the establishment of H3K27me3, we sought to determine mechanisms by which SCML2 is recruited to target sites. To this end, we investigated the genomic features of SCML2 binding sites based on the SCML2 ChIP-seq data from GS cells. Among the 6,561 SCML2 ChIP-seq peaks in GS cells, a major population of SCML2 peaks (46.7%) is enriched in promoters (Fig. 3B), which is consistent with SCML2’s enrichment at guanine-cytosine–rich sequences. These results suggest that SCML2 binds primarily to CpG island promoters.
In ES cells and germ cells, bivalent domains are strongly associated with promoter regions enriched with hypomethylated CpG dinucleotides (10, 12, 29). Therefore, we suspected that SCML2 binds to hypomethylated DNA. To examine the status of DNA methylation at SCML2 binding sites, we performed comparative analyses of our ChIP-seq data with published bisulfite sequencing data in undifferentiated spermatogonia (30). We found that enrichment for SCML2 binding in GS cells tightly correlates with levels of DNA hypomethylation (Fig. 3C). SCML2 enrichment is also correlated with CpG density (Fig. S3B). Since promoters with high CpG density are correlated with hypomethylation status in undifferentiated spermatogonia (Fig. S3C), these results suggest that SCML2 binds to hypomethylated CpG promoters enriched with H3K4me2/3. Indeed, SCML2 binding sites in promoters are hypomethylated for both class I and class II bivalent domains (Fig. S2A).
SCML2 Regulates Bivalent Domains in the Male Germline.
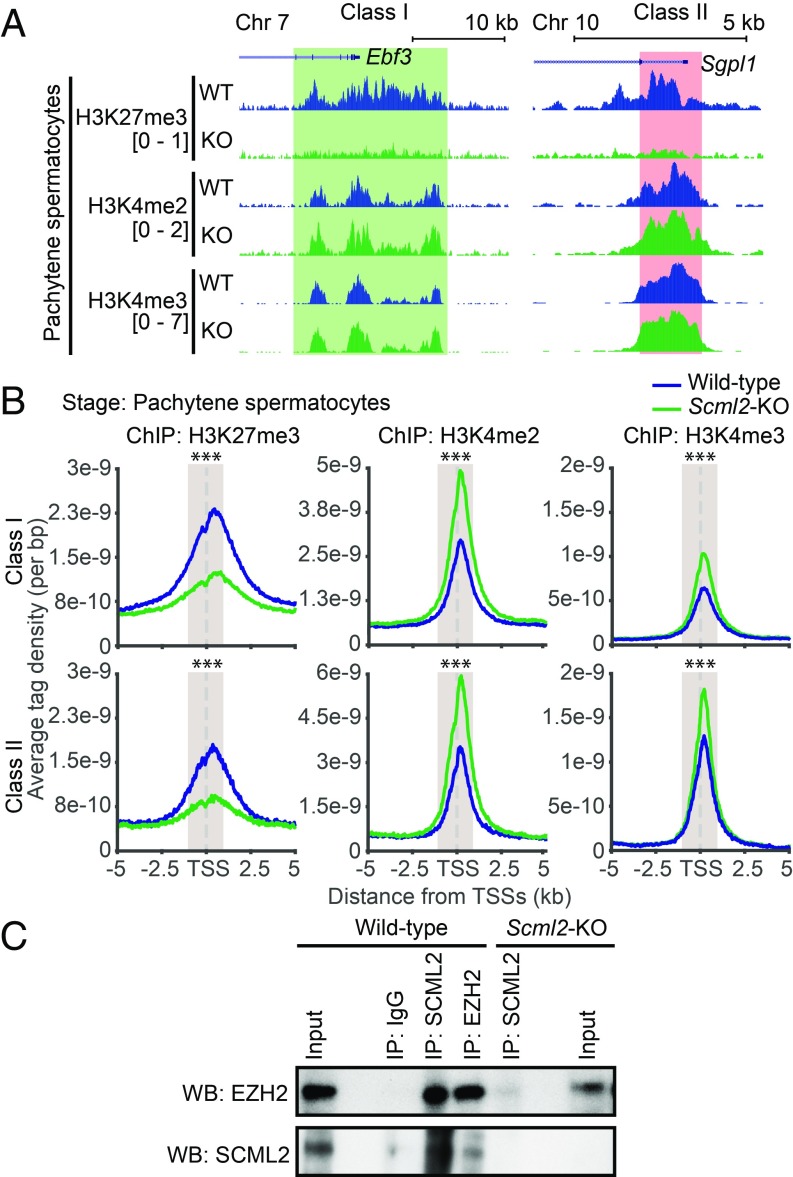
