RNA polymerase II (Pol II) elongation control is utilized during transcription of most metazoan genes (1). Polymerases that successfully initiate must first break contacts with initiation factors and then interact with elongation factors including the DRB sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF). This leads to a reduction of the elongation rate due to an increase in pausing. The stage is now set for positive transcription elongation factor b (P-TEFb) to phosphorylate DSIF, which, in turn, leads to the loss of NELF, association of the PAF1 complex, and the onset of productive elongation. Although it is clear that paused Pol II is found in promoter-proximal regions of most expressed genes in metazoan cells, a controversy has arisen concerning how long paused polymerases remain engaged. The prevailing view is that paused Pol II is relatively stable. However, the findings presented in PNAS by Steurer et al. (2) call this view into question. They generated a human cell line solely expressing GFP-tagged Pol II and then used fluorescence recovery after photobleaching (FRAP) to identify and characterize four distinguishable kinetic states for Pol II. Besides the very rapid recovery due to diffusion of free Pol II, they found three bound states, each with about an order of magnitude slower recovery than the preceding one. These kinetic states were assigned to initiation, pausing, and productive elongation by observing how well-characterized compounds affected the FRAP measurements.
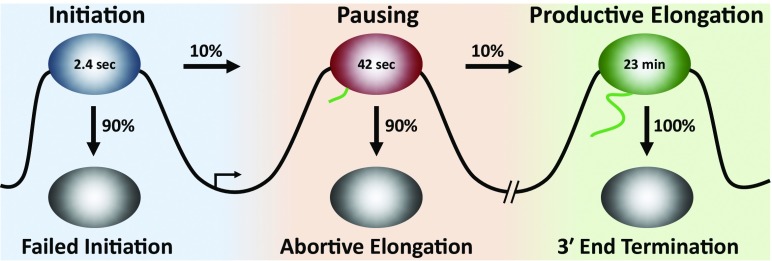
Steurer et al. (2) used computational modeling of the FRAP data to calculate the residence time for Pol II in each of the three bound states, as well as the fraction of each state that progresses into the next state (Fig. 1). They found that in normal untreated cells, Pol II spends, on average, 2.4 s interacting with the promoter. About 90% of these polymerases dissociate because of failed initiation, leaving only 10% engaged in elongation. These polymerases enter into the pausing phase of Pol II elongation control. Critically, Steurer et al. (2) found that the residence time of paused Pol II was, on average, only 42 s and that only 10% those polymerases entered into productive elongation. The remaining paused Pol II was prematurely terminated in a process originally termed abortive elongation (3). Pol II that entered into productive elongation remained bound for an average of 23 min (1). This is the time spent transiting gene bodies at an elongation rate of about 4,000 nt⋅min−1 plus the time spent with Pol II engaged downstream of the 3′ end of the genes before the final termination event.
Fig. 1.
Residence times of Pol II during the three states of Pol II interaction with DNA. The diagram depicts the average time Pol II spends associated with DNA during initiation, pausing, and productive elongation. The fate of Pol II in each of the phases of transcription is indicated as the percentage that dissociates from the DNA versus the percentage that enters the next phase. Values were calculated by Steurer et al. (2).
Support for the findings of Steurer et al. (2) is found in several studies. Darzacq et al. (4) used a similar FRAP method with cells expressing GFP-tagged Pol II and identified three kinetic states for bound Pol II over an engineered gene array. However, without methods to identify the states, they applied the labels “promoter,” “initiating,” and “engaged.” The residence times they calculated were 6 s, 54 s, and 517 s, respectively, which is in reasonable agreement with the three bound states identified by Steurer et al. (2). They found that treatment of cells with a P-TEFb inhibitor blocked entry into the third state, providing evidence that their initiating state was actually paused Pol II (4). They also found the same large loss of polymerases entering downstream states (about 90% each) (4). Recently, a study of the effects of hydrogen peroxide on transcription in human cells found a rapid increase in promoter-proximal paused Pol II (5). Using a nuclear walk-on method in which cells are lysed and Pol II is locked in place within 20 s, paused Pol II was found to increase more than twofold in 1 min and plateaued at about a fourfold increase in 3 min after treatment of the cells with a low dose of peroxide (5). The most plausible model, which was supported by a variety of in vitro experiments, was that termination of paused Pol II was blocked by peroxide. The rapid accumulation of paused Pol II could only arise if the turnover of paused Pol II was normally less than 1 min (5). Rapid termination of paused Pol II has also been observed in vitro using immobilized templates and Drosophila nuclear extract (3). Finally, a recent study using genome-wide, single-molecule footprinting demonstrated a high turnover of paused Pol II in Drosophila cells, although that method did not allow calculation of precise residence times (6).