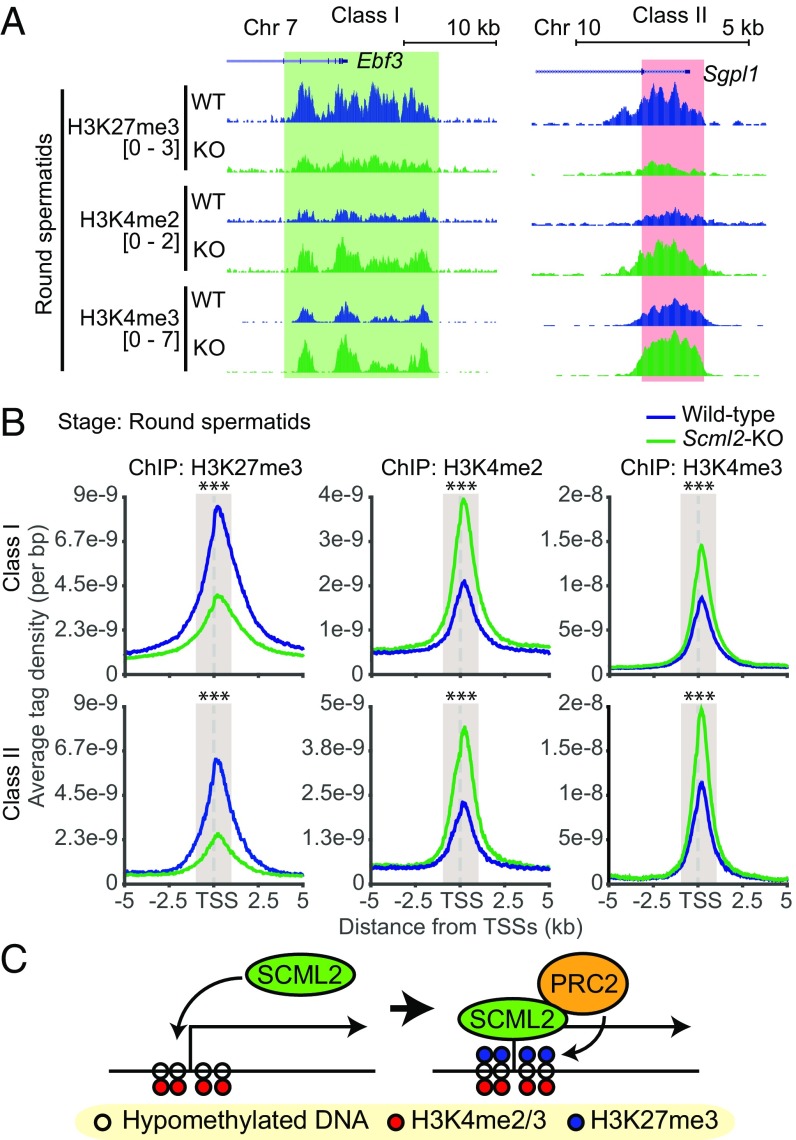
To test the hypothesis that SCML2 regulates the formation of bivalent domains, we performed ChIP-seq using Scml2 knockout (Scml2-KO) mice and examined bivalent domain formation. In PS from Scml2-KO mice, H3K27me3 was markedly decreased around TSSs of both class I and class II genes (Fig. 4A and Fig. S4A). Analyses of average tag densities of H3K27me3 ChIP-seq signals confirmed that H3K27me3 was largely depleted from the TSSs of both class I and class II bivalent domains in Scml2-KO PS (Fig. 4B). On the other hand, average tag densities of H3K4me2 and H3K4me3 ChIP-seq signals were increased in PS from Scml2-KO mice (Fig. 4B). These results suggest that SCML2 promotes H3K27me3 and suppresses H3K4me2 and H3K4me3 on the two major classes of bivalent domains in PS. These results were confirmed with biological replicates of ChIP-seq under cross-linked (fixed) and native conditions (Fig. S4B). A recent study in the fruit fly demonstrated that Sex comb on midleg (Scm), a fly ortholog of mammalian SCM proteins, including SCMH1, SCML1, and SCML2, binds to PRC2 (31). With this finding and our functional data, we reasoned that SCML2 could interact with PRC2 for the regulation of H3K27me3 at bivalent domains. To test this possibility, we performed coimmunoprecipitation using wild-type testis extracts. We found that SCML2 binds to EZH2, a catalytic subunit of PRC2 (Fig. 4C), suggesting that SCML2 interacts with PRC2 for the regulation of H3K27me3 on bivalent domains.
Fig. 4.
SCML2 facilitates H3K27me3 at germline-specific bivalent domains. (A) Distribution of ChIP-seq peaks at a representative gene for each class in PS from wild-type (WT) and Scml2-KO (KO) mice. Chr, Chromosome. (B) Average tag density of ChIP-seq data around TSSs in PSs from wild-type and Scml2-KO mice. A Wilcoxon rank sum test was performed for read counts in the highlighted area (−1,000 to +1,000 bp from TSSs; ***P < 0.001). (C) Coimmunoprecipitation of SCML2 and EZH2 using nuclear extracts from wild-type and Scml2-KO testes. IP, immunoprecipitation; WB, Western blot.
SCML2 Is Required for the Maintenance of Bivalent Domains to RS.
Bivalent domains in the male germline are intriguing for their persistence into sperm, implicating them in the early development of the next generation. To determine the function of SCML2 in the maintenance of bivalent domains into the postmeiotic period of male germline development, we investigated the regulation of bivalent domains in RS. Following the completion of meiosis, H3K27me3 remained depleted around the TSSs of both classes of bivalent domains in RS from Scml2-KO mice (Fig. 5A and Fig. S5A). However, notably, the deposition of H3K4me2 and H3K4me3 was largely increased at both classes of bivalent domains in Scml2-KO mice (Fig. 5A and Fig. S5A). These results were confirmed through analyses of average tag densities for the two classes of bivalent domain genes (Fig. 5B) and independently confirmed with biological replicates using cross-linked (fixed) and native ChIP-seq (Fig. S5B). These results reveal the function of SCML2 in the maintenance of bivalent domains and in counteractive regulation between H3K27me3 and H3K4me2/3.
Fig. 5.
Loss of H3K27me3 leads to an increase of H3K4me2/3 at bivalent domains in RS from Scml2-KO mice. (A) Distribution of ChIP-seq peaks at a representative gene for each class in RS from wild-type (WT) and Scml2-KO (KO) mice. Chr, Chromosome. (B) Average tag density of ChIP-seq data around TSSs (±5 kb) in RS from wild-type and Scml2-KO mice. A Wilcoxon rank sum test was performed for read counts in the highlighted area (−1,000 to +1,000 bp from TSSs; ***P < 0.001). (C) Model of action of SCML2 in the recruitment and induction of H3K27me3.
These results raise the possibility that Scml2-KO mice can serve as a model system to test the impact of decreased H3K27me3 deposition in sperm on embryonic development. To obtain embryos sired by Scml2-KO males, we performed in vitro fertilization (IVF) using Scml2-KO epididymal sperm with wild-type eggs since Scml2-KO males are infertile (21). However, none of Scml2-KO sperm underwent IVF (attempted with sperm from three independent mice). Next, we attempted to perform intracytoplasmic sperm injection (ICSI). However, we were not able to perform ICSI due to a structural abnormality of Scml2-KO sperm: Scml2-KO sperm were abnormally surrounded by their midpieces (Fig. S6), and it was not possible to separate nuclei from their midpieces before ICSI (also attempted with sperm from three independent mice). Therefore, future studies necessitate further technical improvement to generate embryos sired by Scml2-KO males to test paternal epigenetic inheritance.