An alternative view of the stability of paused Pol II has arisen from studies over the past 5 y using methods to examine Pol II occupancy following treatment of cells with triptolide to block initiation. The idea was to block entry to the pause and then follow the decay of Pol II occupancy in the promoter-proximal region. Such triptolide-chase experiments have been carried out in human (7), mouse (8), and Drosophila (9, 10) cells. It was concluded in each of these studies that paused Pol II was either relatively stable or very stable, with residence times between 5 and 15 min (8–10) to longer than 1 h on some genes (7). Another Drosophila study used a photoactivatable GFP-Pol II and fluorescence depletion after photobleaching to examine pausing at the HSP70 locus in Drosophila polytene chromosomes. Those investigators concluded that the half-life was about 5 min (11). It is possible that the kinetics of turnover of Drosophila Pol II are different from mammalian Pol II. This increased temperature experienced by human cells compared with Drosophila cells might increase the termination rate. However, no such correlation was found for Pol II residence times from mammalian and Drosophila studies. In fact, the most stable paused Pol II was found in the human study using HCT116 cells (10).
The discrepancy in residence times of paused Pol II calculated from triptolide-chase experiments and the GFP-Pol II FRAP experiments can be resolved by taking into account the rate of inhibition of initiation by triptolide. Triptolide is a covalent inhibitor of XBP, one of the two helicase subunits found in the initiation factor TFIIH (12). Using an in vitro transcription assay, Nilson et al. (5) demonstrated that inhibition of initiation by triptolide was highly concentration- and time-dependent. Using concentrations chosen in all but one of the triptolide-chase studies, even 30 min of incubation did not achieve complete inhibition (5). Due to kinetics of uptake and competing interactions, the rate of triptolide inhibition is likely slower in cells. The assumption made in the triptolide-chase experiments is that inhibition of initiation is quick, because only then can the rate of depletion of paused Pol II be measured. Unfortunately, this assumption cannot be verified and is likely not the case. Therefore, the actual rate of Pol II turnover would be much quicker than determined by the triptolide-chase experiments. Supporting this idea, there was a rough correlation with the residence time of Pol II calculated with the concentration of triptolide used in the studies. The most stably paused Pol II was found in the study that used 125 nM triptolide (7), which was found to achieve only 40% inhibition of initiation in vitro after a 30-min incubation of the compound with the extract (5).
One common finding among the triptolide-chase experiments was that the rate of depletion of paused Pol II was highly variable even within each group’s experiments. This was likely due, in part, to gene-specific differences in the fraction of paused Pol II that enters into productive elongation. More highly expressed genes would have a higher fraction of depletion caused by entry into productive elongation. Another possibility for variability across genes could be inconsistency in the requirement for XBP. Surprisingly, XBP has been found not to be generally required for transcription in cells (13). When it is depleted, transcription seems to be unaffected and becomes resistant to inhibition by triptolide (13). The mechanism involved in this deserves further investigation. A recent, potentially very informative study by Adelman and coworkers (14) found that the paused Pol II found in active enhancers was more sensitive to triptolide than those in genes. It will be informative to determine the mechanism(s) involved in these differential effects.