Discussion
Unipotent germ cells are unique for their ability to give rise to a totipotent zygote following fertilization. Thus, germ cells have specific mechanisms to ensure the recovery of totipotency. Our study uncovers the regulatory mechanisms of bivalent domains in the male germline, which are proposed to have roles in the preparation of totipotency (15). In this study, we define two major classes of bivalent domains in the germline: one for the suppression of a developmental program (class I) and the other for the suppression of a somatic/progenitor program (class II). We identify SCML2 as a key regulator of H3K27me3 at bivalent domains. Because H3K27me3 remains on promoters and regulatory elements in spermatozoa after the near-global replacement of histones with protamines (6, 7, 9, 20), we propose that these bivalent domains function in two distinct modes to recover totipotency after fertilization. In support of this model, our recent study demonstrated that these bivalent domains are maintained on H3K4me3-enriched promoters into sperm (32). Following fertilization, class I bivalent domains persist into later developmental stages to regulate lineage-specific developmental processes, while class II bivalent domains are required for the recovery of a more general somatic/progenitor program after fertilization. Curiously, H3K27me3 distribution is more broadly dispersed over the regulatory regions of class I genes, while class II bivalent domains tend to be promoter-specific in late spermatogenesis (Fig. S2A). This may possibly reflect the persistence of bivalent domains into embryonic stages since, in ES cells, class II genes are largely active, while class I genes are largely silent and marked by longer-persisting bivalent domains. Thus, the difference between these classes reflects differential developmental programs in embryos. Together, our study provides a possible epigenetic link between gametogenesis and embryogenesis.
Furthermore, our study reveals how Polycomb complexes confer germline-specific functions in addition to the broad functions of core Polycomb proteins in stem cell and developmental contexts. Multiple Polycomb subunits are exchanged to acquire specific functions in different biological contexts (33, 34), and PRC1 and PRC2 are interdependent (35). Although SCML2 was initially identified as a germline-specific subunit of PRC1 (21), our current study demonstrates that SCML2 regulates PRC2 and its associated histone mark H3K27me3. Consistent with the binding of fly Scm to PRC1 and PRC2 (31), mammalian SCML2 could serve as a germline-specific link between PRC1 and PRC2. A previous epigenomic analysis suggested that ∼19% of PRC1 contains SCML2 in GS cells (21), while ∼81% of PRC1 does not contain SCML2. Therefore, it is conceivable that SCML2 links the activities of a portion of PRC1 (∼19%) and the majority of PRC2 since SCML2 substantially regulates H3K27me3 during spermatogenesis.
A previous study demonstrated that PRC2 is required for multiple stages of spermatogenesis, and a PRC2-deficient mouse exhibited a more profound meiotic phenotype than that of the Scml2-KO mouse (21, 26). Thus, PRC2 is likely regulated by SCML2 for germline-specific functions at a specific time point later in spermatogenic differentiation. We found that the genome-wide distribution of H3K27me3 undergoes dynamic changes during the mitosis-to-meiosis transition between c-Kit+ and PS (Fig. 1D), which is concurrent with the de novo formation of class II bivalent domains. Since H3K27me3 at both major classes of bivalent domains was largely diminished in PS, it is possible that SCML2-dependent regulation of PRC2 takes place in the mitosis-to-meiosis transition after the deposition of SCML2 to target loci in undifferentiated spermatogonia. Further investigation is warranted to reveal the interplay and function of Polycomb proteins for key steps of spermatogenesis.
Our study also reveals how specific genes are selected for bivalent domain formation in the germline. Although the mechanism by which Polycomb proteins select target genes remains elusive, one key Polycomb subunit is of particular interest: KDM2B, which recognizes DNA hypomethylation at CpG sites and recruits PRC1 (36–38). Therefore, the function of PRC1 is linked to CpG hypomethylation. Our study of the germline uncovers SCML2 enrichment at hypomethylated CpG sites, thereby providing a mechanistic basis for the recognition of H3K4me2/3-rich hypomethylated promoters and the establishment of H3K27me3 (Fig. 5C). In support of the possible recruitment of SCML2 to hypomethylated DNA, a previous study showed a strong correlation between SCML2 recruitment and DNA hypomethylation: SCML2 was the most enriched protein at hypomethylated chromatin in ES cells when DNA methyltransferases were ablated (39). Intriguingly, ectopic expression of SCML2 is deleterious in methylation-deficient trophoblasts (40). This result could be interpreted such that DNA hypomethylation promotes ectopic SCML2 recruitment to chromatin, leading to deleterious effects. Taken together, we propose that SCML2-dependent mechanisms utilize CpG hypomethylation as molecular landmarks to establish the two major extensive classes of bivalent domains in late spermatogenesis.
We identified counteractive regulation between H3K27me3 and H3K4me2/3 at bivalent domains. One possible explanation is that the suppression of PRC2 increases the activity of MLL, which mediates deposition of H3K4me2/3; this is because Polycomb proteins antagonize transcriptional activation by Trithorax group proteins, including MLL (41). An alternative possibility is that the loss of SCML2 leads to the transcriptional derepression of histone methyltransferases since SCML2 is a suppressor of somatic/progenitor genes. Indeed, Set1a and Mll2 were up-regulated in the Scml2-KO mice (Table S2).
Finally, the SCML2-dependent regulation of germline-specific bivalent domains raises compelling questions about the evolution of bivalent domains. A recent study demonstrated that the bivalent domains in the germline coevolved with bivalent domains involved in the development of somatic lineages (16); this study showed germline-specific bivalent domains diverged on species-specific genes yet are conserved on common regulators of somatic development. An SCML2-dependent mechanism could be a driving force for such an evolutionary trait. This study uncovers fundamental epigenetic mechanisms in the germline and implicates a mechanism for evolution in mammals.
Materials and Methods
Animals.
Scml2-KO mice were previously reported (21). This work was approved by Institutional Animal Care and Use Committee (protocol no. IACUC2015-0032) at Cincinnati Children’s Hospital Medical Center.
Histological Analysis and Germ Cell Slide Preparation.
For preparation of testicular paraffin blocks, testes were fixed with 4% paraformaldehyde at 4 °C overnight. Testes were dehydrated and embedded in paraffin. Chromosome spreads were prepared using hypotonic treatment, modified from an established protocol (42).
ChIP-Seq, RNA-Sequencing, and Data Analysis.
Both cross-linking ChIP-seq and native ChIP-seq were performed for H3K27me3, and cross-linking ChIP-seq was performed for H3K4me2 and H3K4me3. Data analysis for both ChIP-seq and RNA-sequencing was performed using the BioWardrobe Experiment Management System [https://github.com/Barski-lab/biowardrobe (43)]. Briefly, reads were aligned to the mouse genome (mm10) with Bowtie [version 1.0.0 (44)], assigned to RefSeq genes (which have one annotation per gene) using the BioWardrobe algorithm, and displayed on a local mirror of the UCSC Genome Browser as coverage. Islands of H3K27me3, H3K4me2, and H3K4me3 enrichment were identified using MACS2 [version 2.0.10.20130712 (45)]. Average tag density profiles were calculated around TSSs for genes. Resulting graphs were smoothed in 200-bp windows. Enrichment levels for ChIP-seq experiments were calculated for 4-kb windows of ±2 kb surrounding TSSs. To calculate enrichment, total read counts mapping to a coordinate region were normalized to account for the different sizes of the libraries. The Gene Expression Omnibus accession number for ChIP-seq data reported in this paper is GSE89502.