Given the well-designed study from Steurer et al. (2), the reinterpretation of the results from Darzacq et al. (4), the findings
The results from Steurer et al. not only necessitate revising the commonly held conception of Pol II pausing but also provide new insight into the biological role of promoter-proximal pausing.
from Nilson et al. (5), and the problem concerning the rate of inhibition of initiation by triptolide, it seems prudent to ask why promoter-proximal pausing by Pol II is transient. Paused Pol II helps keep promoter regions in an open, accessible chromatin conformation. The histone H3K4 methylation marks found on nucleosomes downstream of the paused Pol II are laid down, in part, by the SET complexes associated with the Ser-5 phosphorylated CTD of Pol II (15). When pausing is disrupted by knockdown of NELF over the course of days, the mark disappears and promoters shut down (16). The critical parameter is the frequency of occupation of each copy of every gene. This is not an issue for highly expressed genes because of their high occupancy. However, the situation with the majority of genes is that they have Pol II occupancies one or two orders of magnitude lower. Stable pausing would keep a small fraction of those promoters open, but across a population of cells, many of those genes might not be occupied often enough to keep promoters open. Rapid flux through the pause alleviates this problem by distributing the paused Pol II across all genes so that no gene has to wait too long. If Pol II pauses for 42 s, a fully occupied gene would be visited about 2,000 times a day. At a gene with 1% occupancy, each copy of that gene across the population of cells would be visited 20 times a day, or about once each hour. In contrast, if Pol II pauses for 20 min, most copies of that low-occupancy gene would be visited less than once per day, and this is not often enough to maintain the H3K4me3 mark, which has a half-life of 6.8 h (17).
In summary, the results from Steurer et al. (2) not only necessitate revising the commonly held conception of Pol II pausing but also provide new insight into the biological role of promoter-proximal pausing. A more integrated view takes into account the rates of initiation, turnover of paused Pol II, and transition into productive elongation (Fig. 1), as well as the relative lifetimes of paused polymerases and the chromatin marks that preserve promoter accessibility. Because the level of paused Pol II occupancy is affected by the rate of turnover, it is logical to assume that it can be regulated. Such regulation may be important during development and during responses to environmental conditions. Consistent with this possibility, oxidative stress has been demonstrated to globally decrease the turnover of paused Pol II (5). These considerations justify a search for novel factors that are able to regulate the residence time of paused Pol II both globally and in gene-specific ways.
Acknowledgments
My research is supported by National Institute of General Medical Science Grants R01-GM35500 and R01-GM113935 (both just replaced by R35-GM126908) and by National Institute of Allergy and Infection Diseases Grant R21 AI130453.
Footnotes
The author declares no conflict of interest.
See companion article on page E4368.
References
- 1.Guo J, Price DH. RNA polymerase II transcription elongation control. Chem Rev. 2013;113:8583–8603. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steurer B, et al. Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proc Natl Acad Sci USA. 2018;115:E4368–E4376. doi: 10.1073/pnas.1717920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilson KA, et al. Oxidative stress rapidly stabilizes promoter-proximal paused Pol II across the human genome. Nucleic Acids Res. 2017;45:11088–11105. doi: 10.1093/nar/gkx724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krebs AR, et al. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol Cell. 2017;67:411–422.e4. doi: 10.1016/j.molcel.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes Dev. 2015;29:39–47. doi: 10.1101/gad.246173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henriques T, et al. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao W, Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet. 2017;49:1045–1051. doi: 10.1038/ng.3867. [DOI] [PubMed] [Google Scholar]
- 11.Buckley MS, Kwak H, Zipfel WR, Lis JT. Kinetics of promoter Pol II on Hsp70 reveal stable pausing and key insights into its regulation. Genes Dev. 2014;28:14–19. doi: 10.1101/gad.231886.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Titov DV, et al. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol. 2011;7:182–188. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alekseev S, et al. Transcription without XPB establishes a unified helicase-independent mechanism of promoter opening in eukaryotic gene expression. Mol Cell. 2017;65:504–514.e4. doi: 10.1016/j.molcel.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Henriques T, et al. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev. 2018;32:26–41. doi: 10.1101/gad.309351.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 16.Gilchrist DA, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Y, Tipton JD, Thomas PM, Kelleher NL, Sweet SM. Site-specific human histone H3 methylation stability: fast K4me3 turnover. Proteomics. 2014;14:2190–2199. doi: 10.1002/pmic.201400060. [DOI] [PMC free article] [PubMed] [Google Scholar]