Supplementary Material
Acknowledgments
We thank Yueh-Chiang Hu, Jérôme Déjardin, Eda Yildrim, and members of the S.H.N. laboratory for discussion and helpful comments regarding the manuscript; Masatoshi Ooga, Yueh-Chiang Hu, Celvie Yuan, and Melissa Scott for technical assistance for IVF and ICSI; and Roberto Bonasio for providing the EZH2 antibody. This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (25112010) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.S.), a Research Grant (FY13-510) from the March of Dimes Foundation (to S.H.N.), NIH Grants GM119134 and HL098691 (to A.B.), and NIH Grants GM098605 and GM122776 (to S.H.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The ChIP-sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE89502).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804512115/-/DCSupplemental.
References
- 1.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 2.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 3.Gill ME, Erkek S, Peters AH. Parental epigenetic control of embryogenesis: A balance between inheritance and reprogramming? Curr Opin Cell Biol. 2012;24:387–396. doi: 10.1016/j.ceb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Kota SK, Feil R. Epigenetic transitions in germ cell development and meiosis. Dev Cell. 2010;19:675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 6.Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 7.Erkek S, et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 8.Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci USA. 2013;110:16061–16066. doi: 10.1073/pnas.1315204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammoud SS, et al. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15:239–253. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Sachs M, et al. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013;3:1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 13.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 14.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesch BJ, Page DC. Poised chromatin in the mammalian germ line. Development. 2014;141:3619–3626. doi: 10.1242/dev.113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesch BJ, Silber SJ, McCarrey JR, Page DC. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet. 2016;48:888–894. doi: 10.1038/ng.3591. [DOI] [PubMed] [Google Scholar]
- 17.Carone BR, et al. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou M, Kurimoto K. Paternal nucleosomes: Are they retained in developmental promoters or gene deserts? Dev Cell. 2014;30:6–8. doi: 10.1016/j.devcel.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 19.Zheng H, et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell. 2016;63:1066–1079. doi: 10.1016/j.molcel.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 20.Jung YH, et al. Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep. 2017;18:1366–1382. doi: 10.1016/j.celrep.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa K, et al. SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell. 2015;32:574–588. doi: 10.1016/j.devcel.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sin HS, Kartashov AV, Hasegawa K, Barski A, Namekawa SH. Poised chromatin and bivalent domains facilitate the mitosis-to-meiosis transition in the male germline. BMC Biol. 2015;13:53. doi: 10.1186/s12915-015-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aranda S, Mas G. Regulation of gene transcription by Polycomb proteins. Sci Adv. 2015;1:e1500737. doi: 10.1126/sciadv.1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisler SJ, Paro R. Trithorax and Polycomb group-dependent regulation: A tale of opposing activities. Development. 2015;142:2876–2887. doi: 10.1242/dev.120030. [DOI] [PubMed] [Google Scholar]
- 25.Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell. 2014;14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Mu W, Starmer J, Fedoriw AM, Yee D, Magnuson T. Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 2014;28:2056–2069. doi: 10.1101/gad.246124.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maezawa S, et al. Polycomb directs timely activation of germline genes in spermatogenesis. Genes Dev. 2017;31:1693–1703. doi: 10.1101/gad.302000.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanatsu-Shinohara M, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 29.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubo N, et al. DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics. 2015;16:624. doi: 10.1186/s12864-015-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang H, et al. Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev. 2015;29:1136–1150. doi: 10.1101/gad.260562.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maezawa S, Yukawa M, Alavattam KG, Barski A, Namekawa SH. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2018;46:593–608. doi: 10.1093/nar/gkx1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farcas AM, et al. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. eLife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J, et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Saksouk N, et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell. 2014;56:580–594, and erratum (2015) 57:202. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Branco MR, et al. Maternal DNA methylation regulates early trophoblast development. Dev Cell. 2016;36:152–163. doi: 10.1016/j.devcel.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 42.Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 43.Kartashov AV, Barski A. BioWardrobe: An integrated platform for analysis of epigenomics and transcriptomics data. Genome Biol. 2015;16:158. doi: 10.1186/s13059-015-0720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.